Abstract
The tomato protein kinase 1 (TPK1b) gene encodes a receptor-like cytoplasmic kinase localized to the plasma membrane. Pathogen infection, mechanical wounding, and oxidative stress induce expression of TPK1b, and reducing TPK1b gene expression through RNA interference (RNAi) increases tomato susceptibility to the necrotrophic fungus Botrytis cinerea and to feeding by larvae of tobacco hornworm (Manduca sexta) but not to the bacterial pathogen Pseudomonas syringae. TPK1b RNAi seedlings are also impaired in ethylene (ET) responses. Notably, susceptibility to Botrytis and insect feeding is correlated with reduced expression of the proteinase inhibitor II gene in response to Botrytis and 1-aminocyclopropane-1-carboxylic acid, the natural precursor of ET, but wild-type expression in response to mechanical wounding and methyl-jasmonate. TPK1b functions independent of JA biosynthesis and response genes required for resistance to Botrytis. TPK1b is a functional kinase with autophosphorylation and Myelin Basis Protein phosphorylation activities. Three residues in the activation segment play a critical role in the kinase activity and in vivo signaling function of TPK1b. In sum, our findings establish a signaling role for TPK1b in an ET-mediated shared defense mechanism for resistance to necrotrophic fungi and herbivorous insects.
INTRODUCTION
Plants recognize attempted pathogen infection and activate defense responses to limit damage caused by disease with the mechanisms of recognition and response depending on the nature of the pathogen encountered. For necrotrophic pathogens, mechanisms of recognition and response are poorly understood. Current molecular models of pathogen response signaling have been derived largely from studies that center on plant interactions with biotrophic pathogens. In general, plants recognize pathogens through race-specific effectors or general elicitors referred to as pathogen-associated molecular patterns (PAMPs) (Jones and Dangl, 2006). Direct or indirect recognition of effectors by plant R proteins triggers a cascade of defense responses culminating in the hypersensitive response (HR) and resistance to specific races of biotrophic pathogens. However, there are no known race-specific effectors or corresponding R proteins described for necrotroph–plant interactions, and the HR may in fact enhance infection by necrotrophic pathogens (Govrin and Levine, 2000). Recently, an R protein was implicated in susceptibility to a necrotrophic pathogen (Lorang et al., 2007). PAMP-triggered basal responses are triggered by diverse pathogens, including necrotrophs, but neither the pattern recognition receptors nor the elicitors are characterized in many plant–necrotroph interactions. In the case of necrotrophic pathogens, toxins, damage to the infected plant cell walls by hydrolytic enzymes, and the oligosaccharides released are all likely to play important roles in perception and response signaling. Botrytis cinerea and other necrotrophs produce toxins and cell wall degrading enzymes, including pathogen polygalacturonases (Prins et al., 2000). Oligogalacturonide (OG) fragments released from cell walls during pathogen attack or insect herbivory induce plant defense responses (Bishop et al., 1981; Doares et al., 1995; Ferrari et al., 2007). Consistent with this, plants may sense damage to the cell wall or recognize OGs through pattern recognition receptors. Additionally, PAMPs and race-specific elicitors induce systemic resistance against biotrophic pathogens (Dempsey et al., 1999; Mishina and Zeier, 2007), although the extent of resistance against necrotrophic pathogens is unclear. Thus, plant responses to necrotrophs and biotrophs are regulated through distinct and overlapping recognition and response mechanisms.
Networks of regulatory factors and defense molecules mediate responses to pathogens. Receptor-like protein kinases (RLKs) regulate recognition and early responses to diverse internal and external signals and frequently play critical roles in defense and symbiosis. RLKs are structurally diverse and constitute a large gene family in many plants with >600 members in Arabidopsis thaliana and ∼1130 in rice (Oryza sativa) (Shiu et al., 2004). The majority of RLKs contain transmembrane and extracellular domains for sensing extracellular signals such as pathogen-derived molecules (Torii, 2000). Xa21 encodes a rice leucine-rich repeat receptor-like kinase (LRR-RLK) that confers resistance to the bacterial blight pathogen Xanthomonas oryzae pv oryzae (Song et al., 1995). The Arabidopsis FLS2 and BAK1 proteins are both LRR-RLKs involved in recognition and signaling of flagellin (Chinchilla et al., 2007). Interestingly, BAK1 is also implicated in cell death control and resistance to necrotrophic pathogens (He et al., 2007; Kemmerling et al., 2007). LysM receptor-like kinase1 (LysM RLK1) from Arabidopsis is required for responses to chitin (Miya et al., 2007; Wan et al., 2008), a component of fungal cell walls recognized as a PAMP by plant cells. Disruption of LysM RLK1 impairs resistance to Alternaria brassicicola and Erysiphe cichoracearum, indicating its role in defense signaling. The tomato (Solanum lycopersicum) RLK SR160 is a receptor for systemin, whereas the SYMRK and HAR1 RLKs are involved in fungal and/or bacterial symbiosis (Krusell et al., 2002; Stracke et al., 2002). Receptor-like cytoplasmic kinases (RLCKs) are a subgroup of RLKs that lack transmembrane and extracellular domains but share a common monophyletic origin with RLKs (Shiu and Bleecker, 2001) and have also been implicated in defense responses. Pto and PBS1 are well-studied RLCKs involved in race-specific resistance to bacterial pathogens in tomato and Arabidopsis, respectively (Martin et al., 1993; Swiderski and Innes, 2001), whereas Arabidopsis BIK1 is a RLCK that mediates crosstalk between defense pathways in Arabidopsis (Veronese et al., 2006). All of these RLCKs act early in recognition or signaling of pathogen defense pathways.
Perception of pathogen-derived molecules initiates a variety of downstream events, including activation of mitogen-activated protein kinase (MAPK) cascades, phosphorylation of downstream targets, and activation of general and specific defense responses (Asai et al., 2002). FLS2-mediated MAPK activation regulates defense gene transcription and resistance to Pseudomonas syringae and Botrytis (Asai et al., 2002), suggesting that defense initiated by a bacterial PAMP can confer resistance to necrotrophic fungi. Perception of systemin by the membrane-bound receptor SR160 activates MPK1 and MPK2 and initiates the synthesis of jasmonate (JA) leading to the induction of defense gene expression, suggesting a role for MAPKs downstream of recognition in insect defense (Kandoth et al., 2007). MAPKs, wound-induced protein kinase, and salicylic acid–induced protein kinase are activated during Cf-9/Avr9 interaction in tobacco (Nicotiana tabacum; Ludwig et al., 2005), and their Arabidopsis orthologs MPK3/6 function in activation of PAMP (Asai et al., 2002) and effector-triggered responses (Menke et al., 2004) in addition to signaling of reactive oxygen intermediates (Rentel et al., 2004). Other studies have demonstrated the activation of MAPKs by wounding, low temperature, drought, salinity, and reactive oxygen intermediates, suggesting extensive crosstalk in plant responses to external cues (Zhang et al., 1998; Romeis, 2001; Zhang and Klessig, 2001). Thus, signal transduction components may be shared among defense responses induced by effectors, PAMPs, insect herbivory, and even abiotic stress factors, and signals initiated by diverse pathogens may converge into conserved MAPK cascades. Downstream from MAPKs are transcription factors and defense molecules. In Arabidopsis, ERF, WRKY, and MYB transcription factor genes have been implicated in plant defense to necrotrophic pathogens (Berrocal-Lobo et al., 2002; Mengiste et al., 2003; Coego et al., 2005; Zheng et al., 2006). The functions of some, but not all, of these transcription factor genes are associated with the expression of PDF1.2, a plant defensin molecule with antifungal activities implicated in resistance to necrotrophic pathogens.
We studied the function of tomato protein kinase 1b (TPK1b) in basal resistance of tomato. We show that TPK1b encodes a membrane associated functional kinase required for resistance against Botrytis and insect herbivory in tomato. TPK1b is a homolog of the Arabidopsis protein kinase 1b (APK1b) (Hirayama and Oka, 1992) that has no known biological function and the Brassicia M locus protein kinase (MLPK) implicated in self-incompatibility (Murase et al., 2004). Interestingly, TPK1b cDNA rescues the disease susceptibility of the Arabidopsis bik1 mutant, suggesting that TPK1b and Arabidopsis BIK1 perform overlapping or redundant functions. Furthermore, TPK1b is required for ethylene (ET) responses of tomato seedlings, suggesting that the impaired defense against insects and Botrytis is due to attenuated activation of ET-dependent defense. These altered pathogen and ET responses in TPK1b RNA interference (RNAi) plants correlated with reduced expression of the tomato proteinase inhibitor II (PI-II) gene in response to Botrytis and 1-aminocyclopropane-1-carboxylic acid (ACC), but expression of PI-II in response to mechanical wounding and methyl-jasmonate (MeJA) was unaffected. Our results demonstrate that TPK1b is involved in ET-dependent but JA-independent defense, thus establishing TPK1b and ET as key regulators of insect and pathogen defense responses.
RESULTS
Cloning and Characterization of TPK1b
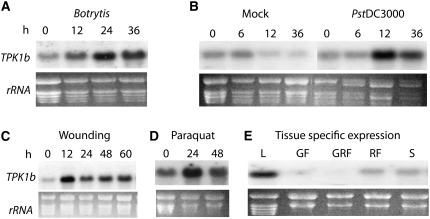
The TPK1b gene was identified during a screen for the tomato homolog of the Arabidopsis Botrytis Induced Kinase1 (BIK1) gene (Veronese et al., 2006). We cloned several cDNAs that are predicted to encode receptor-like cytoplasmic protein kinases (RLCKs) sharing various levels of sequence similarity to Arabidopsis BIK1. Among these, TPK1b (SGN-U319927, SGN-E344457) is induced early during Botrytis infection, consistent with possible function early in a defense response pathway. TPK1b is induced as early as 12 h after Botrytis inoculation in tomato (Figure 1A). In addition, P. syringae, mechanical wounding, and oxidative stress caused by the herbicide paraquat strongly induce TPK1b gene expression (Figures 1B to 1D). TPK1b transcripts are abundant in leaves and are moderately expressed in stems (Figure 1E). Basal expression is low in green fruits but increases as the fruit ripens. Thus, TPK1b is transcriptionally regulated by biotic and abiotic agents, which suggests its role in plant responses to diverse signals.
Figure 1.
Basal and Induced Expression of the TPK1b Gene.
RNA gel blot analysis showing upregulation of TPK1b gene expression by
(A) B. cinerea.
(B) P. syringae.
(C) Mechanical wounding.
(D) Oxidative stress (paraquat, methyl viologen).
(E) Basal expression in fruit and leaf tissues.
Total RNA (15 μg) was loaded per lane. rRNA staining is shown as a loading control. The experiments were repeated at least three times with similar results. Wild-type plants of the tomato cultivar CastlemartII were inoculated with pathogens or were treated with chemicals as described in Methods. h, h after inoculation or chemical treatment; L, leaf; GF, green fruit; GFR, green–red fruit; RF, red fruit; S, stem.
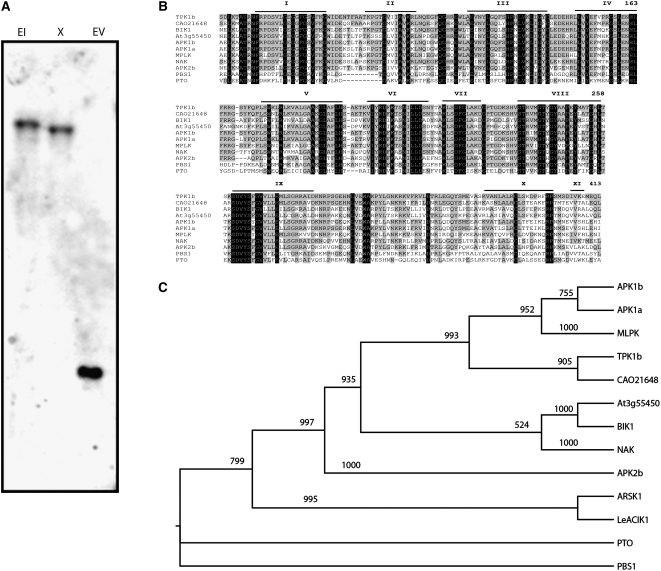
The full-length TPK1b cDNA was cloned by 5′ and 3′ random amplification of cDNA ends (RACE). The gene-specific region of TPK1b was hybridized, under high-stringency conditions, to a DNA blot of tomato genomic DNA digested with different restriction enzymes (Figure 2A). A single hybridizing band was observed for TPK1b consistent with a single-copy gene in the genome. The TPK1b cDNA contains an open reading frame of 404 amino acids encoding an RLCK (Shiu and Bleecker, 2001) with an estimated molecular mass of 44.6 kD. TPK1b contains a consensus plant N-myristoylation motif [MGXXXS/T(K)] (Boisson et al., 2003). Protein N-myristoylation is the covalent attachment of myristic acid to the N-terminal Gly residue of a nascent polypeptide that suggests potential membrane localization (Johnson et al., 1994). In addition, TPK1b has a protein kinase catalytic domain (KD, residues 68 to 353) that includes the protein kinase ATP binding region l and a Ser/Thr kinase active-site signature (VIYRDFKTSNILL, residues 197 to 209) (Figure 2B). The KD occupies the central part of TPK1b with all 11 conserved subdomains of protein kinases (Hardie, 1999). Similar to other Ser/Thr kinases, TPK1b subdomain VI contains a Lys residue in the DFKTSN motif, and subdomain VIII contains a G-T/S-XX-Y/F-X-APE motif (Hardie, 1999), suggesting that TPK1b is a Ser/Thr kinase (Stone and Walker, 1995). TPK1b KD contains a structural element known as the activation segment that lies between the conserved sequence motifs DFG (domain VII) and APE (domain VIII) (Johnson et al., 1996). Also, TPK1b is an RD-type protein kinase because it has the Asn residue in subdomain VIb (D-203) and an Arg immediately preceding this Asn residue (Dardick and Ronald, 2006).
Figure 2.
TPK1b Is a Single-Copy Gene in the Tomato Genome Encoding a Receptor-Like Protein Kinase.
(A) DNA gel blot showing the presence of a single copy of TPK1b in the tomato genome.
(B) Sequence alignment between TPK1b and related protein kinases.
(C) Phylogenetic relationship between TPK1b and related protein kinases.
In (A), tomato genomic DNA was digested with EcoRI (EI), Xba I (X), and EcoRV (EV), resolved on agarose gel, transferred to membrane for hybridization, and probed with 5′ end of the TPK1b gene lacking the kinase domain. In (B), the kinase subdomains are marked with a bar underlined and numbered (I to XI). Residues that are conserved in all TPK1b-related kinases are shaded in black, and amino acids that are shared between TPK1b and the other entries are shaded in gray. Sequences were aligned using ClustalW (Thompson et al., 1994). Numbers above the alignment correspond to amino acid positions in TPK1b. The alignment used for the phylogentic tree in (C) is provided as Supplemental Data Set 1 online. Sequences and phylogenic data are from tomato (TPK1b, GenBank accession number EU555286) CAO21648 (Vitis vinifera), MLPK (Brassica rapa), PTO (Lycopersicum esculentum), and Arabidopsis (BIK1/At2g39660, APK1b/At2g28930, APK1a/At1g07570, BIK1-like/At3g55450, APK2b/At2g02800, NAK/At5g02290, and PBS1).
Sequence and phylogenetic analyses were performed to decipher the exact relationship between TPK1b and related RLCKs. TPK1b is closely related to RLCKs from different plant species with overall sequence identities ranging from 60 to 80%. A predicted RLCK (CAO21648) from grapevine (Vitis vinifera) (Jaillon et al., 2007) shows the highest sequence identity (80%) to TPK1b. Two related Arabidopsis kinases, APK1b (At2g28930) and APK1a (At5g02290), are also closely related to TPK1b, sharing 75 and 68% sequence identities, respectively, to TPK1b. TPK1b is also highly similar to the Brassica MLPK (68% identity), the Arabidopsis proteins NAK (70%, At5g02290) and BIK1 (66%, At2g39660), and the BIK1-like protein (61%, At3g55450) (Figure 2B).
The highest conservation between TPK1b and related RLCK sequences is in the KD with 80 to 87% identities to the related RLCKs. TPK1b KD is flanked by two short nonkinase domains that have no obvious similarities to proteins of known function. The N-terminal region shows a sequence identity of 40 to 54% to related RLCKs. By contrast, the TPK1b C-terminal region consisting of 50 amino acids appears to be specific and shows no homology to any protein in the database, including Arabidopsis APK1b, MPLK, and CAO21648.
Next, we examined the phylogenetic relationship between TPK1b and other RLCKs (Figure 2C). A rooted phylogenetic tree was constructed using the neighbor-joining phylogeny (Saitou and Nei, 1987) based on the full KD of TPK1b and related kinases aligned with ClustalW (Thompson et al., 1994). The tomato PTO kinase or the PBS1 kinases were used as an outgroup. PTO and PBS1 are RLCKs and yet are not closely related to the TPK1b based on sequence analysis. The phylogenetic analysis clusters TPK1b, APK1b, APK1a, grapevine CA02648, and MLPK kinases in the same clade, consistent with the sequence analysis above (Figure 2B). The next related clade contains Arabidopsis NAK1, BIK1, and the BIK1-like proteins (Figure 2C). The functions of most of these related RLCKs have not been determined. APK1a and APK1b show kinase activities in vitro, but their biological function is not determined (Hirayama and Oka, 1992), whereas MLPK is involved in self-incompatibility in Brassicia rapa (Murase et al., 2004).
TPK1b Is Required for Resistance to Botrytis
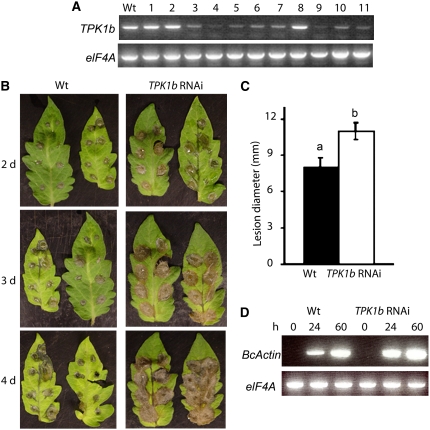
To study the biological function of TPK1b, we generated tomato TPK1b RNAi plants that are reduced in TPK1b gene expression. We cloned a gene-specific fragment of TPK1b into an RNAi vector (http://www.chromdb.org/info/plasmids/pGSA1165.html) and generated transgenic plants through Agrobacterium tumefaciens–mediated transformation of tomato cotyledon explants (McCormick, 1991). RT-PCR analysis using TPK1b gene-specific primers reveals that TPK1b expression is substantially reduced in several lines relative to wild-type plants (Figure 3A). Transgenic TPK1b RNAi lines #4 and #9 showed the greatest reduction of TPK1b gene expression and were used for most studies described in this article.
Figure 3.
TPK1b Is Required for Resistance to Botrytis.
(A) RT-PCR from wild-type and TPK1b RNAi lines showing TPK1b transcript levels.
(B) TPK1b RNAi plants show increased susceptibility to Botrytis.
(C) Disease lesion size in Botrytis-inoculated leaves at 2 DAI.
(D) Accumulation of Botrytis ActinA mRNA as a measure of fungal growth in drop-inoculated leaves.
In (A), numbers above the lanes indicate independent RNAi transgenic lines, and the RNA used for the RT-PCR was extracted from Botrytis-inoculated leaves. In (C), the values represent the mean ± se from a minimum of 60 lesions. Analysis of variance and Duncan's multiple range test were performed to determine the statistical significance of the differences in the mean disease lesion sizes using SAS software (SAS Institute, 1999). Bars with different letters are significantly different from each other at P = 0.05. In (D), RT-PCR was performed using the Botrytis ActinA gene-specific primers. Experiments were repeated at least three times with similar results. h, h after inoculation; d, days after inoculation; BcActin, B. cinerea ActinA gene.
TPK1b RNAi plants showed increased susceptibility to Botrytis with larger disease lesions than wild-type plants at 2 to 4 d after inoculation (DAI) (Figures 3B and 3C). At 3 to 4 DAI, the lesions expanded rapidly and coalesced in TPK1b RNAi leaves, whereas lesions expanded more slowly and remained discrete at 4 DAI in wild-type leaves. The TPK1b RNAi plants also supported more Botrytis growth as measured by the amount of the fungal ActinA mRNA in the inoculated plants (Figure 3D; Benito et al., 1998). Similarly, when we suppressed TPK1b gene expression in the Micro-Tom cultivar using virus-induced gene silencing (VIGS) (Liu et al., 2002), we observed increased susceptibility to Botrytis (see Supplemental Figure 1A online). In the silenced plants, disease developed faster and resulted in tissue collapse in leaves that emerged after the VIGS treatment, consistent with the pattern of silencing in control plants silenced for the tomato phytotene desaturase gene (see Supplemental Figure 1B online, right). TPK1b RNAi plants inoculated with the virulent strain of the bacterial pathogen P. syringae showed no altered responses (see Supplemental Figure 2 online). Thus, our data suggest that TPK1b plays an important role in limiting both disease symptoms and fungal growth in tomato plants inoculated with Botrytis but plays no role in resistance to P. syringae.
TPK1b Is Required for Resistance to Insect Feeding
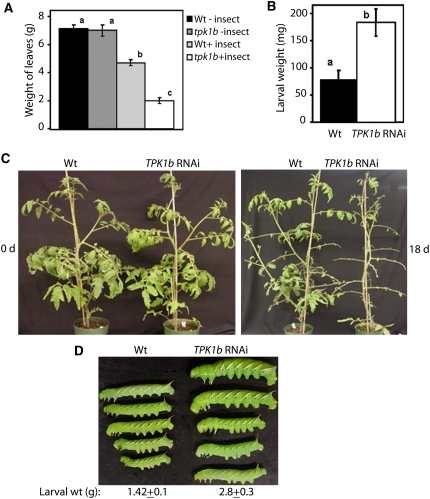
Induction of TPK1b gene expression by wounding and previous reports on tomato defense response to Botrytis (Diaz et al., 2002) suggest a mechanistic link between insect and necrotrophic pathogen defense. Therefore, we tested tomato TPK1b RNAi lines for susceptibility to feeding by tobacco hornworm (Manduca sexta) larvae. In a detached leaf assay, newly hatched tobacco hornworm larvae were placed on leaves of 8-week-old tomato plants to initiate a feeding trial. Larvae and leaf weights were determined when the foliage of TPK1b RNAi plants was ∼90% consumed. In parallel, control detached leaves were kept without larvae to determine leaf weight due to desiccation. The larvae consumed significantly more foliage of TPK1b RNAi than from the wild type (see Supplemental Figure 3A online). After 5 d of larval feeding, 72% of TPK1b RNAi leaves were consumed relative to only 35% of the wild-type plants (Figure 4A). Losses of leaf weight due to desiccation were relatively minor and comparable in both genotypes. The average weights of larvae feeding on TPK1b RNAi plants were 2.3-fold more than larvae recovered from wild-type leaves (Figure 4B; see Supplemental Figure 3B online). In an independent assay, larvae were weighed and placed on whole plants growing in the greenhouse (Figure 4C, left). The TPK1b RNAi plants were defoliated due to the larval feeding (Figure 4C, right). The average weight gain of larva feeding on TPK1b RNAi plants was ∼2.4-fold more than those feeding on wild-type plants regardless of the duration of feeding (Figure 4D), suggesting a clear loss of resistance to larval feeding in the TPK1b RNAi plants. Thus, TPK1b functions in tomato resistance to insects and suggests a shared mechanism against pathogens and insect pests.
Figure 4.
TPK1b RNAi Plants Show Reduced Resistance to Tobacco Hornworm.
(A) Leaf weight recovered at the end of 5 d of feeding trial on detached leaves (n = 25).
(B) Larval weight recovered at the end of 5 d of feeding trial on detached leaves (n = 10).
(C) Wild-type and TPK1b RNAi plants at the beginning (left) and end (right) of tobacco hornworm feeding trial on whole plants.
(D) Size of larvae recovered at the end of tobacco hornworm feeding trial on whole plants.
The feeding trails on detached leaf or whole plants were each repeated three times with similar results. In each experiment of detached leaf and whole-plant assays, six and 10 newly hatched larvae (∼9 to 11 mg each), respectively, were placed on at least five 8-week-old plants of each genotype. Larvae were allowed to feed on the same plant for the duration of the trial. In (A) and (B), the data represent the mean ± se from three different experiments. Bars with different letters are significantly different from each other (P = 0.05). Analysis of variance and Duncan's multiple range test were performed as described in the legend for Figure 3.
TPK1b Functions Independent of Tomato JA Synthesis and Responses
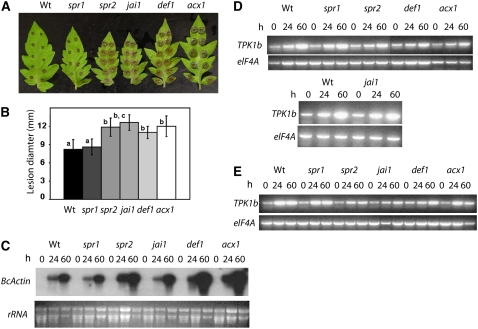
To establish the relationship between TPK1b and genes regulating tomato wound and insect responses, we first determined the Botrytis resistance of the tomato mutants suppressor of prosystemin-mediated response1 (spr1; Lee and Howe, 2003), spr2 (Li et al., 2003), jasmonate-insensitive1 (jai1; Li et al., 2004), defenseless1 (def1; Howe et al., 1996), and acyl-CoA oxidase (acx1; Li et al., 2005), which are impaired in JA synthesis and wound responses. All of the tomato mutants except spr1 showed increased susceptibility to Botrytis compared with the CasltemartII wild type (Figure 5A). In the susceptible mutants, the disease lesions were significantly larger than in wild-type plants at 2 DAI (Figures 5A and 5B). In addition, the mutants supported higher levels of Botrytis growth as measured by the increased accumulation of Botrytis ActinA RNA (Figure 5C). Interestingly, basal resistance of spr1 against Botrytis was comparable to the wild type. Spr1 is an upstream regulator of JA and systemin responses, and the mutant's basal resistance to Botrytis implies the existence of another factor upstream of JA that controls responses to Botrytis. Thus, in tomato, JA levels and responses are required for resistance to Botrytis, but deficiency in systemin responses appears to have no effect. However, the Botrytis and wound-induced expression of TPK1b is largely independent of JA functions defined by SPR2, JAI1, DEF1, and ACX1 or acts upstream of these genes (Figures 5D and 5E).
Figure 5.
Tomato JA and Wound Response Genes Are Required for Botrytis Resistance.
(A) Botrytis disease symptoms on leaves of indicated genotypes.
(B) Disease lesion size.
(C) Fungal growth.
(D) Botrytis-induced TPK1b gene expression.
(E) Wound-induced TPK1b gene expression in the tomato JA and/or wound response mutants.
The pictures in (A) and lesion sizes in (B) are from 3 DAI with Botrytis (3 × 105 spores/mL). Fungal growth in (C) is based on the accumulation of Botrytis ActinA RNA as detected by RNA gel blot hybridization. In (C), total RNA (15 μg) was loaded per lane. In (D) and (E), RT-PCR was performed as described in Methods. The tomato translation initiation factor (eIF4A) gene was amplified in parallel to demonstrate relative quality and quantity of the cDNA. Experiments were repeated at least three times with similar results. h, h after inoculation.
TPK1b Mediates Tomato Responses to ET
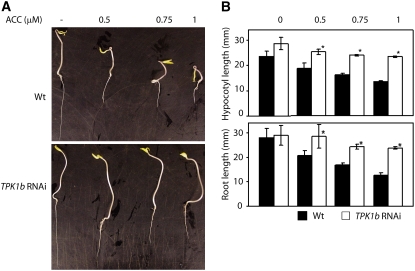
To identify possible links between the function of TPK1b and plant hormones mediating defense and stress responses, we tested TPK1b RNAi plants for hormone sensitivity of seed germination or seedling growth. The TPK1b RNAi plants showed wild-type responses to abscisic acid and MeJA (see Supplemental Figure 4 online). However, TPK1b RNAi seedlings exhibit altered ET responses as measured by the absence of the triple response (Figure 6A). Wild-type tomato seedlings grown in the dark in the presence of ACC displayed a typical triple response, consisting of shortened hypocotyls, reduced root length, and enhanced curvature of the apical hook compared with those grown without ACC (Figures 6A and 6B) (Bleecker et al., 1988). However, in the presence of increasing concentrations of ACC in the dark, TPK1b RNAi seedlings retained elongated hypocotyls with no enhancement of the apical hook and no significant reduction in root length, all features of seedlings not responding to ET (Figures 6A and 6B). TPK1b RNAi seedlings grown in the dark without ACC have longer hypocotyls with no apical hook and longer roots consistent with the loss of ET response (Klee, 2004). Thus, tomato TPK1b has a regulatory function in ET responses of tomato plants.
Figure 6.
ET Responses of TPK1b RNAi and Wild-Type Tomato Seedlings.
(A) The triple response in the wild type but its absence in TPK1b RNAi seedlings grown in the dark in the presence of increasing concentrations of ACC.
(B) Hypocotyl (top) and root (bottom) length of wild-type (black) and TPK1b RNAi (white) seedlings grown in the dark at varying concentrations of ACC.
The pictures in (A) and the measurements in (B) were taken at 6 d after plating. Pictures are representative of seedlings of the corresponding treatments. These experiments were repeated at least five times. The asterisk indicates that the data are statistically significant from the corresponding wild-type control (P = 0.05).
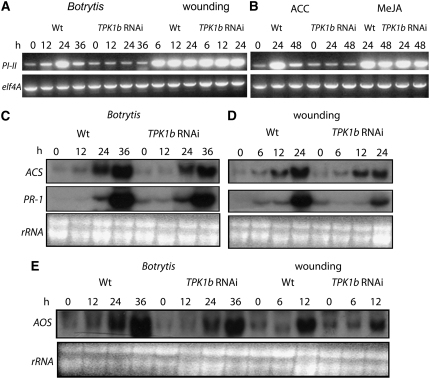
TPK1b Regulates ET and Botrytis-Induced Defense Gene Expression
To determine the molecular basis of TPK1b function, we studied the expression of tomato defense-related genes. Uninfected wild-type plants show low constitutive expression of PI-II but increased expression following Botrytis infection (Figure 7A). In TPK1b RNAi plants, the Botrytis-induced expression of PI-II gene was abolished. After wounding, the expression of PI-II increased starting at 6 h after wounding with comparable patterns of expression in both wild-type and TPK1b RNAi plants. Also, PI-II was induced by MeJA in wild-type and TPK1b RNAi plants to roughly similar levels (Figure 7B). Thus, TPK1b RNAi plants respond normally to JA and wounding, further supporting the hypothesis that TPK1b functions independent of JA and wound response pathways. By contrast, PI-II gene expression was induced by ACC in wild-type plants, but its induction by ACC was abolished in TPK1b RNAi plants, confirming our observation on the altered responses of TPK1b RNAi seedlings to ACC (Figure 7B). The Botrytis and wound-induced expression of ACC synthase showed similar profiles in TPK1b RNAi compared with wild-type plants (Figures 7C and 7D, top rows). Thus, the increased susceptibility of TPK1b RNAi plants to Botrytis and tobacco hornworm correlates with reduced PI-II expression after exogenous ACC application and Botrytis infection.
Figure 7.
Expression of Tomato Defense-Related Genes in Wild-Type and TPK1b RNAi Tomato Plants.
(A) and (B) RT-PCR showing the expression of tomato PI-II during Botrytis infection or mechanical wounding (A) and ACC or MeJA treatment (B).
(C) and (D) RNA gel blot showing the expression of tomato ACC synthase and PR-1 during Botrytis infection (C) and mechanical wounding (D).
(E) Expression of allene oxide synthase2 during Botrytis infection or mechanical wounding.
In (C) to (E), total RNA (15 μg) was loaded per lane, and rRNA is shown as a loading control. Experiments were repeated at least three times with similar results. h, h after Botrytis inoculation or wounding treatment; ACS, ACC synthase; AOS2, allene oxide synthase2.
Basal and induced expression of tomato PR-1, a salicylic acid (SA)–regulated defense gene, was not altered in response to Botrytis in TPK1b RNAi plants but showed a slight reduction of expression in response to wounding (Figures 7C and 7D, middle rows). These results indicate that TPK1b does not mediate SA responses and that TPK1b-mediated Botrytis responses have little effect on Botrytis-induced PR-1 gene expression. However, TPK1b is required for full wound-induced PR-1 expression. Allene oxide synthase 2 (Howe et al., 2000), a late JA-induced gene in tomato, was also induced by Botrytis and wounding independent of TPK1b (Figure 7E).
TPK1b Rescues the Phenotype of the Arabidopsis bik1 Mutant
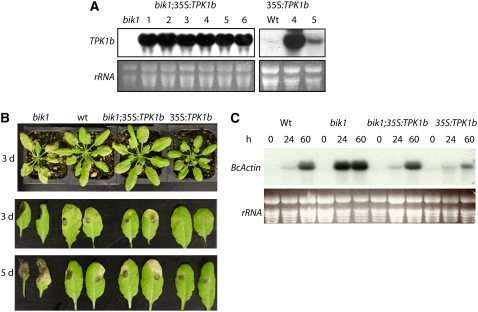
TPK1b and BIK1 are not orthologs, but both are positive regulators of Botrytis resistance and share similarities in sequence, structure, and patterns of pathogen-induced gene expression. To determine whether TPK1b can functionally substitute for BIK1, we generated Arabidopsis bik1 plants expressing 35S:TPK1b (Figure 8A). We also expressed the TPK1b gene in the wild-type plants. Arabidopsis bik1;35S:TPK1b #1 and #2 and BIK1:35S:TPK1b (35S:TPK1b) line #4 were chosen for further experiments due to their high levels of TPK1b gene expression. After spray inoculation with Botrytis, the bik1 mutant was highly susceptible (Veronese et al., 2006), but bik1;35S:TPK1b plants showed levels of resistance comparable to the wild type (Figure 8B). Additionally, the 35S:TPK1b (line #4) plants were more resistant to Botrytis than the wild-type plants, showing restricted disease symptoms (Figure 8B, top panel). After drop inoculation with Botrytis, disease lesions on bik1 expanded and completely macerated the entire leaf at 5 DAI (Figure 8B, bottom panels). The 35S:TPK1b transgenic plants were resistant with smaller disease lesions and with no significant lesion growth between 3 and 5 DAI. Botrytis growth was significantly lower in the 35S:TPK1b plants, further confirming the increased Botrytis resistance (Figure 8C). However, the increased resistance of 35S:TPK1b relative to the bik1;35S:TPK1b plants could result from higher levels of TPK1b expression in the former (Figures 8A and 8B).
Figure 8.
Ectopic Expression of Tomato TPK1b Suppresses the Arabidopsis bik1 Mutant Phenotypes and Confers Increased Resistance to Botrytis.
(A) RNA blot showing expression of 35S:TPK1b in bik1 and wild-type plants.
(B) Botrytis resistance 3 d after spray-inoculation (top panel) and 3 and 5 d after drop inoculation (bottom panels).
(C) Accumulation of the Botrytis ActinA mRNA in spray-inoculated plants.
In (A) and (C), total RNA (15 μg) was loaded per lane, and rRNA is shown as a loading control. Experiments were repeated at least three times with similar results. h, h after inoculation; d, days after inoculation.
Similarly, the bik1;35S:TPK1b plants rescued the A. brassicicola susceptibility of the bik1 mutant to the wild-type level of resistance (see Supplemental Figure 5 online). As expected, the bik1 plants were susceptible to A. brassicicola, with larger disease lesions and increased pathogen reproduction. The 35S:TPK1b transgenic plants show very limited disease symptoms with smaller disease lesions and lower spore counts at 3 and 6 DAI (see Supplemental Figure 5 online). In addition, the shorter primary root length observed in the bik1 mutant was restored to the wild-type level by 35S:TPK1b (see Supplemental Figure 6 online). Thus, ectopic expression of TPK1b rescued the bik1 phenotypes, and TPK1b was sufficient to confer increased resistance to two necrotrophic fungi. These data indicate that TPK1b and Arabidopsis BIK1 may perform similar functions in their respective species.
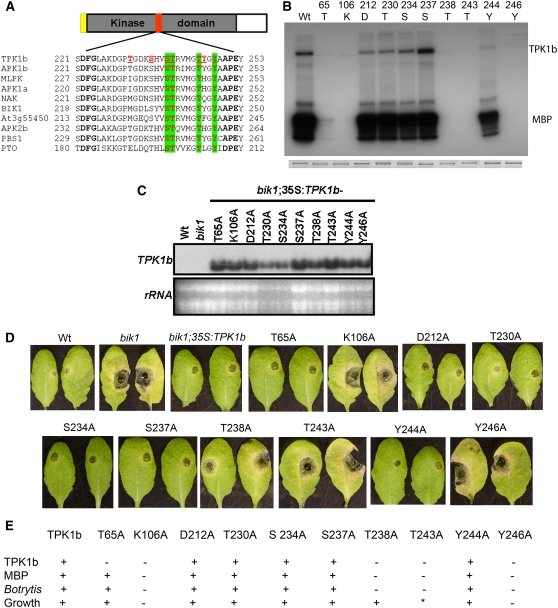
TPK1b Is a Functional Protein Kinase, and Specific Activation Domain Residues Are Important for Kinase Activity and Biological Function
TPK1b is a predicted protein with a large catalytic KD. TPK1b and closely related kinases share particularly high sequence identities in the activation domain with many highly conserved phosphorylatable residues (Figure 9A). Four out of the seven phosphorylatable residues are invariant in RLCKs related to TPK1b. To determine whether TPK1b is a functional protein kinase, we expressed TPK1b as a glutathione S-transferase (GST) fusion protein in Escherichia coli and affinity purified the fusion protein using glutathione resins. The purified protein was incubated in a reaction buffer containing (γ-32P) with or without Myelin Basic Protein (MBP), an artificial kinase substrate, separated by SDS-PAGE, and autoradiographed. The recombinant TPK1b wild-type protein showed autophosphorylation that was visible as a band of ∼70 kD corresponding to the combined molecular mass of TPK1b and GST and also phosphorylated MBP, demonstrating that TPK1b is a functional kinase (Figure 9B). GST alone did not show any kinase activity (see Supplemental Figure 7 online).
Figure 9.
TPK1b Is a Functional Protein Kinase That Localizes to the Plasma Membrane.
(A) Alignment of the activation segment of TPK1b and related kinases.
(B) Autophosphorylation and MBP phosphorylation activities of GST-TPK1b and its mutant derivatives in vitro. Top panel, autoradiogram of the gel; bottom panel, Coomassie blue staining.
(C) RNA blot showing transgenic expression of TPK1b substitution mutants in the Arabidopsis bik1 mutant.
(D) Responses of bik1 plants expressing TPK1b and its substitution mutants to A. brassicicola inoculation.
(E) Summary of kinase activity, Botrytis response, and growth-related phenotypes of the Arabidopsis bik1 expressing TPK1b wild type and substitution mutants.
In (A), TPK1b activation segment phosphorylatable residues are in red and underlined. Residues that are phosphorylatable and conserved in TPK1b-related kinases are shaded green. In (D), A. brassicicola disease symptoms are from 4 DAI. In (E), the asterisk indicates partial complementation; +, rescues phenotypes or shows auto (TPK1b) or MBP phosphorylation activity; −, fails to rescue the bik1 growth phenotype or shows no TPK1b or MBP phosphorylation activity.
We also purified TPK1b proteins substituted at selected residues to determine sites that regulate kinase activity and gain insight into how TPK1b functions. A total of 10 TPK1b residues were mutated, and the recombinant proteins were purified. Seven phosphorylatable residues, Ser, Thr, and Tyr, within the activation segment of TPK1b and three residues in the N-terminal region of the KD were individually substituted to Ala and assayed for kinase activity (Figure 9B). Coomassie blue staining after electrophoresis shows that the TPK1b recombinant proteins with substitutions were expressed at comparable levels (Figure 9B, bottom panel). However, the various mutants affected autophosphorylation and/or phosphorylation of MBP (Figure 9B). The TPK1bK106A mutation outside the activation domain eliminated kinase activity. The Lys residue at 106 is invariant in all kinases (Hanks and Hunter, 1995). The TPK1bT65A mutation completely eliminated autophosphorylation and significantly reduced phosphorylation of MBP. In the activation segment, mutation of three residues, TPK1bT238, TPK1bT243, and TPK1bY246, abolished autophosphorylation and phosphorylation of MBP. Thus, consistent with other kinases, three activation segment residues are required for TPK1b kinase activity (Rathjen et al., 1999; Wang et al., 2005). Kinase activity is frequently controlled by the activation domain, defined by the conserved amino acids D221FG and APE252 in TPK1b (Johnson et al., 1996). In many RD-type RLKs, autophosphorylation of the activation loop is required for general kinase activity. Consistent with this notion, three TPK1b activation segment residues required for autophosphorylation also eliminated phospohrylation of MPB. By contrast, autophosphorylation of N- and C-terminal residues in the KD generates docking sites for specific downstream substrates and affects catalytic activity toward those substrates (Wang et al., 2005).
We took advantage of suppression of Arabidopsis bik1 disease susceptibility by 35S:TPK1b to determine the biological function of TPK1b substitution mutants in vivo. The site-directed TPK1b mutants above were cloned into a plant expression vector and transformed into Arabidopsis bik1. The expression of all TPK1b constructs were verified by RNA gel blots (Figure 9C). The untransformed wild-type, bik1, bik1;35S:TPK1b, and the 10 TPK1b substitution lines were compared for disease resistance and changes in growth related traits (Figures 9D and 9E). In the case of the TPK1bK106A mutant, no rescue of the bik1 disease resistance or growth-related phenotypes were observed. The three mutations in the activation loop (TPK1bT238A, TPK1bT243A, and TPK1bY246A) that eliminated kinase activity of TPK1b failed to rescue bik1 susceptibility to A. brassicicola and Botrytis, indicating these residues are essential for TPK1b signaling function in planta (Figures 9D and 9E). Individual TPK1b mutants, TPK1bD212A, TPK1bT230A, TPK1bS234A, TPK1bS237A, and TPK1bY244A, had no effect on kinase activity and also suppressed the disease phenotype of bik1. Interestingly, the TPK1bY243A mutations that failed to rescue the disease phenotype partially rescued the bik1 growth defects. Expression of these constructs resulted in plants that are intermediate between the wild-type and bik1 plants (Figure 9E; see Supplemental Figure 8 online). The T238A substitution fully restored the growth phenotype of bik1, although it was fully susceptible to disease. These data demonstrate that some residues are essential for disease resistance but are dispensable for growth and that the two functions of TPK1b can be partially uncoupled. Consistent with the recent characterization of Arabidopsis BIR1 (Wang et al., 2005), the correlation between the in vitro kinase data and the biological function of TPK1b suggests that in vitro kinase activity is a good indicator for in vivo kinase activity and function. Interestingly, TPK1bT65A abolished kinase activity but suppressed the disease susceptibility of the bik1 mutant. The residues in the activation loop that are required for TPK1b kinase activity and disease resistance are also invariant in all RLCKs related to TPK1b, possibly indicative of a common regulatory mechanism among these RLCKs (Figure 9A). The TPK1bY246 substitution falls in the region of the activation segment referred to as the P+1 loop, which has been implicated in recognition and binding of protein substrates (Johnson et al., 1996).
The N-Myristoylation Motif Is Required for Disease Resistance Function of TPK1b but Not for Its Association with the Plasma Membrane
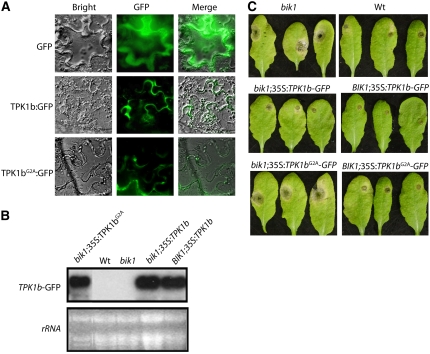
To determine the subcellular localization of TPK1b, the full-length TPK1b was translationally fused with green fluorescent protein (GFP), and the chimeric protein was transiently expressed in Nicotiana benthamiana leaf tissues through Agrobacterium infiltration. The TPK1b-GFP fusion protein was observed at the plasma membrane, whereas cells expressing GFP alone (control, top row) exhibited signal around the nucleus and in the cytosol (Figure 10A). We also generated TPK1bG2A constructs where the invariant penultimate Gly residue that is essential for myristoylation was substituted with Ala. The TPK1bG2A-GFP localized to the plasma membrane similar to the wild-type TPK1b-GFP (Figure 10A, bottom), indicating that TPK1b membrane localization is independent of the N-terminal myristoylation motif. N-terminal myristoylation motifs have been implicated in membrane localization, stabilization of protein structure, and protein–protein interactions (Johnson et al., 1994). The N-terminal myristoylation motif is also found in all of the TPK1b-related RLCKs (Kakita et al., 2007; see Supplemental Figure 9 online). BIK1 and MLPK localize to the plasma membrane (Murase et al., 2004; Veronese et al., 2006).
Figure 10.
The N-Myristoylation Motif Is Not Required for TPK1b Subcellular Localization but Is Required for Its Disease Resistance Function.
(A) Subcellular localization of the GFP control, TPK1b-GFP, and TPK1bG2A-GFP fusion proteins in N. benthamiana cells.
(B) RNA blot showing transgenic expression of TPK1b-GFP and TPK1bG2A-GFP.
(C) Resistance of Arabidopsis plants expressing TPK1b and TPK1bG2A to A. brassicicola.
The experiments in (A) and (C) were repeated at least three times with similar results.
To determine whether the N-myristoylation signal in TPK1b is required for TPK1b disease resistance function, we transformed 35S:TPK1bG2A-GFP constructs into bik1. All of the constructs were expressed based on RNA blot analysis (Figure 10B). However, the expression of 35S:TPK1bG2A-GFP failed to complement the bik1 disease susceptibility (Figure 10C, bottom), although 35S:TPK1b-GFP rescued the A. brassicicola susceptibility of bik1 to wild-type levels. 35S:TPK1bG2A-GFP also failed to confer increased resistance to A. brassicicola when expressed in the wild-type plants, as expected for 35S:TPK1b (Figure 10C, bottom). Thus, TPK1b N-myristoylation signal is required for the disease resistance function but not for localization to the plasma membrane.
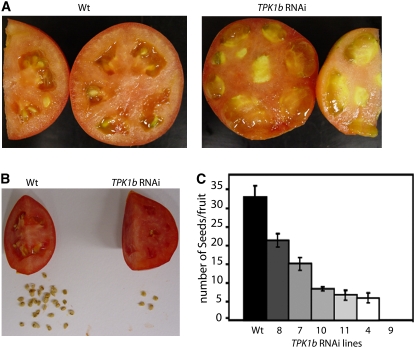
TPK1b Is Required for Normal Fruit Structure and Seed Set
Tomato TPK1b RNAi plants show normal vegetative growth under our growth conditions (see Methods). However, we observed altered fruit structure and reduced number of seeds when we examined six TPK1b RNAi transgenic lines with varying levels of TPK1b expression (Figures 3A and 11A to 11C). The TPK1b RNAi lines produced reduced numbers of seeds in a ratio that correlated with the reduced levels of TPK1b gene expression (Figure 11C). The RNAi line #9 has the greatest reduction in TPK1b expression, and its fruits were parthenocarpic, containing no developed seeds, contained almost no jelly, and were completely filled with flesh compared with CastlemartII wild-type fruits (Figure 11A). TPK1b RNAi line #4 also showed a significant reduction in number of seeds/fruit, which correlated with its level of TPK1b gene expression (Figures 11B and 11C). Thus, TPK1b is required for normal fruit structure and seed set in tomato, consistent with the role of ET in seed and fruit development (Gillaspy et al., 1993).
Figure 11.
TPK1b Is Required for Normal Fruit Structure and Seed Set.
(A) TPK1b RNAi plants show defects in fruit structure.
(B) TPK1b RNAi plants produce fewer seeds.
(C) A comparison of number of seeds/fruit between the wild type and TPK1b RNAi.
Representative fruit tissue and seeds from wild-type and TPK1b RNAi plants are shown. In (B), the picture shows part of the fruit and the entire amount of seed recovered from the whole fruit of the corresponding genotype. At least 10 fruits were examined per plant.
DISCUSSION
This study presents data on the function of TPK1b in defense against Botrytis and the herbivorous insect tobacco hornworm. Reduction of TPK1b gene expression resulted in increased susceptibility to Botrytis. Ectopic expression of TPK1b in Arabidopsis conferred increased resistance to Botrytis and A. brassicicola. These data, coupled with an increased susceptibility of TPK1b RNAi plants to insect feeding, suggest a clear physiological role for TPK1b in resistance against both necrotrophic fungi and insect pests. Interestingly, TPK1b is also required for ET responses of tomato, suggesting that TPK1b regulates ET-dependent defense. The impaired disease and insect resistance in TPK1b RNAi plants is accompanied by altered ET- and Botrytis-induced defense gene expression, further supporting the role of TPK1b in ET-mediated defenses. ET integrates plant responses to developmental and environmental signals, including responses to pathogens (Klee, 2004). Our data on the role of TPK1b in ET responses and insect resistance corroborate the function of ET as an important regulator of insect and pathogen defense in tomato. TPK1b is a functional kinase that localizes to the plasma membrane, suggesting that TPK1b acts early in pathogen and insect response pathway and functions in recognition and/or signaling. Our biochemical and genetic data demonstrate an important role for specific TPK1b KD residues for in planta signaling function. We also present data supporting the function of TPK1b in insect and pathogen defense independent of genes regulating JA biosynthesis and response, which are also required for resistance to Botrytis. Thus, plants deploy multiple and overlapping mechanisms of defense against necrotrophic pathogens and chewing insects, despite the disparate strategies of these organisms in deriving nutrition from plants.
Multiple lines of evidence suggest that responses to insects and necrotrophic pathogens are mechanistically linked in tomato (McCormick, 1991; Ryan, 1992). First, Botrytis induces wound-like responses, including the expression of the wound response gene PI-II, possibly through the actions of OGs released from plant cell walls by the action of fungal endopolygalacturonases (Bishop et al., 1981; Prins et al., 2000). Second, the susceptibility of JA and wound response mutants to Botrytis presented in this article and their impaired resistance to tobacco hornworm (M. sexta) or spider mites (Tetranychus urticae) (Schilmiller and Howe, 2005) demonstrate a clear overlap in the mechanisms of resistance to necrotrophic fungi and chewing insect pests. These overlapping responses are mediated in part by JA levels and signaling (Howe and Jander, 2007). JAI1, SPR2, ACX1, and DEF1 are required for basal resistance of tomato to Botrytis. The acx1 mutant is JA deficient and lacks local and systemic expression of defensive PI genes in response to wounding due to a defect in the first step of β-oxidation in the octadecanoid pathway (Li et al., 2005). SPR2 is a fatty acid desaturase required for the synthesis of JA and the generation of a systemic wound signal mediating defense gene expression in tomato (Li et al., 2003). The def1 and jai1 mutants are impaired in JA responses (Li et al., 2002). JAI1 is the homolog of Arabidopsis COI1 (Li et al., 2004); the coi1 mutant also shows susceptibility to necrotrophic pathogens (Thomma et al., 1998; van Wees et al., 2003). Previous reports on Botrytis responses of def1 were contradictory, with both increased susceptibility and wild-type resistance observed (Audenaert et al., 2002; Diaz et al., 2002). Our data confirm the susceptibility of def1 based on enhanced disease symptoms and fungal growth. Thus, these genes in the JA pathway are part of the defense against Botrytis, and our findings confirm that the Botrytis resistance mechanisms regulated by JA are generally conserved between Arabidopsis and tomato. The spr1 mutation abolishes JA accumulation in response to exogenous systemin and reduces JA accumulation in wounded leaves (Lee and Howe, 2003). However, the spr1 mutant mounts wild-type resistance to Botrytis. SPR1 is an upstream regulator of JA synthesis and systemin signaling (Schilmiller and Howe, 2005), and our data support the hypothesis that an additional factor upstream of JA acts to specify responses to Botrytis. Consistent with this, TPK1b is expressed normally in jai1, spr1, spr2, acx1, and def1 mutants, which demonstrates that TPK1b functions independent of or upstream of the JA and wound response pathways to specify responses to Botrytis. This is consistent with previous observations that JA and wound signaling act independent of ET in resistance to Botrytis in tomato (Diaz et al., 2002). However, the same authors using tomato plants that have increased or decreased prosystemin expression observed that systemin is important for resistance to Botrytis. This appears to contradict our observation in the spr1 mutant that shows normal resistance to Botrytis but is impaired in systemin responses.
The functions of a large number of RLCKs in Arabidopsis and other species have not been determined. In the Arabidopsis genome, there are 46 genes encoding proteins that belong to the RLCK VII subfamily of plant RLCKs that share overall structural and sequence similarity to TPK1b. The TPK1b kinase domain is most closely related to the Arabidopsis APK1b, Brassica MLPK, and the grapevine predicted protein CAO21648 based on sequence and phylogenetic analysis. Arabidopsis APK1a and APK1b are 84% identical and exhibit kinase activities in vitro (Hirayama and Oka, 1992). MLPK shares 76% sequence identity with APK1b and is involved in Brassica self-incompatibility (Murase et al., 2004). The T-DNA insertion alleles of APK1b show no fertility or self-incompatibility related phenotypes (Kakita et al., 2007). Whether MLPK and APK1b perform defense functions is not yet known. Interestingly, ectopic expression of TPK1b rescues the phenotype of the Arabidopsis bik1 mutant, indicating that the two kinases perform similar functions in tomato and Arabidopsis: both TPK1b and Arabidopsis BIK1 are positive regulators of resistance to Botrytis. However, the TPK1b RNAi plants show wild-type responses to P. syringae in contrast with the Arabidopsis bik1 mutant, which shows increased resistance. In addition, the role of the two proteins in plant growth and development appears to be distinct. Although TPK1b affects fruit development and seed set, it has no apparent effect on root growth and leaf shape. The Arabidopsis bik1 mutation has significant effect on leaf shape, primary root, and root hair growth (Veronese et al., 2006). These data further corroborate that TPK1b and BIK1 perform similar yet distinct roles in plant defense and other functions. It will also be interesting to determine the function of Arabidopsis RLCKs that are in the TPK1b clade and tomato BIK1 to establish the extent of functional redundancy. Mutations in Arabidopsis BIK1, Brassica MLPK, and TPK1b all result in defects in plant fertility, further suggesting that members of this protein family perform similar functions. In tomato, diverse processes control parthenocarpic fruit development (Johnson et al., 1996). The molecular mechanisms underlying TPK1b-mediated seed set need further investigation but may relate to the function of ET in fruit development or the function of TPK1b in self-fertility similar to the function of MLPK (Murase et al., 2004). One intriguing possibility is mechanisms underlying pathogen recognition and response and pollen recognition leading to self-incompatibility may share similar molecular mechanisms. In both interactions, mechanisms of self- and non-self recognition play an important role. Indeed, recently, R proteins were implicated in hybrid-induced necrosis resulting in self-incompatibility (Bomblies et al., 2007).
Based on the altered gene expression in response to ACC and impaired triple response in TPK1b RNAi seedlings, TPK1b plays a role in ET signaling. The susceptibility of TPK1b RNAi plants to tobacco hornworm coupled with the lack of ET responses suggest that TPK1b-mediated ET responses are required for defense against pathogens and insect pests. The attenuated PI-II gene expression in TPK1b RNAi plants is also consistent with the action of ET in parallel with the octadecanoid/wounding pathway for full PI-II gene expression (O'Donnell et al., 1996). Mutations causing ET insensitivity in soybean (Glycine max) and Arabidopsis and inhibition of ET perception in tomato result in increased susceptibility to necrotrophic pathogens (Hoffman et al., 1999; Thomma et al., 1999; Diaz et al., 2002). ET also functions as a negative regulator of defense against some pathogens (Lund et al., 1998; Ciardi et al., 2000), indicating that its role varies among different pathosystems.
Insect herbivory is known to promote ET emission (Lund et al., 1998; Ciardi et al., 2000), but ET has been implicated as both a negative and positive factor in insect–plant interactions, similar to its role in pathogen defense. In maize (Zea mays), ET is required for defense against insect herbivory (Harfouche et al., 2006). The bacterial elicitor harpin induces EIN2-dependent resistance to green peach aphid (Myzus persicae) in Arabidopsis (Dong et al., 2004). On the other hand, the Arabidopsis ET response mutants hookless1 and ein2 were more resistant to the generalist insect Egyptian cotton worm (Spodoptera littorali) than wild-type plants, suggesting a negative effect of ET signaling on insect resistance (Bodenhausen and Reymond, 2007). The same mutants show no effect on the specialist herbivore diamondback moth (Plutella xylostella) (Stotz et al., 2000). ET suppresses defense against M. sexta by lowering the putresine amino transferase transcripts in the root and nicotine levels in the shoot in Nicotiana attenuate (Winz and Baldwin, 2001). ET signaling regulates insect oral secretion, which in turn induces nicotine levels and floral longevity, two traits implicated in interaction between N. attenuate and M. sexta (von Dahl et al., 2007). Our data provide evidence for the role of TPK1b in ET signaling and insect resistance in tomato and differ from some of the previous observations, possibly because the role of ET in different plants may be different, involving diverse traits, some of which contribute directly or indirectly.
TPK1b has two major regions, a conserved catalytic KD and a nonconserved flanking region, that are important for the biochemical activities and biological function of TPK1b. The activation segment of the KD controls kinase activity (Johnson et al., 1996), and phosphorylation of one to three residues in the activation loop of KDs is often required for ligand-induced kinase activation (Wang et al., 2005). In the activation segment of TPK1b, three residues (T238, T243, and Y246) are important for in vitro kinase activity and for disease response signaling in planta. These activation segment residues are highly conserved in TPK1b and related RLCKs and are all located in the C-terminal subregion of the activation segment. How TPK1b gets activated is not yet known but may follow the same general trend observed for other RD-type RLKs, in which the activation segments play important roles for general kinase activity and biological function (Sessa et al., 2000; Shah et al., 2001; Guo et al., 2004; Yoshida and Parniske, 2005). The activation segment occupies the catalytic cleft of protein kinases and is composed of smaller regulatory elements, including the T-loop, where activating phosphorylation events often occur, and the C-terminal P+1 loop, which plays a role in recognition and binding of protein substrates (Johnson et al., 1996). Comparisons with other protein kinases show that TPK1b Y246 falls in the P+1 loop of the activation segment. The C-terminal region of the activation domain is highly conserved, suggesting that the tertiary structure of TPK1b is important and may be required for interaction with substrates. The TPK1b residues T238 and T243, located in the T-loop region, are critical for TPK1b function most likely due to their role in general kinase activity, consistent with the function of homologous regions of other kinases. In the activation segment of tomato PTO kinase, S198 (equivalent to TPK1bS237) is required for elicitation of HR (Sessa et al., 2000), T204 (TPK1bT243) is required for recognition specificity (Rathjen et al., 1999), Y205 (TPK1bY244) plays a subsidiary role in recognition (Frederick et al., 1998), and Y207 (TPK1bY246) influences binding properties (Rathjen et al., 1999). BAK1 was autophosphorylated in vitro on residues equivalent to TPK1bT230A and TPK1bS234A. Interestingly, BAK1 was recently implicated as a central player in plant immunity and resistance to Botrytis (Chinchilla et al., 2007; Heese et al., 2007; Kemmerling et al., 2007). These observations indicate that different activation segment residues play different roles among various RLKs. RLKs are predicted to share some common regulatory mechanisms but also have specific mechanisms that control certain physiological processes. Autophosphorylation of less-conserved N-terminal and C-terminal KD regions may help create docking sites for kinase substrates to achieve specificity. Outside the KD, there is significantly less sequence conservation between TPK1b and related RLCKs.
Outside the activation segment, the single amino acid mutation TPK1bT65A was dispensable for disease resistance function, although it eliminated autophosphorylation and severely reduced phosphorylation of MBP. The equivalent residue in PTO is the main autophosphorylation site and is required for HR (Sessa et al., 2000). Currently, we have no plausible explanation for the apparent lack of correlation between the kinase activity and disease resistance function of TPK1b Thr-65. In contrast with TPK1b Thr-65, Lys-106 is important for TPK1b phosphorylation activities and biological function. This residue is the ATP binding site and is frequently mutated to generate kinase dead mutants, suggesting that it may be important for maintaining the tertiary structure of the kinase molecules and thus influence interaction with other proteins (Hanks and Hunter, 1995). Three residues in the TPK1b activation segment are required for autophosphorylation. Some of these residues are likely to be TPK1b autophosphorylation sites. Protein kinases that require autophosphorylation of the activation segment for kinase activity are of the RD type, consistent with TPK1b being an RD-type protein kinase. In sum, specific TPK1b residues that affect tertiary structure and phosphorylation sites are required for function by interfering with kinase activation and interaction with other proteins.
Substitution of the penultimate Gly, a residue essential for myristylation in other proteins, did not affect TPK1b localization but affected its disease resistance function. Consistent with this, the myristylation motif was required for Fen kinase-mediated fenthion sensitivity (Rommens et al., 1995). Myristylation was also required for salt tolerance function of SOS3 but not for its membrane association (Ishitani et al., 2000). It is possible that TPK1b is not myristylated but TPK1b N-terminal conserved sequence affects TPK1b function through an unknown mechanism. Thus, the penultimate Gly residue in TPK1b is required for disease resistance possibly through a myristylation-independent mechanism, such as conferring structural stability or protein–protein interactions.
TPK1b functions early in pathogen and insect response pathways. Future research should focus on the regulation of in vivo kinase activity of TPK1b and its impact on disease resistance and early events in defense against insects and pathogens.
METHODS
Plant Growth
Tomato (Solanum lycopersicum) cultivars CastlemartII and Micro-Tom were grown in plastic pots containing compost soil mix in a greenhouse with a photoperiod extended to 15 h under fluorescent lights (160 W mol−1 m−2 s−1) at a temperature of 24 ± 4°C. The tomato mutants spr1, spr2, def1, acx1, and jai1 were from Gregg Howe (Michigan State University). All plants were fertilized twice weekly.
Fungal Culture and Disease Assays
The Botrytis cinerea strain BO5-10 and Alternaria brassicicola strain MUCL 20297 were used for disease assays. Fungal culture and preparation of conidial spore suspension were as described previously (AbuQamar et al., 2006). Botrytis disease assays were done on whole plants or detached leaves by spray or drop inoculation of a conidial suspension on Arabidopsis thaliana (2.5 × 105 spores/mL) and tomato (5 × 105 spores/mL) unless stated otherwise in the Results.
Bacterial Disease Assay
Bacterial disease assays were done essentially as described (Mengiste et al., 2003). Leaves of 6-week-old tomato plants were infiltrated with suspensions (OD600 = 0.001 in 10 mM MgCl2) of the bacterial strain Pseudomonas syringae pv tomato DC3000 (Generous gift of Greg Martin, Cornell University). To determine bacterial growth, leaf discs from infected leaves were collected at 0, 2, and 4 DAI. Each experiment for bacterial growth assay was performed in three replicates. At each time point, two leaf discs were collected from wild-type and TPK1b RNAi plants for each replicate. Leaf discs of the same size were made using a hole puncher, and bacterial titer per leaf area was determined.
Insect Feeding Trials
Eggs of tobacco hornworm (Manduca sexta) and an artificial diet for the larvae were purchased from Carolina Biological Supply Company. Eggs were hatched by incubation at 26°C as recommended by the supplier. Hatched larvae were kept on the artificial diet for 3 d before transfer to whole tomato plants or detached leaves. The average larval weight at the beginning of the feeding trial was ∼9 to 11 mg.
Generating Constructs for Expression in Tomato and Arabidopsis Transformation
To generate 35S:TPK1b constructs, full-length cDNA was amplified by PCR from the RACE-Ready cDNA with primers TPK1b/LP (5′-TCCCCGCGGCTAATGGGGATATGTTTGAGTG-3′, the SacII site is underlined) and TPK1b/RP (5′-CGCGGATCCTAATTATTTAGCGTAAGGGGGAG-3′, the BamHI site is underlined). Restriction-digested PCR products were cloned into a modified version of the binary vector pCAMBIA1200 behind the cauliflower mosaic virus 35S promoters between the SacII and BamHI sites. Agrobacterium tumefaciens strain GV3101 was used for transformation. Hygromycin-resistant homozygous plants were identified from progenies of primary transformants, and lines that express TPK1b cDNA were identified. Transgenic tomato lines were selected on hygromycin and homozygous transgenic lines identified from selfed progenies of the subsequent generations and used for further experiments.
To generate a TPK1b RNAi construct, 250 bp from the 3′ end (162 bp) and the 3′ untranslated region (88 bp) of TPK1b were amplified by PCR from the RACE-Ready cDNA with primers TPK1bRNAi-LP (5′-GCACTAGTCCATGGGAAATGGAGCAACTTTAT-3′, the SpeI and NcoI sites are underlined) and TPK1bRNAi-RP (5′-CGGGATCCGGCGCGCCATCAGCCTCAACTACTTAT-3′, BamHI and AscI sites are underlined). The inverted repeat is assembled directly in the binary vector by a two-step cloning process using the introduced restriction enzyme sites. In the first cloning step, the PCR product is cleaved at the inner restriction sites, NcoI and AscI, and ligated to NcoI and AscI sites in pGSA1165 (http://www.chromdb.org/info/plasmids/pGSA1165.html). For the second cloning step, the plasmid resulting from the first cloning step serves as a template for a second amplification using the original set of primers. The resulting PCR product is cleaved with BamHI and SpeI and inserted into the BamHI- and SpeI-cleaved template plasmid. This second ligation inserts the PCR product in inverted orientation with respect to first cloned fragment, yielding an inverted repeat separated by the β-glucuronidase fragment.
Plant Transformation and Regeneration
In Arabidopsis, Agrobacterium strain GV3101 was used for transformation through the floral dip method (Clough and Bent, 1998). Hygromycin-resistant homozygous T2 plants were identified from progenies of primary transformants and selfed to produce the T3 homozygous lines.
Tomato transformation was done as described with some modification (Howe et al., 1996). For tomato transformation, S. lycopersicum cv CastlemartII and Micro-Tom were used for RNAi and overexpression experiments, respectively. Seeds were surface-sterilized by immersion for 2 min in 70% ethanol and 30 min in 35% commercial bleach solution (5.25% [w/v] sodium hypoclorite) plus 0.1% Tween 20. Treated seeds were then rinsed five to six times with sterile deionized-distilled water, and 12 seeds per Magenta box were placed on germination medium (GM) (4.3 g L−1 Murashige and Skoog [MS] salts, 30 g L−1 sucrose, 1 mL L−1 Nitsch and Nitsch's Vitamin mix, and 0.8% agar). Seedlings were grown at 25°C and 70% relative humidity for 10 d under fluorescent light (90 μmol m−2 s−1), 8 h dark and 16 h light. Cotyledon explants of 10- to 14-d-old seedlings were cut off, and one to four holes were made on the cotyledon pieces to increase Agrobacterium transformation efficiency. Cotyledon pieces were placed upside down in 90 × 15-mm Petri dishes containing preculture medium (PM = GM + 3 mg L−1 2,4-D) and incubated for 24 h at 25°C.
A single colony from Agrobacterium strain LBA4404 containing desired construct was grown in YEP media (10 g L−1 yeast extract, 10 g L−1 peptone, and 5 g L−1 NaCl) and 50 mL L−1 rifampicin with appropriate antibiotics (40 mg L−1 chloramphenicol) and 300 μM acetosyringone at 28°C to OD600 = 0.5 to 0.8. The culture was centrifuged at low speed for 5 min and diluted by liquid 2% MSO (4.3 g L−1 MS salts, 100 mg L−1 myo-inositol, 0.4 mg L−1 thiamine-HCl, and 20 g L−1 sucrose). Tomato cotyledon explants were removed from the PM plates and transferred to the bacterial suspension for 30 min and then placed on cocultivation culture medium (PM + 150 μM acetosyringone) for 2 d (24 h in dark, 24 h in light). After 2 d, the infected tissue was transferred to shoot induction and regeneration medium containing 2 mg L−1 zeatin with 300 mg L−1 timentin and 40 mg L−1 hygromycin or 50 mg L−1 kanamycin. Explants were subcultured into fresh medium containing 1 mg L−1 zeatin and 1 mg L−1 GA every 2 weeks. After 2 to 4 weeks, initial calli were transferred (with shoot primodia) into regeneration (GM) including desired antibiotics. The 2- to 4-cm-long excised shoots were transferred from calli to rooting media containing 2 mg L−1 indole butyric acid and desired antibiotics, and 10- to 15-cm plants were transferred to the greenhouse after 4 to 6 weeks.
DNA and RNA Extractions, cDNA Synthesis, and RT-PCR
For DNA blots, genomic DNA was extracted from tomato leaves according to the method described (Dellaporta et al., 1983). Total RNA from tomato and Arabidopsis was extracted from tissues frozen in liquid nitrogen as described (Lagrimini et al., 1987). RNA was separated on 1.2% formaldehyde agarose gels. The gels were then blotted onto Hybond N+ nylon membranes (Amersham Pharmacia Biotech). Probes were labeled with 32P by random priming using a commercial kit (Sigma-Aldrich). Hybridization of probe and subsequent washings were performed as described (Church and Gilbert, 1984).
For RT-PCR, cDNA was synthesized from both control and treated samples using equal amounts of total RNA (2 μg), AMV reverse transcriptase (Promega), and oligo (dT15) primers according to standard protocols. The PCR was performed for 35 cycles using 2.5 μL cDNA as a template and specific primer pairs (94°C 30 s, 52°C 30 s, 72°C 1 min). The tomato translation initiation factor (eIF4A) gene was amplified as a control to demonstrate relative quantity of the cDNA. The amplified products were separated on 1.5% agarose gels and visualized under UV light after staining with ethidium bromide. The following primer pairs were used for RT-PCR: TPK1b forward (5′-ATGGGGATATGTTTGAGTGCTAGAA-3′), reverse (5′-GAACGTGTTCTCGTCGATCCACCCT-3′); PI-II forward (5′-ATGGCTGTTCACAAGGAAGTTAATTTTG-3′), reverse (5′-TCACATTACAGGGTACATATTTGCCTTG-3′); B. cinerea Actin A forward (5′-ACTCATATGTTGGAGATGAAGCGCAA-3′), reverse (5′-AATGTTACCATACAAATCCTTACGGACA-3′); eIF4A forward (5′-CAAATCTTTCAGATCTTTCTTTGAC-3′) and reverse (5′-GTGCAAGCTCACGGGTTGGTGCAAG-3′).
VIGS
The TPK1b silencing was done using the TRV vector system essentially as described (Liu et al., 2002).
Subcellular Localization
For the subcellular localization, the TPK1b coding sequence was translationally fused into an N-terminal GFP fusion vector. TPK1b-LP or TPK1b-G2A-LP coding sequence was amplified using the primers 5′-GGATCCATGGG(C)GATATGTTTGAGTGCTAGA-3′ (C was PCR mutated to replace Gly with Ala) and TPK1b-RP 5′-TCTAGATTTAGCGTAAGGGGGAGAAGCAGA-3′ (the BamHI and XbaI sites are underlined, respectively). The correct amplification was verified by sequencing.
Expression and Purification of Recombinant Proteins and in Vitro Kinase Assay
The open reading frame of TPK1b was cloned into GST fusion protein expression vector pGEX4T-1 (Pharmacia). Expression of GST-fusion protein and affinity purification were performed as described previously (Cardinale et al., 2002). The protein concentration of the recombinant proteins were determined with the Bio-Rad detection system using BSA as a standard. Kinase reactions were performed in 15 μL of kinase buffer (20 mM HEPES, pH 7.4, 10 mM MgCl2, 5 mM EGTA, and 1 mM DTT) containing 5 μg of GST-fusion protein, 5 μg of myelin basic protein, 0.1 mM ATP, and 2 μCi [γ-32P]ATP. The protein kinase reactions were performed at room temperature for 30 min, and the reactions were stopped by adding 4× SDS loading buffer. The phosphorylation of MBP was analyzed by autoradiography after separation on 12.5% SDS-PAGE.
Site-Directed Mutagenesis
Mutagenesis of TPK1b residues was performed in the plasmid pGEX4T, containing GST-TPK1b fusion protein. Site-specific mutations were introduced into TPK1b using the Quickchange kit from Stratagene, according to the manufacturer's protocols. The presence of the desired mutations was confirmed by sequencing.
Phylogenetic Analysis
The KD amino acid sequences were used for building the phylogenetic tree comparing TPK1b and related kinases. Sequences were aligned using ClustalW (Thompson et al., 1994) with default gap penalties, and the alignment was manually adjusted where necessary. Mean character distances were used to construct a rooted neighbor-joining phylogeny (Saitou and Nei, 1987) from the PHYLP version 3.67 package (Felsenstein, 1993). Statistical support of the branches was tested with 1000 bootstrap resamples. The tomato PTO and PBS1 kinase were used as outgroups. These two genes are divergent from the other members of the TPK1b-related proteins
Seedling ET Response Assays
Tomato seeds were surface sterilized with 70% ethanol for 2 min and 30 min in 35% commercial bleach solution (5.25% [w/v] sodium hypochlorite) plus 0.1% Tween 20, rinsed in sterile water, and then plated on appropriate media and incubated for 6 d in the dark. In vitro, tomato seeds were cultured on MS medium and 0.8% agar with or without ACC (A0430; Sigma-Aldrich) as described.
Induction Treatments
Induction treatments were performed by spraying 4-weak-old soil-grown tomato plants with 5 × 105 Botrytis spores/mL, 100 μM SA (Sigma-Aldrich), 100 μM abscisic acid (Sigma-Aldrich), 100 μM ACC (Sigma-Aldrich), 100 μM paraquat (methyl viologen; Sigma-Aldrich), and 100 μM MeJA (Sigma-Aldrich). In the wounding experiment, the main veins of apical leaflets of compound leaves were wounded with dented forceps.
Accession Numbers
Sequence data for the genes described in this study or genes that were used in the phylogentic analysis can be found in the GenBank/EMBL data libraries under the following accession numbers: TPK1b (GenBank accession number EU555286), CAO21648, BIK1 (At2g39660), NAK (At5g02290), PBS1 (At5g13160), APK1b (At2g28930), APK1a (At1g07570), APK2b (At2g02800), MPLKe (BAD12263), PTO (A49332), Arabidopsis putative protein kinase (At3g55450), Le ACIK1 (AF332960), and ARSK1 (ROOT-SPECIFIC KINASE1, At2G26290).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Suppression of TPK1b through Virus-Induced Gene Silencing Causes Susceptibility to Botrytis.
Supplemental Figure 2. TPK1b RNAi Plants Show No Altered Responses to the Bacterial Pathogen P. syringae pv Tomato.
Supplemental Figure 3. Feeding Trial on Detached Leaves Showing Reduced Resistance of TPK1b RNAi Plants to Tobacco Hornworm (Manduca sexta).
Supplemental Figure 4. TPK1b RNAi Tomato Plants Do Not Show Any Altered Responses to the Plant Hhormones Abscisic Acid and Methyl-Jasmonate.
Supplemental Figure 5. Tomato TPK1b Suppresses the Arabidopsis bik1 Susceptibility to A. brassicicola.
Supplemental Figure 6. 35S:TPK1b Rescues the Short Root Growth of Arabidopsis bik1 Mutant to Wild-Type Levels.
Supplemental Figure 7. The Recombinant GST-TPK1b Fusion Protein Autophosphorylates and Phosphorylates the Myelin Basic Protein in Vitro.
Supplemental Figure 8. 35S:TPK1bT243A Partially Rescues the Growth-Related Phenotypes of the Arabidopsis bik1 Mutant.
Supplemental Figure 9. Alignment of N-Terminal Amino Acid Sequences of TPK1b and Closely Related Proteins.
Supplemental Data Set 1. Multiple Sequence Alignment (PHYLIP) Used as Input for the Phylogentic Tree Presented in Figure 2C.
Supplementary Material
Acknowledgments
We thank Gregg Howe for the tomato mutant and wild-type seeds used in this study. We thank Brahm Dhillon for his help with the phylogenetic analysis, S.P. Dinesh-Kumar for the VIGS vectors, and Robert Dietrich, Burton Bluhm, and Kristin Laluk for their comments on the manuscript. This research was funded by Grant IOB-0618897 provided to T. Mengiste by the National Science Foundation. This is Purdue University Agricultural Program paper number 2008-18338.
The author responsible for the distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Tesfaye Mengiste (mengiste@purdue.edu).
Online version contains Web-only data.
References
- AbuQamar, S., Chen, X., Dahwan, R., Bluhm, B., Salmeron, J., Lam, S., Dietrich, R.A., and Mengiste, T. (2006). Expression profiling and mutant analysis reveals complex regulatory networks involved in Arabidopsis response to Botrytis infection. Plant J. 48 28–44. [DOI] [PubMed] [Google Scholar]
- Asai, T., Tena, G., Plotnikova, J., Willmann, M.R., Chiu, W.L., Gomez-Gomez, L., Boller, T., Ausubel, F.M., and Sheen, J. (2002). MAP kinase signaling cascade in Arabidopsis innate immunity. Nature 415 977–983. [DOI] [PubMed] [Google Scholar]
- Audenaert, K., De Meyer, G.B., and Hofte, M.M. (2002). Abscisic acid determines basal susceptibility of tomato to Botrytis cinerea and suppresses salicylic acid-dependent signaling mechanisms. Plant Physiol. 128 491–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benito, E.P., ten Have, A., Van Klooster, J.W., and van Kan, J.A.L. (1998). Fungal and plant gene expression during synchronized infection of tomato leaves by Botrytis cinerea. Eur. J. Plant Pathol. 104 207–220. [Google Scholar]
- Berrocal-Lobo, M., Molina, A., and Solano, R. (2002). Constitutive expression of ETHYLENE-RESPONSE-FACTOR 1 in Arabidopsis confers resistance to several necrotrophic fungi. Plant J. 29 23–32. [DOI] [PubMed] [Google Scholar]
- Bishop, P.D., Makus, D.J., Pearce, G., and Ryan, C.A. (1981). Proteinase inhibitor-inducing factor activity in tomato leaves resides in oligosaccharides enzymically released from cell walls. Proc. Natl. Acad. Sci. USA 78 3536–3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleecker, A., Estelle, M., Somerville, C., and Kende, H. (1988). Insensitivity to ethylene conferred by a dominant mutation in Arabidopsis thaliana. Science 241 1086–1089. [DOI] [PubMed] [Google Scholar]
- Bodenhausen, N., and Reymond, P. (2007). Signaling pathways controlling induced resistance to insect herbivores in Arabidopsis. Mol. Plant Microbe Interact. 20 1406–1420. [DOI] [PubMed] [Google Scholar]
- Boisson, B., Giglione, C., and Meinnel, T. (2003). Unexpected protein families including cell defense components feature in the N-myristoylome of a higher eukaryote. J. Biol. Chem. 278 43418–43429. [DOI] [PubMed] [Google Scholar]
- Bomblies, K., Lempe, J., Epple, P., Warthmann, N., Lanz, C., Dangl, J.L., and Weigel, D. (2007). Autoimmune response as a mechanism for a Dobzhansky-Muller-type incompatibility syndrome in plants. PLoS Biol. 5 e236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardinale, F., Meskiene, I., Ouaked, F., and Hirt, H. (2002). Convergence and divergence of stress-induced mitogen-activated protein kinase signaling pathways at the level of two distinct mitogen-activated protein kinase kinases. Plant Cell 14 703–711. [PMC free article] [PubMed] [Google Scholar]
- Chinchilla, D., Zipfel, C., Robatzek, S., Kemmerling, B., Nurnberger, T., Jones, J.D., Felix, G., and Boller, T. (2007). A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448 497–500. [DOI] [PubMed] [Google Scholar]
- Church, G.M., and Gilbert, W. (1984). Genomic sequencing. Proc. Natl. Acad. Sci. USA 81 1991–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciardi, J.A., Tieman, D.M., Lund, S.T., Jones, J.B., Stall, R.E., and Klee, H.J. (2000). Response to Xanthomonas campestris pv. vesicatoria in tomato involves regulation of ethylene receptor gene expression. Plant Physiol. 123 81–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Coego, A., Ramirez, V., Gil, M.J., Flors, V., Mauch-Mani, B., and Vera, P. (2005). An Arabidopsis homeodomain transcription factor, OVEREXPRESSOR OF CATIONIC PEROXIDASE 3, mediates resistance to infection by necrotrophic pathogens. Plant Cell 17 2123–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dardick, C., and Ronald, P. (2006). Plant and animal pathogen recognition receptors signal through non-RD kinases. PLoS Pathog. 2 e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellaporta, S.L., Wood, J., and Hicks, J.B. (1983). A plant DNA mini-preparation: Version II. Plant Mol. Biol. Rep. 1 19–21. [Google Scholar]
- Dempsey, D.A., Shah, J., and Klessig, D.F. (1999). Salicylic acid and disease resistance in plants. Crit. Rev. Plant Sci. 18 547–575. [Google Scholar]
- Diaz, J., ten Have, A., and van Kan, J.A. (2002). The role of ethylene and wound signaling in resistance of tomato to Botrytis cinerea. Plant Physiol. 129 1341–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doares, S.H., Syrovets, T., Weiler, E.W., and Ryan, C.A. (1995). Oligogalacturonides and chitosan activate plant defensive genes through the octadecanoid pathway. Proc. Natl. Acad. Sci. USA 92 4095–4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, H.P., Peng, J., Bao, Z., Meng, X., Bonasera, J.M., Chen, G., Beer, S.V., and Dong, H. (2004). Downstream divergence of the ethylene signaling pathway for harpin-stimulated Arabidopsis growth and insect defense. Plant Physiol. 136 3628–3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein, J. (1993). PHYLIP (Phylogeny Inference Package) Version 3.5c. (Seattle, WA: University of Washington).
- Ferrari, S., Galletti, R., Denoux, C., De Lorenzo, G., Ausubel, F.M., and Dewdney, J. (2007). Resistance to Botrytis cinerea induced in Arabidopsis by elicitors is independent of salicylic acid, ethylene, or jasmonate signaling but requires PHYTOALEXIN DEFICIENT3. Plant Physiol. 144 367–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederick, R.D., Thilmony, R.L., Sessa, G., and Martin, G.B. (1998). Recognition specificity for the bacterial avirulence protein AvrPto is determined by Thr-204 in the activation loop of the tomato Pto kinase. Mol. Cell 2 241–245. [DOI] [PubMed] [Google Scholar]
- Gillaspy, G., Ben-David, H., and Gruissem, W. (1993). Fruits: A developmental perspective. Plant Cell 5 1439–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govrin, E.M., and Levine, A. (2000). The hypersensitive response facilitates plant infection by the necrotrophic pathogen Botrytis cinerea. Curr. Biol. 10 751–757. [DOI] [PubMed] [Google Scholar]
- Guo, Y., Qiu, Q.S., Quintero, F.J., Pardo, J.M., Ohta, M., Zhang, C., Schumaker, K.S., and Zhu, J.K. (2004). Transgenic evaluation of activated mutant alleles of SOS2 reveals a critical requirement for its kinase activity and C-terminal regulatory domain for salt tolerance in Arabidopsis thaliana. Plant Cell 16 435–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanks, S.K., and Hunter, T. (1995). Protein kinases 6. The eukaryotic protein kinase superfamily: Kinase (catalytic) domain structure and classification. FASEB J. 9 576–596. [PubMed] [Google Scholar]
- Hardie, D.G. (1999). Plant protein serine/threonine kinases: Classification and function. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50 97–131. [DOI] [PubMed] [Google Scholar]
- Harfouche, A.L., Shivaji, R., Stocker, R., Williams, P.W., and Luthe, D.S. (2006). Ethylene signaling mediates a maize defense response to insect herbivory. Mol. Plant Microbe Interact. 19 189–199. [DOI] [PubMed] [Google Scholar]
- He, K., Gou, X., Yuan, T., Lin, H., Asami, T., Yoshida, S., Russell, S.D., and Li, J. (2007). BAK1 and BKK1 regulate brassinosteroid-dependent growth and brassinosteroid-independent cell-death pathways. Curr. Biol. 17 1109–1115. [DOI] [PubMed] [Google Scholar]
- Heese, A., Hann, D.R., Gimenez-Ibanez, S., Jones, A.M., He, K., Li, J., Schroeder, J.I., Peck, S.C., and Rathjen, J.P. (2007). The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc. Natl. Acad. Sci. USA 104 12217–12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama, T., and Oka, A. (1992). Novel protein kinase of Arabidopsis thaliana (APK1) that phosphorylates tyrosine, serine and threonine. Plant Mol. Biol. 20 653–662. [DOI] [PubMed] [Google Scholar]
- Hoffman, T., Schmidt, J.S., Zheng, X., and Bent, A.F. (1999). Isolation of ethylene-insensitive soybean mutants that are altered in pathogen susceptibility and gene-for-gene disease resistance. Plant Physiol. 119 935–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe, G.A., and Jander, G. (2007). Plant immunity to insect herbivores. Annu. Rev. Plant Biol. 59 41–66. [DOI] [PubMed] [Google Scholar]
- Howe, G.A., Lee, G.I., Itoh, A., Li, L., and DeRocher, A.E. (2000). Cytochrome P450-dependent metabolism of oxylipins in tomato. Cloning and expression of allene oxide synthase and fatty acid hydroperoxide lyase. Plant Physiol. 123 711–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe, G.A., Lightner, J., Browse, J., and Ryan, C.A. (1996). An octadecanoid pathway mutant (JL5) of tomato is compromised in signaling for defense against insect attack. Plant Cell 8 2067–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishitani, M., Liu, J., Halfter, U., Kim, C.S., Shi, W., and Zhu, J.K. (2000). SOS3 function in plant salt tolerance requires N-myristoylation and calcium binding. Plant Cell 12 1667–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillon, O., et al. (2007). The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449 463–467. [DOI] [PubMed] [Google Scholar]
- Johnson, D.R., Bhatnagar, R.S., Knoll, L.J., and Gordon, J.I. (1994). Genetic and biochemical studies of protein N-myristoylation. Annu. Rev. Biochem. 63 869–914. [DOI] [PubMed] [Google Scholar]
- Johnson, L.N., Noble, M.E., and Owen, D.J. (1996). Active and inactive protein kinases: structural basis for regulation. Cell 85 149–158. [DOI] [PubMed] [Google Scholar]
- Jones, J.D., and Dangl, J.L. (2006). The plant immune system. Nature 444 323–329. [DOI] [PubMed] [Google Scholar]
- Kakita, M., Murase, K., Iwano, M., Matsumoto, T., Watanabe, M., Shiba, H., Isogai, A., and Takayama, S. (2007). Two distinct forms of M-locus protein kinase localize to the plasma membrane and interact directly with S-locus receptor kinase to transduce self-incompatibility signaling in Brassica rapa. Plant Cell 19 3961–3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandoth, P.K., Ranf, S., Pancholi, S.S., Jayanty, S., Walla, M.D., Miller, W., Howe, G.A., Lincoln, D.E., and Stratmann, J.W. (2007). Tomato MAPKs LeMPK1, LeMPK2, and LeMPK3 function in the systemin-mediated defense response against herbivorous insects. Proc. Natl. Acad. Sci. USA 104 12205–12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemmerling, B., et al. (2007). The BRI1-associated kinase 1, BAK1, has a brassinolide-independent role in plant cell-death control. Curr. Biol. 17 1116–1122. [DOI] [PubMed] [Google Scholar]
- Klee, H.J. (2004). Ethylene signal transduction. Moving beyond Arabidopsis. Plant Physiol. 135 660–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krusell, L., et al. (2002). Shoot control of root development and nodulation is mediated by a receptor-like kinase. Nature 420 422–426. [DOI] [PubMed] [Google Scholar]
- Lagrimini, L.M., Burkhart, W., Moyer, M., and Rothstein, S. (1987). Molecular cloning of complementary DNA encoding the liginin-forming peroxidasew from tobacco: Molecular analysis and tissue-specific expression. Proc. Natl. Acad. Sci. USA 84 7542–7546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, G.I., and Howe, G.A. (2003). The tomato mutant spr1 is defective in systemin perception and the production of a systemic wound signal for defense gene expression. Plant J. 33 567–576. [DOI] [PubMed] [Google Scholar]
- Li, C., Liu, G., Xu, C., Lee, G.I., Bauer, P., Ling, H.Q., Ganal, M.W., and Howe, G.A. (2003). The tomato suppressor of prosystemin-mediated responses2 gene encodes a fatty acid desaturase required for the biosynthesis of jasmonic acid and the production of a systemic wound signal for defense gene expression. Plant Cell 15 1646–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, C., Schilmiller, A.L., Liu, G., Lee, G.I., Jayanty, S., Sageman, C., Vrebalov, J., Giovannoni, J.J., Yagi, K., Kobayashi, Y., and Howe, G.A. (2005). Role of beta-oxidation in jasmonate biosynthesis and systemic wound signaling in tomato. Plant Cell 17 971–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, C., Williams, M.M., Loh, Y.T., Lee, G.I., and Howe, G.A. (2002). Resistance of cultivated tomato to cell content-feeding herbivores is regulated by the octadecanoid-signaling pathway. Plant Physiol. 130 494–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, L., Zhao, Y., McCaig, B.C., Wingerd, B.A., Wang, J., Whalon, M.E., Pichersky, E., and Howe, G.A. (2004). The tomato homolog of CORONATINE-INSENSITIVE1 is required for the maternal control of seed maturation, jasmonate-signaled defense responses, and glandular trichome development. Plant Cell 16 126–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y., Schiff, M., and Dinesh-Kumar, S.P. (2002). Virus-induced gene silencing in tomato. Plant J. 31 777–786. [DOI] [PubMed] [Google Scholar]
- Lorang, J.M., Sweat, T.A., and Wolpert, T.J. (2007). Plant disease susceptibility conferred by a “resistance” gene. Proc. Natl. Acad. Sci. USA 104 14861–14866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig, A.A., Saitoh, H., Felix, G., Freymark, G., Miersch, O., Wasternack, C., Boller, T., Jones, J.D., and Romeis, T. (2005). Ethylene-mediated cross-talk between calcium-dependent protein kinase and MAPK signaling controls stress responses in plants. Proc. Natl. Acad. Sci. USA 102 10736–10741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund, S.T., Stall, R.E., and Klee, H.J. (1998). Ethylene regulates the susceptible response to pathogen infection in tomato. Plant Cell 10 371–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, G.B., Brommonschenkel, S.H., Chunwongse, J., Frary, A., Ganal, M.W., Spivey, R., Wu, T., Earle, E.D., and Tanksley, S.D. (1993). Map-based cloning of a protein kinase gene conferring disease resistance in tomato. Science 262 1432–1436. [DOI] [PubMed] [Google Scholar]
- McCormick, S. (1991). Transformation of tomato with Agrobacterium tumefaciens. Plant Tiss. Cult. Man. 6 1–9. [Google Scholar]
- Mengiste, T., Chen, X., Salmeron, J.M., and Dietrich, R.A. (2003). The BOS1 gene encodes an R2R3MYB transcription factor protein that is required for biotic and abiotic stress responses in Arabidopsis. Plant Cell 15 2551–2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menke, F.L., van Pelt, J.A., Pieterse, C.M., and Klessig, D.F. (2004). Silencing of the mitogen-activated protein kinase MPK6 compromises disease resistance in Arabidopsis. Plant Cell 16 897–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishina, T.E., and Zeier, J. (2007). Pathogen-associated molecular pattern recognition rather than development of tissue necrosis contributes to bacterial induction of systemic acquired resistance in Arabidopsis. Plant J. 50 500–513. [DOI] [PubMed] [Google Scholar]
- Miya, A., Albert, P., Shinya, T., Desaki, Y., Ichimura, K., Shirasu, K., Narusaka, Y., Kawakami, N., Kaku, H., and Shibuya, N. (2007). CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 104 19613–19618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murase, K., Shiba, H., Iwano, M., Che, F.S., Watanabe, M., Isogai, A., and Takayama, S. (2004). A membrane-anchored protein kinase involved in Brassica self-incompatibility signaling. Science 303 1516–1519. [DOI] [PubMed] [Google Scholar]
- O'Donnell, P.J., Calvert, C., Atzorn, R., Wasternack, C., Leyser, H.M.O., and Bowles, D.J. (1996). Ethylene as a signal mediating the wound response of tomato plants. Science 274 1914–1917. [DOI] [PubMed] [Google Scholar]
- Prins, T.W., Tudzynski, P., Tiedemann, A.V., Tudzynski, B., ten Have, A., Hansen, M.E., Tenberge, K., and van Kan, J.A.L. (2000). Infection strategies of Botrytis cinerea and related necrotrophic pathogens. In Fungal Pathology, J.W. Kronstad, ed (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 33–64.
- Rathjen, J.P., Chang, J.H., Staskawicz, B.J., and Michelmore, R.W. (1999). Constitutively active Pto induces a Prf-dependent hypersensitive response in the absence of avrPto. EMBO J. 18 3232–3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rentel, M.C., Lecourieux, D., Ouaked, F., Usher, S.L., Petersen, L., Okamoto, H., Knight, H., Peck, S.C., Grierson, C.S., Hirt, H., and Knight, M.R. (2004). OXI1 kinase is necessary for oxidative burst-mediated signalling in Arabidopsis. Nature 427 858–861. [DOI] [PubMed] [Google Scholar]
- Romeis, T. (2001). Protein kinases in the plant defence response. Curr. Opin. Plant Biol. 4 407–414. [DOI] [PubMed] [Google Scholar]
- Rommens, C.M., Salmeron, J.M., Baulcombe, D.C., and Staskawicz, B.J. (1995). Use of a gene expression system based on potato virus X to rapidly identify and characterize a tomato Pto homolog that controls fenthion sensitivity. Plant Cell 7 249–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan, C.A. (1992). The search for the proteinase inhibitor-inducing factor, PIIF. Plant Mol. Biol. 19 123–133. [DOI] [PubMed] [Google Scholar]
- Saitou, N., and Nei, M. (1987). The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4 406–425. [DOI] [PubMed] [Google Scholar]
- SAS Institute (1999). The SAS System for Windows, Release 8.0. (Cary, NC: SAS Institute).
- Schilmiller, A.L., and Howe, G.A. (2005). Systemic signaling in the wound response. Curr. Opin. Plant Biol. 8 369–377. [DOI] [PubMed] [Google Scholar]
- Sessa, G., D'Ascenzo, M., and Martin, G.B. (2000). Thr38 and Ser198 are Pto autophosphorylation sites required for the AvrPto-Pto-mediated hypersensitive response. EMBO J. 19 2257–2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah, K., Vervoort, J., and de Vries, S.C. (2001). Role of threonines in the Arabidopsis thaliana somatic embryogenesis receptor kinase 1 activation loop in phosphorylation. J. Biol. Chem. 276 41263–41269. [DOI] [PubMed] [Google Scholar]
- Shiu, S.H., and Bleecker, A.B. (2001). Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc. Natl. Acad. Sci. USA 98 10763–10768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu, S.H., Karlowski, W.M., Pan, R., Tzeng, Y.H., Mayer, K.F., and Li, W.H. (2004). Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell 16 1220–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, W.Y., Wang, G.L., Chen, L.L., Kim, H.S., Pi, L.Y., Holsten, T., Gardner, J., Wang, B., Zhai, W.X., Zhu, L.H., Fauquet, C., and Ronald, P. (1995). A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science 270 1804–1806. [DOI] [PubMed] [Google Scholar]
- Stone, J.M., and Walker, J.C. (1995). Plant protein kinase families and signal transduction. Plant Physiol. 108 451–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stotz, H.U., Pittendrigh, B.R., Kroymann, J., Weniger, K., Fritsche, J., Bauke, A., and Mitchell-Olds, T. (2000). Induced plant defense responses against chewing insects. Ethylene signaling reduces resistance of Arabidopsis against Egyptian cotton worm but not diamondback moth. Plant Physiol. 124 1007–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke, S., Kistner, C., Yoshida, S., Mulder, L., Sato, S., Kaneko, T., Tabata, S., Sandal, N., Stougaard, J., Szczyglowski, K., and Parniske, M. (2002). A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature 417 959–962. [DOI] [PubMed] [Google Scholar]
- Swiderski, M.R., and Innes, R.W. (2001). The Arabidopsis PBS1 resistance gene encodes a member of a novel protein kinase subfamily. Plant J. 26 101–112. [DOI] [PubMed] [Google Scholar]
- Thomma, B., Eggermont, K., Penninckx, I., Mauch-Mani, B., Vogelsang, R., Cammue, B.P.A., and Broekaert, W.F. (1998). Separate jasmonate-dependent and salicylate-dependent defense-response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc. Natl. Acad. Sci. USA 95 15107–15111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma, B.P., Eggermont, K., Tierens, K.F., and Broekaert, W.F. (1999). Requirement of functional ethylene-insensitive 2 gene for efficient resistance of Arabidopsis to infection by Botrytis cinerea. Plant Physiol. 121 1093–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torii, K.U. (2000). Receptor kinase activation and signal transduction in plants: an emerging picture. Curr. Opin. Plant Biol. 3 361–367. [DOI] [PubMed] [Google Scholar]
- van Wees, S., Chang, H.-S., Zhu, T., and Glazebrook, J. (2003). Characterization of the early response of Arabidopsis to Alternaria brassicicola infection using expression profiling. Plant Physiol. 132 606–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veronese, P., Nakagami, H., Bluhm, B., Abuqamar, S., Chen, X., Salmeron, J., Dietrich, R.A., Hirt, H., and Mengiste, T. (2006). The membrane-anchored BOTRYTIS-INDUCED KINASE1 plays distinct roles in Arabidopsis resistance to necrotrophic and biotrophic pathogens. Plant Cell 18 257–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Dahl, C.C., Winz, R.A., Halitschke, R., Kuhnemann, F., Gase, K., and Baldwin, I.T. (2007). Tuning the herbivore-induced ethylene burst: The role of transcript accumulation and ethylene perception in Nicotiana attenuata. Plant J. 51 293–307. [DOI] [PubMed] [Google Scholar]
- Wan, J., Zhang, X.C., Neece, D., Ramonell, K.M., Clough, S., Kim, S.Y., Stacey, M.G., and Stacey, G. (2008). A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. Plant Cell 20 471–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X., Goshe, M.B., Soderblom, E.J., Phinney, B.S., Kuchar, J.A., Li, J., Asami, T., Yoshida, S., Huber, S.C., and Clouse, S.D. (2005). Identification and functional analysis of in vivo phosphorylation sites of the Arabidopsis BRASSINOSTEROID-INSENSITIVE1 receptor kinase. Plant Cell 17 1685–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winz, R.A., and Baldwin, I.T. (2001). Molecular interactions between the specialist herbivore Manduca sexta (Lepidoptera, Sphingidae) and its natural host Nicotiana attenuata. IV. Insect-induced ethylene reduces jasmonate-induced nicotine accumulation by regulating putrescine N-methyltransferase transcripts. Plant Physiol. 125 2189–2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, S., and Parniske, M. (2005). Regulation of plant symbiosis receptor kinase through serine and threonine phosphorylation. J. Biol. Chem. 280 9203–9209. [DOI] [PubMed] [Google Scholar]
- Zhang, S., Du, H., and Klessig, D.F. (1998). Activation of the tobacco SIP kinase by both a cell wall-derived carbohydrate elicitor and purified proteinaceous elicitins from Phytophthora spp. Plant Cell 10 435–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, S., and Klessig, D.F. (2001). MAPK cascades in plant defense signaling. Trends Plant Sci. 6 520–527. [DOI] [PubMed] [Google Scholar]
- Zheng, Z., Qamar, S.A., Chen, Z., and Mengiste, T. (2006). Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J. 48 592–605. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.