Abstract
Sphingolipids are structural components of endomembranes and function through their metabolites as bioactive regulators of cellular processes such as programmed cell death. A characteristic feature of plant sphingolipids is their high content of trihydroxy long-chain bases (LCBs) that are produced by the LCB C-4 hydroxylase. To determine the functional significance of trihydroxy LCBs in plants, T-DNA double mutants and RNA interference suppression lines were generated for the two Arabidopsis thaliana LCB C-4 hydroxylase genes Sphingoid Base Hydroxylase1 (SBH1) and SBH2. These plants displayed reductions in growth that were dependent on the content of trihydroxy LCBs in sphingolipids. Double sbh1 sbh2 mutants, which completely lacked trihydroxy LCBs, were severely dwarfed, did not progress from vegetative to reproductive growth, and had enhanced expression of programmed cell death associated–genes. Furthermore, the total content of sphingolipids on a dry weight basis increased as the relative amounts of trihydroxy LCBs decreased. In trihydroxy LCB–null mutants, sphingolipid content was ∼2.5-fold higher than that in wild-type plants. Increases in sphingolipid content resulted from the accumulation of molecular species with C16 fatty acids rather than with very-long-chain fatty acids, which are more commonly enriched in plant sphingolipids, and were accompanied by decreases in amounts of C16-containing species of chloroplast lipids. Overall, these results indicate that trihydroxy LCB synthesis plays a central role in maintaining growth and mediating the total content and fatty acid composition of sphingolipids in plants.
INTRODUCTION
Sphingolipids are structurally diverse molecules that are major components of the endomembrane system and have recently been estimated to compose >40% of plasma membrane lipids in plants (Sperling et al., 2005). Not only do sphingolipids provide structural integrity to membranes, they are also believed to contribute to the organization of membrane microdomains or lipid rafts that contain proteins such as glycosylphosphatidylinositol-anchored proteins, which function in cell surface–related activities including cell wall deposition (Mongrand et al., 2004; Borner et al., 2005). Furthermore, sphingolipids function through their metabolites as regulators of key cellular and physiological processes in plants. For example, the ceramide component of sphingolipids has been shown to function as a mediator of programmed cell death (PCD) (Liang et al., 2003), and the sphingolipid long-chain base (LCB) phosphates sphingosine- and phytosphingosine-1-phosphate have been implicated as signaling molecules in abscisic acid–dependent guard cell closure (Ng et al., 2001; Coursol et al., 2003, 2005).
More than 200 different sphingolipid molecules occur in plants (Markham et al., 2006; Markham and Jaworski, 2007). These molecules can differ in the composition of their head groups, degree of hydroxylation, and numbers and positions of double bonds. The functional significance of this immense structural heterogeneity of plant sphingolipids has only begun to be addressed. For example, increases in relative amounts of cis C-8 unsaturated LCBs have been shown to enhance aluminum tolerance in Arabidopsis thaliana (Ryan et al., 2007). One characteristic feature of sphingolipids is the high degree of hydroxylation of the fatty acids and LCBs that compose their ceramide backbone (Lynch and Dunn, 2004). The fatty acids of plant sphingolipids typically are a mixture of C16 and very-long-chain (C20 to C26) moieties that contain an α- or C-2 hydroxy group (Lynch and Dunn, 2004). LCBs, which are derived from the condensation of palmitoyl-CoA and Ser, contain 18 carbon atoms and up to three hydroxyl groups. The C-1 and C-3 hydroxyl groups arise from the Ser and palmitoyl-CoA precursors, respectively, whereas the third hydroxyl group at the C-4 position is added following synthesis of the LCB. The C-1 hydroxyl group can be substituted with polar residues to form complex sphingolipids. In plants, these include glucosylceramides (GlcCers) and the more abundant glycosylinositolphosphoceramides (GIPCs) (Kaul and Lester, 1978). While dihydroxy LCBs, which lack the C-4 hydroxyl group, are typically more enriched in GlcCers than in GIPCs, trihydroxy LCBs predominate in both classes in Arabidopsis and compose nearly 90% of the LCBs in the total sphingolipid extract in Arabidopsis leaves (Chen et al., 2006; Markham et al., 2006).
In Saccharomyces cerevisiae, the C-4 hydroxyl group is introduced by the activity of a diiron-oxo–type hydroxylase encoded by the SUR2 (also known as SYR2) gene (Cliften et al., 1996; Haak et al., 1997; Grilley et al., 1998). A similar enzyme occurs in plants, and the two LCB C-4 hydroxylase genes (At1g69640 and At1g14290; Figure 1) in Arabidopsis have been shown to restore trihydroxy LCB biosynthetic ability to S. cerevisiae sur2Δ mutants (Sperling et al., 2001). In mammals, the C-4 hydroxyl group in LCBs arises from the bifunctional desaturase/hydroxylase activity of the DES2 (Degenerative Spermatocyte2) polypeptide, which is more distantly related to the S. cerevisiae and plant C-4 hydroxylases (Ternes et al., 2002; Omae et al., 2004).
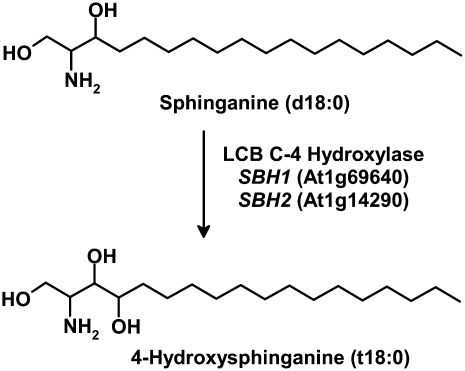
Figure 1.
Example of a Reaction Catalyzed by LCB C-4 Hydroxylase.
The conversion of dihydroxy LCB sphinganine (d18:0) to trihydroxy LCB 4-OH-sphinganine (or phytosphingosine; t18:0) by the activity of LCB C-4 hydroxylase. Arabidopsis contains two genes, SBH1 (At1g69640) and SBH2 (At1g14290), that encode distinct LCB C-4 hydroxylase isoforms.
The significance of sphingolipid LCB C-4 hydroxylation has been studied in S. cerevisiae and more recently in the filamentous fungus Aspergillus nidulans (Li et al., 2007). S. cerevisiae mutants that lack the LCB C-4 hydroxyl group do not display growth phenotypes, indicating that this hydroxyl moiety is not an essential structural feature of S. cerevisiae sphingolipids (Haak et al., 1997). Loss of LCB C-4 hydroxylation, however, does confer resistance to the Pseudomonas syringae toxin syringomycin (Cliften et al., 1996; Grilley et al., 1998). This resistance apparently arises from alterations in the physical properties of the plasma membrane that mitigate the channel-forming activity of syringomycin (Idkowiak-Baldys et al., 2004). In contrast with S. cerevisiae, LCB C-4 hydroxylation is essential for the viability of A. nidulans, and trihydroxy LCBs have been shown to contribute to cell wall formation and the transition from asexual to sexual growth in this organism (Li et al., 2007). In mammals, trihydroxy LCBs are relatively minor components of sphingolipids that are formed by the dual activity of DES2 (Ternes et al., 2002; Omae et al., 2004). As a result, the function of trihydroxy LCBs cannot be dissected from that of C-4 unsaturated LCBs in mammals.
Prior to the studies described here, the functional significance of LCB C-4 hydroxylation in plants had not been well characterized. A recent report, for example, showed that RNA interference (RNAi) and antisense downregulation of one of the five LCB C-4 hydroxylase genes of rice (Oryza sativa) result in reduced fertility (Imamura et al., 2007). However, it was not possible to link this phenotype with LCB C-4 hydroxylation, because no change in sphingolipid LCB composition was detected in these plants (Imamura et al., 2007). To determine the function of LCB C-4 hydroxylation in plants, we have generated double mutants for the two C-4 hydroxylase genes in Arabidopsis that completely lack trihydroxy LCBs and have also generated RNAi lines with intermediate reductions in trihydroxy LCBs. The results presented here show that LCB C-4 hydroxylation is critical for the growth and viability of Arabidopsis. The results also show that synthesis of trihydroxy LCBs plays a central role in the maintenance of sphingolipid fatty acid composition and total sphingolipid content in Arabidopsis. A model is presented that rationalizes the contributions of trihydroxy LCBs to growth and the regulation of sphingolipid biosynthesis.
RESULTS
Expression of SBH1 and SBH2 and Subcellular Localization of the SBH1 and SBH2 Polypeptides
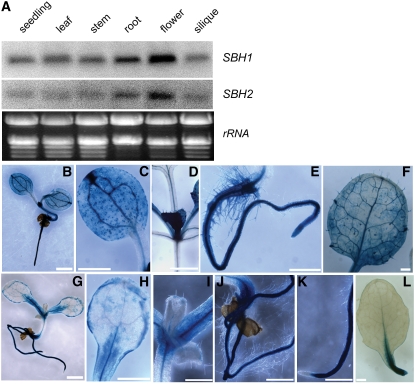
Arabidopsis contains two homologs of the S. cerevisiae LCB C-4 hydroxylase, At1g69640 and At1g14290, designated Sphingoid Base Hydroxylase1 (SBH1) and SBH2, respectively. Each gene has been shown previously to restore trihydroxy LCB synthesis when expressed in S. cerevisiae LCB C-4 hydroxylase mutants (Sperling et al., 2001). The organ-specific expression patterns of SBH1 and SBH2 were examined as a first step toward addressing the possible functional redundancy of these genes. RNA gel blot analyses indicated that both SBH1 and SBH2 are ubiquitously expressed in Arabidopsis and most highly expressed in flowers and roots (Figure 2A). Of the two genes, SBH1 is more highly expressed throughout the Arabidopsis plant. Consistent with this, higher β-glucuronidase (GUS) activity was detected in leaves and cotyledons of plants transformed with an SBH1 promoter–GUS fusion compared with those transformed with an SBH2 promoter–GUS fusion (Figures 2B to 2L).
Figure 2.
Gene Expression Analyses of SBH1 and SBH2.
(A) Gene expression of SBH1 and SBH2 in different organs from a wild-type plant (Col-0) as revealed by RNA gel blot analysis. RNA loading in each lane is indicated by ethidium bromide–stained rRNA. Shown are representative blots from three technical replicates.
(B) to (L) Localization of GUS activity in Arabidopsis plants transformed with SBH1 and SBH2 promoter-GUS fusions.
(B) to (F) Promoter-GUS activities for SBH1.
(B) Ten-day-old seedling grown on MS plates.
(C) Cotyledon.
(D) Emerging young leaf.
(E) Root.
(F) Rosette leaf from a 4-week-old soil-grown plant.
(G) to (L) Promoter-GUS activities for SBH2.
(G) Ten-day-old seedling grown on MS plates.
(H) Cotyledon.
(I) Emerging young leaf.
(J) and (K) Roots.
(L) Rosette leaf from a 4-week-old soil-grown plant.
Bars = 1 mm for (B), (G), (F), and (L) and 500 μm for (C) to (E) and (H) to (K).
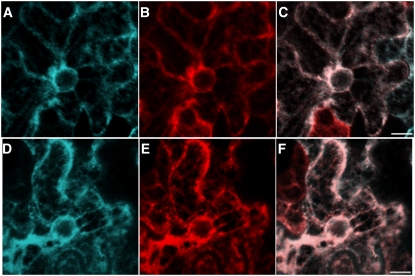
Although the similarity of expression patterns of SBH1 and SBH2 suggests redundancy at the organ level, it is possible that the SBH1 and SBH2 polypeptides reside in different locations within the cell and contribute, for example, to the hydroxylation of LCBs for different sphingolipid classes. To address this possibility, SBH1 and SBH2 were fused at their C termini with cyan fluorescent protein (CFP), and each was transiently expressed in tobacco (Nicotiana tabacum) leaves. Analyses of the Agrobacterium tumefaciens–infiltrated leaves by confocal microscopy revealed that SBH1 and SBH2 colocalize with a fluorescent protein marker containing an endoplasmic reticulum (ER) chitinase signal peptide (CSP) and the HDEL ER retention sequence (CSP-YFP-HDEL) (Chen et al., 2006), but not with the plasma membrane dye FM4-64. This indicates that both Arabidopsis LCB C-4 hydroxylases are ER-localized (Figure 3; see Supplemental Figure 1 online). Both N- and C-terminal fusions of SBH1 and SBH2 with yellow fluorescent protein (YFP) or CFP were able to restore C-4 hydroxylation to a S. cerevisiae sur2Δ mutant containing a knockout of the LCB C-4 hydroxylase gene, indicating that the hydroxylase fusions retain their enzymatic activities (see Supplemental Figure 2 online).
Figure 3.
Subcellular Localization of SBH1 and SBH2.
(A) Distribution of SBH1-CFP.
(B) Distribution of the ER marker CSP-YFP-HDEL.
(C) Merge of images in (A) and (B), which shows the colocalization of SBH1-CFP with the ER marker.
(D) Distribution of SBH2-CFP.
(E) Distribution of the ER marker CSP-YFP-HDEL.
(F) Merge of images in (D) and (E), which shows the colocalization of SBH2-CFP with the ER marker.
Images with additional marker controls are shown in Supplemental Figure 1 online. Bars = 10 μm.
Overall, the constitutive expression of both LCB C-4 hydroxylase genes in Arabidopsis and the localization of both peptides in the ER suggest that SBH1 and SBH2 are functionally redundant.
Relative Contributions of SBH1 and SBH2 to LCB C-4 Hydroxylation
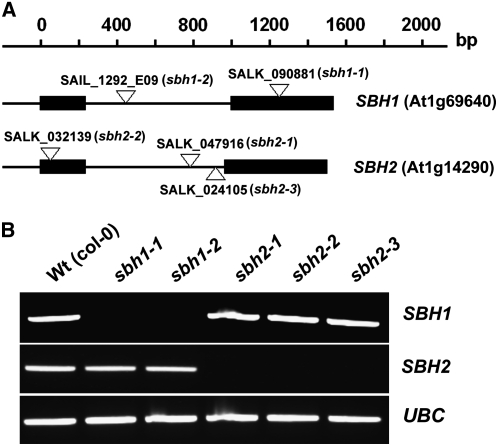
The LCB composition of T-DNA mutants for SBH1 and SBH2 was measured to assess the relative contributions of each gene to C-4 hydroxylation in Arabidopsis plants. For these studies, multiple independent T-DNA mutants were obtained for each gene: (1) for SBH1, SALK_090881, designated sbh1-1, and SAIL_1292_E09, designated sbh1-2; and (2) for SBH2, SALK_047916, designated sbh2-1, SALK_032139, designated sbh2-2, and SALK_024105, designated sbh2-3 (Figure 4A). Each mutant was determined to be a null allele based on RT-PCR (Figure 4B). Double mutants for the two SBH genes were obtained by crossing of sbh1-1 and sbh2-1 and crossing of sbh1-2 and sbh2-2, followed by self-fertilization of the progeny from the crosses. As expected, expression of SBH1 and SBH2 was not detected in the double mutants, as determined by RT-PCR analysis (see Supplemental Figure 3 online).
Figure 4.
Gene Structures and Characterization of Mutant Alleles for SBH1 and SBH2.
(A) Gene structures of SBH1 and SBH2 and T-DNA insertion sites in mutants obtained from the SALK and SAIL collections. Solid bars represent exons, and lines indicate introns. The open triangles indicate the T-DNA insertion sites.
(B) RT-PCR analyses using gene-specific primers show that the T-DNA lines used are null mutants for SBH1 or SBH2. The UBC gene (At5g25760) was used as an internal control. Data shown are representative of three independent analyses.
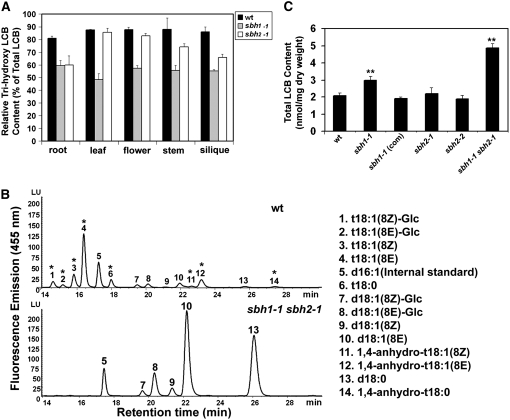
LCB compositions were measured by HPLC following hydrolysis of the total sphingolipid extract and fluorescent derivatization of the released LCBs. Initially, the relative content of trihydroxy and dihydroxy LCBs in the total sphingolipid extract from different organs of 8-week-old wild-type, sbh1, and sbh2 plants was compared. Trihydroxy LCBs, principally in the form of cis/trans isomers of 4-hydroxy-8-sphingenine (t18:1) and 4 hydroxysphinganine (t18:0), accounted for 80 to 90% of the total LCBs of sphingolipids in roots, leaves, flowers, stems, and siliques of wild-type plants (Figure 5A). In the sbh1-1 mutant, the relative content of trihydroxy LCBs was reduced to 50 to 60% of the LCBs in sphingolipids in each of the organs analyzed (Figure 5A). By contrast, reductions in the relative content of trihydroxy LCBs in the sbh2-1 mutant were limited primarily to roots, stems, and siliques, with little or no decrease in leaves compared with wild-type plants (Figure 5A). A similar pattern of alterations in trihydroxy LCB content was also observed in the sbh1-2, sbh2-2, and sbh2-3 mutants. The larger contribution of SBH1 to LCB C-4 hydroxylation in the different organs of Arabidopsis is consistent with its higher expression throughout Arabidopsis, as demonstrated by RNA gel blot analyses (Figure 2A). The sbh1-1 sbh2-1 double mutant contained no detectable amounts of trihydroxy LCBs (Figure 5B) and instead was enriched in cis/trans isomers of Δ8-dihydrosphingenine (d18:1) and sphinganine (d18:0). This result indicates that the combined expression of SBH1 and SBH2 accounts for all LCB C-4 hydroxylation in Arabidopsis.
Figure 5.
Sphingolipid LCB Content and Composition of sbh1 and sbh2 Knockout Mutants.
(A) The relative content of trihydroxy LCBs in total sphingolipid extracts from different organs of wild-type plants and sbh1-1 and sbh2-1 mutants. Values are means ± sd (n = 5).
(B) Profiles of LCBs in the total sphingolipid extracts of wild-type plants and sbh1-1 sbh2-1 double mutant plants obtained by C18 reverse-phase HPLC analysis of fluorescent derivatives of LCBs following barium hydroxide/dioxane hydrolysis of sphingolipids. Trihydroxy LCBs are labeled with asterisks. The identities of the labeled peaks are as follows: 1, t18:1(8Z)-Glc (psychosine form); 2, t18:1(8E)-Glc; 3, t18:1(8Z); 4, t18:1(8E); 5, d16:1 (internal standard); 6, t18:0; 7, d18:1(8Z)-Glc; 8, d18:1(8E)-Glc; 9, d18:1(8Z); 10, d18:1(8E); 11, 1,4-anhydro-t18:1(8Z); 12, 1,4-anhydro-t18:1(8E); 13, d18:0; 14, 1,4-anhydro-t18:0. d, dihydroxy; t, trihydroxy; x:y, x, number of carbon atoms, y, number of double bonds.
(C) Total content of LCBs in wild-type (Col-0) plants, single and double mutants of sbh1-1 and sbh2-1, and the sbh1-1 mutant complemented with a wild-type copy of SBH1 [sbh1-1(com)]. Sphingolipids were extracted from rosettes of 14-d-old plants grown on MS agar plates. Data shown are averages of five independent extractions and analyses ± sd. A Student's t test indicated that the total LCB content of both sbh1-1 and sbh1-1 sbh2-1 was significantly increased relative to the wild-type control (** P < 0.01).
Growth Phenotypes of Arabidopsis sbh Mutants
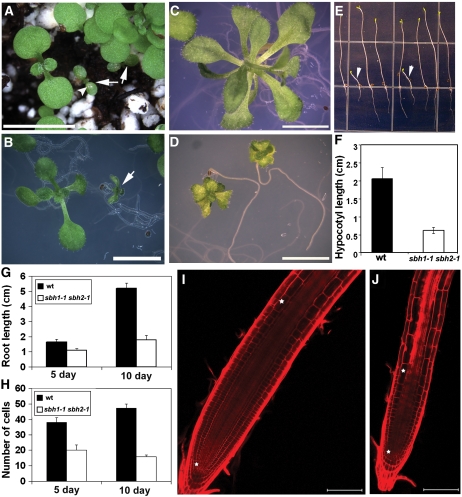
No obvious growth defects were observed when sbh1-1 or sbh2-1 mutant plants were grown in soil under our standard growth conditions. However, sbh1-1 sbh2-1 double mutants displayed severe alterations in growth. Approximately 1/16th of the F2 progeny obtained by self-fertilization of a sbh1-1+/− sbh2-1+/− plant were markedly smaller than their siblings. Similar alterations in growth were also observed in a segregating F2 population from crosses of the sbh1-1 or sbh1-2 allele with the sbh2-1, sbh2-2, or sbh2-3 allele. Double mutants displayed reduced vigor and usually died before expansion of the true leaves when grown in soil. In addition to the reduced size, these plants also developed lesions near the center of their cotyledons (Figure 6A, arrowhead).
Figure 6.
Growth Phenotypes of sbh1 sbh2 Double Mutants.
(A) Ten-day-old seedlings in soil. Arrows indicate the sbh1-1 sbh2-1 mutant segregated from the sbh1-1+/− sbh2-1−/− parent. Necrotic lesions are indicated by the arrowhead.
(B) One week-old seedling grown on 1× MS plates. The arrow indicates the sbh1-1 sbh2-1 double mutant.
(C) and (D) Three-week-old seedlings on MS plates: wild type (C) and sbh1-1 sbh2-1 double mutant (D). Bars = 0.5 cm in (A) to (D).
(E) and (F) Reduced hypocotyl growth in 7-d-old etiolated sbh1-1 sbh2-1 plants relative to wild-type plants. Plants with shorter hypocotyls were confirmed to be sbh1-1 sbh2-1 mutants (arrowheads) by use of PCR genotyping. The data shown are from one of three experiments.
(G) Root length of the wild type and the sbh1-1 sbh2-1 double mutant at 5 and 10 d after germination.
(H) Number of cells in the root meristem zone of wild-type and sbh1-1 sbh2-1 plants. Values are means ± sd (n = 20) for (F) to (H).
(I) and (J) Meristem size of wild-type plants (I) and sbh1-1 sbh2-1 double mutants (J). Stars indicate the upper and lower ends of the meristem zone (see text for definition of the root meristem zone). Bars = 100 μm.
Despite their inability to transition to reproductive growth, double mutants could be obtained from segregating populations of progeny from a sbh1-1+/− sbh2-1−/− parental line. These plants displayed extended viability when maintained on agar plates but were nonetheless reduced in size relative to their siblings, and necrotic lesions were again observed on cotyledons (Figure 6B, arrow). These plants had small, curled leaves and short petioles that resulted in a compact, epinastic morphology of the rosette (Figures 6C and 6D). The inability to bolt was also observed in double mutants maintained on plates for 6 weeks or more, whereas wild-type plants typically bolted within 4 weeks under these growth conditions. The addition of trihydroxy LCB phytosphingosine (t18:0) to the medium at concentrations of up to 200 μM was unable to restore the growth of sbh1-1 sbh2-1 plants, and higher concentrations of t18:0 were not used because of its adverse effects on the growth and viability of wild-type plants (Abbas et al., 1994). However, the growth of sbh1-1 sbh2-1 mutants appeared to be restored to that of wild-type plants by genetic complementation with a wild-type copy of SBH1.
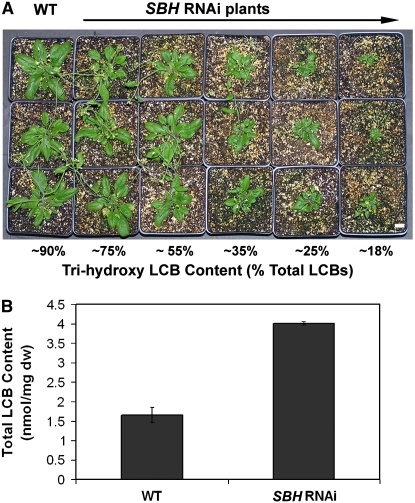
In addition, RNAi suppression lines that targeted both SBH genes were generated to examine the effects of a range of trihydroxy LCB contents on growth and sphingolipid content. As shown in Figure 7A, reduced growth was observed in plants with a relative content of trihydroxy LCBs ≤ ∼35% of the total LCBs in leaves, and growth was progressively reduced in plants with lower relative amounts of trihydroxy LCBs. Plants containing as little as 18% trihydroxy LCBs, although dwarfed, retained the ability to bolt and set seed.
Figure 7.
Growth and LCB Content of SBH RNAi Lines.
(A) Four-week-old wild-type seedlings and T2 seedlings of independent SBH RNAi lines with varying levels of trihydroxy LCB content. Bar = 1 cm.
(B) An RNAi line (SBH RNAi) with a trihydroxy LCB content of ∼20% total sphingolipids was grown in soil, and total LCBs were measured from leaves of 6-week-old plants by HPLC analysis of o-phthaldialdehyde derivatives of LCBs following barium hydroxide/dioxane hydrolysis of total sphingolipids. The RNAi-suppressed plants showed increased levels of total LCBs (4.02 ± 0.04 nmol/mg dry weight [dw]) compared with wild-type plants grown under the same conditions (1.65 ± 0.20 nmol/mg dry weight). Results are averages of five independent extractions and analyses ± sd.
Additional studies were conducted to determine whether the reduced growth of sbh1-1 sbh2-1 plants was due to defects in cell division or cell elongation. When plants were grown vertically on agar plates under conditions of etiolation, the hypocotyl length of the sbh1-1 sbh2-1 mutant was only ∼25% of that of wild-type plants (Figures 6E and 6F). In addition, the length of epidermal cells in the middle part of the hypocotyls of the sbh1-1 sbh2-1 mutant was shorter than that of cells in the corresponding part of wild-type plants (see Supplemental Figures 4A and 4B online). When grown vertically in the light, roots of sbh1-1 and sbh2-1 single mutants were approximately the same length as those of wild-type plants. However, roots of 5-d-old sbh1-1 sbh2-1 double mutant seedlings were ∼66% the length of roots of wild-type seedlings of the same age, and at 10 d, the roots of sbh1-1 sbh2-1 seedlings were ∼34% the length of wild-type seedling roots (Figure 6G). Microscopic analyses of roots revealed that sbh1-1 sbh2-1 plants had shorter and thinner meristematic zones than those of roots from wild-type plants (Figures 6I and 6J). To compare meristem growth between wild-type and sbh1-1 sbh2-1 plants, the number of cortical cells from the initial cell to the first rapidly elongating cell (indicated by stars in Figures 6I and 6J) was counted using a previously described methodology (Casamitjana-Martinez et al., 2003). According to this criterion, root meristems of 5- and 10-d-old sbh1-1 sbh2-1 seedlings contained ∼20 and 16 cortical cells, respectively, while wild-type 5- and 10-d-old seedlings had ∼38 and 47 cortical cells, respectively (Figure 6H). These data suggest that cell division is reduced in the root meristem of the sbh1-1 sbh2-1 mutant. Collectively, these results indicate that defects in both cell elongation and division contribute to the reduced size of the sbh1-1 sbh2-1 mutant.
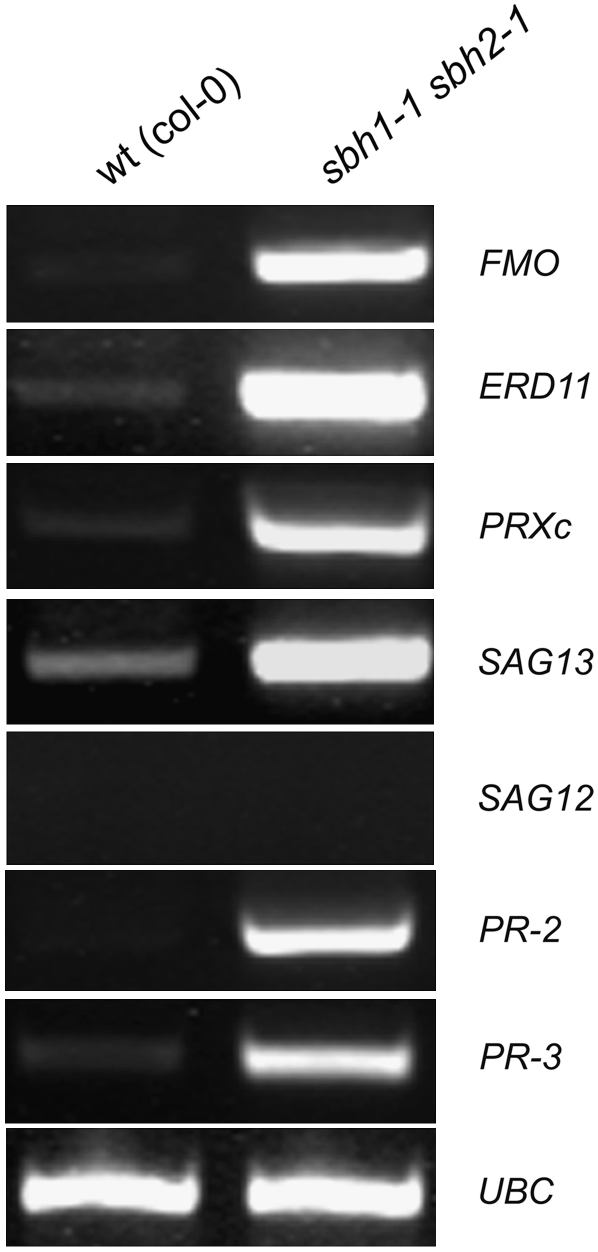
The presence of necrotic lesions on cotyledons of double mutant plants and the premature death of these plants are reminiscent of phenotypes associated with accelerated cell death (acd) Arabidopsis mutants, including the sphingolipid-related acd11 and acd5 mutants (Brodersen et al., 2002; Liang et al., 2003). To gain clues about the type of cell death present in sbh1 sbh2 double mutants, the expression of PCD marker genes was examined. For these studies, gene expression was assessed by RT-PCR using leaves from 2-week-old plants maintained on Murashige and Skoog (MS) plates. The marker genes chosen were those previously used to examine cell death in acd11 mutants (Brodersen et al., 2002). Similar to previous observations for acd11 plants (Brodersen et al., 2002), expression of PCD and related hypersensitive response marker genes was detected at substantially higher levels in the hydroxylase double mutants relative to wild-type plants (Figure 8). These included SENESCENCE-ASSOCIATED GENE13 (SAG13; At2g29350), FLAVIN-CONTAINING MONOXYGENASE (FMO; At1g19250), PEROXIDASE C (PRXc; At3g49120), PATHOGENESIS-RELATED PROTEIN2 (PR-2; At2g14610), and PR-3 (At3g57260). By contrast, no expression of the senescence-associated gene SAG12 (At5g45890) was detected in the LCB C-4 hydroxylase double mutant, as was also observed with acd11 (Brodersen et al., 2002). These results suggest that, like acd11, a hypersensitive response–type PCD is activated in sbh1 sbh2 double mutants.
Figure 8.
RT-PCR Analysis of the Expression of PCD– and Hypersensitive Response (HR)–Related Genes in Wild-Type (Col-0) and sbh1-1 sbh2-1 Double Mutant Plants.
The genes chosen are FMO (At1g19250), ERD11 (At1g02930), PRXc (At3g49120), SAG13 (At2g29350), SAG12 (At5g45890), PR2 (At3g57260), and PR3 (At3g12500), as reported previously (Brodersen et al., 2002). UBC (At5g25760) was used as an internal positive control. Shown are representative results from three biological replicates. Oligonucleotide sequences used for these analyses are provided in Supplemental Table 3 online.
Effects of Altered LCB C-4 Hydroxylation on Sphingolipid Content
The effects of altered trihydroxy LCB composition on the sphingolipid content of mutants and RNAi suppression lines were examined by analysis of LCBs following hydrolysis of the total sphingolipid extract. Quantitative measurements of sphingolipids from leaves of 2-week-old agar-grown seedlings revealed an ∼1.5-fold increase in the total LCB content in sbh1-1 relative to the wild-type control (Figure 5C). This increase was reversed by complementation of sbh1-1 with a wild-type copy of the SBH1 gene (Figure 5C). No change in sphingolipid content was detected in sbh2-1 and sbh2-2 seedlings, which also do not have detectable alterations in total trihydroxy LCB composition relative to wild-type seedlings (Figure 5C). Most strikingly, the sbh1-1 sbh2-1 double mutant, which lacks detectable amounts of trihydroxy LCBs, displayed twofold to threefold increases in the total LCB content of leaves relative to that of the wild-type control (Figure 5C). In addition, soil-grown RNAi suppression lines with ∼20% trihydroxy LCBs in total sphingolipid extract contained nearly twofold higher levels of sphingolipids than wild-type plants grown under similar conditions (Figure 7B). These results indicate that reductions in LCB C-4 hydroxylation not only affect the relative content of trihydroxy LCBs but also lead to increases in total sphingolipid content.
Sphingolipidomic Analyses of Mutant and RNAi Suppression Lines
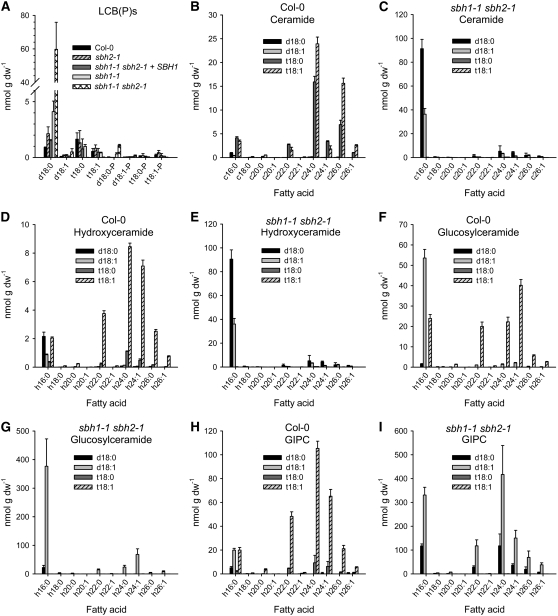
Comprehensive analyses of the complete complement of sphingolipids (sphingolipidomic analyses) in T-DNA mutants and RNAi suppression lines were conducted using the recently developed HPLC coupled to electrospray ionization-tandem mass spectrometry (ESI-MS/MS) methods that allow for the detection and quantification of the molecular species (or exact pairings of fatty acids and LCBs) of different sphingolipid classes (Markham and Jaworski, 2007). The goals of these studies were to determine the effects of partial and complete loss of LCB C-4 hydroxylation on sphingolipid metabolism and to examine whether SBH1 and SBH2 contribute preferentially to the hydroxylation of different sphingolipid classes. Four major classes of sphingolipid (ceramides, hydroxyceramides, GlcCers, and GIPCs) were analyzed along with free LCBs and phosphorylated LCBs (LCBPs) in 2-week-old T-DNA mutant seedlings maintained on MS plates.
Free LCBs and LCBPs, which are normally present at low levels, were twofold to fourfold higher in the sbh1-1 and sbh2-1 mutants and >60-fold higher in the sbh1-1 sbh2-1 double mutant than in wild-type plants (Figure 9A; see Supplemental Figure 5A online). The increase in free LCBs and LCBPs in the double mutant was due largely to the accumulation of free d18:0 (60 ± 16 nmol/g dry weight). This substantial increase in free d18:0 in the double mutant is not explained simply by reduced C-4 hydroxylation, as wild-type plants contained only 1.6 ± 0.6 nmol/g dry weight free t18:0. Instead, accumulation of d18:0 is more likely due to a bottleneck in the incorporation of this LCB into ceramides. For example, enhanced production of d18:0 in sbh1-1 sbh2-1 plants may exceed the capacity of ceramide synthase to incorporate these LCBs into ceramides.
Figure 9.
Measurements of Free LCBs and LCBPs [LCB(P)s]) and Molecular Species Composition of Sphingolipid Classes in Extracts from Leaves of Wild-Type (Col-0) and sbh1-1 sbh2-1 Plants Determined by HPLC-ESI-MS/MS.
(B), (D), (F), and (H) show measurements obtained from wild-type plants, and (C), (E), (G), and (I) show measurements obtained from sbh1-1 sbh2-1 double mutants. The measurements shown are of LCBPs (A), ceramide ([B] and [C]), hydroxyceramide ([D] and [E]), glucosylceramide ([F] and [G]), and GIPC ([H] and [I]). Results are averages from analyses of three independent samples ± sd.
Increases in the content of substrate pools of LCBs for ceramide synthases may also lead to higher levels of ceramides and ultimately to increased amounts of complex sphingolipids. Consistent with this, the ceramide content of sbh1-1 sbh2-1 mutants (150 ± 12 nmol/g dry weight) was nearly twofold higher than that of wild-type plants (86 ± 4 nmol/g dry weight) (see Supplemental Figure 5A online). In addition, the fatty acid composition of the ceramides of the double mutant was distinctly different from that of wild-type plants. Most notably, palmitic acid (16:0) accounted for ∼85% of the fatty acids in ceramides of the double mutant but only 13% of the fatty acids in ceramides of wild-type plants (Figures 9B and 9C). Accumulation of ceramides in single mutants of SBH1 and SBH2 was much less pronounced; however, the palmitic acid content of sbh1-1 ceramides was elevated relative to that of wild-type ceramides (44 versus 13%) (see Supplemental Figures 6A and 6B online).
Ceramides containing 2- (or α-) hydroxy fatty acids (hydroxyceramides) were similarly affected. The hydroxyceramide content of the sbh1 sbh2 double mutant (150 ± 12 nmol/g dry weight) was nearly fivefold higher than that in wild-type plants (31 ± 1 nmol/g dry weight) (Figures 9D and 9E). In addition, the relative content of 2OH-palmitic acid was increased from 18% in hydroxyceramides of wild-type plants to 89% in hydroxyceramides of sbh1-1 sbh2-1 plants (Figures 9D and 9E). Again, like ceramides in the single mutants, hydroxyceramides were unaffected in the sbh2-1 mutant. Increases in the total content of hydroxyceramides as well as in the relative content of 2OH-palmitic acid were detectable in the sbh1-1 mutant but were less severe than those in the double mutant (see Supplemental Figures 6D and 6F online).
Examination of the GlcCers from the sbh1-1 sbh2-1 double mutant showed that, as expected, they contained exclusively dihydroxy-LCBs, and the 2OH-palmitic acid content of their ceramide backbones was increased to 76 ± 1% from the 45 ± 1% found in GlcCers from wild-type plants (Figures 9F and 9G). Similarly, GIPCs from the sbh1-1 sbh2-1 double mutant contained only dihydroxy-LCBs and more than twice as much 2OH-palmitic acid than GIPCs from wild-type plants (Figures 9H and 9I). Consistent with enhanced pool sizes of ceramides in the double mutant, the total content of GlcCers was increased by nearly threefold and that of GIPCs was increased by nearly fivefold in the sbh1-1 sbh2-1 plants relative to wild-type plants (see Supplemental Figure 5A online). Overall, the increase in total sphingolipid content detected in sbh1-1 sbh2-1 double mutants was similar in magnitude (twofold to threefold) to those detected by HPLC analysis of LCBs from the hydrolyzed total complement of sphingolipids described above (Figure 5C).
GlcCer and GIPC levels in the sbh1-1 single mutant were increased and contained higher levels of 2OH-palmitic acid than in the wild-type controls, but the extent of these changes was considerably less than that detected in the sbh1-1 sbh2-1 mutants (Figures 9F to 9I; see Supplemental Figures 6G, 6H, 6J, and 6K online). Modest increases in the content of 2OH-palmitic acid were detected in GlcCers and GIPCs in the sbh2-1 mutant (Figures 9F to 9I; see Supplemental Figures 6H and 6K online), but little change in the total content of these lipids was measured. The observation that SBH1 mutants have at least small alterations in the composition of all sphingolipid classes suggests that the corresponding hydroxylase contributes globally to the synthesis of trihydroxy LCBs rather than to the synthesis of trihydroxy LCBs for specific sphingolipid classes.
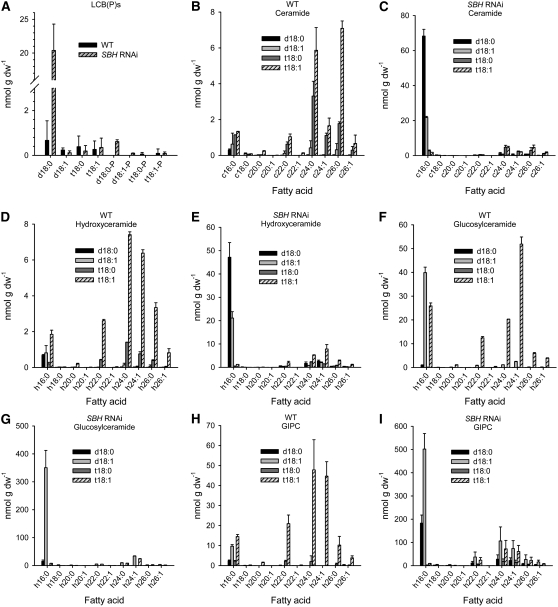
Analyses of soil-grown RNAi lines with 20% of the total sphingolipid LCBs in the trihydroxy form revealed similar phenotypes as those of the double mutant, including increased total content of each sphingolipid class and the accumulation of C16 fatty acid–containing sphingolipids (Figure 10; see Supplemental Figure 5B online).
Figure 10.
HPLC-ESI-MS/MS Analysis of the Sphingolipid Composition of Leaves from 6-Week-Old Soil-Grown Wild-Type and SBH RNAi Suppression Plants.
Approximately 20% of the total LCBs in the RNAi line used for these analyses was in the trihydroxy form. Panels show measurements from wild-type plants ([B], [D], [F], and [H]) and the SBH RNAi suppression line ([C], [E], [G], and [I]). Each panel shows measurements of individual molecular species for different sphingolipid classes, as follows: LCBPs (A), ceramides ([B] and [C]), hydroxyceramides ([D] and [E]), glucosylceramides ([F] and [G]), and GIPCs ([H] and [I]). Values are means ± sd (n = 3).
Overall, these results demonstrate a profound redirection of sphingolipid metabolism accompanying reductions in LCB C-4 hydroxylation of sphingolipids, leading to increased sphingolipid content and enhanced C16 fatty acid content of sphingolipids.
Glycerolipid Analyses of Double Mutant and RNAi Suppression Lines
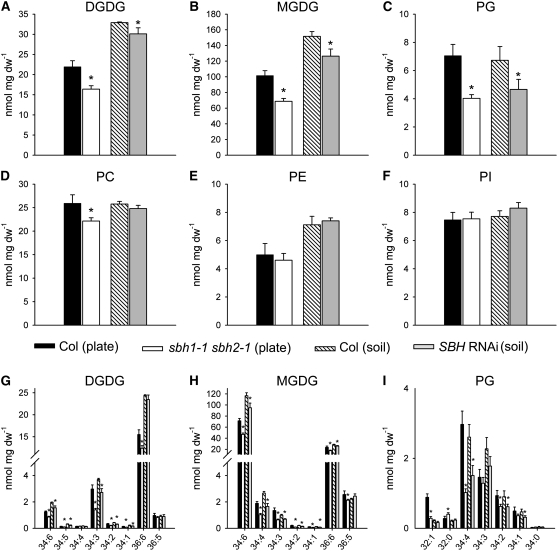
The content and molecular species composition of glycerolipids in a sbh1-1 sbh2-1 double mutant and an SBH RNAi line were analyzed by ESI-MS/MS. The purpose of these analyses was to determine if the large increases in the total content of sphingolipids and in the content of C16 fatty acid–containing species resulted in changes in glycerolipids. These studies were performed using plants grown under the same conditions and of similar age as those used for the sphingolipidomic analyses described above. The analytical technique used for these analyses allows for the measurement of 144 molecular species and has now been widely applied for the comprehensive measurement of glycerolipids in plants (Devaiah et al., 2006; Fritz et al., 2007; Welti et al., 2007). Using these methods, only a small reduction was detected in the content of phosphatidylcholine in the sbh1-1 sbh2-1 double mutant relative to the wild-type control (Figure 11D). Otherwise, little difference was measured in amounts of the major extraplastidic glycerolipids, including phosphatidylethanolamine and phosphatidylinositol, between wild-type plants, the C-4 hydroxylase double mutant, and the RNAi suppression line (Figures 11E and 11F). In addition, the molecular species compositions of these glycerolipids were similar among these lines (see Supplemental Data Set 1). By contrast, significant differences were detected in the content and composition of chloroplast-type lipids. In particular, amounts of the major leaf lipid monogalactosyldiacylglycerol (MGDG) were ∼33% lower in the double mutant than in the wild-type control and 17% lower in the RNAi line than in wild-type plants (Figure 11B). In both cases, most of these differences were the result of reductions in the C18/C16-containing molecular species 34:4 and 34:6 (Figure 11H). In addition, ∼25 and 9% less digalactosyldiacylglycerol (DGDG) and 43 and 30% less phosphatidylglycerol (PG) were detected in the sbh1-1 sbh2-1 double mutant and RNAi lines, respectively, compared with wild-type control plants (Figures 11A and 11C). As observed for MGDG, the lower levels of PG and DGDG were due primarily to reductions in amounts of the C18/C16-containing species 34:4 for PG and 34:3 and 34:6 for DGDG (Figures 11G and 11I). Overall, these results point to the possibility that the increased demand for palmitic acid (16:0) to support sphingolipid biosynthesis in LCB C-4 hydroxylase mutants is met by reducing the synthesis of chloroplast glycerolipids, particularly molecular species containing C16 fatty acids.
Figure 11.
Glycerolipid Content and Composition of Leaves from Wild-Type Plants, the sbh1-1 sbh2-1 Double Mutant, and SBH RNAi Lines.
(A) to (F) Content of major glycerolipids in leaves from 2-week-old seedlings of the wild type (Col-0), the sbh1-1 sbh2-1 double mutant, and SBH RNAi lines, as determined by ESI-MS/MS analysis. As indicated, double mutant and the corresponding wild-type control seedlings were maintained on MS plates, and the RNAi and control plants were grown in soil. Values are averages ± sd (n = 5).
(G) to (I) Lipid species of DGDG, MGDG, and PG. Values are averages ± sd (n = 5).
* Significant at P < 0.01 compared with the wild type based on Student's t test.
DISCUSSION
Plant sphingolipids are enriched in trihydroxy LCBs, and trihydroxy LCBs compose nearly 90% of the sphingolipids of Arabidopsis leaves(Markham et al., 2006). In this report, the physiological and metabolic significance of sphingolipid trihydroxy LCBs was examined by characterization of LCB C-4 hydroxylase mutants or RNAi suppression lines that have no detectable trihydroxy LCBs or have intermediate levels of LCB C-4 hydroxylation. We show that complete loss of trihydroxy LCBs results in severe reductions in the size of plants due to defects in both cell expansion and division, and trihydroxy LCB–deficient plants are unable to transition from vegetative to reproductive growth. In addition, examination of RNAi suppression lines revealed that the degree of dwarfing is correlated with the loss of trihydroxy LCBs, and reduced growth was observed in lines with trihydroxy LCB content levels ≤ ∼35% of the total LCBs in leaf extracts. By use of the recently developed ESI-MS/MS protocols for sphingolipid molecular species analysis (Markham and Jaworski, 2007), it was determined that reductions in trihydroxy LCBs are accompanied by the accumulation of sphingolipids with C16 fatty acids rather than the more typical very-long-chain fatty acids. In addition, the total content of sphingolipids in all classes was found to increase as the relative content of trihydroxy LCBs decreased. In mutants that completely lack trihydroxy LCBs, the total content of sphingolipids in leaves was twofold to threefold higher than that in leaves of wild-type plants. Overall, these results demonstrate that C-4 hydroxylation of LCBs is a critical structural modification for the maintenance of sphingolipid content and fatty acid composition and ultimately for the growth and reproductive potential of Arabidopsis.
Several formal explanations may account for the growth phenotypes associated with the loss of LCB C-4 hydroxylation in Arabidopsis seen in our studies. Among the possibilities is that the altered growth of the LCB C-4 hydroxylase mutants is the direct result of the shift in LCB composition from trihydroxy to dihydroxy moieties. Alternatively, growth defects may be due to secondary alterations in sphingolipid profiles, including increases in total sphingolipid content or the accumulation of sphingolipids containing fatty acids of shorter chain lengths. Notably, Arabidopsis cer10 mutants, which have reduced enoyl-CoA reductase activity, contain sphingolipids with increased relative amounts of C16 fatty acids and corresponding decreases in relative amounts of very-long-chain fatty acids (Zheng et al., 2005). These mutants display reduced growth, although not as severe as the reductions in growth in the LCB C-4 hydroxylase null mutants (Zheng et al., 2005). The growth phenotype of cer10 mutants was attributed to defects in sphingolipid-mediated endocytic protein trafficking (Zheng et al., 2005). Indeed, sphingolipids are major structural components of plasma membrane and tonoplast of plant cells (Verhoek et al., 1983; Yoshida and Uemura, 1986), and alterations in sphingolipid composition undoubtedly affect the physical properties of these membranes and perhaps the organization of membrane microdomains or lipid rafts, which have been linked to cellular functions such as cell wall formation and cell–cell recognition (Borner et al., 2005). In addition, sphingolipid metabolites are believed to be involved in the regulation of certain physiological processes in plant cells. For example, the LCB-1-phosphate derivative of phytosphingosine, a trihydroxy LCB, appears to be important in the abscisic acid–dependent signaling events leading to guard cell closure (Coursol et al., 2005). As such, it is possible that the loss of phytosphingosine-1-phosphate in the LCB C-4 hydroxylase mutants affects these and other physiological and cellular processes. Furthermore, ceramides are known to serve as components of glycosylphosphatidylinositol-type anchors for some cell surface proteins, including arabinogalactan proteins (Svetek et al., 1999). Although the quantitative significance of proteins with ceramide-containing anchors in plant cells is not known, the functions of these proteins and their transport to the plasma membrane may be altered in plants deficient in trihydroxy LCBs.
An additional phenotype observed in sbh1 sbh2 double mutants is the presence of necrotic lesions on cotyledons and premature death (Figure 6A). Indeed, the accumulation of ceramides and free LCBs, as observed in the hydroxylase double mutant, has been shown to induce PCD (Abbas et al., 1994; Spassieva et al., 2002; Liang et al., 2003; Townley et al., 2005). In addition, the acd11 mutant, which is defective in a putative LCB transport protein, has been shown to have constitutively upregulated marker genes for hypersensitive response–type PCD (Brodersen et al., 2002). Although the in vivo role of the putative LCB transport protein in sphingolipid metabolism has yet to be demonstrated, one possibility is that lesions in the ACD11 gene result in increased levels of LCBs or ceramides in plant cells that trigger PCD (Brodersen et al., 2002). Interestingly, we observed that a number of the PCD-associated genes that are expressed at high levels in acd11 mutants are also upregulated in the sbh1-1 sbh2-1 double mutant (Figure 8). This finding suggests that the necrotic lesions and premature death of the LCB C-4 hydroxylase null mutant arise from PCD, likely caused by the buildup of sphingolipids, including free LCBs and ceramides.
The observation that the LCB C-4 hydroxylase null mutants and RNAi suppression lines have twofold to threefold higher amounts of sphingolipids, primarily as C16 fatty acid–containing species, raises the question of whether these large alterations in sphingolipid production affect the synthesis of other lipids in plant cells. Comprehensive analyses of glycerolipids by ESI-MS/MS were conducted to address this question. These analyses revealed that amounts of chloroplast lipids, particularly MGDG and PG, were significantly lower in the sbh1-1 sbh2-1 double mutant and the RNAi suppression line relative to wild-type control plants. The decreased amounts of MGDG and PG were due largely to the reduced content of C16-containing molecular species. Conversely, only small changes or no significant changes were detected in the content and composition of the major ER-type lipids (e.g., phosphatidylcholine and phosphatidylethanolamine). Although a number of explanations could be evoked from these data, one possibility is that plant cells reduce the synthesis of chloroplast lipids to meet the increased requirement for 16:0 to support enhanced sphingolipid production in plants defective in LCB C-4 hydroxylation. This implies metabolic coordination between the ER and chloroplasts for regulation of the biosynthesis of sphingolipids and chloroplast-specific lipids. The existence of such a metabolic network awaits further investigation. It should be noted that ESI-MS/MS analyses of sphingolipids and glycerolipids were conducted using independent methodologies, which limits direct comparisons of absolute amounts of these lipids between data sets.
Our results indicate that the combined activities of SBH1 and SBH2 account for all detectable synthesis of trihydroxy LCBs in Arabidopsis leaves. However, it cannot be ruled out that sphingolipid Δ4 desaturases (Ternes et al., 2002), which are distantly related to LCB C-4 hydroxylases, also introduce some portion of the C-4 hydroxy groups found in trihydroxy LCBs, particularly in plants such as tomato (Solanum lycopersicum) and maize (Zea mays), which contain high levels of Δ4 unsaturated LCBs (Dunn et al., 2004; Markham et al., 2006). It is notable that in mammals and certain fungi, the Δ4 desaturase homologs of DES2 also display some level of LCB C-4 hydroxylase activity (Ternes et al., 2002; Omae et al., 2004). The contributions of the sphingolipid Δ4 desaturase to LCB hydroxylation is difficult to assess in Arabidopsis. The Arabidopsis sphingolipid Δ4 desaturase gene is expressed only in flowers, as indicated by publicly available microarray data (Zimmermann et al., 2004), and LCB products of Δ4 desaturation are not detectable in nonfloral tissues of Arabidopsis (Columbia; Sperling et al., 2005; Markham et al., 2006). It has been reported previously that that LCB C-4 hydroxylation in crude microsomes from maize leaves can use free LCBs or LCB components of ceramides as substrates (Wright et al., 2003). It was suggested that discrete hydroxylases catalyze these reactions, because free LCBs and ceramides do not act as competitors in these assays (Wright et al., 2003). It is possible that these different activities arise from the LCB C-4 hydroxylase and the sphingolipid Δ4 desaturase. In this regard, Δ4 unsaturated LCBs are found almost entirely in GlcCers rather than in GIPCs (Markham et al., 2006; Markham and Jaworski, 2007), suggesting that Δ4 desaturases function directly on ceramides in GlcCers or on a separate pool of ceramides used in the synthesis of GlcCers. Conversely, consistent with the activity on free LCBs, C-4 hydroxylase–derived trihydroxy LCBs are found in all sphingolipid classes, including pools of free LCBs and LCB-1 phosphates (Markham and Jaworski, 2007).
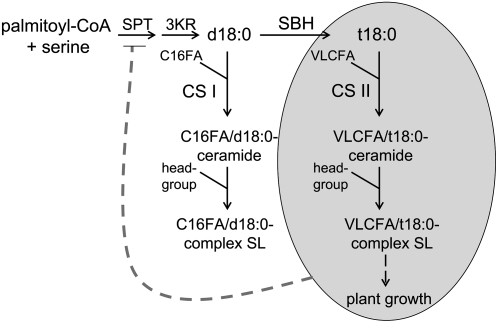
The results presented here indicate that trihydroxy LCBs resulting from LCB C-4 hydroxylation play a central role in the maintenance of growth, the fatty acid composition, and the total content of sphingolipids. A model that rationalizes these observations is proposed in Figure 12. As indicated, the accumulation of sphingolipid molecular species enriched in C16 fatty acids is likely a reflection of the substrate specificity of acyl-CoA–dependent ceramide synthases (or sphinganine N-acyltransferases), which catalyze the condensation of LCBs with fatty acyl-CoAs. The data presented here are most consistent with the occurrence of two classes of acyl-CoA–dependent ceramide synthases (designated CS I and CS II in Figure 12) in Arabidopsis. The putative CS I class preferentially links dihydroxy LCBs with C16 fatty acyl-CoAs, while the CS II class preferentially links trihydroxy LCBs with very-long-chain fatty acyl-CoAs. Indeed, in wild-type Arabidopsis, dihydroxy LCBs are most frequently bound to C16 fatty acids (Markham and Jaworski, 2007), which is accentuated in the LCB C-4 hydroxylase mutants. In addition, acyl-CoA–dependent ceramide synthases with distinct specificities for acyl-CoAs of different chain lengths have been described in mouse and human cells (Guillas et al., 2003; Mizutani et al., 2005; Pewzner-Jung et al., 2006). For example, the mouse LASS6 ceramide synthase is most active, with acyl-CoAs containing fatty acid chains with 12 to 18 carbon atoms, but it displays little activity with C20 to C26 acyl-CoAs (Mizutani et al., 2005). Conversely, the mouse LASS2 ceramide synthase is most active, with acyl-CoAs containing fatty acids with 22, 24, and 26 carbon atoms, but it has only low activity with shorter chain acyl-CoAs (Mizutani et al., 2005). The occurrence in plants of ceramide synthases with differing substrate specificities has also been suggested from research conducted on the Alternaria stem canker-1 (asc-1) mutant of tomato (Spassieva et al., 2002). In these studies, expression of the Asc-1–encoded ceramide synthase in yeast lacking the endogenous genes for ceramide synthase created new sphingolipid species, suggesting a different substrate specificity for the tomato ceramide synthase (Spassieva et al., 2002).
Figure 12.
Model Showing the Central Role of Trihydroxy LCB Synthesis in Growth and Sphingolipid Metabolism.
The accumulation of dihydroxy LCBs (d18:0) resulting from impaired C-4 hydroxylation causes sphingolipids (SL) to accumulate that are enriched in C16 fatty acids (C16FA). This can be rationalized by the presence of two classes of acyl-CoA–dependent ceramide synthases: CS I, which preferentially uses dihydroxy LCBs and CoA esters of C16 fatty acids (FA) as substrates, and CS II, which preferentially uses trihydroxy LCBs (t18:0) and CoA esters of very-long-chain (≥C20) fatty acids (VLCFA) as substrates. As revealed by the phenotypes of sbh1-1 sbh2-1 mutants and RNAi suppression lines, LCB C-4 hydroxylation (as indicated by SBH) is critical for growth (as indicated in the shaded oval), and the synthesis of trihydroxy LCB–containing sphingolipids is likely regulated, perhaps through Ser palmitoyltransferase (SPT), to meet the demands of growth. Loss of LCB C-4 hydroxylation appears to uncouple this regulation, resulting in the accumulation of sphingolipids with dihydroxy LCB/C16 fatty acid–containing ceramide backbones. 3KR, 3-ketosphinganine reductase.
As indicated in the model in Figure 12 and supported by the phenotypes of C-4 hydroxylase mutants and RNAi suppression lines, trihydroxy LCB production is required for growth. The model does not distinguish between the contributions of trihydroxy LCBs and very-long-chain fatty acids to growth. Regardless, LCB C-4 hydroxylation is important for mediating the incorporation of very-long-chain fatty acids into the ceramides of sphingolipids. In the absence of LCB C-4 hydroxylation, a preponderance of C16 fatty acid–containing sphingolipids accumulates. It is likely that the regulation of sphingolipid synthesis and growth are tightly intertwined. Consistent with this, we recently showed that plants adjust growth to compensate for reduced sphingolipid synthesis in Arabidopsis RNAi suppression lines for the LCB1 subunit of Ser palmitoyltransferase (Chen et al., 2006). We propose that reduction or elimination of LCB C-4 hydroxylation uncouples the regulation of sphingolipid synthesis from the demands for growth, resulting in enhanced flux through the sphingolipid biosynthetic pathway and the increased accumulation of sphingolipids, as observed in mutants and RNAi suppression lines. Regulation of sphingolipid synthesis in yeast and animals is generally believed to be mediated through Ser palmitoyltransferase (designated SPT in Figure 12), although the regulation of sphingolipid synthesis in these systems is still not well defined (Hanada, 2003). The regulatory interactions between the formation of trihydroxy LCBs and SPT is likely to be a fruitful topic for research aimed at understanding how sphingolipid synthesis is controlled in plants.
Overall, the characterization of T-DNA mutants and RNAi suppression lines for C-4 hydroxylase genes in these studies has provided unexpected insights into the significance of sphingolipid structure for plant growth and development. These results also highlight how structural modifications, such as C-4 hydroxylation, of LCBs can dictate metabolic flow in the sphingolipid biosynthetic pathway in Arabidopsis. It is expected that the results presented here will provide direction for future studies of sphingolipid function and metabolism in plants.
METHODS
Plant Material and Growth Conditions
Wild-type and mutant Arabidopsis thaliana lines used for these studies were of the Columbia (Col-0) ecotype. Multiple T-DNA insertion mutants for SBH1 and SBH2 (Figure 4A) were obtained from the ABRC (Alonso et al., 2003). Primers used for PCR genotyping of these mutants are provided in Supplemental Table 1 online.
Plants were grown on soil or MS agar as described previously (Chen et al., 2006). For growth under long-day conditions, plants were maintained at 22°C and 50% humidity with a 16-h-light (100 μmol·m−2·s−1)/8-h-dark cycle. Root elongation measurements were conducted on plants grown on plates in a vertical orientation. To measure elongation of hypocotyls in response to etiolation, seeds were sown on MS agar plates and exposed to light for 6 to 8 h after stratification to promote germination. Plates were then wrapped in aluminum foil and maintained for 7 d at 22°C in a vertical orientation. The ability of exogenous LCBs to complement C-4 hydroxylase sbh1-1 sbh2-1 double mutants was examined by the addition of phytosphingosine (t18:0; Matreya) in methanol at varying concentrations up to 200 μM to MS medium supplemented with 0.2% (w/v) Tergitol Nonidet P-40 (Sigma-Aldrich) (Abbas et al., 1994).
RNA Isolation and RNA Gel Blotting
Total RNA was isolated from 10-d-old soil-grown seedlings. For analyses of organ-specific expression of SBH1 and SBH2, 6- to 8-week-old wild-type plants were used as sources of plant material. RNA extraction was performed using the RNeasy Plant Kit (Qiagen) according to the manufacturer's protocol. Total RNA (15 μg) was treated with glyoxal (Ambion) and electrophoresed on a 1% (w/v) agarose gel and then transferred to Hybond nylon membranes (GE Bioscience) using the reagents and protocol supplied with the NorthernMax kit (Ambion). Hybridization and detection were conducted according to the protocol for the DIG High Primer DNA Labeling and Detection Starter Kit II (Roche). The DNA probe was labeled with the PCR DIG Probe Synthesis Kit (Roche). Gene-specific probes were obtained from PCR products corresponding to the 3′-untranslated region of SBH1 or SBH2. Oligonucleotides P9 and P10 were used for generation of the SBH1-specific probe, and P11 and P12 were used to generate the SBH2-specific probe (see Supplemental Table 2 online for the sequences of oligonucleotides).
RT-PCR Verification of T-DNA Mutants
Total RNA was isolated from 10-d-old wild-type and mutant seedlings grown on 1× MS plates. Total RNA (1 μg) was first treated with DNase (Roche), and first-strand cDNA was subsequently synthesized using SuperScript III reverse transcriptase (Invitrogen) and oligo(dT) primer, according to the manufacturer's instructions. A 2-μL aliquot of first-strand cDNA was used as template to amplify SBH1, SBH2, or the gene for the ubiquitin-conjugating enzyme (UBC; At5g25760) in a 20-μL reaction with 30 cycles of amplification and 55°C annealing temperature. PCR amplification was conducted with Taq DNA polymerase (New England Biolabs). Primers used for amplification of SBH1-derived cDNA were P9 and P10, and those used for SBH2 were P11 and P12 (see Supplemental Table 2 online). Amplification products were analyzed by electrophoresis in 1% agarose (w/v) and detected by ethidium bromide staining. The gene for the UBC (At5g25760) was used as an internal control (Czechowski et al., 2005). Primers P13 and P14 (see Supplemental Table 2 online) were used for amplification of the UBC-derived cDNA.
Expression Analysis of PCD Marker Genes
Total RNA was extracted from 2-week-old control and sbh1-1 sbh2-1 double mutant plants grown on MS plates, and first-strand cDNA was prepared as described above. RT-PCR analysis was conducted with equal amounts of first-strand cDNA as template. Oligonucleotides and the numbers of PCR cycles used for each target gene are provided in Supplemental Table 3 online. UBC (At5g25760) was amplified and used as an internal positive control.
Promoter-GUS Assay of SBH1 and SBH2
A SBH1 promoter–GUS reporter construct was generated by amplification of an ∼2.0-kb sequence upstream of the start codon of SBH1 (At1g69640) using the sense and antisense oligonucleotides P19 and P20 (see Supplemental Table 2 online). The product was digested with HindIII and XbaI and cloned into the corresponding sites of binary vector pBI121 (Clontech) to generate a transcriptional fusion with the GUS coding region. The resulting plasmid was designated ProSBH1:GUS. ProSBH2:GUS was generated in a similar way using primer pair P21 and P22 (see Supplemental Table 2 online). Homozygous lines containing both ProSBH1:GUS and ProSBH2:GUS constructs were isolated from the T3 generation. Histochemical assay for GUS activity was performed as described previously (Gallagher, 1992).
Subcellular Localization of SBH1 and SBH2
CFP fusion proteins with SBH1 and SBH2 were prepared by amplification of the SBH1 and SBH2 open reading frames with oligonucleotides P3/P4 and P6/P7 (see Supplemental Table 2 online), respectively. PCR products were first cloned into the pENTR/D-TOPO vector (Invitrogen). The resulting plasmids were combined with the destination vector pEarleygate 102 (Earley et al., 2006) in an attL × attR recombination reaction to generate the CFP C-terminal fusion constructs pSBH1-CFP and pSBH2-CFP. Similarly, YFP-SBH1 and YFP-SBH2 fusion proteins with YFP at their N termini were generated using the primer pair P3/P5 and P6/P8 (see Supplemental Table 2 online), respectively, and the destination vector pEarleygate 104.
Agrobacterium tumefaciens–mediated infiltration of tobacco (Nicotiana tabacum) leaves with pSBH1-CFP and pSBH2-CFP and confocal laser scanning microscopy were conducted using a Carl Zeiss LSM 510 laser scanning microscope as described previously (Chen et al., 2006). The ER-specific marker used for these studies was YFP fused to the ER signal peptide of basic chitinase at its N terminus and to the HDEL ER retention sequence at its C terminus (Chen et al., 2006).
The localizations of SBH1– and SBH2–fluorescent protein fusions were also examined relative to a plasma membrane marker dye. For these studies, leaves of tobacco transiently expressing SBH1-CFP or SBH2-CFP were immersed in 20 μM FM4-64 (Invitrogen) for 10 min to stain the plasma membrane of cells. CFP was excited with a 458-nm argon laser line and a 480- to 520-nm band-pass emission filter, while FM4-64 was excited with a 543-nm argon laser line and a 565- to 615-nm band-pass emission filter.
Complementation of a Yeast sur2Δ Mutant with SBH–Fluorescent Protein Fusions
An LCB C-4 hydroxylase mutant (sur2Δ) of Saccharomyces cerevisiae strain BY4741 was prepared by disruption of the SUR2 gene with a LEU2 marker using methods described previously (Haak et al., 1997). The coding sequences of N-terminal fusions of SBH1 or SBH2 with YFP and C-terminal fusions of these proteins with CFP were amplified by PCR from plant expression vectors used in transient localization studies. The oligonucleotides used for amplification of SBH1- or SBH2-CFP nucleotide sequences were P23/P24 and P24/P25, respectively, and those used for amplification of YFP-SBH1 or YFP-SBH2 sequences were P26/P27 and P26/P28, respectively (see Supplemental Table 2 online). PCR products were digested with HindIII and XbaI and cloned into the yeast expression vector pYES2 (Invitrogen). The resulting plasmids were transformed into the sur2Δ yeast mutant, and the fusion proteins were expressed with galactose induction of 3-mL cultures using previously described methods (Cahoon and Kinney, 2004). The total LCB composition of the yeast was determined by heating of cell pellets in barium hydroxide/dioxane followed by HPLC analysis of fluorescent derivatives of released LCBs, as described previously (Morrison and Hay, 1970; Chen et al., 2006).
Genetic Complementation of sbh1-1
For genomic complementation of sbh1-1 or sbh1-1 sbh2-1 plants, an ∼2.7-kb fragment of SBH1 was amplified from Arabidopsis (Col-0) genomic DNA using the oligonucleotides P1 and P2 (for primer sequences, see Supplemental Table 2 online). The amplified product was then digested with AscI and PacI and cloned into the corresponding restriction sites of the binary vector pMDC123 (Curtis and Grossniklaus, 2003) in place of the cloning cassette to produce pMDC123_SBH1g. Transformation of sbh1-1 or sbh1-1−/+ sbh2-1−/− with pMDC123_SBH1g was performed by the floral dip method (Clough and Bent, 1998), and transformants were selected by resistance to glufosinate (10 mg/L; Sigma-Aldrich).
Generation of SBH RNAi Suppression Plants
A SBH1 RNAi suppression construct was generated using the Kannibal RNAi vector system (Helliwell and Waterhouse, 2003). A 356-bp fragment containing base pairs 25 to 380 of the SBH1 open reading frame was amplified using two pairs of oligonucleotides, P15/P16 and P17/P18 (see Supplemental Table 2 online), and the products of the two reactions were cloned sequentially into the pKannibal vector. The amplified portion of SBH1 contains regions of absolute homology with the open reading frame of SBH2. The resulting hairpin construct together with the cauliflower mosaic virus 35S promoter and the ocs terminator were released and inserted into the NotI site of binary vector pART27 to generate pKanSBH1i. Binary vectors were introduced into Agrobacterium GV3101 by electroporation, and transgenic plants were generated by floral dip of Arabidopsis (Col-0) (Clough and Bent, 1998) and screened on MS plates containing 50 mg/L kanamycin monosulfate. Four- to 6-week-old T2 plants grown in soil were used to analyze total LCBs and sphingolipid molecular species.
Microscopy
Imaging of wild-type and mutant plants was performed using a Nikon SMZ1500 dissection microscope attached to a digital camera (Retiga 1300; Qimaging), and images were processed with IPLab software. Hypocotyls were cleared with Hoyer's solution and observed with a Nikon Eclipse E800 microscope equipped with a differential interference contrast apparatus (Nomarski optics) as described previously (Chen et al., 2006). Observations of root meristem zones were conducted with whole seedlings that were stained with propidium iodide (10 μg/mL [w/v]; Sigma-Aldrich) for 2 to 5 min and then rinsed with water. Stained roots were analyzed with a Carl Zeiss LSM 510 laser scanning microscope with excitation at 543 nm and emission at 590 nm.
Analysis of Sphingolipid LCBs
The total content and composition of sphingolipid LCBs were determined following strong alkaline hydrolysis of extracted complex sphingolipids, fluorescent derivatization of released LCBs, and subsequent reverse-phase HPLC analysis as described previously (Morrison and Hay, 1970; Chen et al., 2006). For analysis of the total content of sphingolipid, plants were grown for 14 d on the MS plates supplied with 1% (w/v) sucrose. For analysis of the sphingolipid LCB composition of different organs, plants were grown under long-day conditions (see above) and harvested at 8 weeks of age.
Sphingolipidomic Analyses
Wild-type or mutant plants were grown on 1× MS agar plates supplied with 1% sucrose for 14 d, and aerial portions of the seedlings were collected and lyophilized. For RNAi lines and wild-type controls, plants were maintained on soil using the growth conditions described above, and leaves were collected from 6-week-old plants and lyophilized. Global analyses of molecular species of different sphingolipid classes were conducted with reverse-phase HPLC coupled to ESI-MS/MS as described by Markham and Jaworski (2007).
Glycerolipid Analyses
Comprehensive glycerolipid analyses were conducted on sbh1-1 sbh2-1 double mutant and RNAi suppression lines grown and harvested as described above for sphingolipidomic analyses. Lipid extractions, ESI-MS/MS analyses, and lipid quantification were conducted as described (Devaiah et al., 2006) with minor modifications. For analysis of DGDG, 341.11 [M + NH4]+ and collision energy of 24 V were used. For analysis of MGDG, 179.08 [M + NH4]+ and collision energy of 21 V were used. The ESI-MS/MS analyses were performed by the Kansas Lipidomics Research Center.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: SBH1, NM_105632; SBH2, NM_101295.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Comparison of the ER Localization of SBH1-CFP and SBH2-CFP Versus a Plasma Membrane Marker Dye.
Supplemental Figure 2. LCB C-4 Hydroxylase Activity of Fluorescent Protein Fusions of SBH1 and SBH2.
Supplemental Figure 3. Expression Analysis of SBH1 and SBH2 in sbh1 sbh2 Double Mutants.
Supplemental Figure 4. Epidermal Cell Length in Arabidopsis Col-0 Hypocotyls Versus That in the sbh1-1 sbh2-1 Double Mutant.
Supplemental Figure 5. Content of Sphingolipid Classes in sbh1-1 sbh2-1 Double Mutant and RNAi Suppression Lines.
Supplemental Figure 6. Sphingolipid Composition of Leaves from sbh1-1, sbh2-1, and the SBH1-Complemented sbh1-1 sbh2-1 Double Mutant.
Supplemental Table 1. Oligonucleotide Primer Sequences Used for Confirmation of T-DNA Insertion Sites of sbh1 and sbh2 Mutants.
Supplemental Table 2. Oligonucleotide Primer Sequences Used for RT-PCR, Complementation, Subcellular Localizations, Promoter-GUS Assays, and RNA Gel Blot Analyses.
Supplemental Table 3. Oligonucleotides and Numbers of PCR Cycles Used for Expression Analysis of Cell Death and Hypersensitive Response–Related Genes.
Supplemental Data Set 1. ESI-MS/MS Analysis of Glycerolipid Molecular Species in Mutant and RNAi Lines.
Supplementary Material
Acknowledgments
We thank our colleagues Daniel Lynch and Teresa Dunn for helpful comments, Jia Li for technical assistance, R. Howard Berg for assistance with confocal laser microscopy, and the ABRC for providing seeds for SALK and SAIL mutants. We are also indebted to Mary Roth and Ruth Welti of the Kansas Lipidomics Research Center for their expert profiling of glycerolipids. The Kansas Lipidomics Research Center is supported by the National Science Foundation (Grants EPS 0236913, MCB 0455318, and DBI 0521587), the Kansas Technology Enterprise Corporation, the K-IDeA Networks of Biomedical Research Excellence of the National Institutes of Health (Grant P20 RR-16475), and Kansas State University. This work was supported by a National Science Foundation Arabidopsis 2010 Grant to J.G.J. and E.B.C. (Grant MCB 0312559).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Edgar B. Cahoon (ecahoon@danforthcenter.org).
Online version contains Web-only data.
References
- Abbas, H.K., Tanaka, T., Duke, S.O., Porter, J.K., Wray, E.M., Hodges, L., Sessions, A.E., Wang, E., Merrill, A.H., Jr., and Riley, R.T. (1994). Fumonisin- and AAL-toxin-induced disruption of sphingolipid metabolism with accumulation of free sphingoid bases. Plant Physiol. 106 1085–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Borner, G.H., Sherrier, D.J., Weimar, T., Michaelson, L.V., Hawkins, N.D., Macaskill, A., Napier, J.A., Beale, M.H., Lilley, K.S., and Dupree, P. (2005). Analysis of detergent-resistant membranes in Arabidopsis. Evidence for plasma membrane lipid rafts. Plant Physiol. 137 104–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen, P., Petersen, M., Pike, H.M., Olszak, B., Skov, S., Odum, N., Jorgensen, L.B., Brown, R.E., and Mundy, J. (2002). Knockout of Arabidopsis ACCELERATED-CELL-DEATH11 encoding a sphingosine transfer protein causes activation of programmed cell death and defense. Genes Dev. 16 490–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoon, E.B., and Kinney, A.J. (2004). Dimorphecolic acid is synthesized by the coordinate activities of two divergent Δ12-oleic acid desaturases. J. Biol. Chem. 279 12495–12502. [DOI] [PubMed] [Google Scholar]
- Casamitjana-Martinez, E., Hofhuis, H.F., Xu, J., Liu, C.M., Heidstra, R., and Scheres, B. (2003). Root-specific CLE19 overexpression and the sol1/2 suppressors implicate a CLV-like pathway in the control of Arabidopsis root meristem maintenance. Curr. Biol. 13 1435–1441. [DOI] [PubMed] [Google Scholar]
- Chen, M., Han, G., Dietrich, C.R., Dunn, T.M., and Cahoon, E.B. (2006). The essential nature of sphingolipids in plants as revealed by the functional identification and characterization of the Arabidopsis LCB1 subunit of serine palmitoyltransferase. Plant Cell 12 3576–3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cliften, P., Wang, Y., Mochizuki, D., Miyakawa, T., Wangspa, R., Hughes, J., and Takemoto, J.Y. (1996). SYR2, a gene necessary for syringomycin growth inhibition of Saccharomyces cerevisiae. Microbiology 142 477–484. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Coursol, S., Fan, L.M., Le Stunff, H., Spiegel, S., Gilroy, S., and Assmann, S.M. (2003). Sphingolipid signalling in Arabidopsis guard cells involves heterotrimeric G proteins. Nature 423 651–654. [DOI] [PubMed] [Google Scholar]
- Coursol, S., Le Stunff, H., Lynch, D.V., Gilroy, S., Assmann, S.M., and Spiegel, S. (2005). Arabidopsis sphingosine kinase and the effects of phytosphingosine-1-phosphate on stomatal aperture. Plant Physiol. 137 724–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis, M.D., and Grossniklaus, U. (2003). A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski, T., Stitt, M., Altmann, T., Udvardi, M.K., and Scheible, W.R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaiah, S.P., Roth, M.R., Baughman, E., Li, M., Tamura, P., Jeannotte, R., Welti, R., and Wang, X. (2006). Quantitative profiling of polar glycerolipid species from organs of wild-type Arabidopsis and a PHOSPHOLIPASE Dα1 knockout mutant. Phytochemistry 67 1907–1924. [DOI] [PubMed] [Google Scholar]
- Dunn, T.M., Lynch, D.V., Michaelson, L.V., and Napier, J.A. (2004). A post-genomic approach to understanding sphingolipid metabolism in Arabidopsis thaliana. Ann. Bot. (Lond.) 93 483–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley, K.W., Haag, J.R., Pontes, O., Opper, K., Juehne, T., Song, K., and Pikaard, C.S. (2006). Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45 616–629. [DOI] [PubMed] [Google Scholar]
- Fritz, M., Lokstein, H., Hackenberg, D., Welti, R., Roth, M., Zahringer, U., Fulda, M., Hellmeyer, W., Ott, C., Wolter, F.P., and Heinz, E. (2007). Channeling of eukaryotic diacylglycerol into the biosynthesis of plastidial phosphatidylglycerol. J. Biol. Chem. 282 4613–4625. [DOI] [PubMed] [Google Scholar]
- Gallagher, S.R., ed (1992). GUS Protocols: Using the GUS Gene as a Reporter of Gene Expression. (San Diego, CA: Academic Press).
- Grilley, M.M., Stock, S.D., Dickson, R.C., Lester, R.L., and Takemoto, J.Y. (1998). Syringomycin action gene SYR2 is essential for sphingolipid 4-hydroxylation in Saccharomyces cerevisiae. J. Biol. Chem. 273 11062–11068. [DOI] [PubMed] [Google Scholar]
- Guillas, I., Jiang, J.C., Vionnet, C., Roubaty, C., Uldry, D., Chuard, R., Wang, J., Jazwinski, S.M., and Conzelmann, A. (2003). Human homologues of LAG1 reconstitute acyl-CoA-dependent ceramide synthesis in yeast. J. Biol. Chem. 278 37083–37091. [DOI] [PubMed] [Google Scholar]
- Haak, D., Gable, K., Beeler, T., and Dunn, T. (1997). Hydroxylation of Saccharomyces cerevisiae ceramides requires Sur2p and Scs7p. J. Biol. Chem. 272 29704–29710. [DOI] [PubMed] [Google Scholar]
- Hanada, K. (2003). Serine palmitoyltransferase, a key enzyme of sphingolipid metabolism. Biochim. Biophys. Acta 1632 16–30. [DOI] [PubMed] [Google Scholar]
- Helliwell, C., and Waterhouse, P. (2003). Constructs and methods for high-throughput gene silencing in plants. Methods 30 289–295. [DOI] [PubMed] [Google Scholar]
- Idkowiak-Baldys, J., Grilley, M.M., and Takemoto, J.Y. (2004). Sphingolipid C4 hydroxylation influences properties of yeast detergent-insoluble glycolipid-enriched membranes. FEBS Lett. 569 272–276. [DOI] [PubMed] [Google Scholar]
- Imamura, T., Kusano, H., Kajigaya, Y., Ichikawa, M., and Shimada, H. (2007). A rice dihydrosphingosine C4 hydroxylase (DSH1) gene, which is abundantly expressed in stigmas, vascular cells and apical meristem, may be involved in fertility. Plant Cell Physiol. 48 1108–1120. [DOI] [PubMed] [Google Scholar]
- Kaul, K., and Lester, R.L. (1978). Isolation of six novel phosphoinositol-containing sphingolipids from tobacco leaves. Biochemistry 17 3569–3575. [DOI] [PubMed] [Google Scholar]
- Li, S., Bao, D., Yuen, G., Harris, S.D., and Calvo, A.M. (2007). basA regulates cell wall organization and asexual/sexual sporulation ratio in Aspergillus nidulans. Genetics 176 243–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, H., Yao, N., Song, J.T., Luo, S., Lu, H., and Greenberg, J.T. (2003). Ceramides modulate programmed cell death in plants. Genes Dev. 17 2636–2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch, D.V., and Dunn, T.M. (2004). An introduction to plant sphingolipids and a review of recent advances in understanding their metabolism and function. New Phytol. 161 677–702. [DOI] [PubMed] [Google Scholar]
- Markham, J.E., and Jaworski, J.G. (2007). Rapid measurement of sphingolipids from Arabidopsis thaliana by reversed-phase high-performance liquid chromatography coupled to electrospray ionization tandem mass spectrometry. Rapid Commun. Mass Spectrom. 21 1304–1314. [DOI] [PubMed] [Google Scholar]
- Markham, J.E., Li, J., Cahoon, E.B., and Jaworski, J.G. (2006). Separation and identification of major plant sphingolipid classes from leaves. J. Biol. Chem. 281 22684–22694. [DOI] [PubMed] [Google Scholar]
- Mizutani, Y., Kihara, A., and Igarashi, Y. (2005). Mammalian Lass6 and its related family members regulate synthesis of specific ceramides. Biochem. J. 390 263–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongrand, S., Morel, J., Laroche, J., Claverol, S., Carde, J.P., Hartmann, M.A., Bonneu, M., Simon-Plas, F., Lessire, R., and Bessoule, J.J. (2004). Lipid rafts in higher plant cells: Purification and characterization of Triton X-100-insoluble microdomains from tobacco plasma membrane. J. Biol. Chem. 279 36277–36286. [DOI] [PubMed] [Google Scholar]
- Morrison, W.R., and Hay, J.D. (1970). Polar lipids in bovine milk. II. Long-chain bases, normal and 2-hydroxy fatty acids, and isomeric cis and trans monoenoic fatty acids in the sphingolipids. Biochim. Biophys. Acta 202 460–467. [DOI] [PubMed] [Google Scholar]
- Ng, C.K., Carr, K., McAinsh, M.R., Powell, B., and Hetherington, A.M. (2001). Drought-induced guard cell signal transduction involves sphingosine-1-phosphate. Nature 410 596–599. [DOI] [PubMed] [Google Scholar]
- Omae, F., Miyazaki, M., Enomoto, A., Suzuki, M., Suzuki, Y., and Suzuki, A. (2004). DES2 protein is responsible for phytoceramide biosynthesis in the mouse small intestine. Biochem. J. 379 687–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pewzner-Jung, Y., Ben-Dor, S., and Futerman, A.H. (2006). When do Lasses (longevity assurance genes) become CerS (ceramide synthases)? Insights into the regulation of ceramide synthesis. J. Biol. Chem. 281 25001–25005. [DOI] [PubMed] [Google Scholar]
- Ryan, P.R., Liu, Q., Sperling, P., Dong, B., Franke, S., and Delhaize, E. (2007). A higher plant Δ8 sphingolipid desaturase with a preference for (Z)-isomer formation confers aluminum tolerance to yeast and plants. Plant Physiol. 144 1968–1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spassieva, S.D., Markham, J.E., and Hille, J. (2002). The plant disease resistance gene Asc-1 prevents disruption of sphingolipid metabolism during AAL-toxin-induced programmed cell death. Plant J. 32 561–572. [DOI] [PubMed] [Google Scholar]
- Sperling, P., Franke, S., Luthje, S., and Heinz, E. (2005). Are glucocerebrosides the predominant sphingolipids in plant plasma membranes? Plant Physiol. Biochem. 43 1031–1038. [DOI] [PubMed] [Google Scholar]
- Sperling, P., Ternes, P., Moll, H., Franke, S., Zahringer, U., and Heinz, E. (2001). Functional characterization of sphingolipid C4-hydroxylase genes from Arabidopsis thaliana. FEBS Lett. 494 90–94. [DOI] [PubMed] [Google Scholar]
- Svetek, J., Yadav, M.P., and Nothnagel, E.A. (1999). Presence of a glycosylphosphatidylinositol lipid anchor on rose arabinogalactan proteins. J. Biol. Chem. 274 14724–14733. [DOI] [PubMed] [Google Scholar]
- Ternes, P., Franke, S., Zahringer, U., Sperling, P., and Heinz, E. (2002). Identification and characterization of a sphingolipid Δ4-desaturase family. J. Biol. Chem. 277 25512–25518. [DOI] [PubMed] [Google Scholar]
- Townley, H.E., McDonald, K., Jenkins, G.I., Knight, M.R., and Leaver, C.J. (2005). Ceramides induce programmed cell death in Arabidopsis cells in a calcium-dependent manner. Biol. Chem. 386 161–166. [DOI] [PubMed] [Google Scholar]
- Verhoek, B., Haas, R., Wrage, K., Linscheid, M., and Heinz, E. (1983). Lipids and enzymatic activities in vacuolar membranes isolated via protoplasts from oat primary leaves. Z Naturforsch 38c 770–777. [Google Scholar]
- Welti, R., Shah, J., Li, W., Li, M., Chen, J., Burke, J.J., Fauconnier, M.L., Chapman, K., Chye, M.L., and Wang, X. (2007). Plant lipidomics: Discerning biological function by profiling plant complex lipids using mass spectrometry. Front. Biosci. 12 2494–2506. [DOI] [PubMed] [Google Scholar]
- Wright, B.S., Snow, J.W., O'Brien, T.C., and Lynch, D.V. (2003). Synthesis of 4-hydroxysphinganine and characterization of sphinganine hydroxylase activity in corn. Arch. Biochem. Biophys. 415 184–192. [DOI] [PubMed] [Google Scholar]
- Yoshida, S., and Uemura, M. (1986). Lipid composition of plasma membranes and tonoplasts isolated from etiolated mung bean (Vigna radiata L.). Plant Physiol. 82 807–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, H., Rowland, O., and Kunst, L. (2005). Disruptions of the Arabidopsis enoyl-CoA reductase gene reveal an essential role for very-long-chain fatty acid synthesis in cell expansion during plant morphogenesis. Plant Cell 17 1467–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann, P., Hirsch-Hoffmann, M., Hennig, L., and Gruissem, W. (2004). GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136 2621–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.