Abstract
Autophagy is the engulfment of cytosol and organelles by double-membrane vesicles termed autophagosomes. Autophagosome formation is known to require phosphatidylinositol 3-phosphate (PI(3)P) and occurs near the endoplasmic reticulum (ER), but the exact mechanisms are unknown. We show that double FYVE domain–containing protein 1, a PI(3)P-binding protein with unusual localization on ER and Golgi membranes, translocates in response to amino acid starvation to a punctate compartment partially colocalized with autophagosomal proteins. Translocation is dependent on Vps34 and beclin function. Other PI(3)P-binding probes targeted to the ER show the same starvation-induced translocation that is dependent on PI(3)P formation and recognition. Live imaging experiments show that this punctate compartment forms near Vps34-containing vesicles, is in dynamic equilibrium with the ER, and provides a membrane platform for accumulation of autophagosomal proteins, expansion of autophagosomal membranes, and emergence of fully formed autophagosomes. This PI(3)P-enriched compartment may be involved in autophagosome biogenesis. Its dynamic relationship with the ER is consistent with the idea that the ER may provide important components for autophagosome formation.
Introduction
Autophagy is a mechanism for the degradation of cellular material either as a way to provide nutrients during times of starvation or as a quality control mechanism that eliminates unneeded proteins and/or organelles during normal growth and development (Klionsky, 2005; Lum et al., 2005). It was described morphologically more than 50 years ago but its complex molecular mechanism is only now beginning to be elucidated in some detail. Morphologically, autophagy is characterized by formation of double-membrane vesicles termed autophagosomes, which engulf cytoplasmic material and organelles and deliver them to lysosomes for degradation. More than 31 autophagy-related (ATG) genes whose products regulate autophagy have been identified, primarily through the use of yeast genetics (Klionsky et al., 2003; Yorimitsu and Klionsky, 2005). Half of the yeast genes, comprising the machinery for selective autophagy, do not have a mammalian counterpart. The rest, comprising the basic machinery for nonselective autophagy, are also found in higher eukaryotes, which suggests that the mammalian and yeast pathways for autophagosome formation are broadly similar (Reggiori and Klionsky, 2005; Suzuki and Ohsumi, 2007).
Despite much progress, two critical questions in autophagy remain unanswered: what is the origin of the autophagosomal membrane and how is it formed? In yeast, the earliest discernible autophagy precursor is a proteinaceous aggregate assembled in the vicinity of the vacuole and termed preautophagosomal structure (PAS). As the PAS components assemble, they somehow acquire a double membrane, leading to formation of the autophagosome (Reggiori and Klionsky, 2005). Several gene products are required for this induction step in yeast (Atg1, Atg9, Atg13, Atg17, and Atg23) in addition to the type III phosphoinositide 3-kinase (PI 3-kinase) complex (Vps34, Vps15, Atg6, and Atg14) that produces phosphatidylinositol 3-phosphate (PI(3)P; Kihara et al., 2001b; Suzuki et al., 2007). Analogous to the PAS structure in mammals is a small PAS termed the isolation membrane or phagophore, whose formation also requires type III PI 3-kinase activity (Blommaart et al., 1997; Mizushima et al., 2001, 2003; Lindmo and Stenmark, 2006).
The debate on the origin of the autophagosomal membrane, first reviewed in 1966 (De Duve and Wattiaux, 1966), continues to this date (Juhasz and Neufeld, 2006). Two general models (maturation vs. assembly) have been considered. According to the maturation model, the origin of the autophagosomal membrane is the ER, but direct evidence for this is lacking. The assembly model postulates that autophagosomal membranes assemble de novo from localized lipid synthesis or transport. As discussed in Juhasz and Neufeld (2006), support for this model comes mostly from an inability to directly implicate the ER in autophagosome biogenesis.
We and others recently described the identification of a novel protein termed double FYVE domain–containing protein 1 (DFCP1) based on its ability to bind PI(3)P in vitro (Derubeis et al., 2000; Cheung et al., 2001; Ridley et al., 2001). In those earlier studies, it was shown that DFCP1 contains two FYVE domains (thus explaining its PI(3)P binding) but is unusual among FYVE domain–containing proteins because it localizes to ER and Golgi membranes instead of endosomes. In this work we exploit the localization and movement of DFCP1 during amino acid starvation to identify a PI(3)P-enriched compartment dynamically connected to the ER and important in autophagosome formation.
Results
Identification of a DFCP1 domain necessary and sufficient for ER targeting
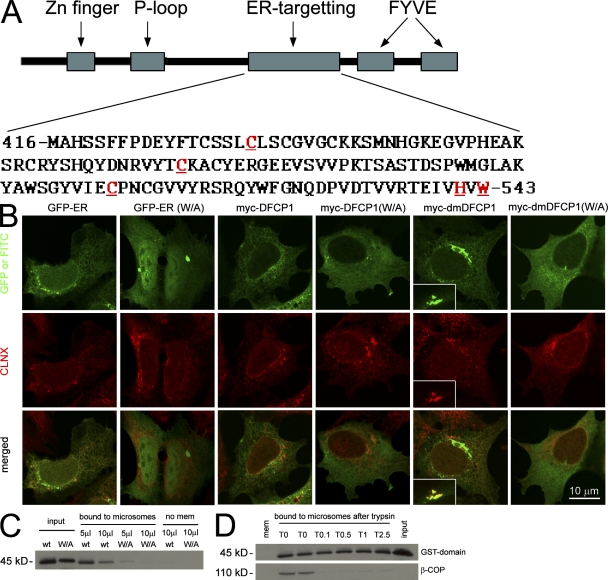
To account for its unusual localization, we hypothesized that DFCP1 may contain a novel domain that allows it to target the ER/Golgi independently of its FYVE domains. Previous work had also suggested the existence of such a domain within residues 416–602 (Cheung et al., 2001). GFP-tagged fragments of DFCP1 around this region identified a domain between amino acids 416–543 (Fig. 1 A), which, when expressed on its own, localized to the ER (Fig. 1 B, GFP-ER). Several residues within this domain were targeted for further mutagenesis (Fig. 1 A, red and underlined) based on their conservation across species and on fine mapping of the C terminus (not depicted). Mutagenesis of W543, H541, or the three indicated cysteine residues resulted in a protein with cytoplasmic localization (Fig. 1 B, GFP-ER(W/A) for W543A, and others not depicted). These residues were also mutagenized in the full-length protein either in the wild type (WT) or the double FYVE domain mutant (DM) background (the DM contains two point mutations inactivating both FYVE domains; described in Ridley et al., 2001). Although the WT and the DM are primarily localized to the ER/Golgi (Fig. 1 B, myc-DFCP1 and myc-dmDFCP1; Ridley et al., 2001), mutagenesis of the critical residues resulted in cytoplasmic localization (Fig. 1 B, myc-DFCP1(W/A) and myc-dmDFCP1(W/A) for W543A, and others not depicted). Thus, the reason that DFCP1 resides on ER/Golgi membranes and not on endosomes is that it contains an ER-targeting domain that is dominant over the two FYVE domains. To provide biochemical data on the binding characteristics of this domain, we created GST-tagged constructs and expressed them in bacteria. The domain derived from the WT protein bound to microsomes isolated from rat kidney, whereas the W543A point mutant had reduced binding (Fig. 1 C). Interestingly, trypsin digestion of the microsomal membranes did not inhibit this binding, indicating that the domain binds either to lipids or to a trypsin-insensitive protein epitope. Work to identify the binding partner of this domain is in progress.
Figure 1.
Identification and partial characterization of an ER-targeting domain within DFCP1. (A) Sequential truncations of DFCP1 identified an internal domain necessary and sufficient to specify localization in the ER. Residues in red and underlined are required for ER localization. (B) The GFP-[416–543] construct localized to the ER in HEK-293 cells, whereas mutants within this domain (shown here is a W543A construct) were cytosolic. Selected residues important in ER targeting were also mutagenized in the full-length protein as indicated. The full-length DFCP1 (myc-DFCP1) localized to the ER, whereas the DM (myc-dmDFCP1) had ER/Golgi localization (insets in myc-dmDFCP1 panels show colocalization with giantin). Mutagenesis of critical residues resulted in cytoplasmic redistribution (shown here as W/A mutant). All samples were costained with antibodies to calnexin (CLNX), an ER protein. (C) The WT or the W/A mutant of the 416–543 domain were purified as GST fusion proteins (input) and mixed with rat kidney microsomes. Bound material was recovered by centrifugation. Note that the WT-derived protein bound to microsomes, whereas the W/A mutant did not. (D) Microsomes as in C were treated on ice with the indicated units of trypsin before incubation with the GST-[416–543] domain and centrifugation. Note that β-COP, a peripheral protein found on microsomes, was almost completely digested by trypsin; under these conditions, binding of the DFCP1 fragment to microsomes was not changed.
DFCP1 translocates to a punctate compartment during amino acid starvation
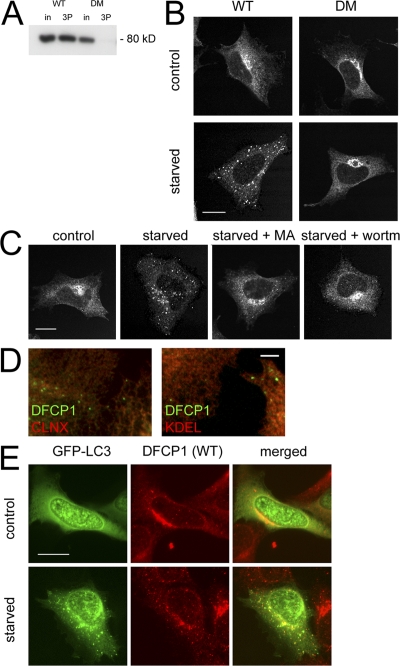
To study the function of DFCP1, we created clonal HEK-293 cell lines expressing myc-tagged versions of the protein. In the stable cell lines, WT DFCP1 and the DM localized to the ER/Golgi (Fig. 2 B, control), although only WT protein bound PI(3)P (Fig. 2 A). Because PI(3)P has been shown to be involved in autophagy (Lindmo and Stenmark, 2006), we examined DFCP1 localization during amino acid starvation and found that WT DFCP1 translocated to a punctate compartment, whereas the DM (Fig. 2 B, bottom) and the W/A mutant did not (not depicted). Translocation was inhibited by known inhibitors of autophagy such as 3-methyladenine and wortmannin (Fig. 2 C). Immunostaining of DFCP1 after starvation showed that the punctate structures did not correspond to any known organelles (ER exit sites, Golgi, early/late endosomes, lysosomes, mitochondria, and not depicted) but appeared to be adjacent to the ER (Fig. 2 D). The punctate compartment colocalized partially with autophagosomes, as defined by transient expression of GFP-tagged MAP-LC3 (Fig. 2 E; Kabeya et al., 2000). In general, 60% of myc-DFCP1 puncta colocalized with GFP-MAPLC3 in fixed cells, but we believe that this is an underestimate based on the dynamic interaction between the two proteins (see below).
Figure 2.
Starvation-induced and PI(3)P-dependent translocation of DFCP1 to punctate structures partially colocalizing with autophagosomes. (A) Lysates from cells expressing WT DFCP1 or the DM were incubated with Affigel beads coupled to PI(3)P; WT DFCP1 bound to PI(3)P, whereas the DM did not. in, 5% of input. (B) Stable HEK-293 cell lines expressing WT DFCP1 or the DM were left untreated or starved of amino acids for 90 min. Cells were then fixed and stained for DFCP1. Both WT and DM DFCP1 under normal conditions localized to the ER/Golgi; after starvation, only the WT translocated to punctate structures. (C) HEK-293 cells stably expressing WT DFCP1 were starved alone or in the presence of 3-methyladenine (MA) or wortmannin (wortm) as indicated before fixation and staining for DFCP1. (D) Starved HEK-293 cells expressing DFCP1 were costained with markers to the ER such as calnexin and KDEL. (E) HEK-293 cells stably expressing WT DFCP1 were transfected with GFP-MAP-LC3 and starved for 90 min as indicated. Note that in cells expressing WT DFCP1, both MAP-LC3 and DFCP1 translocated to punctate structures upon starvation, some of which colocalized. Bars: (A–C and E) 20 μm; (D) 0.5 μm.
A starvation-induced PI(3)P-enriched compartment related to the ER and to autophagosomes
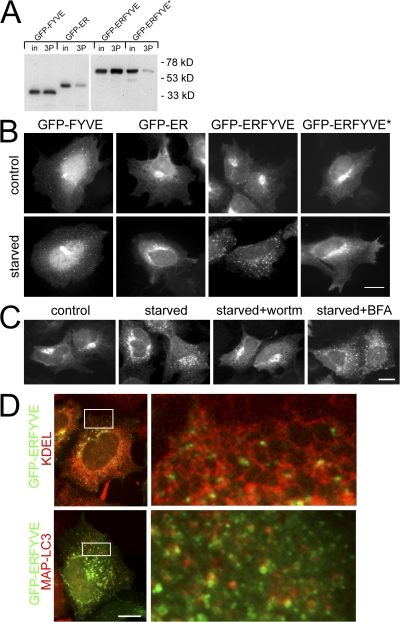
Because starvation-induced translocation of DFCP1 required functional FYVE domains and was sensitive to inhibitors of PI(3)P formation, we hypothesized that this movement reflected formation and recognition of a pool of PI(3)P during starvation. Two additional PI(3)P-specific probes were used to explore this further: an isolated FYVE domain from FENS-1 (Ridley et al., 2001) or an isolated PHOX homology (PX) domain from p40 PHOX (Bravo et al., 2001). Both GFP-FYVE and GFP-PX partially colocalized with early endosomal markers (such as early endosome antigen 1 [EEA1], unpublished data) unlike DFCP1, which was on the ER/Golgi. Upon amino acid starvation, all three proteins became more punctate (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200803137/DC1), but MAP-LC3 colocalized primarily with DFCP1 and much less with the other two PI(3)P probes (Fig. S1). These results suggest that multiple pools of PI(3)P are formed during amino acid starvation and that DFCP1 may be the most appropriate reporter for a pool related to the ER. To address this directly, we fused the ER-targeting domain of DFCP1 to the FYVE domain of FENS-1 to generate a PI(3)P probe tethered peripherally to the ER. This probe (GFP-ERFYVE) bound to PI(3)P as well as the isolated FYVE domain, whereas a point mutant inactivating the FENS-1 FYVE domain (Ridley et al., 2001) showed 10-fold less binding (Fig. 3 A). The localization of the four proteins under normal conditions was as expected: GFP-FYVE was cytosolic and partially punctuate, whereas the other three constructs were found on ER/Golgi membranes (Fig. 3 B, top). Upon amino acid starvation, GFP-FYVE became more punctate (as shown above), whereas GFP-ER and GFP-ERFYVE* did not change. Significantly, GFP-ERFYVE redistributed completely to a punctate compartment (Fig. 3 B, bottom). Like DFCP1, redistribution of the GFP-ERFYVE construct was inhibited by wortmannin (and 3-MA, not depicted) but not affected by brefeldin A (BFA; Fig. 3 C). In addition, the GFP-ERFYVE punctate compartment was adjacent to the ER and colocalized partially with MAP-LC3 (Fig. 3 D).
Figure 3.
FYVE domain tethered peripherally to the ER translocates to starvation-induced punctate structures in a pathway dependent on PI(3)P. (A) GFP-FYVE domain from FENS-1 bound to PI(3)P-coupled lipid beads, whereas the GFP-ER–targeting domain of DFCP1 showed little binding (first four lanes; in, 5% of input; 3P, the faction bound to PI(3)P-coupled beads). A construct with the FENS-1 FYVE domain fused to the ER domain of DFCP1 (GFP-ERFYVE) maintained PI(3)P binding, whereas a construct with a point mutation in the FYVE domain (GFP-ERFYVE*) showed much-reduced binding. (B) Stable cell lines expressing the four constructs shown in A were left untreated or starved for 45 min before fixation and fluorescence microscopy. Note that the GFP-ERFYVE construct translocated to punctate structures upon starvation. (C) Cells expressing GFP-ERFYVE were starved alone or in the presence of wortmannin or BFA as indicated. (D) Cells expressing GFP-ERFYVE and starved for 45 min were either counterstained for the ER using antibodies to KDEL (top) or cotransfected with mRFP-MAP-LC3 (bottom). Note that the GFP-ERFYVE punctate structures frequently localized on the ER and showed partial colocalization with LC3. The panels on the right show an enlarged view of the boxed regions on the left. Bars, 10 μm.
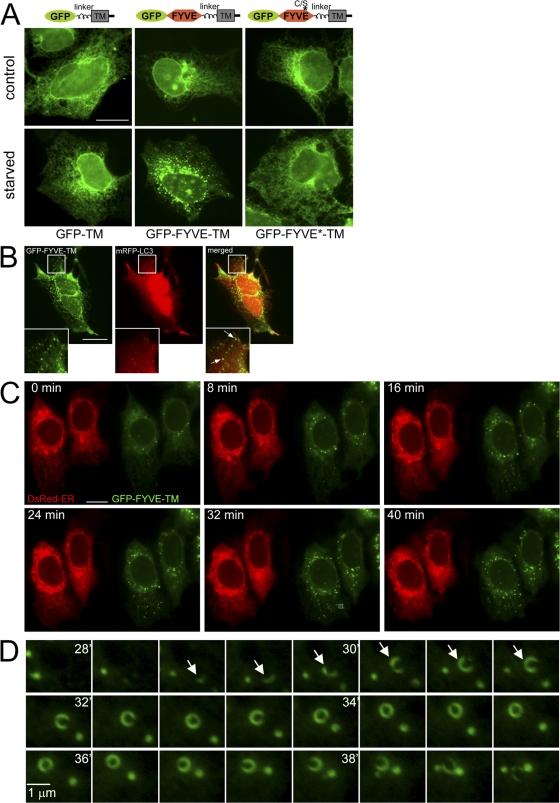
To move away from DFCP1 completely, we constructed a novel reporter consisting of the FYVE domain from FENS-1 anchored on the ER membrane by a transmembrane domain. Such tail-anchored proteins have been extensively studied previously (Bulbarelli et al., 2002; Borgese et al., 2007). As previously reported, GFP with such an anchor derived from cytochrome b5 localized to the ER (Bulbarelli et al., 2002) and did not translocate during amino acid starvation (Fig. 4 A). In contrast, the GFP-FYVE-TM protein, which under normal growth conditions was on the ER (Fig. 4 A) and in a small percentage of cells on punctate structures (not depicted), translocated to punctate structures upon starvation (Fig. 4 A). A construct with a single point mutation inactivating the FYVE domain did not translocate (Fig. 4 A), which indicates that punctate localization requires PI(3)P recognition. In support of this, localization of the GFP-FYVE-TM construct on punctate structures was completely inhibited by wortmannin (not depicted). These GFP-TM-FYVE punctate structures colocalized partially with MAP-LC3 (Fig. 4 B) and frequently encircled RFP-positive structures (Fig. 4 B, arrows). This localization gives a clue on the function of this compartment and will be discussed extensively later. In live imaging experiments, translocation of GFP-FYVE-TM occurred within 30 min, whereas in the same cells, the localization of dsRED-ER (a dsRED protein with an ER retention signal) did not change (Fig. 4 C and Video 1, available at http://www.jcb.org/cgi/content/full/jcb.200803137/DC1). Finally, we frequently saw in these cells, the formation and collapse of GFP-FYVE-TM rings such as shown in Fig. 4 D (arrows mark the particle at early stages). These structures were also seen during live imaging of DFCP1 and will be discussed in detail later.
Figure 4.
ER-anchored FYVE domain translocates to starvation-induced punctate structures in a pathway dependent on PI(3)P. (A) Three GFP-based constructs were fitted with a linker region, a transmembrane domain (TM), and a short cytoplasmic tail: GFP alone (GFP-TM), GFP fused to the WT FYVE domain of FENS-1 (GFP-TM-FYVE), and GFP fused to a point mutant of the FENS-1 FYVE domain unable to bind PI(3)P (GFP-FYVE*-TM). All three constructs were expressed transiently in HEK-293 cells, and their localization was examined with or without amino acid starvation for 60 min as indicated. Note that GFP-TM-FYVE translocated to punctate structures during starvation. (B) HEK-293 cells were transfected with GFP-FYVE-TM and mRFP-LC3 and starved for 60 min. Note that puncta of the two reporters colocalize, with the GFP construct frequently encircling mRFP-LC3 membranes (arrows). In general, cells had many more GFP-FYVE-TM than mRFP-LC3 puncta, and on average, 80% of LC3 puncta colocalized with a GFP-FYVE-TM particle. Insets show enlarged views of the boxed regions. (C) Live imaging of HEK-293 cells coexpressing GFP-FYVE-TM and dsRED-ER and starved for 60 min. Also see Video 1 (available at http://www.jcb.org/cgi/content/full/jcb.200803137/DC1). (D) A region indicated in C (bottom middle, box) is expanded and shown for 28–40 min during starvation. Note the formation and collapse of a ringlike particle. Arrows mark the particle at early stages. Bars: (A–C) 20 μm; (D) 1 μm.
Live imaging of the PI(3)P compartment during starvation
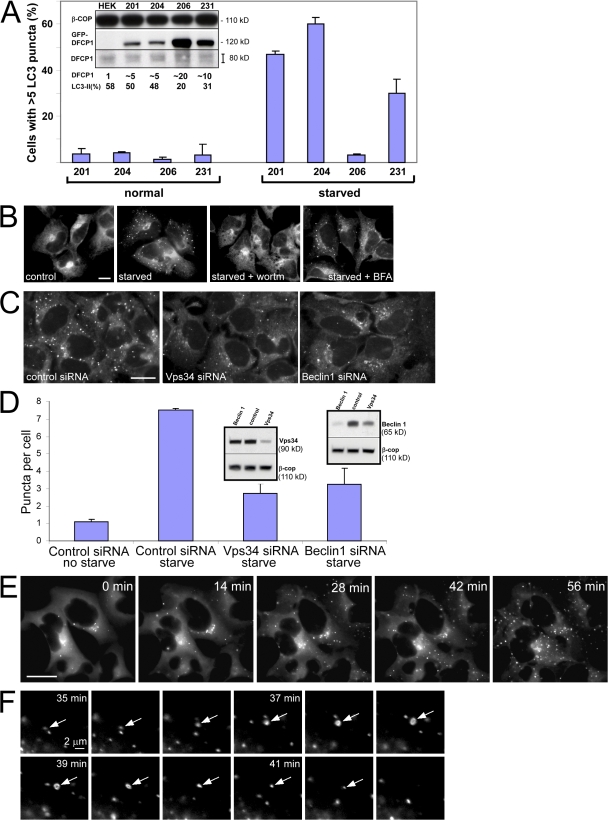
To study the dynamics of this punctate compartment during autophagy, we established HEK-293 stable cell lines expressing GFP-tagged DFCP1. We had noted from work with myc-DFCP1 stable cell lines that high levels of DFCP1 (but not of the DM) suppressed specifically autophagosome formation (Fig. S2, A–D, available at http://www.jcb.org/cgi/content/full/jcb.200803137/DC1, for overexpressed GFP-MAP-LC3; and not depicted for endogenous MAP-LC3). For live-imaging studies, several clones were selected and examined. In analogy to myc-DFCP1 (Fig. S2, A–D), high expression of GFP-DFCP1 inhibited autophagosome formation as measured by the translocation of MAP-LC3 to punctate structures and formation of the type II form during electrophoresis (Fig. 5 A, clones 206 and 231). For the remainder of this work, we chose clone 201, which showed a good response to starvation, and is inhibitable by wortmannin treatment but insensitive to BFA (Fig. 5 B).
Figure 5.
Isolation and characterization of stable HEK-293 clones expressing GFP-DFCP1. (A) Four clones were screened for GFP-DFCP1 expression (inset shows blots of endogenous DFCP1 and GFP-DFCP1 with β-COP as loading control; the fold overexpression of tagged DFCP1 over endogenous DFCP1 is also indicated) and for a good response to starvation, as measured by translocation of GFP-MAP-LC3 to punctate structures (shown in the graph) or acquisition of endogenous LC3-II form (last lane of inset). High levels of DFCP1 inhibited starvation responses (especially evident for clone 206). Error bars show the standard deviation from three independent experiments. (B) Clone 201 cells were left untreated or starved for 45 min in the absence or presence of wortmannin (wortm) or BFA as indicated, and the distribution of GFP-DFCP1 was examined by fluorescence microscopy. Note that wortmannin inhibited translocation, whereas BFA was without effect. (C) Clone 201 cells were treated with siRNA against Vps34 or beclin-1, or with a control siRNA as shown, starved for 60 min, and examined by fluorescence microscopy. Note that the level of punctate structures is reduced in Vps34- and beclin-1–reduced cells; this is analyzed for three independent experiments in D (error bars show standard deviation). (D, inset) The levels of Vps34 and beclin-1 after siRNA treatments. Of peripheral interest for this work is that the reduction of Vps34 during treatment with siRNA for beclin-1 is reproducible. (E) Clone 201 cells were starved for 60 min. Imaging was at 1 frame per 20 s, and selected frames throughout the sequence are shown. Also see Video 2 (available at http://www.jcb.org/cgi/content/full/jcb.200803137/DC1). (F) For a selected time interval (starting approximately at 35 min after starvation), a sequence showing formation and collapse of an omegasome (arrows) is shown. Bars (A–C and E) 20 μm; (F) 2 μm.
Autophagy in yeast and mammals is known to depend on the function of Vps34 and its adaptor beclin. We used siRNA against both genes to determine whether the DFCP1 response depends on the function of these proteins. Under conditions where Vps34 and beclin levels were reduced by 50–80% (Fig. 5 D, inset), we observed a significant 50–60% reduction on the level of DFCP1 punctate structures (Fig. 5, C and D).
Live imaging during starvation showed that DFCP1 became more punctate as a function of time, peaking at ∼50 min after starvation (Fig. 5 E and Video 2, available at http://www.jcb.org/cgi/content/full/jcb.200803137/DC1). Although the overall pattern was toward a more punctate morphology, the individual ringlike punctate structures formed and collapsed continuously as a function of time (Fig. 5 F). These structures were also seen with the GFP-FYVE-TM construct (Fig. 4 D). They were on average 1 μm in maximum diameter, and their lifetime (from formation to collapse) was ∼6–7 min. Because they were frequently seen in association with the underlying ER (see the following section) forming an Ω-like shape, we term them “omegasomes.”
Interaction of omegasomes with autophagosomes and the ER
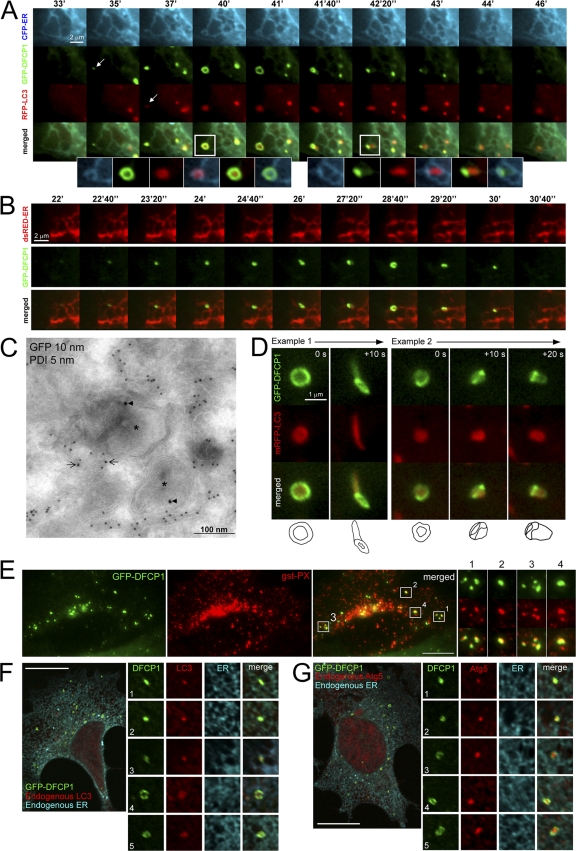
The DFCP1-specific punctate structures (omegasomes) were very dynamic and colocalized partially with endogenous autophagy-specific proteins (such as MAP-LC3 and ATG-5; Fig. S3, A–C). Therefore, we examined by live imaging the impact of amino acid starvation on the dynamic distribution of coexpressed GFP-DFCP1, mRFP-MAP-LC3, and CFP-ER (a CFP protein containing an ER localization signal). It was apparent from these videos that autophagosomes form within omegasomes that are in turn in dynamic equilibrium with the ER. A characteristic sequence for a time interval of 10 min during starvation is shown in Fig. 6 A (and Video 3, available at http://www.jcb.org/cgi/content/full/jcb.200803137/DC1). At the beginning, a small amount of DFCP1 concentrated at the edge of an ER strand (Fig. 6 A, arrow on GFP-DFCP1 panel). As a function of time, DFCP1 enlarged, and at some point (Fig. 6 A, 37′), MAP-LC3 began to accumulate in the same region (arrow on RFP-LC3 panel). Expansion of DFCP1 and MAP-LC3 occurred for the next 5 min (Fig. 6 A, 40′–41′) until a DFCP1 omegasome fully encircled a MAP-LC3 particle. It is of note that all of this expansion corresponded to changes in the underlying ER strand but that the three domains (ER, DFCP1, and MAP-LC3) remained distinct (Fig. 6 A, inset for 40′ time point). When the DFCP1 omegasome reached its maximum diameter, the LC3 particle began to exit it (Fig. 6 A, 41′40′′–44′). This was a very complicated movement, and it appeared to involve some acquisition (“coating”) of DFCP1-staining membrane by the LC3 particle as it exited the omegasome (Fig. 6 A, inset for 42′20′′ time point; note the faint green color outlining the red particle). Finally, late in the sequence, the LC3 particle was almost entirely free of DFCP1 staining and all of DFCP1 had been absorbed back in the ER (Fig. 6 A, 46′ onwards). Several aspects of these results were explored further.
Figure 6.
Relationship of PI(3)P-containing omegasomes with the ER and autophagosomes by live imaging and in fixed cells. (A) Clone 201 cells were transfected with mRFP-MAP-LC3 (red) and CFP-ER (blue) and starved for 60 min. Imaging was performed at 1 frame per 10 s, and a selected interval within this sequence is shown, starting at 33 min after starvation. Arrows indicate the first discernible omegasome (green) and autophagosome (red) occurrences. The enlarged panels are from the two boxed areas and represent views of (1) ER, (2) DFCP1, (3) MAP-LC3, (4) ER-MAPLC3, (5) DFCP1-MAPLC3, and (6) MAPLC3-ER, in that order. Also see Video 3 (available at http://www.jcb.org/cgi/content/full/jcb.200803137/DC1). (B) Dynamic relation of omegasomes and the ER using TIRFM imaging. Note that omegasomes form in regions containing ER and collapse there. Also see Video 4. (C) Thawed cryosection of clone 201 cells 45 min after starvation and double-labeled for anti–GFP-DFCPI (arrowheads, 10 nm gold) and anti-PDI (arrows, 5 nm gold). Note the putative autophagic-like vacuoles (asterisks) labeled for DFCPI-GFP adjacent to PDI-labeled ER membranes. This is one panel of a multipanel supplemental figure (Fig. S4). (D) Selected frames from live imaging of GFP-DFCP1 and mRFP-MAP-LC3 during starvation, whereby an intermediate is formed in which MAP-LC3 appears to bud from the omegasome while still being outlined with a DFCP1-staining membrane. At the bottom of each panel are line drawings of these structures using magnified photographs of the relevant frames. Also see Video 6. (E) Colocalization of omegasomes with a PI(3)P-binding protein applied exogenously. Clone 201 cells were starved for 60 min, perforated using nitrocellulose, and stained with purified GST-PX domain from p40phox. The majority of GSP-PX domain stains early endosomes (not depicted) but a substantial amount of the protein also binds to DFCP1 omegasomes (F and G). (F and G) Relationship of omegasomes to autophagy proteins and the ER. Clone 201 cells counterstained with antibodies to endogenous ER and endogenous MAP-LC3 or Atg5 as indicated. Selected examples from such cells (in addition to the one shown) where the ER was in a single layer and well resolved from cytosol, to allow evaluation of colocalizations, are shown in magnified panels 1–5. Bars: (A, B, and D) 1 μm; (E–G) 20 μm.
Relationship of omegasomes with the ER
To obtain an alternative view of the omegasome–ER dynamics, we used total internal reflection fluorescence microscopy (TIRFM), which can give a clearer view of the ER (Fig. 6 B and Video 4, available at http://www.jcb.org/cgi/content/full/jcb.200803137/DC1). These movies showed that omegasomes formed de novo and that their expansion and collapse were accompanied by changes of the underlying ER. A similar spatial relationship between ER and omegasomes was seen during confocal live imaging, where the DFCP1 rings were seen on the same plane as equivalent ER rings (Fig. S3 D). To obtain a higher resolution view of the omegasomes and the ER, we used immuno-EM. When thawed, cryosections of aldehyde-fixed cells were double-labeled with anti-GFP and anti–protein disulfide isomerase (PDI; an ER marker); the GFP-DFCP1–labeled structures were in most cases in close contiguity with the ER membranes (Fig. 6 C, and Fig. S4).
Omegasomes and MAP-LC3
To obtain a 3D view of the omegasome-MAP-LC3 structure, we used serial reconstruction (Video 5, available at http://www.jcb.org/cgi/content/full/jcb.200803137/DC1). In this view, the omegasome appeared as a torus, i.e., a donut-shaped ring, which enclosed the MAP-LC3 particle with the DFCP1- and MAP-LC3–staining domains, which were apparently distinct but had some overlap at the edges. This structure was very similar to what was seen with the GFP-FYVE-TM construct (Fig. 4 B) and consistent with experiments staining for endogenous LC3 and Atg5 (see Fig. S3, A and C). We also sought to obtain additional views of the LC3 “coating” step, which appears to be the exit step before omegasomes collapse back to the ER and autophagosomes are fully formed. Two types of movement were evident: either the MAP-LC3 particle exited the omegasome smoothly (Fig. 6 D, example 1) or it was possible to discern several discrete steps as the DFCP1 membranes “zipped” along the LC3 particle (Fig. 6 D, example 2). Video 6 shows five examples of this process. This type of sequential interaction was also noted when cells expressing GFP-DFCP1 were starved and stained for endogenous MAP-LC3 (Fig. S3 A, enlarged panels).
Staining of omegasomes with exogenous PI(3)P probe
To ensure that omegasomes were indeed PI(3)P-enriched membranes, we perforated starved cells and stained them with purified GSP-PX domain known to bind PI(3)P (Ellson et al., 2001). We saw that a subset of the GST-PX signal was on omegasomes (Fig. 6 E), whereas the remaining was on early endosomes.
Omegasomes, endogenous autophagosomes, and ER
Because all of these results were obtained with live imaging of overexpressed protein, we sought to verify them using staining for endogenous proteins. Our antibodies against DFCP1 could not detect the endogenous protein. Therefore, cells expressing GFP-DFCP1 were starved and stained for endogenous ER and endogenous MAP-LC3 (Fig. 6 F) or endogenous Atg5 (Fig. 6 G). Fig. 6 (F and G) shows a single cell from such a staining, whereas the enlarged panels are enlarged examples representative of 100 such cells. We saw that the colocalization of DFCP1 omegasomes with MAP-LC3– or Atg-5–positive structures frequently occurred on the underlying ER. Finally, we also ensured that starvation responses in these cells were normal, as shown by three independent assays: formation of punctate MAL-LC3, formation of punctate Atg5, and formation of the type II MAP-LC3 form during electrophoresis (Fig. S5, available at http://www.jcb.org/cgi/content/full/jcb.200803137/DC1).
The main parameters of the results with DFCP1 were also observed during live imaging of the GFP-ERFYVE probe. During starvation, stable cells expressing GFP-ERFYVE displayed many dynamic punctate structures that frequently colocalized with MAP-LC3, and the ERFYVE/LC3 association occurred on ER strands (unpublished data).
Autophagosome maturation after omegasome exit
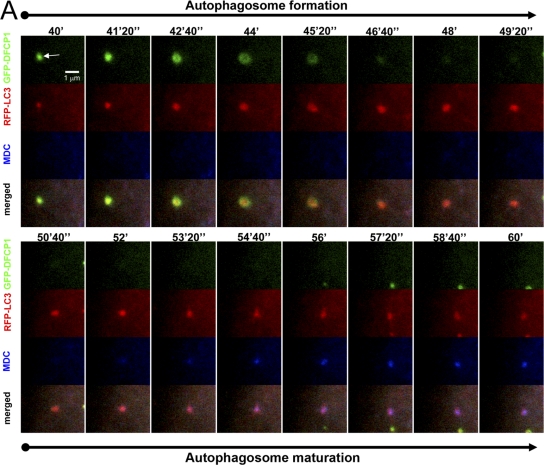
The fate of the LC3 particle upon omegasome exit was followed in live cells by staining with monodansylcadaverine (MDC), which is known to stain acidic compartments including mature autophagosomes (Bampton et al., 2005). We observed a 3–5-min delay between autophagosome formation and its staining with MDC (Fig. 7), which is consistent with the idea that autophagosomes emerging from omegasomes are not fully mature: this occurs at a later time. Interestingly, MDC staining of the autophagosome did not involve fusion with another MDC-positive membrane such as a late endosome/lysosome.
Figure 7.
Autophagosome maturation after omegasome exit. Cells expressing GFP-DFCP1 and mRFP-MAP-LC3 were starved and imaged for the indicated time interval. At 30 min after starvation, the cells were also incubated with 2 μM MDC. Note that an autophagosome emerges first from an omegasome (panels labeled autophagosome formation; arrow indicates the omegasome) and then begins to stain with MDC (panels labeled autophagosome maturation) without appearing to change its appearance or to fuse with another MDC-positive vesicle.
The source of PI(3)P: live imaging of Vps34
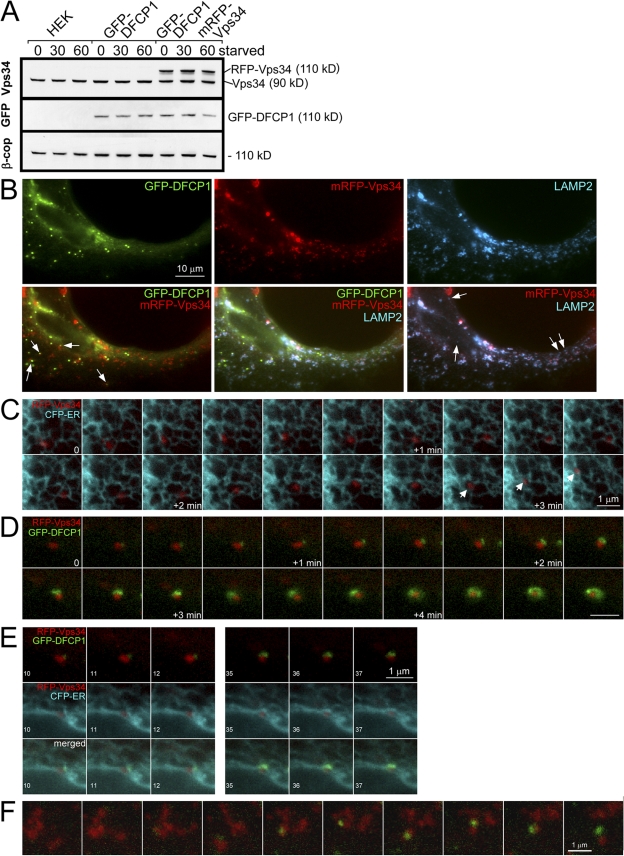
Based on the siRNA data (Fig. 5), we assumed that the source of PI(3)P in these cells is the Vps34 enzyme presumably being delivered to the ER either by translocation or via a vesicular route. To address these possibilities, we introduced mRFP-Vps34 into the stable cell lines expressing GFP-DFCP1 and derived new stable cell lines expressing both reporters. In these cells, mRFP-Vps34 was expressed at levels comparable to the endogenous protein, and levels of both DFCP1 and Vps34 did not change during starvation (Fig. 8 A). In cells with moderate expression of Vps34, starvation responses were normal (Fig. S5). The localization of Vps34 was cytosolic and vesicular in most cells, and staining with various organellar markers showed that Vps34 vesicles corresponded to Lamp-2– and Lamp-1–positive late endosomes/lysosomes (Fig. 8 B), whereas staining for EEA1 (an early endosome protein) showed no colocalization (not depicted). Interestingly, these Vps34 vesicles were always in close proximity to the ER and were frequently seen to move along ER strands (Fig. 8 C) irrespective of starvation status. Having established that Vps34 exists in a cytosolic and a vesicular form, we performed live imaging experiments during amino acid starvation. We did not obtain any evidence that cytosolic Vps34 translocated to ER strands during starvation or that its cytosolic localization changed. In contrast, we saw repeatedly during starvation that omegasomes formed in close association to Vps34-containing vesicles (Fig. 8, D and F; and Video 7, available at http://www.jcb.org/cgi/content/full/jcb.200803137/DC1). In many cases, the earliest visible DFCP1 accumulation occurred near a Vps34 vesicle whereby the Vps34 vesicle remained constant in size, whereas the DFCP1 membranes enlarged and expanded (compare size of Vps34 particle at the beginning and end of sequence in Fig. 8 D). The position of the underlying ER during this interaction is shown in Fig. 8 E for selected frames of Fig. 8 D, and for the whole sequence in Video 7. Omegasome expansion was accompanied by changes in the underlying ER, in tight proximity to the Vps34 particle. We note that in cells expressing very high levels of mRFP-Vps34, autophagy was suppressed both in terms of Atg5 and GFP-DFCP1 accumulation in punctate structures (unpublished data).
Figure 8.
Vps34 dynamics during omegasome formation. (A) Clone 201 cells were transfected with mRFP-Vps34, and a stable population expressing both proteins was selected (last three lanes). Note that exogenous mRFP-Vps34 was comparable to endogenous Vps34 and stable during amino acid starvation. (B) Cells as in A were starved for 60 min, fixed, and stained for Lamp-2. Note that DFCP1 punctate structures are frequently in proximity to Vps34 membranes (arrows on the bottom left) and that the majority but not all of Lamp-2 vesicles (∼80%) colocalize with Vps34, whereas all of the Vps34 vesicles colocalize with Lamp-2 (arrows on the bottom right indicate examples of Lamp-2 vesicles devoid of Vps34). (C) Selected frames from live imaging of mRFP-Vps34 in cells also expressing CFP-ER. Note that the Vps34 vesicle is in constant proximity to the ER and frequently appears to use the ER strands to move long distances (arrows on the bottom). The times refer to the period after amino acid starvation, but similar types of movement are evident without starvation. (D and E) Selected frames from live imaging of mRFP-Vps34, GFP-DFCP1, and CFP-ER during amino-acid starvation. (D) An omegasome being formed in constant and close proximity to a Vps34 particle. Bar, 1 μm. For selected frames of this video, as indicated (10–12 and 35–37), E shows the relationship of Vps34 to DFCP1 (top), Vps34 to ER (middle), and all three (bottom). Also see Video 7 (available at http://www.jcb.org/cgi/content/full/jcb.200803137/DC1). (F) An additional example from live imaging experiments showing an omegasome forming in close proximity to a Vps34 particle.
Down-regulation of DFCP1 does not inhibit autophagy
Endogenous DFCP1 was down-regulated by siRNA in HEK-293 cells, and the resultant autophagy response of endogenous MAP-LC3 was explored. Under conditions where >80% of DFCP1 expression was eliminated, MAP-LC3 type II autophagy-specific form was evident after amino acid starvation and wortmannin still inhibited the response (Fig. S2, E–H).
Discussion
A PI(3)P-enriched compartment dynamically connected to the ER during induction of autophagy
In this work, we have provided evidence that a pool of PI(3)P is formed in a membrane compartment shortly after amino acid starvation and during the early stages of autophagy. Three different reporter proteins translocated to this compartment during amino acid starvation: DFCP1, a known mammalian protein; ER-FYVE, an artificial reporter created by fusing the ER-targeting domain of DFCP1 with a heterologous FYVE domain normally residing in endosomes; and FYVE-TM, a reporter consisting of a heterologous (endosomal) FYVE domain anchored to the ER via a transmembrane domain. In all three cases, translocation was dependent on amino acid starvation and required functional PI(3)P recognition, i.e., a WT FYVE domain. Translocation was blocked by wortmannin and by other inhibitors of PI 3-kinase, and required Vps34 and beclin function. We have termed the membranes of this compartment omegasomes to provide an indication that they are neither autophagosomes nor autophagosomal precursors that mature into autophagosomes.
We propose that these PI(3)P-enriched membranes are dynamically connected to the ER based on several complementary observations: (a) Live imaging data using wide-field, confocal, and TIRFM revealed that omegasomes and the underlying ER frequently coincide; (b) confocal microscopy of endogenous autophagy proteins, omegasomes, and endogenous ER also indicated that there is some colocalization between omegasomes and the ER; (c) this compartment was revealed when an endosomal PI(3)P probe (FENS FYVE domain) was tethered to the ER but not when the same probe was cytosolic; and (d) immuno-EM labeling of the ER and omegasomes showed that the signal was frequently interspersed. The safest interpretation of all these data is that this punctate compartment can be accessed by proteins that localize to the ER. Our data cannot exclude the possibility that Golgi membranes may also give rise to omegasomes, although we would argue that they are not essential, given that disruption of the Golgi with BFA did not affect omegasome formation at all. Finally, we cannot eliminate (and in fact we favor) the possibility that this compartment originates in the ER but that it acquires distinct characteristics once autophagosomal proteins are accumulated there. Future work identifying the components of this compartment biochemically will address this question.
This PI(3)P-enriched compartment is involved in autophagosome biogenesis. We clearly observed continuous and dynamic interaction of omegasomes with forming autophagosomes, and we captured autophagosomes first appearing within the omegasomes before becoming distinct and acquiring mature characteristics such as a low pH. The simplest interpretation is that omegasomes may provide the sites for biogenesis of at least a certain type of autophagosome. Our data cannot exclude the possibility that not all autophagosomes originate in this way because we did not obtain a 100% colocalization between omegasomes and endogenous autophagosome proteins. During live imaging experiments, and in cases where the first appearance of a MAP-LC3–positive punctate structure could be clearly established, we almost always saw it appearing in association with an omegasome. In this sense we believe that omegasome association may account for a significant proportion of autophagosome formation.
As is currently understood, autophagosome biogenesis requires a series of biochemical reactions whereby two conjugation systems converge to create a multimeric protein assembly that nucleates formation of the autophagosome (Yorimitsu and Klionsky, 2005). One conjugation system creates a 4:4:4 Atg16/Atg12/Atg5 multimer (Yorimitsu and Klionsky, 2005), whereas a parallel conjugation system involves an unusual covalent modification of Atg8 (mammalian homologues MAP-LC3, GABARAP, and GATE-16) with phosphatidylethanolamine or phosphatidylserine (Sou et al., 2006). Because omegasomes colocalized with components of both conjugation systems (Atg5 and MAP-LC3), they would be well-suited for nucleating formation of such a particle, and the underlying ER membrane would provide a good source of the lipids that are used in the conjugation step.
PI(3)P enriched in omegasomes would also be important for attracting and localizing effectors such as Atg18 (Guan et al., 2001; Proikas-Cezanne et al., 2004), Atg20 and Atg21 (Stromhaug et al., 2004), Atg24 (Ano et al., 2005), and Alfy1 (Simonsen et al., 2004), all of which bind PI(3)P. DFCP1 is another such effector whose function is currently unknown. Based on the fact that DFCP1 does not have a counterpart in yeast, and especially in Drosophila melanogaster, which exhibits normal autophagic responses (Rusten et al., 2004; Scott et al., 2004), we consider it likely that DFCP1 itself may be involved in some (nonessential) specialized function during autophagy.
Although DFCP1 is not essential for autophagy, the pool of PI(3)P that it binds must be given the genetic and pharmacological data that reveal PI(3)P formation as an essential requirement in autophagosome formation (Blommaart et al., 1997; Kihara et al., 2001b; Mizushima et al., 2001). Our finding that high levels of ER/Golgi-localized WT DFCP1 (but not of the similarly localized DM) inhibit autophagosome formation can be explained by a mechanism involving sequestration of PI(3)P and are analogous with work showing that a double FYVE domain construct (Gillooly et al., 2000) can inhibit endosome function in vitro and in vivo when used at high levels (Boeddinghaus et al., 2002; Petiot et al., 2003; Asano et al., 2004). In particular, overexpression of a FYVE domain has been shown to lead to sequestration of endosomal PI(3)P and redistribution of EEA1 from endosomes to the cytosol (Byfield et al., 2005). In the case of DFCP1, both a myc- and a GFP-tagged construct expressed at levels higher than 10-fold over the endogenous protein inhibited autophagosome formation.
The source of PI(3)P
The ER in eukaryotic cells is highly compartmentalized (Levine and Rabouille, 2005) but contains little PI(3)P under normal conditions (Gillooly et al., 2000). How then is PI(3)P formed on the ER during starvation? We saw that vesicles containing Vps34 were frequently associated with the earliest discernible omegasome spots and continued associating as the omegasomes expanded. These Vps34 vesicles were positive for LAMP-2, which indicates that they are late endosomes or lysosomes, and were always found in the vicinity of the ER, frequently moving long distances along the ER strands. The simplest hypothesis for our observations is that Vps34 is delivered to the ER via some type of vesicular transport step, although we cannot rule out the possibility that Vps34-positive vesicles interact directly with the ER, allowing Vps34 to generate PI(3)P on the ER in trans. Remarkably, this proposed mode of action of Vps34 is analogous to the situation in yeast. The single autophagosome produced in yeast during autophagy localizes near the vacuole, in close proximity to Vps34 protein, which also localizes to the vacuole (Obara et al., 2006). Given that vacuoles are functionally equivalent to mammalian late endosomes/lysosomes, our proposal that autophagosomes arise near Vps34-positive vesicles makes the two systems very similar.
Previous work has shown that Vps34 is essential for initiation of the autophagic response (Kihara et al., 2001b) but, paradoxically, total Vps34 activity is stimulated by amino acids, and can actually drop during amino acid starvation and autophagy (Byfield et al., 2005; Nobukuni et al., 2005). These observations suggest that a subpopulation of Vps34 molecules is activated during amino acid starvation, whereas the majority of the protein is inhibited. In support of this, work in yeast has suggested the existence of two Vps34 complexes, with one involved in endosomal function and the other in autophagy (Kihara et al., 2001b). The complex involved in autophagy contains, in addition to Vps34, the two accessory proteins Vps14 and Vps30/Atg6. The mammalian homologue of Atg6, beclin 1, has been shown to be essential for induction of autophagy (Liang et al., 1999), and recent localization and functional studies have suggested that Beclin 1 is partially targeted to the ER (Pattingre et al., 2005), although the majority of Vps34 is targeted to the trans-Golgi network and to late endosomes/lysosomes (Kihara et al., 2001a). Given that omegasome formation requires Vps34 and beclin function, a possible mechanism for PI(3)P formation is that a Vps34-positive vesicle interacting with the ER delivers Vps34 to its binding partner, beclin, which is already there.
A real-time view of autophagosome formation
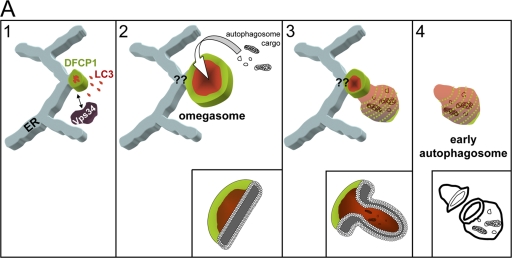
Based on our results and on extensive previous work, we propose the following working model for the biogenesis of at least certain types of autophagosomes (Fig. 9). Early during amino acid starvation, PI(3)P begins to accumulate in a part of a flat ER cisterna (green) in the vicinity of Vps34-containing vesicles. This PI(3)P-enriched membrane helps to localize autophagy-related proteins (red), and a mixed cisterna is created. Topologically, the PI(3)P-enriched membrane (omegasome) is always found encircling the autophagosomal membrane, but its exact connection to the ER is still not known (Fig. 9, question marks). This mixed cisterna continues to expand, with the PI(3)P- and the autophagosomal-enriched membranes remaining spatially distinct but continuous with each other. Our model thus far is similar to the pathway of isolation membrane expansion previously proposed, with the difference being that in our view, all of this takes place on a PI(3)P-enriched membrane platform. The final step in previous models of autophagosome formation involves fusion of the isolation membrane into a double membrane vesicle by an unknown mechanism. Again, our proposal is similar, but we would suggest that fusion of the flat isolation membrane is aided by the PI(3)P outer layer: when omegasomes reach their maximum size, inward budding would generate a prefusion autophagosome intermediate whereby the PI(3)P-enriched membrane marks the point of eventual closure of the double membrane vesicle. Indeed, live imaging showed that the last discernible pool of PI(3)P during omegasome collapse decorates the edge of the autophagosome.
Figure 9.
Potential role of omegasomes in autophagosome biogenesis. Our hypothesis is that during amino acid starvation, Vps34-containing vesicles interact with the ER and form PI(3)P on a membrane connected to the ER (step 1, green). This membrane domain associates with autophagosomal proteins (red) to create a mixed membrane domain that continues to expand but maintains its spatial separation with PI(3)P on the outside, and autophagosomal membranes inside (steps 1 and 2). Note that the connection of this membrane to the ER is not fully shown, thus the question marks at the junction. Once omegasomes reach their maximum size, the autophagosomal membranes bud inwards (steps 2 and 3), giving rise to a fully formed double-membrane autophagosome (step 4). The PI(3)P outer membrane in the intermediate of step 3 may aid the fusion reaction. Inward budding would allow the autophagosome to ingest cytoplasmic material and/or organelles indicated as autophagosome cargo. At the bottom of steps 2, 3, and 4, we have drawn cut-outs of the relevant structures to indicate the geometry of the bilayer.
Our proposed model is not in conflict with previous observations on autophagosome biogenesis, but it does place emphasis on the generation of PI(3)P as a very critical early event. In our view, PI(3)P is a regulator of this pathway by being the determining factor for the localization of autophagosome induction. In addition, our proposal is more consistent with the maturation model of autophagosome biogenesis and suggests that the ER plays an important role in this process both by providing the site for omegasome formation and the membrane used.
Materials and methods
All chemicals were obtained from Sigma-Aldrich unless otherwise stated.
Antibodies
The antibodies used in the course of this work were: mouse anti-GST (GeneTex, Inc.), mouse anti–β-COP (a gift from the late T. Kreis, European Molecular Biology Laboratory, Heidelberg, Germany), rat anti-ER proteins (primarily BiP, a gift from G. Butcher, Babraham Institute, Cambridge, England, UK), rabbit anti-GFP (Invitrogen), 9E10 mouse anti-myc, rabbit anti–MAP-LC3 (Santa Cruz Biotechnology Inc.), rabbit anti–MAP-LC3 (Sigma-Aldrich), mouse anti-EEA1 (BD Biosciences), rabbit anti-calnexin (Assay Designs), rabbit anti-calreticulin (prepared in house), rabbit anti-COPII (Affinity BioReagents), mouse anti-LAMP2 (Developmental Studies Hybridoma Bank), rabbit anti-mannosidase 2 (obtained from K. Moremen, University of Georgia, Athens, Georgia), rabbit anti-Vps34 (Invitrogen), rabbit anti–beclin 1 (Santa Cruz Biotechnology, Inc.), rabbit anti-giantin (Covance), and rabbit anti-Atg5 (Sigma-Aldrich).
Plasmids used in this work
Wild-type and C650S/C770S DFCP1 (DM) were cloned into pEGFPC2 (Invitrogen) and pCMV3myc as described previously (Ridley et al., 2001). Truncated mutants of DFCP1 (residues 416–535, 416–543, 416–549, and 416–576) were cloned into pEGFPC2 using standard techniques. Site-directed mutagenesis to generate point mutants within DFCP1 was performed using standard PCR techniques, and all constructs were verified by DNA sequencing. The pECFP-ER plasmid (which contains CFP flanked by an ER insertion signal and a KDEL C-terminal ER retention signal) was a kind gift of C. Taylor (University of Cambridge, Cambridge, England, UK). The pdsRED-ER plasmid (similar to the CFP one) was obtained from Clontech Laboratories, Inc.
To generate the GFP-ER-FYVE reporters, the FYVE domain of FENS-1 (Ridley et al., 2001) was amplified by PCR and ligated to the pEGFPC2 plasmid encoding the 416–543 fragment of DFCP1 (designated ER). The exact orientation in this reporter is important: GFP-ER-FYVE works, whereas GFP-FYVE-ER does not. To generate a version of this with a mutated FYVE domain, oligonucleotide-based mutagenesis was used to create a C/S mutation as described previously (Ridley et al., 2001).
To generate the GFP-TM and GFP-FYVE-TM constructs, we used oligonucleotides to add the following sequence in frame at the N terminus of GFP or the GFP-FYVE plasmid: N′-GGGSGGGSGGGSPSETLITTVESNSSWWTNWVIPAISALVVALMYRLYMAED-C′
The spacer region in this construct (amino acids starting with GGGS and ending with ESNS) is extremely important for its function. A shorter spacer results in a construct that stays in the ER without translocating to punctate structures.
Truncated DFCP1 constructs (416–543 and W543A 416–543 DFCP1) were also cloned into pGEX 4T-1 (GE Healthcare) using standard techniques.
To produce pSilencer RNAi B and RNAi C specific for DFCP1, the following oligonucleotides were designed: B, 5′-GATCCCACCATGAGCGGATAAGACTTTCAAGAGAAGTCTTATCCGCTCATGGTTTTTTGGAAA-3′; B′, 5′-AGCTTTTCCAAAAAACCATGAGCGGATAAGACTTCTCTTGAAAGTCTTATCCGCTCATGGTGG-3′; C, 5′-GATCCCTGTCAGTAATCTCCAGGAGTTCAAGAGACTCCTGGAGATTACTGACATTTTTGGAAA-3′; C′, 5′-AGCTTTTCCAAAAATGTCAGTAATCTCCAGGAGTCTCTTGAACTCCTGGAGATTACTGACAGG-3′.
The oligonucleotides were annealed and ligated into the pSilencer vector (Ambion) containing either an RNA U6 or H1 promoter using standard techniques. Constructs were verified by DNA sequencing. pEGFPC2 expressing iFYVE of FENS-1 was a gift from P. Hawkins (Babraham Institute). pEGFPC1 LC3 was a gift from T. Yoshimori (Osaka University, Osaka, Japan). To create the RFP version, LC3 was cloned into the pmRFPC1 vector using standard techniques. pEGFPC1 expressing the PX domain from p40phox was a gift from C. Ellson (Babraham Institute).
Cell culture, transfections, and generation of stable cell lines
HEK-293 cell lines and COS-7 cell lines were grown in DME (Invitrogen) containing 100 U/μl penicillin and streptomycin (Invitrogen) and 10% fetal calf serum (Invitrogen). Stable DFCP1-expressing cell lines were selected in 800 μg/ml geneticin (Invitrogen). Stable DFCP1- and Vps34-expressing cell lines were selected in 800 μg/ml geneticin and 100 μg/ml zeocin.
COS-7 cell lines were transiently transfected with diethylaminoethyl-dextran as described previously (Manifava et al., 2001). HEK-293 cell lines were transfected using FuGENE-6 and FuGENE HD transfection reagent (Roche) according to the manufacturer's instructions.
siRNA
We used predesigned oligonucleotides from Thermo Fisher Scientific (SMARTpool) to reduce expression levels of Vps34, beclin, and DFCP1. Cells were transfected using dharmafect-1 and examined 72 h later.
Starvation of HEK-293 cells and inhibition of autophagy
Throughout the paper, we refer to amino acid starvation, although it should be noted that other growth factors normally present in the medium are also absent. However, we determined that the DFCP1 response was only evident during amino acid withdrawal, i.e., when cells were incubated in serum-free DME, DFCP1 remained ER/Golgi associated. For amino acid starvation, cells were washed once with prewarmed PBS and then twice with prewarmed starvation medium (140 mM NaCl, 1 mM CaCl2, 1 mM MgCl2, 5 mM glucose, and 20 mM Hepes, pH 7.4) before incubation with starvation medium plus 1% BSA unless indicated otherwise. Note that this medium lacks amino acids as well as potassium; in preliminary experiments, we found it to give faster starvation responses in HEK-293 cells. The duration of the starvation treatment varied between experiments as indicated. PI 3-kinase was inhibited by the addition of either 10 nM wortmannin or 100 μM LY294002 to the cell medium. Autophagy was inhibited by the addition of 10 mM 3-methyladenine to the cell medium. BFA was used at 5 μg/ml. Duration of the treatments varied between experiments. When MDC was used, a stock was made in starvation medium at 0.05 mM, and this was diluted 1:20–1:40 during experiments.
Immunofluorescence Microscopy
Cells for immunofluorescence were grown on glass coverslips and fixed in 3.7% formaldehyde in 200 mM Hepes, pH 7.2. Staining for immunofluorescence and digital photography were done as described previously (Manifava et al., 1999).
Preparation of rat kidney microsomes and binding of GST-tagged DFCP1 fragments
Rat kidneys (Harlan Sera Laboratory) stored at −80°C were thawed, dissected, and suspended in 4× volume kidney membrane buffer (0.2 M sucrose, 20 mM Hepes-KOH, 2 mM DTT, 1 mM EGTA, and GST protease inhibitor cocktail, pH 7.2 [Roche]). After homogenization and centrifugation at 30,000 rpm, the membrane pellet was washed, resuspended in buffer, and stored at −80°C. Purified GST-tagged DFCP1 proteins were precleared by centrifugation at 14,000 g for 10 min and mixed with microsomes in siliconized Eppendorf tubes in a total volume of 200 μl for 15 min at 37°C. After centrifugation, the pellets were analyzed by SDS-PAGE.
Binding of proteins to PI(3)P-conjugated beads
COS-7 cells transiently transfected with the appropriate constructs were lysed in lysis buffer (50 mM Tris-HCl, pH 8.0, 50 mM KCl, 10 mM EDTA, 0.6 mM phenylmethylsulfonyl fluoride, 1 μg/ml trypsin inhibitor, and 0.5% Nonidet P-40) and centrifuged at 14,000 g to remove cell debris. Binding to PI(3)P-coupled beads was done as described previously (Ridley et al., 2001).
Staining of omegasomes with exogenous GST-PX
Cells expressing GFP-DFCP1 and grown on coverslips were starved for 60 min and washed extensively with PBS. They were then perforated with nitrocellulose as described previously (Simons and Virta, 1987) and fixed with formaldehyde. Staining was done with 50 μg/ml of purified GST-PX domain from p40phox (a gift from C. Ellson) followed by monoclonal anti-GST antibodies and TRITC-conjugated goat anti–mouse secondary.
Confocal imaging
Images were captured with a confocal microscope (FV1000; Olympus) using a 60× 1.4 NA objective (Olympus). Samples triple labeled with GFP, TRITC, and Cy5 were imaged using a sequential scan setting using excitation light at 488, 543, and 633 nm, respectively. Emission was collected at 495–535 nm (GFP), 550–600 nmn (TRITC), and >650 nm (Cy5).
Live cell imaging
Two imaging systems were used to capture images of live cells. Confocal images were taken using a confocal microscope (UltraVIEW LCI; PerkinElmer), whereas wide-field images (including for TIRFM) were taken using a Cell^R imaging system (Olympus). For both systems, cells were plated onto 22-mm-diameter glass coverslips (BDH) and transiently transfected with the relevant constructs, then individual coverslips were secured in an imaging chamber with 2 ml of cell medium or starvation medium added as indicated. The assembled imaging chamber was fitted into a heated stage on the microscope, and cells were maintained at 37°C. The UltraView LCI confocal was equipped with a 100× 1.4 NA objective (Nikon), CSU 10 scan head (Yokogawa), camera (Orca ER; Hamamatsu), sutter filter wheels, and an argon-krypton laser to excite at 488 (GFP) and 568 nm (DsRed). Emission was collected using 497–547 nm (GFP) and 577–622 (DsRed) band-pass filters. The Cell^R imaging system was equipped with 60× 1.45 NA and 100× 1.45 NA objectives, MT-20 illumination unit (150 W xenon/mercury mix bulb), and an Orca ER camera. CFP, GFP, and mRFP were excited using 417–442 nm, 486–498 nm, and 560–583 nm band-pass filters, respectively. Emission was collected using 455–475 nm (CFP), 510–545 nm (GFP), and 600–650 nm (mRFP) band-pass filters. Data were analyzed using UltraView or Cell^R software, with subsequent image processing using ImageJ (National Institutes of Health). Deconvolution was performed using Autodeblur (MediaCybernetics), and 3D reconstructions were made using Volocity software (PerkinElmer) where indicated.
EM
Samples for EM were washed three times in ice-cold EM preparation buffer (PBS, 0.2 M sucrose, 1 mM MgCl2, 0.2 mM CaCl2, and 0.1% BSA, pH 7.5) before addition of 1 ml EM preparation buffer with or without 5 ng/ml digitonin for 5 min on ice. Cells were fixed in 3.7% formaldehyde and 0.1% glutaraldehyde, and quenched in DME. For preembedding labeling, cells were stained with primary antibodies and protein–A gold before a second fixation step and embedding in epon. For cryo-EM, cells were scraped and centrifuged at 800 rpm for 5 min at RT, and the cell pellet was processed for immunogold labeling with rabbit anti-GFP antibodies (Invitrogen) using standard techniques.
Quantitation of puncta
We measured 100 cells in three separate experiments to determine levels of punctate structures or the extent of colocalization. These measurements were done on randomly selected fields of view. In addition, all data reported that show differences in puncta formation were verified qualitatively in blind fashion by an independent observer.
Online supplemental material
Fig. S1 shows the response of three PI(3)P-binding proteins to starvation and concludes that DFCP1 is the most appropriate reporter for colocalization with autophagosomes. Fig. S2 shows that overexpression of WT DFCP1 at high levels inhibits autophagosome formation, whereas its down-regulation by siRNA does not inhibit this response. Fig. S3 shows additional examples of colocalization of GFP-DFCP1 with autophagy-related proteins (MAP-LC3 and Atg5) and the ER. Fig. S4 shows additional examples of localization of DFCP1 by immuno-EM. Fig. S5 shows that autophagy as measured by three independent assays proceeds normally in the cell lines used throughout this work. Video 1 shows double imaging of GFP-TM-FYVE and dsRED-ER during starvation. Video 2 shows single imaging of GFP-DFCP1 during starvation. Video 3 shows triple imaging of GFP-DFCP1, mRFP-MAP-LC3, and CFP-ER during starvation. Video 4 shows double imaging of GFP-DFCP1 and dsRED-ER during starvation using TIRFM imaging. Video 5 shows a 3D reconstruction of GFP-DFCP1 and mRFP-MAP-LC3 at different planes of rotation. Video 6 shows double imaging of GFP-DFCP1 and mRFP-MAP-LC3 during starvation. Several panels from different videos showing the “budding” step are shown. Video 7 shows triple imaging of GFP-DFCP1, mRFP-Vps34, and CFP-ER during starvation. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200803137/DC1.
Supplementary Material
Acknowledgments
We are grateful to many colleagues for supplying reagents as listed in Materials and methods, to John Coadwell for bioinformatics support; to Michael Hinton for help with photography/movies; and to Len Stephens, Phill Hawkins, Jonathan Backer, and Aviva Tolkovsky for useful discussions.
This work was supported by the Biotechnology and Biological Sciences Research Council (BBSRC). E.L. Axe was the recipient of a BBSRC special committee studentship on live imaging. H.L. Roderick is a Royal Society University Research Fellow.
E.L. Axe and S.A. Walker contributed equally to this paper.
Abbreviations used in this paper: ATG, autophagy related; BFA, brefeldin A; DFCP1, double FYVE domain–containing protein 1; DM, domain mutant; EEA1, early endosome antigen 1; MDC, monodansylcadaverine; PAS, preautophagosomal structure; PDI, protein disulfide isomerase; PI 3-kinase, phosphoinositide 3-kinase; PI(3)P, phosphatidylinositol 3-phosphate; TIRFM, total internal reflection fluorescence microscopy; WT, wild type.
References
- Ano, Y., T. Hattori, M. Oku, H. Mukaiyama, M. Baba, Y. Ohsumi, N. Kato, and Y. Sakai. 2005. A sorting nexin PpAtg24 regulates vacuolar membrane dynamics during pexophagy via binding to phosphatidylinositol-3-phosphate. Mol. Biol. Cell. 16:446–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano, Y., H. Ihn, K. Yamane, M. Jinnin, Y. Mimura, and K. Tamaki. 2004. Phosphatidylinositol 3-kinase is involved in alpha2(I) collagen gene expression in normal and scleroderma fibroblasts. J. Immunol. 172:7123–7135. [DOI] [PubMed] [Google Scholar]
- Bampton, E.T., C.G. Goemans, D. Niranjan, N. Mizushima, and A.M. Tolkovsky. 2005. The dynamics of autophagy visualized in live cells: from autophagosome formation to fusion with endo/lysosomes. Autophagy. 1:23–36. [DOI] [PubMed] [Google Scholar]
- Blommaart, E.F., U. Krause, J.P. Schellens, H. Vreeling-Sindelarova, and A.J. Meijer. 1997. The phosphatidylinositol 3-kinase inhibitors wortmannin and LY294002 inhibit autophagy in isolated rat hepatocytes. Eur. J. Biochem. 243:240–246. [DOI] [PubMed] [Google Scholar]
- Boeddinghaus, C., A.J. Merz, R. Laage, and C. Ungermann. 2002. A cycle of Vam7p release from and PtdIns 3-P-dependent rebinding to the yeast vacuole is required for homotypic vacuole fusion. J. Cell Biol. 157:79–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgese, N., S. Brambillasca, and S. Colombo. 2007. How tails guide tail-anchored proteins to their destinations. Curr. Opin. Cell Biol. 19:368–375. [DOI] [PubMed] [Google Scholar]
- Bravo, J., D. Karathanassis, C.M. Pacold, M.E. Pacold, C.D. Ellson, K.E. Anderson, P.J. Butler, I. Lavenir, O. Perisic, P.T. Hawkins, et al. 2001. The crystal structure of the PX domain from p40(phox) bound to phosphatidylinositol 3-phosphate. Mol. Cell. 8:829–839. [DOI] [PubMed] [Google Scholar]
- Bulbarelli, A., T. Sprocati, M. Barberi, E. Pedrazzini, and N. Borgese. 2002. Trafficking of tail-anchored proteins: transport from the endoplasmic reticulum to the plasma membrane and sorting between surface domains in polarised epithelial cells. J. Cell Sci. 115:1689–1702. [DOI] [PubMed] [Google Scholar]
- Byfield, M.P., J.T. Murray, and J.M. Backer. 2005. hVps34 is a nutrient-regulated lipid kinase required for activation of p70 S6-kinase. J. Biol. Chem. 280:33076–33082. [DOI] [PubMed] [Google Scholar]
- Cheung, P.C., L. Trinkle-Mulcahy, P. Cohen, and J.M. Lucocq. 2001. Characterization of a novel phosphatidylinositol 3-phosphate-binding protein containing two FYVE fingers in tandem that is targeted to the Golgi. Biochem. J. 355:113–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Duve, C., and R. Wattiaux. 1966. Functions of lysosomes. Annu. Rev. Physiol. 28:435–492. [DOI] [PubMed] [Google Scholar]
- Derubeis, A.R., M.F. Young, L. Jia, P.G. Robey, and L.W. Fisher. 2000. Double FYVE-containing protein 1 (DFCP1): isolation, cloning and characterization of a novel FYVE finger protein from a human bone marrow cDNA library. Gene. 255:195–203. [DOI] [PubMed] [Google Scholar]
- Ellson, C.D., S. Gobert-Gosse, K.E. Anderson, K. Davidson, H. Erdjument-Bromage, P. Tempst, J.W. Thuring, M.A. Cooper, Z.Y. Lim, A.B. Holmes, et al. 2001. PtdIns(3)P regulates the neutrophil oxidase complex by binding to the PX domain of p40(phox). Nat. Cell Biol. 3:679–682. [DOI] [PubMed] [Google Scholar]
- Gillooly, D.J., I.C. Morrow, M. Lindsay, R. Gould, N.J. Bryant, J.M. Gaullier, R.G. Parton, and H. Stenmark. 2000. Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J. 19:4577–4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan, J., P.E. Stromhaug, M.D. George, P. Habibzadegah-Tari, A. Bevan, W.A. Dunn Jr., and D.J. Klionsky. 2001. Cvt18/Gsa12 is required for cytoplasm-to-vacuole transport, pexophagy, and autophagy in Saccharomyces cerevisiae and Pichia pastoris. Mol. Biol. Cell. 12:3821–3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhasz, G., and T.P. Neufeld. 2006. Autophagy: a forty-year search for a missing membrane source. PLoS Biol. 4:e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya, Y., N. Mizushima, T. Ueno, A. Yamamoto, T. Kirisako, T. Noda, E. Kominami, Y. Ohsumi, and T. Yoshimori. 2000. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19:5720–5728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara, A., Y. Kabeya, Y. Ohsumi, and T. Yoshimori. 2001. a. Beclin-phosphatidylinositol 3-kinase complex functions at the trans-Golgi network. EMBO Rep. 2:330–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara, A., T. Noda, N. Ishihara, and Y. Ohsumi. 2001. b. Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J. Cell Biol. 152:519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky, D.J. 2005. The molecular machinery of autophagy: unanswered questions. J. Cell Sci. 118:7–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky, D.J., J.M. Cregg, W.A. Dunn Jr., S.D. Emr, Y. Sakai, I.V. Sandoval, A. Sibirny, S. Subramani, M. Thumm, M. Veenhuis, and Y. Ohsumi. 2003. A unified nomenclature for yeast autophagy-related genes. Dev. Cell. 5:539–545. [DOI] [PubMed] [Google Scholar]
- Levine, T., and C. Rabouille. 2005. Endoplasmic reticulum: one continuous network compartmentalized by extrinsic cues. Curr. Opin. Cell Biol. 17:362–368. [DOI] [PubMed] [Google Scholar]
- Liang, X.H., S. Jackson, M. Seaman, K. Brown, B. Kempkes, H. Hibshoosh, and B. Levine. 1999. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 402:672–676. [DOI] [PubMed] [Google Scholar]
- Lindmo, K., and H. Stenmark. 2006. Regulation of membrane traffic by phosphoinositide 3-kinases. J. Cell Sci. 119:605–614. [DOI] [PubMed] [Google Scholar]
- Lum, J.J., R.J. DeBerardinis, and C.B. Thompson. 2005. Autophagy in metazoans: cell survival in the land of plenty. Nat. Rev. Mol. Cell Biol. 6:439–448. [DOI] [PubMed] [Google Scholar]
- Manifava, M., J. Sugars, and N.T. Ktistakis. 1999. Modification of catalytically active phospholipase D1 with fatty acid in vivo. J. Biol. Chem. 274:1072–1077. [DOI] [PubMed] [Google Scholar]
- Manifava, M., J.W. Thuring, Z.Y. Lim, L. Packman, A.B. Holmes, and N.T. Ktistakis. 2001. Differential binding of traffic-related proteins to phosphatidic acid- or phosphatidylinositol (4,5)- bisphosphate-coupled affinity reagents. J. Biol. Chem. 276:8987–8994. [DOI] [PubMed] [Google Scholar]
- Mizushima, N., A. Yamamoto, M. Hatano, Y. Kobayashi, Y. Kabeya, K. Suzuki, T. Tokuhisa, Y. Ohsumi, and T. Yoshimori. 2001. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J. Cell Biol. 152:657–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima, N., A. Kuma, Y. Kobayashi, A. Yamamoto, M. Matsubae, T. Takao, T. Natsume, Y. Ohsumi, and T. Yoshimori. 2003. Mouse Apg16L, a novel WD-repeat protein, targets to the autophagic isolation membrane with the Apg12-Apg5 conjugate. J. Cell Sci. 116:1679–1688. [DOI] [PubMed] [Google Scholar]
- Nobukuni, T., M. Joaquin, M. Roccio, S.G. Dann, S.Y. Kim, P. Gulati, M.P. Byfield, J.M. Backer, F. Natt, J.L. Bos, et al. 2005. Amino acids mediate mTOR/raptor signaling through activation of class 3 phosphatidylinositol 3OH-kinase. Proc. Natl. Acad. Sci. USA. 102:14238–14243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obara, K., T. Sekito, and Y. Ohsumi. 2006. Assortment of phosphatidylinositol 3-kinase complexes–Atg14p directs association of complex I to the pre-autophagosomal structure in Saccharomyces cerevisiae. Mol. Biol. Cell. 17:1527–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattingre, S., A. Tassa, X. Qu, R. Garuti, X.H. Liang, N. Mizushima, M. Packer, M.D. Schneider, and B. Levine. 2005. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 122:927–939. [DOI] [PubMed] [Google Scholar]
- Petiot, A., J. Faure, H. Stenmark, and J. Gruenberg. 2003. PI3P signaling regulates receptor sorting but not transport in the endosomal pathway. J. Cell Biol. 162:971–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proikas-Cezanne, T., S. Waddell, A. Gaugel, T. Frickey, A. Lupas, and A. Nordheim. 2004. WIPI-1alpha (WIPI49), a member of the novel 7-bladed WIPI protein family, is aberrantly expressed in human cancer and is linked to starvation-induced autophagy. Oncogene. 23:9314–9325. [DOI] [PubMed] [Google Scholar]
- Reggiori, F., and D.J. Klionsky. 2005. Autophagosomes: biogenesis from scratch? Curr. Opin. Cell Biol. 17:415–422. [DOI] [PubMed] [Google Scholar]
- Ridley, S.H., N. Ktistakis, K. Davidson, K.E. Anderson, M. Manifava, C.D. Ellson, P. Lipp, M. Bootman, J. Coadwell, A. Nazarian, et al. 2001. FENS-1 and DFCP1 are FYVE domain-containing proteins with distinct functions in the endosomal and Golgi compartments. J. Cell Sci. 114:3991–4000. [DOI] [PubMed] [Google Scholar]
- Rusten, T.E., K. Lindmo, G. Juhasz, M. Sass, P.O. Seglen, A. Brech, and H. Stenmark. 2004. Programmed autophagy in the Drosophila fat body is induced by ecdysone through regulation of the PI3K pathway. Dev. Cell. 7:179–192. [DOI] [PubMed] [Google Scholar]
- Scott, R.C., O. Schuldiner, and T.P. Neufeld. 2004. Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev. Cell. 7:167–178. [DOI] [PubMed] [Google Scholar]
- Simons, K., and H. Virta. 1987. Perforated MDCK cells support intracellular transport. EMBO J. 6:2241–2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen, A., H.C. Birkeland, D.J. Gillooly, N. Mizushima, A. Kuma, T. Yoshimori, T. Slagsvold, A. Brech, and H. Stenmark. 2004. Alfy, a novel FYVE-domain-containing protein associated with protein granules and autophagic membranes. J. Cell Sci. 117:4239–4251. [DOI] [PubMed] [Google Scholar]
- Sou, Y.S., I. Tanida, M. Komatsu, T. Ueno, and E. Kominami. 2006. Phosphatidylserine in addition to phosphatidylethanolamine is an in vitro target of the mammalian Atg8 modifiers, LC3, GABARAP, and GATE-16. J. Biol. Chem. 281:3017–3024. [DOI] [PubMed] [Google Scholar]
- Stromhaug, P.E., F. Reggiori, J. Guan, C.W. Wang, and D.J. Klionsky. 2004. Atg21 is a phosphoinositide binding protein required for efficient lipidation and localization of Atg8 during uptake of aminopeptidase I by selective autophagy. Mol. Biol. Cell. 15:3553–3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, K., and Y. Ohsumi. 2007. Molecular machinery of autophagosome formation in yeast, Saccharomyces cerevisiae. FEBS Lett. 581:2156–2161. [DOI] [PubMed] [Google Scholar]
- Suzuki, K., Y. Kubota, T. Sekito, and Y. Ohsumi. 2007. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells. 12:209–218. [DOI] [PubMed] [Google Scholar]
- Yorimitsu, T., and D.J. Klionsky. 2005. Autophagy: molecular machinery for self-eating. Cell Death Differ. 12(Suppl 2):1542–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.