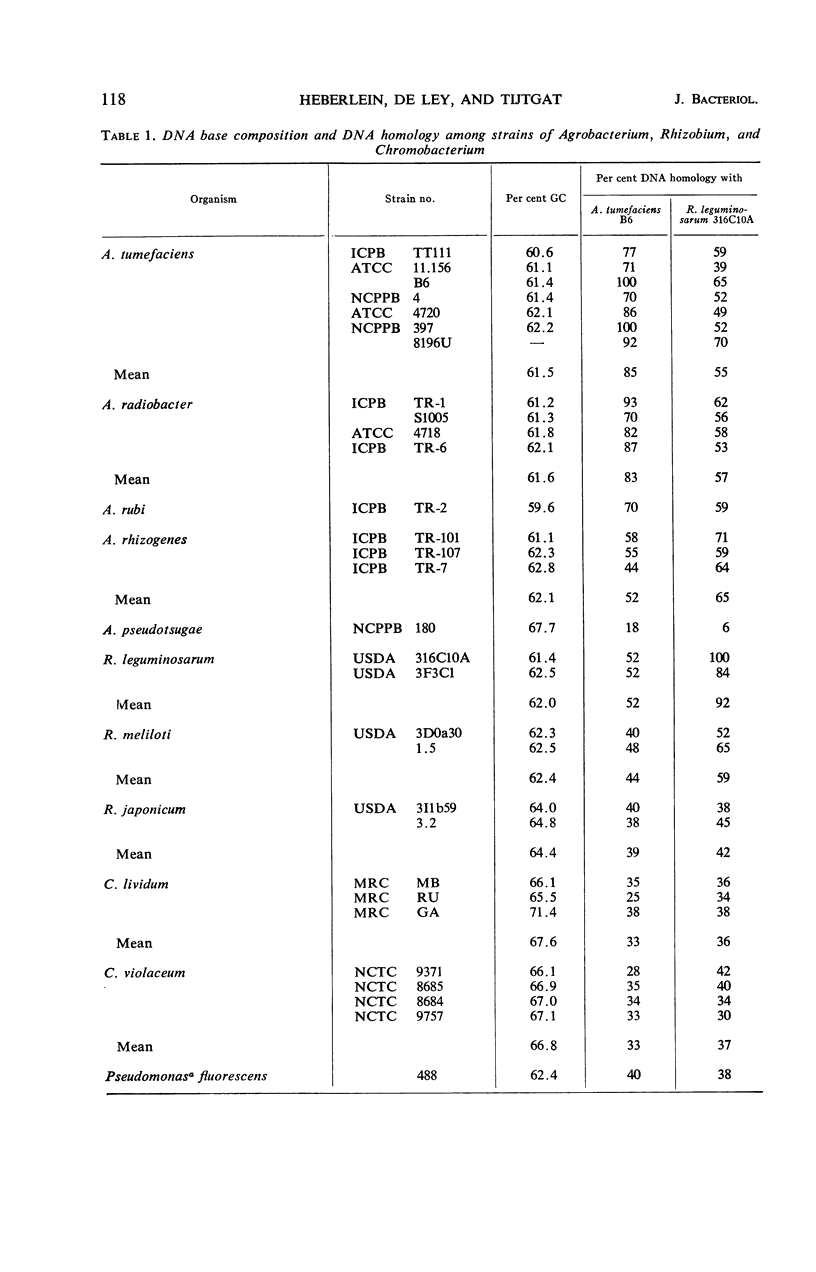
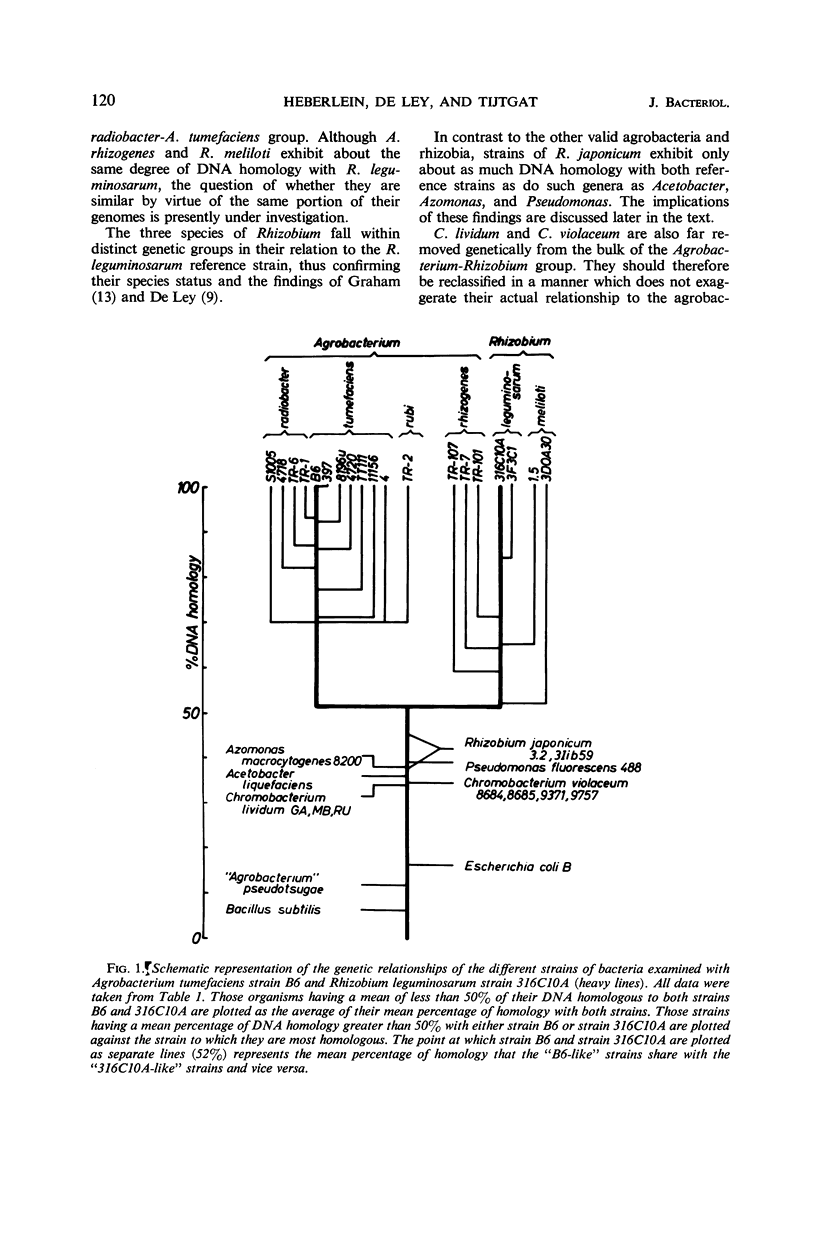
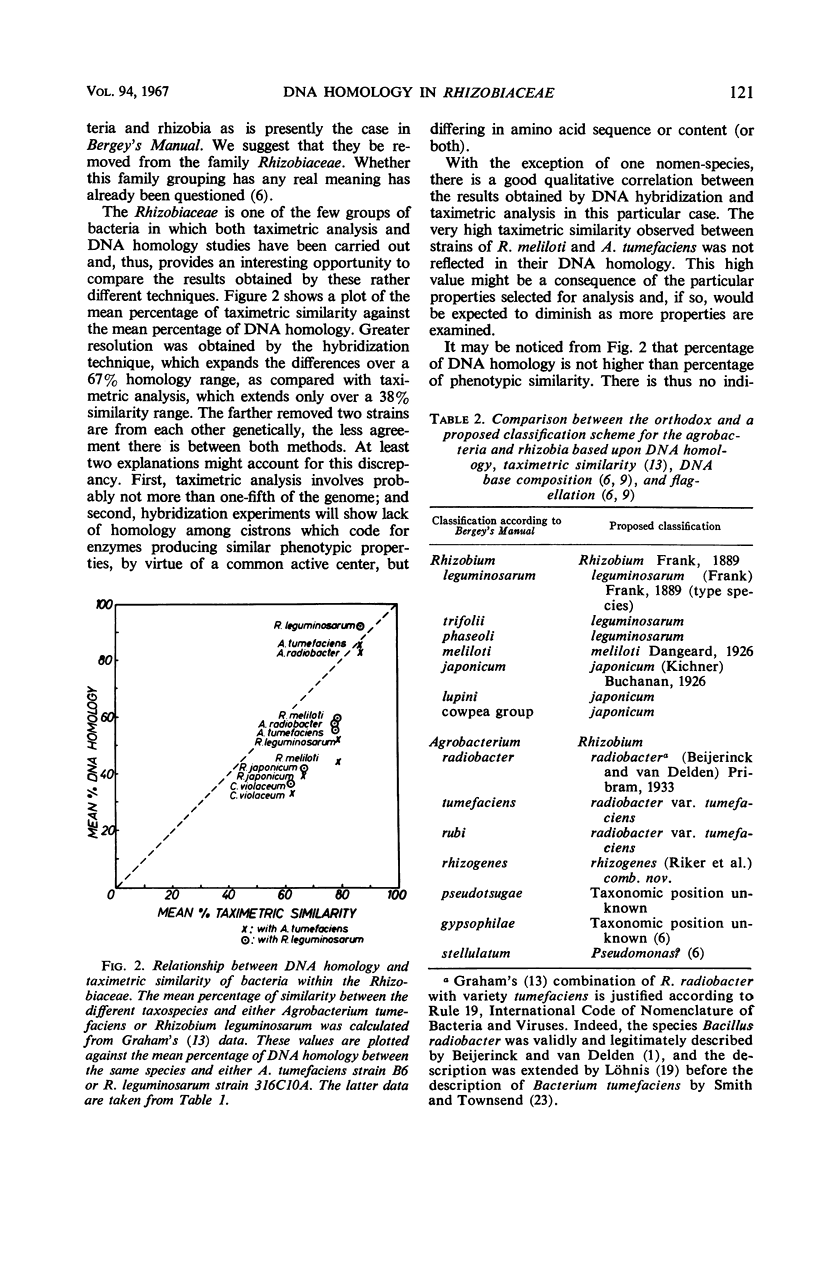
Abstract
Hybridization experiments were carried out between high molecular weight, denatured, agar-embedded deoxyribonucleic acid (DNA) and homologous, nonembedded, sheared, denatured 14C-labeled DNA from a strain of Agrobacterium tumefaciens and Rhizobium leguminosarum (the reference strains) in the presence of sheared, nonembedded, nonlabeled DNA (competing DNA) from the same or different nomen-species of Agrobacterium, Rhizobium, Chromobacterium, and several other organisms. Percentage of DNA homology was calculated from the results. The findings are discussed in relation to previous taximetric studies, present classification schemes, and guanine-cytosine content of the DNA. Strains of A. tumefaciens, A. radiobacter, A. rubi, A. rhizogenes, R. leguminosarum, and R. meliloti exhibited a mean percentage of DNA homology greater than 50 with the two reference strains. A. tumefaciens, A. radiobacter, and A. rubi were indistinguishable on the basis of DNA homology, with strain variations for this group involving up to 30% of their base sequences. The remainder of the organisms studied fall into at least six distinct genetic groups: (i) R. (Agrobacterium) rhizogenes, which is more homologous to R. leguminosarum than to the A. tumefaciens-A. radiobacter group; (ii) R. leguminosarum; (iii) R. meliloti; (iv) R. japonicum, which has a mean DNA homology of some 38 to 45% with the reference strains; (v) Chromobacterium, which is as genetically remote from the reference strains as, for example, Pseudomonas; and (vi) A. pseudotsugae strain 180, which has a DNA homology with A. tumefaciens and R. leguminosarum of only about 10%. Since this latter homology value is similar to what was found after hybridizations between the reference strains and organisms such as Escherichia coli and Bacillus subtilis, A. pseudotsugae should definitely be removed from the genus.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- BOLTON E. T., McCARTHY B. J. A general method for the isolation of RNA complementary to DNA. Proc Natl Acad Sci U S A. 1962 Aug;48:1390–1397. doi: 10.1073/pnas.48.8.1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DELEY J. EFFECT OF MUTATION ON DNA-COMPOSITION OF SOME BACTERIA. Antonie Van Leeuwenhoek. 1964;30:281–288. doi: 10.1007/BF02046734. [DOI] [PubMed] [Google Scholar]
- DELEY J., FRIEDMAN S. SIMILARITY OF XANTHOMONAS AND PSEUDOMONAS DEOXYRIBONUCLEIC ACID. J Bacteriol. 1965 May;89:1306–1309. doi: 10.1128/jb.89.5.1306-1309.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DELEY J., VANMUYLEM J. SOME APPLICATIONS OF DEOXYRIBONUCLEIC ACID BASE COMPOSITION IN BACTERIAL TAXONOMY. Antonie Van Leeuwenhoek. 1963;29:344–358. doi: 10.1007/BF02046087. [DOI] [PubMed] [Google Scholar]
- De Ley J., Bernaerts M., Rassel A., Guilmot J. Approach to an improved taxonomy of the genus Agrobacterium. J Gen Microbiol. 1966 Apr;43(1):7–17. doi: 10.1099/00221287-43-1-7. [DOI] [PubMed] [Google Scholar]
- De Ley J., Park I. W., Tijtgat R., Van Ermengem J. DNA homology and taxonomy of Pseudomonas and Xanthomonas. J Gen Microbiol. 1966 Jan;42(1):43–56. doi: 10.1099/00221287-42-1-43. [DOI] [PubMed] [Google Scholar]
- De Ley J., Rassel A. DNA base composition, flagellation and taxonomy of the genus Rhizobium. J Gen Microbiol. 1965 Oct;41(1):85–91. doi: 10.1099/00221287-41-1-85. [DOI] [PubMed] [Google Scholar]
- HYER B. H., MCCARTHY B. J., BOLTON E. T. A MOLECULAR APPROACH IN THE SYSTEMATICS OF HIGHER ORGANISMS. DNA INTERACTIONS PROVIDE A BASIS FOR DETECTING COMMON POLYNUCLEOTIDE SEQUENCES AMONG DIVERSE ORGANISMS. Science. 1964 May 22;144(3621):959–967. doi: 10.1126/science.144.3621.959. [DOI] [PubMed] [Google Scholar]
- Lippincott J. A., Heberlein G. T. The quantitative determination of the infectivity of Agrobacterium tumefaciens. Am J Bot. 1965 Sep;52(8):856–863. [PubMed] [Google Scholar]
- MCCARTHY B. J., BOLTON E. T. An approach to the measurement of genetic relatedness among organisms. Proc Natl Acad Sci U S A. 1963 Jul;50:156–164. doi: 10.1073/pnas.50.1.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SNEATH P. H. The application of computers to taxonomy. J Gen Microbiol. 1957 Aug;17(1):201–226. doi: 10.1099/00221287-17-1-201. [DOI] [PubMed] [Google Scholar]
- Smith E. F., Townsend C. O. A PLANT-TUMOR OF BACTERIAL ORIGIN. Science. 1907 Apr 26;25(643):671–673. doi: 10.1126/science.25.643.671. [DOI] [PubMed] [Google Scholar]
- Starr M. P. The Nutrition of Phytopathogenic Bacteria: II. The Genus Agrobacterium. J Bacteriol. 1946 Aug;52(2):187–194. [PMC free article] [PubMed] [Google Scholar]
- VALERA C. L., ALEXANDER M. NODULATION FACTOR FOR RHIZOBIUM-LEGUME SYMBIOSIS. J Bacteriol. 1965 Apr;89:1134–1139. doi: 10.1128/jb.89.4.1134-1139.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]


