Abstract
Dynamic organization of the cell interior, which is crucial for cell function, largely depends on the microtubule cytoskeleton. Microtubules move and position organelles by pushing, pulling, or sliding. Pushing forces can be generated by microtubule polymerization, whereas pulling typically involves microtubule depolymerization or molecular motors, or both. Sliding between a microtubule and another microtubule, an organelle, or the cell cortex is also powered by molecular motors. Although numerous examples of microtubule-based pushing and pulling in living cells have been observed, it is not clear why different cell types and processes employ different mechanisms. This review introduces a classification of microtubule-based positioning strategies and discusses the efficacy of pushing and pulling. The positioning mechanisms based on microtubule pushing are efficient for movements over small distances, and for centering of organelles in symmetric geometries. Mechanisms based on pulling, on the other hand, are typically more elaborate, but are necessary when the distances to be covered by the organelles are large, and when the geometry is asymmetric and complex. Thus, taking into account cell geometry and the length scale of the movements helps to identify general principles of the intracellular layout based on microtubule forces.
Keywords: Cytoskeleton, Microtubules, Force, Positioning, Mitotic spindle, Cell biophysics
Introduction
Cells are basic units of life, performing a multitude of complex functions and readily changing their program in response to environmental changes. Much is known about the intracellular elements, from large organelles to minute molecules, but how they interact and how these interactions are regulated to sustain an organized and functional cell is largely unknown. Microtubules are key organizers of the cell interior. These stiff hollow 25-nm wide tubes made of tubulin dimers (Alberts et al. 2008; Bouchet-Marquis et al. 2007) arrange into supramolecular structures with diverse functions such as the mitotic spindle, which separates the chromosomes during cell division, and microtubule bundles in axons, which serve as roads for intracellular traffic. Microtubules are dynamic polymers: phases of growth and shrinkage typically alternate (Mitchison and Kirschner 1984). This dynamic instability allows microtubules to interact temporarily with cellular components, to search the intracellular space, to disassemble and assemble into different arrangements, and to dynamically position cell organelles (Howard 2006).
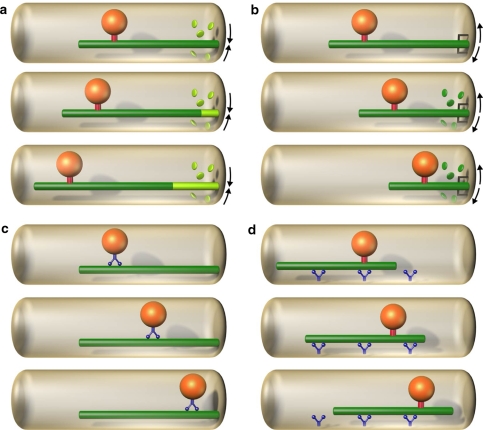
Microtubule-based positioning mechanisms can be divided into two classes. In class 1 the organelle is firmly bound to, and moves together with, the microtubule (Fig. 1a, b, d). In class 2 the organelle slides along the microtubule (Fig. 1c). The class 1 movements can be further divided according to the site of force generation, which is either at the microtubule end as in Fig. 1a, b, or along the lateral sides of the microtubule as in Fig. 1d. With respect to the force direction, the movements can be driven either by pushing as in Fig. 1a or pulling as in Fig. 1b. A pushing force generated by the microtubule end (Fig. 1a) is typically based on microtubule polymerization (Dogterom and Yurke 1997). As a consequence of pushing, the microtubule is under compression, which often leads to microtubule buckling. A pulling force (Fig. 1b) is generated by motor proteins (Howard 2001) and/or microtubule depolymerization. In the case of pulling the microtubule is under tension. Microtubule sliding (Fig. 1d) is powered by motor proteins, and can be regarded as either pushing or pulling, depending on the direction of motor motion. At a higher level of complexity, organelles can be bound to a set of overlapping microtubules that pull them together or push them apart, according to the motor activity in the overlap zone.
Fig. 1.
Basic types of microtubule force generation. a Pushing, b pulling; c, d sliding. a, b The organelle (orange) is bound to the microtubule (green) by a fixed link (red). a The organelle is being pushed away from the cell edge by a microtubule polymerization force. The microtubule polymerizes by addition of new subunits (light green discs) at its end (arrows). b A depolymerizing microtubule, which is connected to the cell edge by an active motor or a passive “adaptor” (dark grey), is pulling the organelle towards the cell edge. Depolymerization is accompanied by a loss of old subunits (dark green discs and arrows). c A motor protein (blue) walks along the microtubule and carries the organelle. d Motor proteins (blue) are anchored at the cell cortex and walk along the microtubule, thus translating the microtubule together with the bound organelle
Example of microtubule push–pull mechanisms: the mitotic spindle
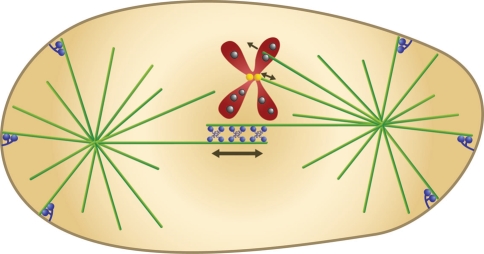
The mitotic spindle in animal cells consists of the central spindle, i.e., the microtubules connecting the spindle poles, and two asters (Fig. 2). Both single and overlapping microtubules can be found in the spindle. The center of the spindle is the meeting area for the anti-parallel microtubules that grow from each spindle pole, whereas asters contain microtubules growing from a single pole outwards in all directions. The main task of the mitotic spindle is to segregate sister chromatids: first, to separate them from each other, and then to move them across the cleavage plane, one set into each of the two future daughter cells. A key question is how these chromosome movements are achieved.
Fig. 2.
The mitotic spindle. Microtubules (green) form two asters and the central spindle, where the microtubules growing from the two spindle poles meet. Microtubule “plus-ends” (the more dynamic ends) are in the center of the spindle and at the cell periphery, while the “minus-ends” (the less dynamic ends) are anchored at the spindle poles. Motor proteins (blue, at the cell center) cross-link the overlapping anti-parallel microtubules in the central spindle and slide them past each other. Chromosomes (red; only one is shown for clarity) are attached to microtubules at the kinetochore (yellow), where forces towards and away from the spindle pole are generated. Chromokinesins (grey) reside at the chromosome arms and push the chromosomes away from the spindle pole by walking towards the microtubule plus-end. Minus-end directed motors (blue, at the cell periphery) are anchored at the cell cortex and pull on astral microtubules, thereby elongating the spindle
The separation of sister chromatids is lead by the kinetochore microtubules, which interact end-on with a kinetochore, a structure formed from proteins on the centromere of each chromosome (Fig. 2). The attachment of microtubules to the kinetochores is a dynamic process where the microtubules growing from the spindle pole towards the central region of the spindle search the spindle space for kinetochores and become stabilized when the search is successful. Unsuccessful microtubules undergo a transition from growth to shrinkage (termed “catastrophe”) and shorten towards the spindle poles, allowing other microtubules to do their search. Microtubules that interact with the chromosomes can push them away from the spindle pole from which the microtubule grows, or pull them towards the pole, depending on the type of interaction.
Spindle elongation, i.e., the movement of the spindle poles, plus the attached chromosomes, away from each other can be accomplished by two mechanisms: (1) the overlap microtubules of the central spindle interact via molecular motors which slide them apart; (2) astral microtubules interact with the cell cortex via molecular motors, which pull the spindle poles apart (Fig. 2).
Laser-cutting as a method to test the microtubule forces
A direct way to distinguish between pushing and pulling mechanisms in the cell is to mechanically perturb the force balance in the structure of interest, and to observe its subsequent movements. For example, in order to identify the role of the forces exerted by astral microtubules and those by the central spindle on the separation of the spindle poles, one can cut the central spindle using a laser microbeam or a microneedle (Aist and Berns 1981). The model in which astral forces dominate, hence, the spindle is under tension, predicts that the poles of the cut spindle will continue moving apart after the cut. On the contrary, the model in which the major forces driving spindle elongation are those generating compression in the central spindle predicts that the spindle poles of a centrally severed spindle will stop moving apart. Micro-cutting techniques have been applied to spindles and other microtubule-based structures in a variety of cell systems. The results of these and related experiments are summarized in the following section according to models of microtubule-based pushing, pulling, and sliding.
Classification of microtubule-based positioning strategies
Microtubule pushing on an organelle
Pushing forces on a non-dividing nucleus
Aster. During interphase in the budding yeast Saccharomyces cerevisiae, microtubules are organized in an aster or a conical array radiating from the spindle pole body (a yeast centrosome). The microtubules of the aster exhibit dynamic instability and push against the cell cortex, thus propelling the nucleus in the opposite direction (Shaw et al. 1997).
Parallel array. In cylindrically shaped cells of the fission yeast Schizosaccharomyces pombe, interphase microtubules lie parallel with the cell axis, surrounding the nucleus like a barrel. They are attached to the nuclear envelope at several points, including the spindle pole body and interphase microtubule-organizing centers. These microtubules grow from the nuclear region towards the cell tips, and exert a transient pushing force on the nucleus upon reaching the cell tips (Tran et al. 2001). Microtubule pushing keeps the nucleus at the cell center and can even re-center a displaced nucleus. This was shown previously by experiments where the nucleus was moved away from the cell center using optical tweezers (Tolić-Nørrelykke et al. 2005) or cell centrifugation (Daga et al. 2006). Interestingly, when the microtubule organization was changed from a parallel array into an aster by ectopically expressing the meiosis-specific spindle pole body component Hrs1p/Mcp6p, pushing forces by astrally arranged microtubules were not able to center the nucleus, but generated large nuclear excursions instead (Tanaka et al. 2005). Thus, the ability of microtubule pushing forces to center the nucleus depends on the arrangement of microtubules, besides its more obvious dependence on microtubule dynamics and cell geometry.
Pushing forces on a dividing nucleus
Pushing on the spindle. In S. pombe, spindle movements involve microtubule-pushing forces. At the transition from interphase to mitosis, interphase microtubules are attached to the duplicated spindle pole body. Pushing forces exerted by these microtubules against the cell tips position the duplicated spindle pole body in the cell center, thereby setting the central location and the alignment of the future spindle (Vogel et al. 2007). Later in mitosis, during anaphase B, astral microtubules grow from the spindle pole bodies in a direction nearly perpendicular to the central spindle. Without contributing to spindle elongation, these astral microtubules help to align the spindle with the cell axis by pushing against the cell sides (Tolić-Nørrelykke et al. 2004).
Pushing on chromosomes. Experiments, where a prometaphase chromosome was cut by a laser microbeam, demonstrated that microtubules can push on the chromosomes in the spindle. The fragment without the kinetochore moved rapidly away from the spindle pole, suggesting that “polar ejection forces” push chromosome arms away from the proximal spindle pole (Rieder et al. 1986). Recent studies have revealed the molecular entity generating ejection forces to be the chromokinesin Kid (Antonio et al. 2000; Brouhard and Hunt 2005; Funabiki and Murray 2000). This motor is bound to chromosome arms and “walks” towards the microtubule plus-end, which points away from the spindle pole.
An additional motor protein contributes to chromosome movements away from the spindle pole. The plus-end directed motor CENP-E binds the chromosome at the kinetochore and slides it along another microtubule bound to the kinetochore of another chromosome (Kapoor et al. 2006). This movement is not only microtubule-based “pushing” of an organelle away from the spindle pole, but it also involves the idea of “sliding” of an organelle along a microtubule (see below).
Microtubule pulling on an organelle
Pulling forces on a non-dividing nucleus
In meiotic prophase of S. pombe, the nucleus shows a peculiar movement: it oscillates from one end of the cell to the other with a period of 5–10 min repeated over 2–3 h. The oscillatory movement of the nucleus is lead by the spindle pole body and depends on microtubules growing from the spindle pole body, and on the motor protein cytoplasmic dynein (Yamamoto et al. 1999). Observation of the spindle pole body movements in relation to the microtubule position (Yamamoto et al. 2001), as well as laser-cutting of microtubules (Vogel et al. 2008), indicated that the driving force for nuclear oscillations is pulling by astral microtubules. This pulling force is generated by cortically anchored dynein motors that walk along the microtubules towards microtubule minus-ends, which are focused at the spindle pole body.
After fertilization in multicellular organisms, the male pronucleus migrates towards the center of the egg to reach the female pronucleus. This movement depends on the aster of microtubules that grow from two centrosomes attached to the male pronucleus. Minus-end directed motors, anchored at organelles in the cytoplasm, move the aster and the associated male pronucleus by walking along the astral microtubules (Hamaguchi and Hiramoto 1986; Kimura and Onami 2005). Simultaneously, the female pronucleus does not passively wait, but moves towards the male pronucleus by sliding along the same astral microtubules (see below).
Pulling forces on a dividing nucleus
Pulling on the spindle. Since the pioneering experiments of Aist and Berns (1981), forces exerted by astral microtubules on the spindle poles have been explored by laser-cutting of the spindle in a variety of cell types. Cutting of the spindle in the fungus Fusarium solani showed that extranuclear forces, presumably involving astral microtubules, pull on the spindle poles and that the central spindle limits the separation rate (Aist and Berns 1981). Similar conclusions were obtained by laser ablation of centrosomes in rat kangaroo kidney epithelium (PtK2) cells (Aist et al. 1993). In the single-cell stage C. elegans embryo, cutting of the central spindle revealed that pulling forces external to the spindle act on the two spindle poles, a stronger net force acting on the posterior pole. The asymmetry in the pulling force results in an asymmetric spindle position, which translates into an asymmetric cell division (Grill et al. 2001; Grill and Hyman 2005). Laser-cutting has identified pulling forces by microtubules and dynein to be responsible for rapid elongation and positioning of the central spindle in the fungus Ustilago maydis (Fink et al. 2006). Observation of astral microtubules and spindle pole body movements have suggested that astral microtubules pull one spindle pole into the bud during budding yeast mitosis (Adames and Cooper 2000; Yeh et al. 2000). In fibroblasts, astral microtubules transiently anchored at the bottom of the cell exert pulling forces to position the spindle (Schultz and Onfelt 2001). These mechanisms of pulling the organelles via cortically anchored molecular motors that move along astral microtubules can be described as “pulling” from the organelle viewpoint, and as “sliding” from the cortex perspective.
Pulling on chromosomes. Experiments with laser-cutting of prometaphase chromosomes, mentioned above in the context of pushing forces, have also revealed a pulling force on chromosomes. Whereas the chromosome fragment without the kinetochore moved away from the proximal spindle pole, the fragment with the kinetochore moved towards the pole, suggesting that kinetochore microtubules pull on the kinetochore (Rieder et al. 1986). Recent work has shown that the Dam1 ring complex sliding along the microtubule translates the force generated by microtubule depolymerization into a poleward kinetochore movement along the microtubule lattice to drive chromosome segregation (Westermann et al. 2006).
Organelle-microtubule sliding
In this class of intracellular movements the organelle is loosely bound to, and moves with respect to, the microtubule, which serves as a track for the movement. The movement is driven by molecular motors. Plus-end directed motors, such as kinesins, distribute the endoplasmic reticulum and the Golgi complex along microtubules (Lippincott-Schwartz et al. 1995). Kinesins and other proteins from the kinesin-superfamily transport mitochondria along microtubules (Fujita et al. 2007), as well as precursors of synaptic vesicles and axonal membranes in neurons (Kondo et al. 1994). The minus-end directed motor dynein moves the endoplasmic reticulum in U. maydis (Wedlich-Soldner et al. 2002). After fertilization in multicellular organisms, the female pronucleus moves towards the male pronucleus along the astral microtubules extending from the centrosome associated with the male pronucleus (Hamaguchi and Hiramoto 1986; Schatten 1981).
Similar movement of membrane-bound organelles along microtubules contributes to skin pigmentation. In keratinocytes, a “microparasol” made of phagocytosed melanosomes filled with the dark pigment melanin protects the nucleus from UV-induced DNA damage. Cytoplasmic dynein is the motor driving the perinuclear-directed aggregation of melanosomes along microtubules (Byers et al. 2003).
Microtubule-microtubule sliding
Central spindle
Microtubules growing from the two spindle poles meet in an anti-parallel arrangement in the central part of the spindle. The spindle length is nearly constant during metaphase while the kinetochore microtubules are capturing the chromosomes. In anaphase B, on the other hand, the spindle elongates in order to separate the two sets of chromosomes. The spindle length is controlled by a “push-me-pull-you” mechanism: motors of opposite polarity cross-link the overlapping microtubules in the central spindle. Their opposing forces slide the spindle microtubules with respect to one another. The plus-end directed motors (BimC kinesins, e.g., KLP61F in Drosophila) push the spindle poles apart, whereas minus-end directed motors (kinesin-14 family, e.g., Ncd in Drosophila) pull them together (Sharp et al. 1999). The balance of forces exerted by these two classes of motors helps to set the constant spindle length in metaphase and the elongation onset at the transition to anaphase.
In S. pombe, spindle elongation in anaphase B is driven by pushing forces in the central spindle, as opposed to pulling-apart of the spindle poles by astral microtubules as described above. The role of pushing forces was revealed by laser-cutting experiments where the poles of severed spindles did not continue to move apart, suggesting that the forces external to the spindle do not have a significant impact on spindle elongation. Thus, the elongation forces are instead generated in the central spindle (Khodjakov et al. 2004; Tolić-Nørrelykke et al. 2004).
Spindle pole focusing
Minus-end directed motor proteins, such as dynein, can cross-link and slide single free microtubules with respect to each other. If several motor molecules are connected, their activity can rearrange the microtubules into an aster with a focus of minus-ends (Verde et al. 1991). This mechanism of motor-based microtubule sliding and convergence, together with the nucleation of microtubules from centrosomes, is required for focusing of minus-ends into spindle poles in vertebrate somatic cells (Gaglio et al. 1997).
Cilia and flagella
Another specialized force-generating structure based on microtubules and dynein is the axoneme, the core of motile cilia and flagella. A key structural feature of the axoneme, compared to the central spindle and asters, is that the parallel microtubules in the axoneme are not free to move, but are connected to their neighbors by protein links. This prevents sliding of the microtubules with respect to one another. The motor activity of the axonemal dynein is, therefore, converted into bending of the microtubules, resulting in the beating of cilia and the wave motion of flagella (Lindemann 2003).
Microtubule-actin push–pull mechanism
Besides pushing or pulling on the cell cortex, nucleus, spindle, chromosomes, and various membrane-bound organelles, microtubules exert forces on other cytoskeleton systems such as the actin network. A model of cell mechanics called “tensegrity,” which is a contraction of “tensional integrity,” describes the cell as a structure consisting of a continuous, tensed network of structural elements together with other isolated support elements that resist compression (Ingber 1993). According to this model, which is used to describe cell shape changes, cell deformability depends on the level of tension in the cell. In the simplest embodiment of the cellular tensegrity model, the actin–myosin contractile system is under tension, which is balanced by the compression borne by microtubules and the attachment points to the external substrate. The observed correlation between the extent of microtubule buckling and the level of tension in the actin cytoskeleton supports the idea that microtubules bear compression as they balance a substantial portion of the contractile stress (Wang et al. 2001). Another prediction of the model is that, upon microtubule disruption, the portion of stress balanced by microtubules would shift to the substrate, thereby causing an increase in the forces exerted by the cell at the attachment points to the substrate. This was observed in adherent smooth muscle cells (Stamenovic et al. 2002), providing further evidence in favor of the model of the push–pull relationship between microtubules and actin.
Summary
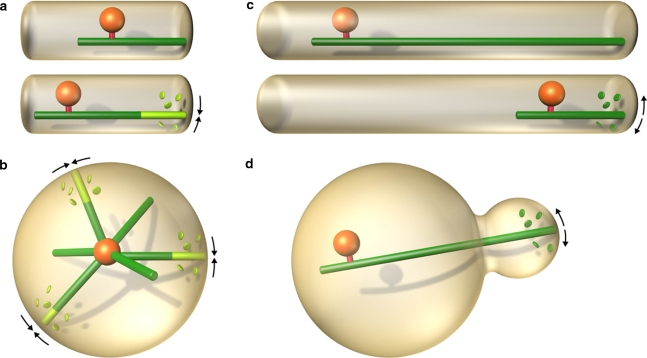
Why do some cell types employ microtubule pushing, whereas other cells use pulling, to position their organelles? Although much is known about examples of microtubule pushing and pulling in living cells, the relation between the cell systems and the positioning mechanisms employed is not clear. Different cell geometries, and distances over which the organelles have to be moved, may require different force generation strategies. The positioning mechanisms based on microtubule pushing are efficient for movements over short distances (up to ∼10 μm; Fig. 3a), where microtubules do not buckle, and for centering of organelles in symmetric geometries (Fig. 3b). These conditions are met, for example, in S. pombe interphase nuclear centering, as well as in mitotic spindle centering by astral microtubules in the same organism. Whereas pushing mechanisms can be based on microtubule polymerization force, without requirements for additional force-generating proteins, mechanisms based on pulling are typically more complex to engineer since they involve accessory proteins such as motors or adaptors. Pulling mechanisms are, however, necessary when the distances to be covered by the organelles are large (Fig. 3c), or when the geometry is asymmetric and complex (Fig. 3d). Spindle elongation in large animal cells, and extensive nuclear movements in S. pombe meiotic prophase, are examples of large movements, spindle positioning in the C. elegans embryo and in S. cerevisiae are examples of asymmetric movements. The cell’s “choice” of the positioning strategy may thus depend on the cell size, shape, and specific requirements for organelle positioning. Focusing on these aspects will help to uncover unifying themes of how microtubule pushing and pulling forces organize the cellular interior.
Fig. 3.
Pushing versus pulling. The positioning mechanisms based on microtubule pushing are efficient for movements over small distances (a), and for centering in symmetric geometries (b). Mechanisms based on pulling are preferred in cases where the distances to be covered by the organelles are large (c), and where the geometry is asymmetric and complex (d)
Acknowledgments
Thanks to Franziska Friedrich and Daniel Schmidt for drawing the figures; Nenad Pavin, Nicola Maghelli, Sven Vogel, Joe Howard, Frank Jülicher, Francesco Pavone, Leonardo Sacconi, and the members of the Tolić-Nørrelykke group for inspiring discussions and collaborations; and Judith Nicholls for proof-reading.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Footnotes
Regional Biophysics Conference of the National Biophysical Societies of Austria, Croatia, Hungary, Italy, Serbia, and Slovenia.
References
- Adames NR, Cooper JA. Microtubule interactions with the cell cortex causing nuclear movements in Saccharomyces cerevisiae. J Cell Biol. 2000;149:863–874. doi: 10.1083/jcb.149.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aist JR, Berns MW. Mechanics of chromosome separation during mitosis in Fusarium (Fungi imperfecti): new evidence from ultrastructural and laser microbeam experiments. J Cell Biol. 1981;91:446–458. doi: 10.1083/jcb.91.2.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aist JR, Liang H, Berns MW. Astral and spindle forces in PtK2 cells during anaphase B: a laser microbeam study. J Cell Sci. 1993;104:1207–1216. doi: 10.1242/jcs.104.4.1207. [DOI] [PubMed] [Google Scholar]
- Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. Molecular biology of the cell. 5th edn. New York: Garland Science; 2008. [Google Scholar]
- Antonio C, Ferby I, Wilhelm H, Jones M, Karsenti E, Nebreda AR, Vernos I. Xkid, a chromokinesin required for chromosome alignment on the metaphase plate. Cell. 2000;102:425–435. doi: 10.1016/S0092-8674(00)00048-9. [DOI] [PubMed] [Google Scholar]
- Bouchet-Marquis C, Zuber B, Glynn AM, Eltsov M, Grabenbauer M, Goldie KN, Thomas D, Frangakis AS, Dubochet J, Chretien D. Visualization of cell microtubules in their native state. Biol Cell. 2007;99:45–53. doi: 10.1042/BC20060081. [DOI] [PubMed] [Google Scholar]
- Brouhard GJ, Hunt AJ. Microtubule movements on the arms of mitotic chromosomes: polar ejection forces quantified in vitro. Proc Natl Acad Sci USA. 2005;102:13903–13908. doi: 10.1073/pnas.0506017102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers HR, Maheshwary S, Amodeo DM, Dykstra SG. Role of cytoplasmic dynein in perinuclear aggregation of phagocytosed melanosomes and supranuclear melanin cap formation in human keratinocytes. J Invest Dermatol. 2003;121:813–820. doi: 10.1046/j.1523-1747.2003.12481.x. [DOI] [PubMed] [Google Scholar]
- Daga RR, Yonetani A, Chang F. Asymmetric microtubule pushing forces in nuclear centering. Curr Biol. 2006;16:1544–1550. doi: 10.1016/j.cub.2006.06.026. [DOI] [PubMed] [Google Scholar]
- Dogterom M, Yurke B. Measurement of the force–velocity relation for growing microtubules. Science. 1997;278:856–860. doi: 10.1126/science.278.5339.856. [DOI] [PubMed] [Google Scholar]
- Fink G, Schuchardt I, Colombelli J, Stelzer E, Steinberg G. Dynein-mediated pulling forces drive rapid mitotic spindle elongation in Ustilago maydis. EMBO J. 2006;25:4897–4908. doi: 10.1038/sj.emboj.7601354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita T, Maturana AD, Ikuta J, Hamada J, Walchli S, Suzuki T, Sawa H, Wooten MW, Okajima T, Tatematsu K, Tanizawa K, Kuroda S. Axonal guidance protein FEZ1 associates with tubulin and kinesin motor protein to transport mitochondria in neurites of NGF-stimulated PC12 cells. Biochem Biophys Res Commun. 2007;361:605–610. doi: 10.1016/j.bbrc.2007.07.050. [DOI] [PubMed] [Google Scholar]
- Funabiki H, Murray AW. The Xenopus chromokinesin Xkid is essential for metaphase chromosome alignment and must be degraded to allow anaphase chromosome movement. Cell. 2000;102:411–424. doi: 10.1016/S0092-8674(00)00047-7. [DOI] [PubMed] [Google Scholar]
- Gaglio T, Dionne MA, Compton DA. Mitotic spindle poles are organized by structural and motor proteins in addition to centrosomes. J Cell Biol. 1997;138:1055–1066. doi: 10.1083/jcb.138.5.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grill SW, Hyman AA. Spindle positioning by cortical pulling forces. Dev Cell. 2005;8:461–465. doi: 10.1016/j.devcel.2005.03.014. [DOI] [PubMed] [Google Scholar]
- Grill SW, Gonczy P, Stelzer EH, Hyman AA. Polarity controls forces governing asymmetric spindle positioning in the Caenorhabditis elegans embryo. Nature. 2001;409:630–633. doi: 10.1038/35054572. [DOI] [PubMed] [Google Scholar]
- Hamaguchi MS, Hiramoto Y. Analysis of the role of astral rays in pronuclear migration by the colcemid-UV method. Dev Growth Differ. 1986;28:143–156. doi: 10.1111/j.1440-169X.1986.00143.x. [DOI] [PubMed] [Google Scholar]
- Howard J. Mechanics of Motor Proteins and the Cytoskeleton. Sunderland: Sinauer Associates; 2001. [Google Scholar]
- Howard J. Elastic and damping forces generated by confined arrays of dynamic microtubules. Phys Biol. 2006;3:54–66. doi: 10.1088/1478-3975/3/1/006. [DOI] [PubMed] [Google Scholar]
- Ingber DE. Cellular tensegrity: defining new rules of biological design that govern the cytoskeleton. J Cell Sci. 1993;104(Pt 3):613–627. doi: 10.1242/jcs.104.3.613. [DOI] [PubMed] [Google Scholar]
- Kapoor TM, Lampson MA, Hergert P, Cameron L, Cimini D, Salmon ED, McEwen BF, Khodjakov A. Chromosomes can congress to the metaphase plate before biorientation. Science. 2006;311:388–391. doi: 10.1126/science.1122142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodjakov A, La Terra S, Chang F. Laser microsurgery in fission yeast; role of the mitotic spindle midzone in anaphase B. Curr Biol. 2004;14:1330–1340. doi: 10.1016/j.cub.2004.07.028. [DOI] [PubMed] [Google Scholar]
- Kimura A, Onami S. Computer simulations and image processing reveal length-dependent pulling force as the primary mechanism for C. elegans male pronuclear migration. Dev Cell. 2005;8:765–775. doi: 10.1016/j.devcel.2005.03.007. [DOI] [PubMed] [Google Scholar]
- Kondo S, Sato-Yoshitake R, Noda Y, Aizawa H, Nakata T, Matsuura Y, Hirokawa N. KIF3A is a new microtubule-based anterograde motor in the nerve axon. J Cell Biol. 1994;125:1095–1107. doi: 10.1083/jcb.125.5.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindemann CB. Structural-functional relationships of the dynein, spokes, and central-pair projections predicted from an analysis of the forces acting within a flagellum. Biophys J. 2003;84:4115–4126. doi: 10.1016/S0006-3495(03)75136-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott-Schwartz J, Cole NB, Marotta A, Conrad PA, Bloom GS. Kinesin is the motor for microtubule-mediated Golgi-to-ER membrane traffic. J Cell Biol. 1995;128:293–306. doi: 10.1083/jcb.128.3.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison T, Kirschner M. Dynamic instability of microtubule growth. Nature. 1984;312:237–242. doi: 10.1038/312237a0. [DOI] [PubMed] [Google Scholar]
- Rieder CL, Davison EA, Jensen LC, Cassimeris L, Salmon ED. Oscillatory movements of monooriented chromosomes and their position relative to the spindle pole result from the ejection properties of the aster and half-spindle. J Cell Biol. 1986;103:581–591. doi: 10.1083/jcb.103.2.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatten G. Sperm incorporation, the pronuclear migrations, and their relation to the establishment of the first embryonic axis: time-lapse video microscopy of the movements during fertilization of the sea urchin Lytechinus variegatus. Dev Biol. 1981;86:426–437. doi: 10.1016/0012-1606(81)90201-3. [DOI] [PubMed] [Google Scholar]
- Schultz N, Onfelt A. Spindle positioning in fibroblasts supports an astral microtubule length dependent force generation at the basal membrane. Cell Motil Cytoskeleton. 2001;50:69–88. doi: 10.1002/cm.1042. [DOI] [PubMed] [Google Scholar]
- Sharp DJ, Yu KR, Sisson JC, Sullivan W, Scholey JM. Antagonistic microtubule-sliding motors position mitotic centrosomes in Drosophila early embryos. Nat Cell Biol. 1999;1:51–54. doi: 10.1038/9025. [DOI] [PubMed] [Google Scholar]
- Shaw SL, Yeh E, Maddox P, Salmon ED, Bloom K. Astral microtubule dynamics in yeast: a microtubule-based searching mechanism for spindle orientation and nuclear migration into the bud. J Cell Biol. 1997;139:985–994. doi: 10.1083/jcb.139.4.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamenovic D, Mijailovich SM, Tolić-Nørrelykke IM, Chen J, Wang N. Cell prestress. II. Contribution of microtubules. Am J Physiol Cell Physiol. 2002;282:C617–C624. doi: 10.1152/ajpcell.00271.2001. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Kohda T, Yamashita A, Nonaka N, Yamamoto M. Hrs1p/Mcp6p on the meiotic SPB organizes astral microtubule arrays for oscillatory nuclear movement. Curr Biol. 2005;15:1479–1486. doi: 10.1016/j.cub.2005.07.058. [DOI] [PubMed] [Google Scholar]
- Tolić-Nørrelykke IM, Sacconi L, Thon G, Pavone FS. Positioning and elongation of the fission yeast spindle by microtubule-based pushing. Curr Biol. 2004;14:1181–1186. doi: 10.1016/j.cub.2004.06.029. [DOI] [PubMed] [Google Scholar]
- Tolić-Nørrelykke IM, Sacconi L, Stringari C, Raabe I, Pavone FS. Nuclear and division-plane positioning revealed by optical micromanipulation. Curr Biol. 2005;15:1212–1216. doi: 10.1016/j.cub.2005.05.052. [DOI] [PubMed] [Google Scholar]
- Tran PT, Marsh L, Doye V, Inoue S, Chang F. A mechanism for nuclear positioning in fission yeast based on microtubule pushing. J Cell Biol. 2001;153:397–411. doi: 10.1083/jcb.153.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verde F, Berrez JM, Antony C, Karsenti E. Taxol-induced microtubule asters in mitotic extracts of Xenopus eggs: requirement for phosphorylated factors and cytoplasmic dynein. J Cell Biol. 1991;112:1177–1187. doi: 10.1083/jcb.112.6.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel SK, Raabe I, Dereli A, Maghelli N, Tolić-Nørrelykke I. Interphase microtubules determine the initial alignment of the mitotic spindle. Curr Biol. 2007;17:438–444. doi: 10.1016/j.cub.2007.01.047. [DOI] [PubMed] [Google Scholar]
- Vogel SK, Pavin N, Maghelli N, Jülicher F, Tolić-Nørrelykke IM (2008) Dynamic self-organization of dynein motors and microtubules generates meiotic nuclear oscillations (submitted) [DOI] [PMC free article] [PubMed]
- Wang N, Naruse K, Stamenovic D, Fredberg JJ, Mijailovich SM, Tolić-Nørrelykke IM, Polte T, Mannix R, Ingber DE. Mechanical behavior in living cells consistent with the tensegrity model. Proc Natl Acad Sci USA. 2001;98:7765–7770. doi: 10.1073/pnas.141199598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedlich-Soldner R, Schulz I, Straube A, Steinberg G. Dynein supports motility of endoplasmic reticulum in the fungus Ustilago maydis. Mol Biol Cell. 2002;13:965–977. doi: 10.1091/mbc.01-10-0475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann S, Wang HW, Avila-Sakar A, Drubin DG, Nogales E, Barnes G. The Dam1 kinetochore ring complex moves processively on depolymerizing microtubule ends. Nature. 2006;440:565–569. doi: 10.1038/nature04409. [DOI] [PubMed] [Google Scholar]
- Yamamoto A, West RR, McIntosh JR, Hiraoka Y. A cytoplasmic dynein heavy chain is required for oscillatory nuclear movement of meiotic prophase and efficient meiotic recombination in fission yeast. J Cell Biol. 1999;145:1233–1249. doi: 10.1083/jcb.145.6.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A, Tsutsumi C, Kojima H, Oiwa K, Hiraoka Y. Dynamic behavior of microtubules during dynein-dependent nuclear migrations of meiotic prophase in fission yeast. Mol Biol Cell. 2001;12:3933–3946. doi: 10.1091/mbc.12.12.3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E, Yang C, Chin E, Maddox P, Salmon ED, Lew DJ, Bloom K. Dynamic positioning of mitotic spindles in yeast: role of microtubule motors and cortical determinants. Mol Biol Cell. 2000;11:3949–3961. doi: 10.1091/mbc.11.11.3949. [DOI] [PMC free article] [PubMed] [Google Scholar]