Abstract
The core of the neurotransmitter release machinery is formed by SNARE complexes, which bring the vesicle and plasma membranes together and are key for fusion, and by Munc18-1, which controls SNARE-complex formation and may also have a direct role in fusion. In addition, SNARE complex assembly is likely orchestrated by Munc13s and RIMs, active-zone proteins that function in vesicle priming and diverse forms of presynaptic plasticity. Synaptotagmin-1 mediates triggering of release by Ca2+, probably through interactions with SNAREs and both membranes, as well as through a tight interplay with complexins. Elucidation of the release mechanism will require a full understanding of the network of interactions among all these proteins and the membranes.
The immense variety and complexity of the functions performed by the nervous system rely on the ability of neurons to communicate with each other in defined and precisely timed patterns. Such precise timing is enabled in part by the fast speed of synaptic transmission. It takes as little as 100 μs from the arrival of an action potential to the release of neurotransmitters by Ca2+-evoked synaptic vesicle exocytosis1. These high speeds arise because, after synaptic vesicles dock onto specialized sites of the plasma membrane called active zones, a priming reaction(s) leaves the vesicles in a metastable state that is ready for fast Ca2+-triggered fusion with the plasma membrane. Neurotransmitter release does not just constitute a means to send signals between neurons; acute, dynamic as well as long-term changes in the efficiency of release during presynaptic plasticity processes shape the properties of neural networks and underlie some forms of information processing in the brain1. Synaptic exocytosis is thus an exquisitely regulated form of intracellular membrane fusion, and understanding the complexity of this process requires characterization of not only the mechanism of membrane fusion but also how fusion is controlled to occur at the right time and with the correct probability.
Intense research that started more than 20 years ago has led to the identification of many proteins involved in neurotransmitter release. Among the components of this machinery are proteins that have homologs in most types of intracellular membrane fusion and include the Sec1/Munc18-1 (SM) protein Munc18-1; N-ethylmaleimide sensitive factor (NSF); soluble NSF attachment proteins (SNAPs); the SNAP receptors (SNAREs) synaptobrevin (also known as VAMP, for vesicle-associated membrane protein), syntaxin-1 and SNAP-25 (no relation to SNAPs); and small GTPases from the Rab3 family. These proteins have conserved properties that underlie general mechanisms of docking, fusion and recycling, but some of them also show unique features that are likely to have evolved to meet the tight regulatory requirements of synaptic exocytosis. In addition, release also depends on diverse specialized proteins that control different steps leading to exocytosis.
The focus of the field changed gradually from identification and initially characterization of components to detailed studies of how they function. This research is increasingly illustrating the complexity of the network of intermolecular interactions that controls synaptic exocytosis, showing that great advances have been made but much remains to be learned in order to reach a true understanding of the mechanism of release. In this review, we primarily discuss mechanistic aspects, with an emphasis on recent data but also describing earlier key results. We focus on a few proteins that have emerged as crucial players in this system, including the SNAREs and Munc18-1 as the core components of the general fusion machinery, and specialized proteins with critical roles in vesicle priming and presynaptic plasticity (Munc13s and RIMs) or in the Ca2+-triggered step of release (synaptotagmin-1 and complexins). Many reviews have recently covered these and other facets of the vast literature that is accumulating in this field1–8, and other reviews in this issue of Nature Structural & Molecular Biology discuss diverse aspects of intracellular membrane fusion.
SNAREs
SNARE proteins govern most types of intracellular membrane traffic and are characterized by sequences called SNARE motifs that have a high propensity to form coiled coils. The key roles in synaptic exocytosis of the synaptic vesicle SNARE synaptobrevin and the plasma membrane SNAREs syntaxin-1 and SNAP-25 (see domain diagrams in Fig. 1a) were demonstrated by the finding that they are the targets of clostridial neurotoxins and by genetic experiments (reviewed in refs. 1,5,8). The three proteins form a highly stable ‘SNARE complex’ that is disassembled upon binding to SNAPs and NSF through the ATPase activity of NSF9,10. The SNARE complex consists of a parallel four-helix bundle formed by the SNARE motifs of the three neuronal SNAREs11,12 (Fig. 1b). Because the SNARE motifs of syntaxin-1 and synaptobrevin are adjacent to their transmembrane regions, the parallel arrangement of the helices indicated that the SNARE complex should bring the vesicle and plasma membranes into close proximity and suggested that complex assembly could provide the energy required for fusion13 (Fig. 1c, left panel). Indeed, reconstitution experiments with synaptobrevin and syntaxin-1–SNAP-25 incorporated into separate populations of liposomes indicated that the SNAREs alone can induce membrane fusion14. Although the relevance of this finding has been debated for diverse reasons (see below and ref. 4.), the reconstitution approach has provided an invaluable tool to study the components of the release machinery in a membrane environment and to correlate their ability to govern membrane merger in vitro with functional studies in vivo. Another strategy that provides complementary information involves monitoring fusion between cells bearing flipped SNAREs on their plasma membranes15.
Figure 1.
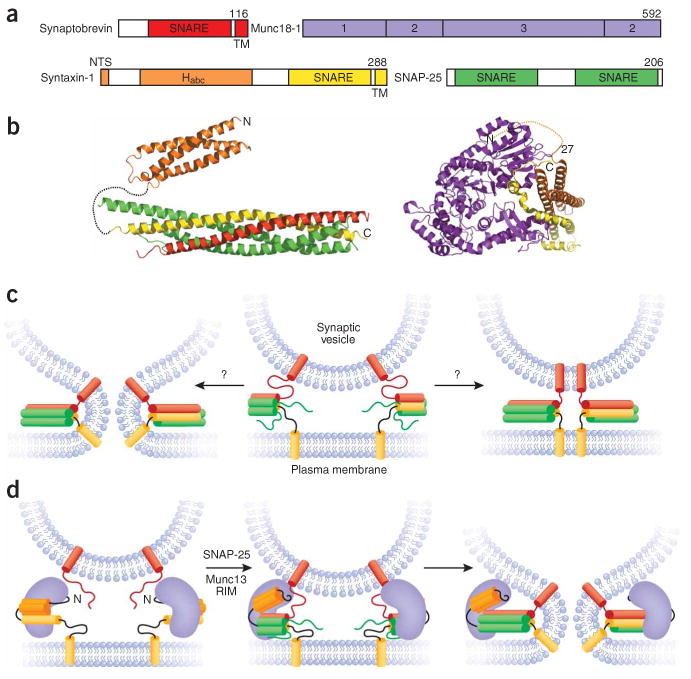
SNAREs and Munc18-1. (a) Domain diagrams of the neuronal SNAREs and Munc18-1. The number of residues of each protein is indicated above each diagram on the right. The same color coding for these proteins is used in all other figures. TM, transmembrane. (b) Ribbon diagrams of the structures of the SNARE complex11 and the syntaxin-1 Habc domain16 (shown connected by a dashed curve that represents the syntaxin-1 sequence linking them) and of the binary complex between Munc18-1 and the closed conformation of syntaxin-142. The orange dashed curve indicates that the syntaxin-1 NTS also participates in Munc18-1 binding52,53, even though it was not observed in the structure of the complex. The N and C termini of syntaxin-1 are indicated in both diagrams. (c) Partially assembled SNARE complexes (center) and how they could induce membrane fusion as they fully assemble (left) or could remained fully assembled between the two membranes (right) if the linkers between the SNARE motifs and transmembrane regions are flexible. (d) The closed conformation of syntaxin-1 bound to Munc18-1 (left) and Munc18-1 bound to partially assembled SNARE complexes (center) that could induce fusion as the SNARE complexes fully assemble (right). Munc13s and RIMs are likely to mediate the transition between the two complexes.
The notion that formation of trans-SNARE complexes between two membranes is key for fusion is now widely accepted, and the activity of SNAPs–NSF is thus believed to be required for disassembly of cis-SNARE complexes to recycle the SNAREs. Much of the recent research on the mechanism of release has thus focused on studying how other critical factors govern SNARE complex assembly and/or cooperate with the SNAREs in membrane fusion (see below), as well as how assembly, fusion and coupling with these factors depend on distinct SNARE sequences. Particularly important for control of SNARE complex assembly is the long N-terminal region of syntaxin-1 containing an autonomously folded three-helix bundle called the Habc domain16 (Fig. 1a,b). In isolated syntaxin-1, this domain folds back onto the SNARE motif, resulting in a so-called ‘closed conformation’ that hinders SNARE complex assembly17 (Fig. 1d, left panel). In addition to mediating coupling with Munc18-1 (see below), the syntaxin-1 closed conformation is likely to be important for preventing reassembly of the SNARE complex after disassembly by SNAPs–NSF. Moreover, removing the syntaxin-1 N-terminal region in coexpressed syntaxin-1–SNAP-25 heterodimers enhances the rate of SNARE-induced liposome fusion18, suggesting that this region may have a further role in controlling SNARE complex formation downstream of heterodimer formation19.
SNARE complex assembly in vitro is also hindered by the tendency of syntaxin-1 and SNAP-25 to form 2:1 heterodimers20 wherein synaptobrevin is replaced by a second copy of syntaxin-1 in the four-helix bundle21. Indeed, syntaxin-1 and SNAP-25 have a natural tendency to associate into diverse four-helix bundles in vitro and are highly promiscuous5. However, coexpression of syntaxin-1 and SNAP-25 does yield 1:1 heterodimers19,22 and, in the original reconstitution assays14, was required to observe liposome fusion, suggesting that coexpression leads to a metastable state that is kinetically unreachable by mixing separately expressed proteins. Conversely, recent data have shown that syntaxin-1 reconstituted into lipid bilayers at very low protein-to-lipid (P/L) ratios can form stable 1:1 heterodimers with SNAP-25 that interconvert between two- and three-helix bundle configurations23. Although the 1:1 heterodimers formed under these conditions or by coexpression may have a different nature, both types of heterodimer bind to Munc18-1 and Munc13-1, among other factors22,23, and these interactions stabilize the three-helix bundle23. Thus, binding of these factors to either type of 1:1 heterodimer is likely to lead to the same three-helix bundle configuration of the syntaxin-1–SNAP-25 SNARE motifs that is expected to serve as the acceptor complex for synaptobrevin. These findings suggest that a crucial task of the release apparatus is to help form this acceptor complex and to prevent unwanted interactions of the promiscuous SNARE motifs of syntaxin-1 and SNAP-25.
Binding of synaptobrevin to pre-formed syntaxin-1–SNAP-25 heterodimers is believed to start at the N terminus of its SNARE motif and then ‘zipper’ toward the C terminus (Fig. 1c). This notion is supported by several lines of evidence, including the differential inhibition of release caused by clostridial neurotoxins8, antibodies24 or mutations at the N- and C-terminal regions of the SNARE complex25. Furthermore, biophysical data showed that the N-terminal half of the SNARE complex is extremely stable26 and can fold independently of the C terminus20,25 and that synaptobrevin and syntaxin-1–SNAP-25 anchored at apposed lipid bilayers form an intermediate state in which about 70% of the SNARE complex is assembled27. This intermediate has a very high stabilization energy (35 kBT), and assembly of the C terminus of the SNARE complex is expected to provide further energy. Because several SNARE complexes (estimated between 3 and 17; ref. 8.) are believed to form a ring around the site of fusion, the bulk energy of formation of these complexes easily exceeds the energy required for membrane fusion28. However, it is still unclear whether all of this energy or only that provided by C-terminal assembly can be transduced to cause fusion, whether more energy is provided by other factors, and how the available energy is transduced into fusion. Note also that the transmembrane regions of synaptobrevin and syntaxin-1 are critical in SNARE-mediated liposome fusion29 and probably lower the energy barrier to fusion by destabilizing the lipid bilayers30, but it is unknown to what extent.
Liposome fusion assays showed that the SNAREs alone could force the merger of two membranes14, but the efficiency of fusion depends highly on the P/L ratios and the homogeneity of the liposomes31,32. The P/L ratios required to observe fusion lead to liposome leakiness33, probably because high densities of the SNARE transmembrane regions compromise membrane integrity. Note, however, that the low fusion efficiency arises in part from inhibition by the syntaxin-1 Habc domain or by formation of 2:1 syntaxin-1–SNAP-25 heterodimers, and peptides spanning the C terminus of the synaptobrevin SNARE motif can release both types of inhibition, facilitating the N- to C-terminal zippering and allowing highly efficient fusion in a few minutes19,20. Still, single-molecule spectroscopy studies showed that multiple SNARE complexes (∼12) can assemble between vesicles containing low synaptobrevin/lipid ratios and supported bilayers containing syntaxin-1–SNAP-25 without inducing membrane fusion34. A plausible rationalization for all these results is that a limited number of assembling SNARE complexes can diffuse to the center of the intermembrane space and remain assembled without inducing fusion (Fig. 1c, right panel); accumulation of many SNARE complexes may cause sufficient strain on the membranes to force them to merge32, but such accumulation may compromise membrane integrity. More efficient fusion mechanisms can be envisaged by cooperation with other factors4 (see below).
The observation of SNARE complex formation without fusion suggests that there is some degree of flexibility in the linker between the SNARE motifs and transmembrane regions of synaptobrevin and syntaxin-1 (ref. 4), in contrast to the assumption in early models of SNARE-mediated fusion that these sequences form continuous helices. It is noteworthy in this context that inclusion of helix-breaking residues in the synaptobrevin linker does not alter Ca2+-evoked exocytosis35 or the rate of liposome fusion36, but the linker sequences contain basic residues that bind to membranes37 and could help in coupling SNARE-complex assembly to mechanical action on the membranes. Such coupling was suggested by the findings that inserting flexible sequences in the synaptobrevin linker strongly inhibits Ca2+-triggered exocytosis38 and that that inhibition depends steeply on the length of the linker35. Interestingly, inhibition of spontaneous release in synapses38 and the slow phase of exocytosis in chromaffin cells35 depend less steeply on the linker length, which correlates with the effects of similar linkers on SNARE-mediated liposome fusion36. These findings suggest that the primed state that is ready for fast Ca2+-evoked exocytosis may involve hemifusion of the vesicle and plasma membranes35. Overall, the results summarized above show that elucidating how SNARE complex assembly is coupled to membrane fusion remains as a key challenge in this field and emphasize the importance of understanding how other factors govern assembly and may cooperate with the SNAREs in fusion.
Munc18-1
Like the SNAREs, SM proteins are crucial for most types of intracellular membrane traffic1,6, as emphatically shown by the total abrogation of neurotransmitter release observed in Munc18-1 knockout mice39. However, the function(s) of SM proteins has been enigmatic. The finding that Munc18-1 binds tightly to syntaxin-1 suggested that Munc18-1 is functionally coupled to the SNAREs and forms part of the fusion machinery40, but this interaction was found to require the closed conformation of syntaxin-1 (ref. 17) and to hinder SNARE complex formation41. Moreover, X-ray crystallography showed that Munc18-1 forms an arch-shaped structure with a cavity in the middle where syntaxin-1 binds (Fig. 1a,b) and that the closed conformation is clearly incompatible with the SNARE complex42. Although it was proposed that Munc18-1 may somehow assist in SNARE-complex assembly to explain its critical function in release17,42, the structural and biochemical data seemed to contradict this notion.
Key insights clarifying SM protein function came from studies of SNAREs and SM proteins from diverse membrane compartments. The yeast SM protein involved in exocytosis, Sec1p, was shown to bind to cognate SNARE complexes rather than to isolated Sso1p, the yeast plasma membrane syntaxin43,44. Although Sso1p forms a closed conformation45 and all syntaxins contain an Habc domain46–48, it became clear that the closed conformation is not a general feature of syntaxins46,48, and the syntaxins from the ER, Golgi, TGN and early endosomes were found to bind tightly to their cognate SM proteins through a short N-terminal sequence (NTS; Fig. 1a), in an interaction that is compatible with the SNARE complex47–49. The apparently confusing picture emerging from these findings has been recently reconciled by several studies showing that many SM proteins, including Munc18-1, bind to SNARE complexes and that the syntaxin NTS most often contributes to binding (refs. 50,51 and references cited therein; note that the recent observation of Munc18-1 binding to the SNARE complex contrasts with results of an earlier study41).
These results suggest that SM-protein binding to SNARE complexes underlies the general mode of coupling between these two critical protein families and most often involves the syntaxin NTS, whereas the binary complex of Munc18-1 with closed syntaxin-1 represents a specialization of regulated exocytosis. Both types of interaction of Munc18-1 with the neuronal SNAREs involve the syntaxin-1 NTS (Fig. 1b,d), which plays a crucial role in exocytosis and was proposed to provide a pivot point for the transition between the two complexes52. Conversely, recent data suggested that the NTS needs to be released from Munc18-1 for SNARE complex assembly to occur53, but it is unclear whether such release is required in the presence of other factors. Indeed, although Munc18-1 binding stabilizes syntaxin-1 in vivo39 and seems to be key for transport of syntaxin-1 to the plasma membrane54, this binary interaction poses an energetic barrier to SNARE complex assembly, and other factors such as Munc13s must be required to promote the transition to the SNARE complex. Despite its apparently inhibitory interaction with closed syntaxin-1, Munc18-1 is likely to play an additional, active role in SNARE complex assembly, as suggested by the Munc18-1–SNARE complex interaction. Such a role may involve providing a template to form the syntaxin-1–SNAP-25 acceptor complex for synaptobrevin, as Munc18-1 also binds to syntaxin-1–SNAP-25 heterodimers22,23,55. This proposal is supported by the finding that Munc18-1 facilitates assembly of SNARE complexes between synaptobrevin and coexpressed syntaxin-1–SNAP-25 heterodimers51. Note also that recent evidence56 suggests multiple roles for Munc18-1 throughout the steps leading to exocytosis, including a function in vesicle priming that could arise from its action in SNARE complex assembly.
As SNAREs alone can form SNARE complexes in vitro, a key question is whether a general function for Munc18-1 and SM proteins in promoting SNARE complex assembly can explain the strong phenotypes observed upon genetic deletion of these proteins; or, perhaps they play a direct, key role in membrane fusion. Genetic experiments indeed suggested a function for Sec1p downstream of SNARE complex formation57, and a model for how Munc18-1 might participate in fusion was proposed considering that diffusion of assembling SNARE complexes to the center of the intermembrane space may hinder application of mechanical force to the membranes (Fig. 1c; see above)4. This model predicts that binding of a bulky protein(s) such as Munc18-1 to the assembling SNARE complexes may prevent such diffusion and provide an asymmetry in the resulting Munc18-1–SNARE complex assemblies to allow energy transduction to the membranes (Fig. 1d)4. Although the findings that Munc18-1 binds to SNARE complexes50,51 and strongly enhances SNARE-mediated liposome fusion51 support this model, it is still unclear whether Munc18-1 stimulates fusion indirectly (that is, by promoting SNARE complex assembly) or directly. Recent evidence favors the later possibility58, but further research will be necessary to demonstrate whether and how Munc18-1 directly contributes to fusion.
Munc13s and RIMs
Presynaptic active zones are closely apposed to postsynaptic densities and are formed by large proteins that facilitate rapid mobilization, fusion and recycling of synaptic vesicles, enabling the precise timing and localization of synaptic transmission2. Among these proteins, Munc13s and RIMs (unc13 and unc10, respectively, in Caenorhabditis elegans) play prominent roles in vesicle priming and diverse forms of presynaptic plasticity. Figure 2a shows the multidomain structures of Munc13-1 and RIM1α, two of the major isoforms of these protein families in brain; other isoforms contain the same or only a subset of these domains. Note the abundance of C2 domains in both Munc13s and RIMs. These widespread protein modules adopt characteristic β-sandwich structures and commonly function as Ca2+ binding modules59, but the Munc13 C2A domain and both RIM C2 domains are Ca2+-independent60–62, and only the Munc13 C2B domain is predicted to bind Ca2+.
Figure 2.
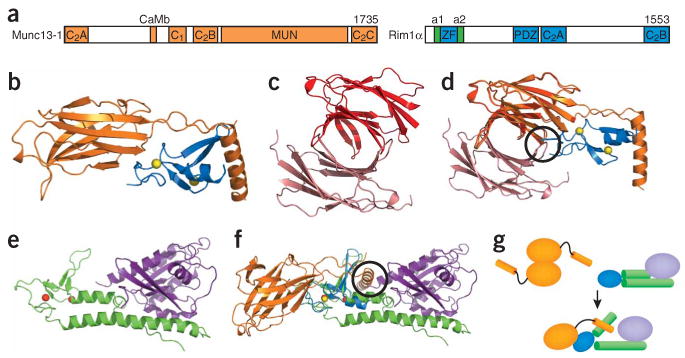
Munc13s and RIMs. (a) Domain diagram of Munc13-1 and RIM1α. The number of residues of each protein is indicated above each diagram on the right. The helices adjacent to the RIM1α ZF domain, which are involved in Rab3 binding, are labeled a1 and a2. The same color-coding is used in b,d–g. (b) Structure of the complex between the RIM2α ZF domain and the N-terminal region of Munc13-1, including the C2A domain60. Yellow spheres, zinc ions. (c,d) Structure of the Munc13-1 C2A domain homodimer60 (c) and superposition with the Munc13-1/RIM2α heterodimer (d). The two protomers of the Munc13-1 C2A domain homodimer are shown in red and salmon to distinguish them from the Munc13-1 C2A domain in the heterodimer. (e) Structure of Rab3A (purple) bound to the rabphilin N-terminal region containing its ZF domain and the two adjacent helices involved in Rab3 binding (green)78. Red spheres, zinc ions. (f) Superposition of the Munc13-1–RIM2α and Rab3A–rabphilin structures shown in b and e using the two ZF domains for the superposition. Black circles, regions of overlap between the RIM2α ZF domain and one Munc13-1 C2A domain protomer of the homodimer in d, and between Rab3A and the Munc13-1 α-helix at the C terminus of the C2A domain in f. (g) Model summarizing the structural rearrangements and changes in protein-protein interfaces proposed to occur during the switch from the Munc13-1 homodimer to the Munc13-1–RIM-Rab3 tripartite complex.
The key role of Munc13s for priming is illustrated by the total abrogation of spontaneous and evoked release observed upon deletion of Munc13-1 and the closely related Munc13-2 in mice, or of the homologs in C. elegans and Drosophila melanogaster (reviewed in ref. 2). Crucial for this function is the MUN domain, which is sufficient to rescue release in Munc13-1/2 double-knockout mice63. The observation that a constitutively open syntaxin mutant17 partially rescues release in unc13 nulls in C. elegans suggested that unc13 and Munc13s play a role in the conformational transition of syntaxin-1 (ref. 64). This role was initially attributed to an interaction with the syntaxin-1 N-terminal region, but the Munc13-1 MUN domain does not bind to isolated syntaxin-1 (refs. 22,63). However, the MUN domain does bind to membrane-anchored SNARE complexes and to syntaxin-1–SNAP-25 heterodimers22,23. These results support the idea that Munc13s function in opening syntaxin-1, but perhaps by a mechanism different from that originally envisioned, acting in concert with Munc18-1 to form the syntaxin-1–SNAP-25 heterodimer and promote vesicle priming. It is also worth noting that the protein CAPS, which also contains a MUN domain63, has also been implicated in vesicle priming, suggesting that priming may depend generally on MUN domains. These bulky domains could also play a direct role in fusion through their interaction with the SNARE complex, as proposed above for Munc18-1 (Fig. 1d), but this notion has not been tested.
A function for Munc13s in presynaptic plasticity in addition to priming was shown by the observation of distinct changes in synaptic amplitudes upon high-frequency stimulation in the presence of either Munc13-1 or Munc13-2, as well as by the functional effects of mutations in the C1 domain and a calmodulin-binding (CaMb) sequence65–68 (see Fig. 2a). Coupling of the Munc13 C2A domain to RIMs and Rab3 may provide a link to further forms of plasticity (see below). These observations led to the hypothesis that various factors involved in distinct presynaptic plasticity processes may regulate release efficiency by altering intramolecular interactions of their receptor domains with the Munc13 MUN domain, thus controlling its priming activity and, as a consequence, the vesicular release probability63.
RIM1α was originally identified as a Rab3 effector, but RIMs have multiple functions beyond this role69, as only a subset of RIMs contain the N-terminal Rab-binding region, and a much stronger impairment of neurotransmitter release is observed upon deletion of RIM1α and the closely related RIM2α in mice or of unc10 in C. elegans than is observed in the absence of Rab3s (reviewed in ref. 69; note that the function of Rab3s is still unclear and that the mild inhibition of release observed in their absence contrasts with the strong blocks in membrane traffic caused by mutations in other Rab proteins1). The phenotype of unc10 nulls in C. elegans and the finding that this phenotype can be partially rescued by the open syntaxin mutant70 suggested that unc10 and RIMs play a role in vesicle priming that is functionally linked to unc13 and Munc13s. Such a link has been demonstrated by the reduction in Munc13-1 levels observed in RIM1α knockout mice71, by the observation of a direct interaction between the N-terminal regions of RIM1α and Munc13-1 (ref. 72), and by the impairment of vesicle priming caused by interference with this interaction72,73. RIM1α shows additional interactions with other active zone proteins that suggest a role in organizing the active zone69,71. Moreover, mossy fiber long-term potentiation is abolished in RIM1α knockout mice74, as observed in Rab3A knockout mice75, and short-term plasticity is altered71,76. The severe deficits in memory and learning observed in RIM1α knockout mice77 emphasize the importance of these varied functions.
The mechanisms of RIM function are poorly understood. The Munc13-1–RIM interaction has been well characterized and shown to involve the Munc13-1 C2A domain and a sequence at its C terminus, which wrap around the RIM ZF domain60 (Fig. 2b). The Munc13-1 C2A domain also forms a tight homodimer (Fig. 2c) that is incompatible with Munc13–RIM heterodimerization, although there is only a slight overlap between the surfaces involved in homo and heterodimerization60 (Fig. 2d). Conversely, the interactions of Munc13 and Rab3 with RIM can occur simultaneously and thus lead to a tripartite complex73. RIM binds to Rab3A through two α-helices adjacent to the ZF finger73, as expected from the crystal structure of a complex between Rab3A and rabphilin, another Rab3 effector78 (Fig. 2e), and from the homology between the rabphilin and RIM N-terminal sequences. Superposition of the Munc13-1–RIM heterodimer with a homology model of the RIM–Rab3A complex suggests that some steric clashes between Munc13-1 and Rab3A occur in the tripartite Munc13-1–RIM–Rab3A complex (Fig. 2f), but they can be relieved by a slight structural rearrangement60 (Fig. 2g). Overall, these results suggest that a switch from a Munc13-1 homodimer to a Munc13-1–RIM heterodimer, perhaps influenced by Rab3s, may regulate the activity of the MUN domain in priming and provide a link between priming and diverse forms of presynaptic plasticity that depend on RIMs and Rab3s (refs. 60,73). However, this model remains to be tested, and it has also been proposed that the Munc13-1–RIM–Rab3A complex may help to recruit Munc13-1 and synaptic vesicles at release sites in the active zone73. Distinguishing these possibilities and characterizing in detail the relevant interactions of the highly conserved C2 domains will be crucial to understanding RIM function.
Synaptotagmin-1 and complexins
Key for the Ca2+-triggering step of release are synaptotagmin-1 and complexins1,3,4,7 (Figs. 3 and 4). Synaptotagmin-1 is a synaptic vesicle protein with two C2 domains, the C2A and C2B domains, that adopt similar β-sandwich structures and bind three and two Ca2+ ions, respectively, through loops at the top of the sandwich79–82 (Fig. 3a,b). These top loops also mediate Ca2+-dependent phospholipid binding to both C2 domains82–84. Mutations that decrease or increase the apparent Ca2+ affinity of synaptotagmin-1 lead to parallel changes in the Ca2+ sensitivity of release85,86, showing that synaptotagmin-1 acts as a Ca2+ sensor in release and that Ca2+-dependent phospholipid binding to both C2 domains is key for this function. However, mutations in the Ca2+-binding ligands of the C2B domain impair release much more severely than analogous mutations in the C2A domain87,88, indicating that Ca2+ binding to the C2B domain is more critical for release. Among several potential explanations for these findings4,7, particularly attractive is the observation that the C2B domain mediates simultaneous binding to two membranes through its Ca2+-binding loops and the abundant basic residues around its surface, thus showing that the C2B domain can bring two membranes together as the SNAREs do, but through a Ca2+-dependent mechanism89 (Fig. 3d).
Figure 3.
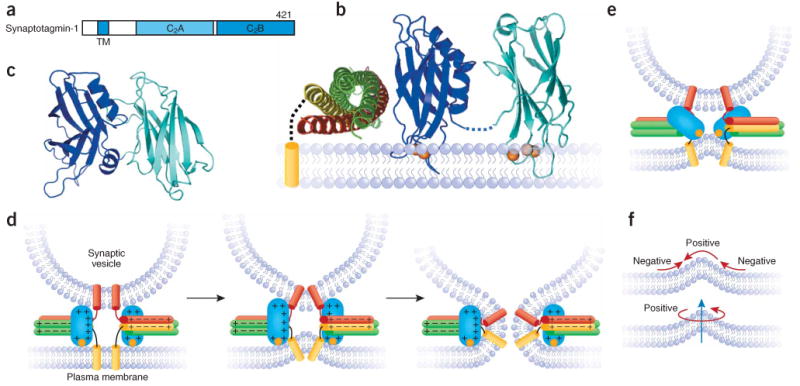
Syntaptotagmin-1 and its coupling to SNAREs and membranes. (a) Domain structure of synaptotagmin-1, with the number of residues indicated in the top right corner. TM, transmembrane. (b) Model of the SSCAP complex built from the structures of the SNARE complex11 and the Ca2+-bound synaptotagmin-1 C2 domains81,82 and a mutagenesis analysis of SSCAP complex formation92. Orange spheres, Ca2+ ions; dashed black curve, the linker between the syntaxin-1 SNARE motif and transmembrane region. Note that it is uncertain whether the synaptotagmin-1 Ca2+-binding loops bind to the plasma membrane or the synaptic vesicle membrane. (c) Crystal structure of the tandem synaptotagmin-1 C2 domains in the absence of Ca2+; the structure involves an antiparallel interaction between the two domains that needs to be disrupted to allow Ca2+ binding to the C2A domain109. (d) Model of how the synaptotagmin-1 C2B domain could cooperate with the SNAREs in triggering membrane fusion upon Ca2+ influx by binding to both membranes and the C terminus of the SNARE complex. Each orange circle represents the two Ca2+ ions bound to the C2B domain. The + and − signs illustrate the electrostatic charge distribution of the C2B domain and the SNARE complex. The model assumes that synaptotagmin-1 interacts with the SNARE complex through the polybasic region on the side of the C2B domain, as in b. This interaction is weak in solution but is likely to be strengthened by colocalization of synaptotagmin-1 and the SNARE complex on one membrane, which at the same time may increase binding specificity by disfavoring irrelevant interactions existing in solution between these highly charged molecules92. The C2A domain is not shown in this model for simplicity, but could play a related role. (e) Model of how the Ca2+ binding loops of the synaptotagmin-1 C2 domains (only the C2B domain is shown for simplicity) could help to cause membrane fusion by inducing positive curvature on the plasma membrane96. (f) Two diagrams showing the types of curvature involved in membrane bending and illustrating that such bending requires both positive and negative curvature.
Figure 4.
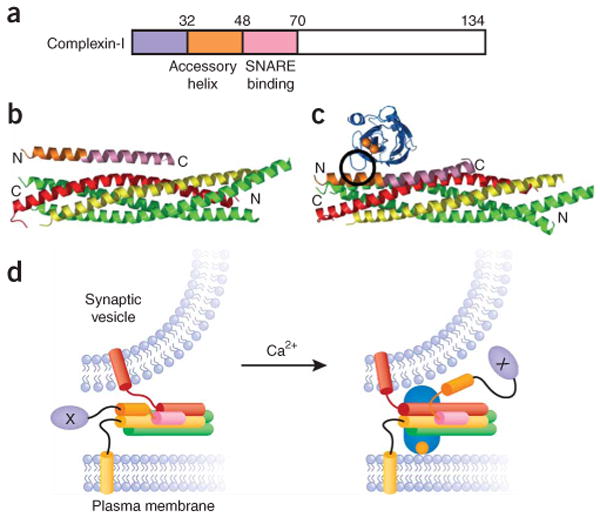
Complexins and their coupling to SNAREs and synaptotagmin-1. (a) Domain diagram of complexin-I. Residue numbers are indicated above the diagram. The same color-coding is used in the remaining panels. (b) Structure of a complexin-I fragment bound to the SNARE complex26. The N and C termini of the complexin-I fragment and the SNARE complex are indicated. (c) Superposition of the crystal structure shown in b with the model of the SCCAP complex shown in Figure 3b (omitting the membrane and the C2A domain). Black circle, the region of overlap between the C2B domain and the accessory helix of complexin-I. (d) Models of how binding of the accessory helix to the C terminus of the SNARE complex could hinder full SNARE complex assembly and thus inhibit fusion (left), and how this inhibition could be released by partial competition between the synaptotagmin-1 C2B domain and complexin for binding to the C terminus of the SNARE complex (right). The N terminus of complexin is shown as a purple ellipse with an X in the center to indicate an as-yet-unidentified interaction of this region that is critical for complexin function. The orientation of this N-terminal region in both diagrams is arbitrary.
In addition to membrane binding, diverse interactions between SNAREs and synaptotagmin-1 have been described (reviewed in refs. 1,3,4,7), and those involving SNARE complexes are the most likely to couple their functions in triggering release. Although the relevance of Ca2+-dependent interactions of synaptotagmin-1 with soluble SNARE complexes was questioned because phospholipids compete with these interactions90, synaptotagmin-1 actually binds simultaneously to membranes and SNARE complexes if they are anchored on the membranes91, resulting in a quaternary SNARE–synaptotagmin-1–Ca2+–phospholipid (SSCAP) complex92. These findings can be rationalized by considering the highly charged nature of these proteins (Fig. 3d) together with a model that was built from a mutagenesis analysis of SSCAP complex formation92 (Fig. 3b) and is compatible with single-molecule spectroscopy data93. The model envisions the relevant synaptotagmin-1/SNARE complex interaction as being mediated by a polybasic region on the side of the C2B domain β-sandwich (Fig. 3b) (but note that this region has also been implicated in diverse interactions including phospholipid binding4,7). An attractive feature of this model of the SSCAP complex is that the top Ca2+-binding loops and the bottom side of the C2B domain can still interact with the apposed membranes to bring them together in cooperation with the SNAREs; moreover, the highly positive electrostatic potential generated by the C2B domain and the C terminus of the SNARE complex could help induce negative curvature on the membranes to bend them and induce fusion89,92 (Fig. 3d).
This overall model has been supported by theoretical calculations94 and is consistent with multiple functional studies, including those showing a predominant role of the C2B domain in release92. Note, however, that basic residues in the C2A domain that are functionally important have also been implicated in SNAP-25 binding95 and that alternative models have also been proposed. Thus, the vesicle and plasma membranes might be hemifused before Ca2+ influx, and the positive electrostatic potential of the C2B domain may help to open the fusion pore92,94. Another model predicts that synaptotagmin-1 causes fusion by inducing positive curvature in the plasma membrane96 (Fig. 3e); this model is also supported by theoretical calculations96 and is compatible with the induction of negative curvature proposed in Figure 3d (membrane bending involves both types of curvature; Fig. 3f). However, the significance of the liposome tubulation experiments supporting the induction of positive membrane curvature by synaptotagmin-1 is unclear because they used high concentrations of negative stain96 and only a small degree of tubulation is observed in aqueous solution89. Regardless of these models, a role for synaptotagmin-1 in fusion has been supported by the observation that Ca2+-bound synaptotagmin-1 can enhance SNARE-mediated liposome fusion97,98.
The function of complexins is intimately coupled to that of the SNAREs and synaptotagmin-1. Complexins are largely unstructured in solution99 and bind tightly to the SNARE complex100, forming an α-helix that interacts in an antiparallel fashion with the synaptobrevin and syntaxin-1 SNARE motifs26 (Fig. 4b). The observations that complexin inhibits SNARE-induced cell-cell or liposome fusion and that Ca2+-bound synaptotagmin-1 releases this inhibition led to the proposal that complexins act as fusion clamps101,102. This model is consistent with biophysical data suggesting that synaptotagmin-1 displaces complexin-I from membrane-anchored SNARE complexes and with the observation that increasing the local concentration of complexin-I by fusing it to the N terminus of synaptobrevin strongly impairs synchronous release103. Furthermore, a marked increase in spontaneous release is observed in Drosophila complexin null mutants104. However, a merely inhibitory role for complexin cannot explain the severe impairment in release observed in complexin-I/II double-knockout mice105 and the finding that the other two mammalian complexin isoforms (III and IV) rescue this phenotype106. Taking into account these findings and the observation that complexin binding stabilizes the C terminus of the SNARE complex26, it was proposed that complexins have dual roles and that complexin binding to the SNARE complex generates a metastable state that serves as a substrate for synaptotagmin-1 to trigger fast release103. Studies of sperm acrosomal fusion indicated a similar synaptotagmin-complexin interplay107, suggesting that this interplay is not limited to neuronal exocytosis.
Studies of the ability of complexin-I mutants to rescue release in complexin-I/II double knockout mice have illustrated the complexity of complexin function and shown that SNARE complex binding is necessary but not sufficient for this function108. Mutations that disrupt binding to the SNARE complex abolish rescue, but rescue is also abrogated by deleting the 26 N-terminal residues of complexin-I, which preserves the SNARE interaction; intriguingly, partial rescue was observed upon deletion of the 46 N-terminal residues. These results led to the proposal that residues 27–46, which form the ‘accessory helix’ preceding the SNARE complex–binding region defined by crystallography (residues 48–70), play an inhibitory role, perhaps by replacing the C terminus of the synaptobrevin SNARE motif in the SNARE complex108 (Fig. 4d; left panel). Recent data showing that complexin-I binds to syntaxin-1–SNAP-25 heterodimers22,23 have provided some support for this proposal. The rescue data also revealed that the N terminus is essential for complexin function and must be involved in an as-yet-unidentified interaction that is critical for release108. This notion suggests that SNARE-complex binding provides an attachment point for complexin so that it may perform this role, but it seems unlikely that complexin is then fully displaced by synaptotagmin-1 from the SNARE complex upon Ca2+ influx. Note that the model of the SSCAP complex built by monitoring complexin-I displacement from SNARE complexes by various synaptotagmin-1 mutants (Fig. 3b) predicts that the C2B domain collides with the accessory helix but not with the SNARE-binding region of complexin-I (ref. 92) (Fig. 4c). Hence, the complexin-I displacement assays may reflect a local competition and weakening of the complexin–SNARE complex interaction due to steric hindrance, rather than a full competition between complexin-I and synaptotagmin-1 for SNARE complex binding (Fig. 4d). Moreover, co-flotation assays have indicated that complexin and synaptotagmin-1 can indeed bind simultaneously to SNARE complexes102.
These observations suggest that both complexin-I and synaptotagmin-1 are bound to the SNAREs after Ca2+ influx and that Ca2+ induces a rearrangement of interactions that leads to fast neurotransmitter release through the concerted action of all these proteins, although the exact nature of this rearrangement is unclear. Note that Ca2+-free synaptotagmin-1 binds to membrane-anchored syntaxin-1–SNAP-25 heterodimers23, but it is unknown whether this interaction is compatible with complexin binding. In addition, X-ray crystallography recently revealed an intramolecular interaction between the C2 domains in Ca2+-free synaptotagmin-1 (ref. 109) (Fig. 3c), which, although not observed in solution89, could influence how Ca2+ rearranges the release machinery. Hence, elucidating the conformational and binding status of synaptotagmin-1 before Ca2+ influx will be as crucial to understand the mechanism of release as testing the emerging ideas about its Ca2+-triggered action.
Perspective
Studies of the neurotransmitter release machinery have led to some well-established conclusions about the functions of its components and to several models for their mechanism of action, some of which are depicted in Figures 1–4. However, many of the ideas discussed above remain as likely but undemonstrated, or they are purely speculative, and critical questions need to be answered before all these ideas can be integrated into a unified model of the mechanism of neurotransmitter release. It seems clear that the SNARE complex is central in membrane fusion and that many factors interact with this complex (Fig. 5) to control its assembly and/or membrane fusion, but most of these interactions are not well-understood, and it is unclear whether they occur simultaneously or sequentially. Perhaps the most crucial question for understanding not only synaptic exocytosis but also intracellular membrane fusion in general is whether SM proteins are directly involved in fusion, in addition to having a role in SNARE-complex assembly. Munc13s and RIMs appear to control the conformational transition of syntaxin-1 and various forms of presynaptic plasticity, but the mechanisms of these functions remain to be defined, and it is plausible that Munc13s also have a direct role in fusion. Synaptotagmin-1 and complexins are clearly involved in the Ca2+-triggered step of release, and both may play a role in fusion, but the exact mechanism of their interplay with the SNAREs is still enigmatic. Also unclear are the functional differences between synaptotagmin-1 and other synaptotagmin isoforms that are likely to play related but slightly different roles in different types of exocytosis1. But perhaps most unclear of all is the mechanism of vesicle docking, since removal of any of these proteins does not impair docking in vertebrate central nervous system synapses, but increasing evidence suggests that Munc18-1, Munc13s and syntaxin-1 are actually key for docking in chromaffin cells and C. elegans synapses110–113. Hence, it appears that the basic mechanism of docking that operates in these other systems may be redundant with vesicle tethering mechanisms in the active zones of vertebrate central nervous system synapses.
Figure 5.
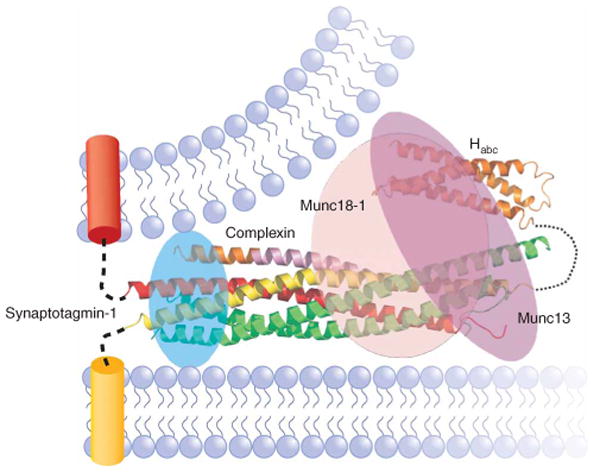
Diagram illustrating the notion that Munc18-1, Munc13, complexin and synaptotagmin-1 bind to the SNARE complex. The ribbon diagram shows the structure of the complexin-I–SNARE complex26, which illustrates the only one among these interactions that has been revealed at atomic resolution. The binding site of synaptotagmin-1 on the four-helix bundle is inferred from mutagenesis92, whereas those of Munc18-1 and Munc13 are unknown. The figure intends to illustrate that all these SNARE complex interactions may be compatible, mutually exclusive, or partially competitive, and that resolving this issue as well as how these interactions occur during the steps that lead to exocytosis will be critical to understand the mechanism of release.
Consideration of three emerging themes will be critical for unraveling the mechanism of release. First, while there is a natural tendency to assign either active or inhibitory roles to the components of the release machinery, it is becoming clear that several components may play dual roles through diverse interactions with other proteins or the membrane. Second, interactions of two components with a common target often involve largely different but slightly overlapping binding sites (Figs. 2d,f and 4c), which may facilitate rapid rearrangements of interactions during docking, priming and fusion60. Third, the influence of membranes on interactions of the SNARE complex with synaptotagmin-1 (refs. 92,103) and the Munc13-1 MUN domain22 underlines the importance of working with reconstituted systems to study the interactions within the release machinery, which is not surprising given the very nature of the biological process controlled by this machinery. Thus, further development of methods for structural studies of protein complexes on a membrane or, ideally, between two membranes will be crucial for reaching a detailed understanding of the complex mechanism of Ca2+-triggered neurotransmitter release.
Acknowledgments
Work in the authors' laboratories is supported by US National Institutes of Health grants NS40944 and NS37200 (to J.R.) and NS050655 and NS051262 (to C.R.).
References
- 1.Sudhof TC. The synaptic vesicle cycle. Annu Rev Neurosci. 2004;27:509–547. doi: 10.1146/annurev.neuro.26.041002.131412. [DOI] [PubMed] [Google Scholar]
- 2.Rosenmund C, Rettig J, Brose N. Molecular mechanisms of active zone function. Curr Opin Neurobiol. 2003;13:509–519. doi: 10.1016/j.conb.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 3.Brunger AT. Structure and function of SNARE and SNARE-interacting proteins. Q Rev Biophys. 2005;38:1–47. doi: 10.1017/S0033583505004051. [DOI] [PubMed] [Google Scholar]
- 4.Rizo J, Chen X, Arac D. Unraveling the mechanisms of synaptotagmin and SNARE function in neurotransmitter release. Trends Cell Biol. 2006;16:339–350. doi: 10.1016/j.tcb.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 5.Jahn R, Scheller RH. SNAREs — engines for membrane fusion. Nat Rev Mol Cell Biol. 2006;7:631–643. doi: 10.1038/nrm2002. [DOI] [PubMed] [Google Scholar]
- 6.Verhage M, Toonen RF. Regulated exocytosis: merging ideas on fusing membranes. Curr Opin Cell Biol. 2007;19:402–408. doi: 10.1016/j.ceb.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 7.Chapman ER. How does synaptotagmin trigger neurotransmitter release? Annu Rev Biochem. 2008;77:615–641. doi: 10.1146/annurev.biochem.77.062005.101135. [DOI] [PubMed] [Google Scholar]
- 8.Montecucco C, Schiavo G, Pantano S. SNARE complexes and neuroexocytosis: how many, how close? Trends Biochem Sci. 2005;30:367–372. doi: 10.1016/j.tibs.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 9.Sollner T, Bennett MK, Whiteheart SW, Scheller RH, Rothman JE. A protein assembly-disassembly pathway in vitro that may correspond to sequential steps of synaptic vesicle docking, activation, and fusion. Cell. 1993;75:409–418. doi: 10.1016/0092-8674(93)90376-2. [DOI] [PubMed] [Google Scholar]
- 10.Sollner T, et al. SNAP receptors implicated in vesicle targeting and fusion. Nature. 1993;362:318–324. doi: 10.1038/362318a0. [DOI] [PubMed] [Google Scholar]; This paper introduces the SNARE hypothesis and postulates that a universal membrane fusion apparatus includes vesicle SNAREs and target-membrane SNAREs that bind to each other and mediate targeting specificity.
- 11.Sutton RB, Fasshauer D, Jahn R, Brunger AT. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 Å resolution. Nature. 1998;395:347–353. doi: 10.1038/26412. [DOI] [PubMed] [Google Scholar]; The crystal structure of the neuronal SNARE complex is described, providing a structural basis for understanding SNARE function at atomic resolution.
- 12.Poirier MA, et al. The synaptic SNARE complex is a parallel four-stranded helical bundle. Nat Struct Biol. 1998;5:765–769. doi: 10.1038/1799. [DOI] [PubMed] [Google Scholar]
- 13.Hanson PI, Roth R, Morisaki H, Jahn R, Heuser JE. Structure and conformational changes in NSF and its membrane receptor complexes visualized by quick-freeze/deep-etch electron microscopy. Cell. 1997;90:523–535. doi: 10.1016/s0092-8674(00)80512-7. [DOI] [PubMed] [Google Scholar]; The SNARE motifs of syntaxin-1 and synaptobrevin are shown to bind in a parallel fashion, leading to the proposal that SNARE complex assembly may provide the energy for membrane fusion.
- 14.Weber T, et al. SNAREpins: minimal machinery for membrane fusion. Cell. 1998;92:759–772. doi: 10.1016/s0092-8674(00)81404-x. [DOI] [PubMed] [Google Scholar]; This paper describes reconstitution experiments that provide a powerful tool to study the role of diverse factors in membrane fusion and suggested that SNAREs alone may be able to induce membrane fusion.
- 15.Hu C, et al. Fusion of cells by flipped SNAREs. Science. 2003;300:1745–1749. doi: 10.1126/science.1084909. [DOI] [PubMed] [Google Scholar]
- 16.Fernandez I, et al. Three-dimensional structure of an evolutionarily conserved N-terminal domain of syntaxin 1A. Cell. 1998;94:841–849. doi: 10.1016/s0092-8674(00)81742-0. [DOI] [PubMed] [Google Scholar]
- 17.Dulubova I, et al. A conformational switch in syntaxin during exocytosis: role of munc18. EMBO J. 1999;18:4372–4382. doi: 10.1093/emboj/18.16.4372. [DOI] [PMC free article] [PubMed] [Google Scholar]; Syntaxin-1 is shown to adopt a closed conformation that binds to Munc18-1 and is incompatible with SNARE complex formation, suggesting that syntaxin-1 undergoes a large conformational change during exocytosis.
- 18.Parlati F, et al. Rapid and efficient fusion of phospholipid vesicles by the alphahelical core of a SNARE complex in the absence of an N-terminal regulatory domain. Proc Natl Acad Sci USA. 1999;96:12565–12570. doi: 10.1073/pnas.96.22.12565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Melia TJ, et al. Regulation of membrane fusion by the membrane-proximal coil of the t-SNARE during zippering of SNAREpins. J Cell Biol. 2002;158:929–940. doi: 10.1083/jcb.200112081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pobbati AV, Stein A, Fasshauer D. N- to C-terminal SNARE complex assembly promotes rapid membrane fusion. Science. 2006;313:673–676. doi: 10.1126/science.1129486. [DOI] [PubMed] [Google Scholar]
- 21.Zhang F, Chen Y, Kweon DH, Kim CS, Shin YK. The four-helix bundle of the neuronal target membrane SNARE complex is neither disordered in the middle nor uncoiled at the C-terminal region. J Biol Chem. 2002;277:24294–24298. doi: 10.1074/jbc.M201200200. [DOI] [PubMed] [Google Scholar]
- 22.Guan R, Dai H, Rizo J. Binding of the Munc13–1 MUN domain to membrane-anchored SNARE complexes. Biochemistry. 2008;47:1474–1481. doi: 10.1021/bi702345m. [DOI] [PubMed] [Google Scholar]
- 23.Weninger K, Bowen ME, Choi UB, Chu S, Brunger AT. Accessory proteins stabilize the acceptor complex for synaptobrevin, the 1:1 syntaxin/SNAP-25 complex. Structure. 2008;16:308–320. doi: 10.1016/j.str.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu T, et al. Inhibition of SNARE complex assembly differentially affects kinetic components of exocytosis. Cell. 1999;99:713–722. doi: 10.1016/s0092-8674(00)81669-4. [DOI] [PubMed] [Google Scholar]
- 25.Sorensen JB, et al. Sequential N- to C-terminal SNARE complex assembly drives priming and fusion of secretory vesicles. EMBO J. 2006;25:955–966. doi: 10.1038/sj.emboj.7601003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen X, et al. Three-dimensional structure of the complexin/SNARE complex. Neuron. 2002;33:397–409. doi: 10.1016/s0896-6273(02)00583-4. [DOI] [PubMed] [Google Scholar]; A combination of NMR spectroscopy and X-ray crystallography reveals the interaction between complexin-I and the SNARE complex at atomic resolution and suggests that complexin binding stabilizes the C-terminus of the SNARE complex.
- 27.Li F, et al. Energetics and dynamics of SNAREpin folding across lipid bilayers. Nat Struct Mol Biol. 2007;14:890–896. doi: 10.1038/nsmb1310. [DOI] [PubMed] [Google Scholar]
- 28.Cohen FS, Melikyan GB. The energetics of membrane fusion from binding, through hemifusion, pore formation, and pore enlargement. J Membr Biol. 2004;199:1–14. doi: 10.1007/s00232-004-0669-8. [DOI] [PubMed] [Google Scholar]
- 29.McNew JA, et al. Close is not enough: SNARE-dependent membrane fusion requires an active mechanism that transduces force to membrane anchors. J Cell Biol. 2000;150:105–117. doi: 10.1083/jcb.150.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Langosch D, et al. Peptide mimics of SNARE transmembrane segments drive membrane fusion depending on their conformational plasticity. J Mol Biol. 2001;311:709–721. doi: 10.1006/jmbi.2001.4889. [DOI] [PubMed] [Google Scholar]
- 31.Kweon DH, Kim CS, Shin YK. Regulation of neuronal SNARE assembly by the membrane. Nat Struct Biol. 2003;10:440–447. doi: 10.1038/nsb928. [DOI] [PubMed] [Google Scholar]
- 32.Chen X, et al. SNARE-mediated lipid mixing depends on the physical state of the vesicles. Biophys J. 2006;90:2062–2074. doi: 10.1529/biophysj.105.071415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dennison SM, Bowen ME, Brunger AT, Lentz BR. Neuronal SNAREs do not trigger fusion between synthetic membranes but do promote PEG-mediated membrane fusion. Biophys J. 2006;90:1661–1675. doi: 10.1529/biophysj.105.069617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bowen ME, Weninger K, Brunger AT, Chu S. Single molecule observation of liposome-bilayer fusion thermally induced by soluble N-ethyl maleimide sensitive-factor attachment protein receptors (SNAREs) Biophys J. 2004;87:3569–3584. doi: 10.1529/biophysj.104.048637. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper shows that multiple SNARE complexes bridging a vesicle and a planar membrane can form without inducing membrane fusion, suggesting that SNARE complex formation is not sufficient for membrane fusion.
- 35.Kesavan J, Borisovska M, Bruns D. v-SNARE actions during Ca2+-triggered exocytosis Cell. 2007;131:351–363. doi: 10.1016/j.cell.2007.09.025. [DOI] [PubMed] [Google Scholar]
- 36.McNew JA, Weber T, Engelman DM, Sollner TH, Rothman JE. The length of the flexible SNAREpin juxtamembrane region is a critical determinant of SNARE-dependent fusion. Mol Cell. 1999;4:415–421. doi: 10.1016/s1097-2765(00)80343-3. [DOI] [PubMed] [Google Scholar]
- 37.Kim CS, Kweon DH, Shin YK. Membrane topologies of neuronal SNARE folding intermediates. Biochemistry. 2002;41:10928–10933. doi: 10.1021/bi026266v. [DOI] [PubMed] [Google Scholar]
- 38.Deak F, Shin OH, Kavalali ET, Sudhof TC. Structural determinants of synaptobrevin 2 function in synaptic vesicle fusion. J Neurosci. 2006;26:6668–6676. doi: 10.1523/JNEUROSCI.5272-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Verhage M, et al. Synaptic assembly of the brain in the absence of neurotransmitter secretion. Science. 2000;287:864–869. doi: 10.1126/science.287.5454.864. [DOI] [PubMed] [Google Scholar]; This paper shows that neurotransmitter secretion from synaptic vesicles is completely abrogated in Munc18-1 knockout mice and that the brain can still assemble in the absence of neurotransmitter release.
- 40.Hata Y, Slaughter CA, Sudhof TC. Synaptic vesicle fusion complex contains unc-18 homologue bound to syntaxin. Nature. 1993;366:347–351. doi: 10.1038/366347a0. [DOI] [PubMed] [Google Scholar]
- 41.Yang B, Steegmaier M, Gonzalez LC, Jr, Scheller RH. nSec1 binds a closed conformation of syntaxin1A. J Cell Biol. 2000;148:247–252. doi: 10.1083/jcb.148.2.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Misura KM, Scheller RH, Weis WI. Three-dimensional structure of the neuronal-Sec1-syntaxin 1a complex. Nature. 2000;404:355–362. doi: 10.1038/35006120. [DOI] [PubMed] [Google Scholar]; The crystal structure of syntaxin-1 bound to Munc18-1 is described, providing the first atomic resolution structure of a SM protein and revealing how Munc18-1 binds to the syntaxin-1 closed conformation.
- 43.Carr CM, Grote E, Munson M, Hughson FM, Novick PJ. Sec1p binds to SNARE complexes and concentrates at sites of secretion. J Cell Biol. 1999;146:333–344. doi: 10.1083/jcb.146.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]; Sec1p is shown to bind to its cognate SNARE complex, providing the first evidence for formation of the SM protein–SNARE complex assemblies that may form the core of intracellular membrane fusion machineries.
- 44.Togneri J, Cheng YS, Munson M, Hughson FM, Carr CM. Specific SNARE complex binding mode of the Sec1/Munc-18 protein, Sec1p. Proc Natl Acad Sci USA. 2006;103:17730–17735. doi: 10.1073/pnas.0605448103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nicholson KL, et al. Regulation of SNARE complex assembly by an N-terminal domain of the t-SNARE Sso1p. Nat Struct Biol. 1998;5:793–802. doi: 10.1038/1834. [DOI] [PubMed] [Google Scholar]
- 46.Dulubova I, Yamaguchi T, Wang Y, Sudhof TC, Rizo J. Vam3p structure reveals conserved and divergent properties of syntaxins. Nat Struct Biol. 2001;8:258–264. doi: 10.1038/85012. [DOI] [PubMed] [Google Scholar]
- 47.Yamaguchi T, et al. Sly1 binds to Golgi and ER syntaxins via a conserved N-terminal peptide motif. Dev Cell. 2002;2:295–305. doi: 10.1016/s1534-5807(02)00125-9. [DOI] [PubMed] [Google Scholar]
- 48.Dulubova I, et al. How Tlg2p/Syntaxin 16 “snares” Vps45. EMBO J. 2002;21:3620–3631. doi: 10.1093/emboj/cdf381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bracher A, Weissenhorn W. Structural basis for the Golgi membrane recruitment of Sly1p by Sed5p. EMBO J. 2002;21:6114–6124. doi: 10.1093/emboj/cdf608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dulubova I, et al. Munc18–1 binds directly to the neuronal SNARE complex. Proc Natl Acad Sci USA. 2007;104:2697–2702. doi: 10.1073/pnas.0611318104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shen J, Tareste DC, Paumet F, Rothman JE, Melia TJ. Selective activation of cognate SNAREpins by Sec1/Munc18 proteins. Cell. 2007;128:183–195. doi: 10.1016/j.cell.2006.12.016. [DOI] [PubMed] [Google Scholar]
- 52.Khvotchev M, et al. Dual modes of Munc18–1/SNARE interactions are coupled by functionally critical binding to syntaxin-1 N terminus. J Neurosci. 2007;27:12147–12155. doi: 10.1523/JNEUROSCI.3655-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Burkhardt P, Hattendorf DA, Weis WI, Fasshauer D. Munc18a controls SNARE assembly through its interaction with the syntaxin N-peptide. EMBO J. 2008;27:923–933. doi: 10.1038/emboj.2008.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Medine CN, Rickman C, Chamberlain LH, Duncan RR. Munc18–1 prevents the formation of ectopic SNARE complexes in living cells. J Cell Sci. 2007;120:4407–4415. doi: 10.1242/jcs.020230. [DOI] [PubMed] [Google Scholar]
- 55.Zilly FE, Sorensen JB, Jahn R, Lang T. Munc18-bound syntaxin readily forms snare complexes with synaptobrevin in native plasma membranes. PLoS Biol. 2006;4:e330. doi: 10.1371/journal.pbio.0040330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gulyas-Kovacs A, et al. Munc18–1: sequential interactions with the fusion machinery stimulate vesicle docking and priming. J Neurosci. 2007;27:8676–8686. doi: 10.1523/JNEUROSCI.0658-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Grote E, Carr CM, Novick PJ. Ordering the final events in yeast exocytosis. J Cell Biol. 2000;151:439–452. doi: 10.1083/jcb.151.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tareste D, Shen J, Melia TJ, Rothman JE. SNAREpin/Munc18 promotes adhesion and fusion of large vesicles to giant membranes. Proc Natl Acad Sci USA. 2008;105:2380–2385. doi: 10.1073/pnas.0712125105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rizo J, Sudhof TC. C2-domains, structure and function of a universal Ca2+-binding domain. J Biol Chem. 1998;273:15879–15882. doi: 10.1074/jbc.273.26.15879. [DOI] [PubMed] [Google Scholar]
- 60.Lu J, et al. Structural basis for a Munc13–1 homodimer to Munc13–1/RIM heterodimer switch. PLoS Biol. 2006;4:e192. doi: 10.1371/journal.pbio.0040192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dai H, et al. Crystal structure of the RIM2 C2A-domain at 1.4 Å resolution. Biochemistry. 2005;44:13533–13542. doi: 10.1021/bi0513608. [DOI] [PubMed] [Google Scholar]
- 62.Guan R, et al. Crystal structure of the RIM1α C2B domain at 1.7 Å resolution. Biochemistry. 2007;46:8988–8998. doi: 10.1021/bi700698a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Basu J, et al. A minimal domain responsible for Munc13 activity. Nat Struct Mol Biol. 2005;12:1017–1018. doi: 10.1038/nsmb1001. [DOI] [PubMed] [Google Scholar]
- 64.Richmond JE, Weimer RM, Jorgensen EM. An open form of syntaxin bypasses the requirement for UNC-13 in vesicle priming. Nature. 2001;412:338–341. doi: 10.1038/35085583. [DOI] [PMC free article] [PubMed] [Google Scholar]; Rescue experiments suggest that the function of Unc13/Munc13s in vesicle priming is at least in part to facilitate opening of the syntaxin-1 conformation.
- 65.Rosenmund C, et al. Differential control of vesicle priming and short-term plasticity by Munc13 isoforms. Neuron. 2002;33:411–424. doi: 10.1016/s0896-6273(02)00568-8. [DOI] [PubMed] [Google Scholar]; This paper clearly establishes a role for Munc13s in short-term presynaptic plasticity.
- 66.Rhee JS, et al. Beta phorbol ester- and diacylglycerol-induced augmentation of transmitter release is mediated by Munc13s and not by PKCs. Cell. 2002;108:121–133. doi: 10.1016/s0092-8674(01)00635-3. [DOI] [PubMed] [Google Scholar]
- 67.Basu J, Betz A, Brose N, Rosenmund C. Munc13–1 C1 domain activation lowers the energy barrier for synaptic vesicle fusion. J Neurosci. 2007;27:1200–1210. doi: 10.1523/JNEUROSCI.4908-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Junge HJ, et al. Calmodulin and Munc13 form a Ca2+ sensor/effector complex that controls short-term synaptic plasticity. Cell. 2004;118:389–401. doi: 10.1016/j.cell.2004.06.029. [DOI] [PubMed] [Google Scholar]
- 69.Kaeser PS, Sudhof TC. RIM function in short- and long-term synaptic plasticity. Biochem Soc Trans. 2005;33:1345–1349. doi: 10.1042/BST0331345. [DOI] [PubMed] [Google Scholar]
- 70.Koushika SP, et al. A post-docking role for active zone protein Rim. Nat Neurosci. 2001;4:997–1005. doi: 10.1038/nn732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schoch S, et al. RIM1α forms a protein scaffold for regulating neurotransmitter release at the active zone. Nature. 2002;415:321–326. doi: 10.1038/415321a. [DOI] [PubMed] [Google Scholar]; Biochemical and functional experiments suggest that RIM1α provides a scaffold to organize the active zone and plays a role in short-term presynaptic plasticity in addition to controlling vesicle priming.
- 72.Betz A, et al. Functional interaction of the active zone proteins Munc13–1 and RIM1 in synaptic vesicle priming. Neuron. 2001;30:183–196. doi: 10.1016/s0896-6273(01)00272-0. [DOI] [PubMed] [Google Scholar]
- 73.Dulubova I, et al. A Munc13/RIM/Rab3 tripartite complex: from priming to plasticity? EMBO J. 2005;24:2839–2850. doi: 10.1038/sj.emboj.7600753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Castillo PE, Schoch S, Schmitz F, Sudhof TC, Malenka RC. RIM1α is required for presynaptic long-term potentiation. Nature. 2002;415:327–330. doi: 10.1038/415327a. [DOI] [PubMed] [Google Scholar]
- 75.Castillo PE, et al. Rab3A is essential for mossy fibre long-term potentiation in the hippocampus. Nature. 1997;388:590–593. doi: 10.1038/41574. [DOI] [PubMed] [Google Scholar]
- 76.Calakos N, Schoch S, Südhof TC, Malenka RC. Multiple roles for the active zone protein RIM1α in late stages of neurotransmitter release. Neuron. 2004;42:889–896. doi: 10.1016/j.neuron.2004.05.014. [DOI] [PubMed] [Google Scholar]
- 77.Powell CM, et al. The presynaptic active zone protein RIM1α is critical for normal learning and memory. Neuron. 2004;42:143–153. doi: 10.1016/s0896-6273(04)00146-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ostermeier C, Brunger AT. Structural basis of Rab effector specificity: crystal structure of the small G protein Rab3A complexed with the effector domain of rabphilin-3A. Cell. 1999;96:363–374. doi: 10.1016/s0092-8674(00)80549-8. [DOI] [PubMed] [Google Scholar]
- 79.Sutton RB, Davletov BA, Berghuis AM, Sudhof TC, Sprang SR. Structure of the first C2 domain of synaptotagmin I: a novel Ca2+/phospholipid-binding fold. Cell. 1995;80:929–938. doi: 10.1016/0092-8674(95)90296-1. [DOI] [PubMed] [Google Scholar]
- 80.Ubach J, Zhang X, Shao X, Sudhof TC, Rizo J. Ca2+ binding to synaptotagmin: how many Ca2+ ions bind to the tip of a C2-domain? EMBO J. 1998;17:3921–3930. doi: 10.1093/emboj/17.14.3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shao X, Fernandez I, Sudhof TC, Rizo J. Solution structures of the Ca2+-free and Ca2+-bound C2A domain of synaptotagmin I: does Ca2+ induce a conformational change? Biochemistry. 1998;37:16106–16115. doi: 10.1021/bi981789h. [DOI] [PubMed] [Google Scholar]
- 82.Fernandez I, et al. Three-dimensional structure of the synaptotagmin 1 C2B-domain: synaptotagmin 1 as a phospholipid binding machine. Neuron. 2001;32:1057–1069. doi: 10.1016/s0896-6273(01)00548-7. [DOI] [PubMed] [Google Scholar]
- 83.Chapman ER, Davis AF. Direct interaction of a Ca2+-binding loop of synaptotagmin with lipid bilayers. J Biol Chem. 1998;273:13995–14001. doi: 10.1074/jbc.273.22.13995. [DOI] [PubMed] [Google Scholar]
- 84.Zhang X, Rizo J, Sudhof TC. Mechanism of phospholipid binding by the C2A-domain of synaptotagmin I. Biochemistry. 1998;37:12395–12403. doi: 10.1021/bi9807512. [DOI] [PubMed] [Google Scholar]
- 85.Fernandez-Chacon R, et al. Synaptotagmin I functions as a calcium regulator of release probability. Nature. 2001;410:41–49. doi: 10.1038/35065004. [DOI] [PubMed] [Google Scholar]; A point mutation in synaptotagmin-1 causes a parallel decrease in its apparent Ca2+ affinity and in the Ca2+ sensitivity of release, leaving little doubt for the proposed role of synaptotagmin-1 as a Ca2+ sensor in release.
- 86.Rhee JS, et al. Augmenting neurotransmitter release by enhancing the apparent Ca2+ affinity of synaptotagmin 1. Proc Natl Acad Sci USA. 2005;102:18664–18669. doi: 10.1073/pnas.0509153102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Robinson IM, Ranjan R, Schwarz TL. Synaptotagmins I and IV promote transmitter release independently of Ca2+ binding in the C2A domain. Nature. 2002;418:336–340. doi: 10.1038/nature00915. [DOI] [PubMed] [Google Scholar]
- 88.Mackler JM, Drummond JA, Loewen CA, Robinson IM, Reist NE. The C2B Ca2+-binding motif of synaptotagmin is required for synaptic transmission in vivo. Nature. 2002;418:340–344. doi: 10.1038/nature00846. [DOI] [PubMed] [Google Scholar]; A mutation in the Ca2+ binding sites of the synaptotagmin-1 C2B domain causes a dramatic impairment in neurotransmitter release, showing the crucial importance of Ca2+ binding to the C2B domain for release.
- 89.Arac D, et al. Close membrane-membrane proximity induced by Ca(2+)-dependent multivalent binding of synaptotagmin-1 to phospholipids. Nat Struct Mol Biol. 2006;13:209–217. doi: 10.1038/nsmb1056. [DOI] [PubMed] [Google Scholar]; The observation of simultaneous binding of synaptotagmin-1 to two membranes suggests that this Ca2+ sensor functions by bringing the vesicle and plasma membranes into close proximity, similar to the SNAREs but in a Ca2+-dependent manner.
- 90.Arac D, Murphy T, Rizo J. Facile detection of protein-protein interactions by one-dimensional NMR spectroscopy. Biochemistry. 2003;42:2774–2780. doi: 10.1021/bi0272050. [DOI] [PubMed] [Google Scholar]
- 91.Bhalla A, Chicka MC, Tucker WC, Chapman ER. Ca2+-synaptotagmin directly regulates t-SNARE function during reconstituted membrane fusion. Nat Struct Mol Biol. 2006;13:323–330. doi: 10.1038/nsmb1076. [DOI] [PubMed] [Google Scholar]
- 92.Dai H, Shen N, Arac D, Rizo JA. Quaternary SNARE-synaptotagmin-Ca(2+)-phospholipid complex in neurotransmitter release. J Mol Biol. 2007;367:848–863. doi: 10.1016/j.jmb.2007.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bowen ME, Weninger K, Ernst J, Chu S, Brunger AT. Single-molecule studies of synaptotagmin and complexin binding to the SNARE complex. Biophys J. 2005;89:690–702. doi: 10.1529/biophysj.104.054064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zimmerberg J, Akimov SA, Frolov V. Synaptotagmin: fusogenic role for calcium sensor? Nat Struct Mol Biol. 2006;13:301–303. doi: 10.1038/nsmb0406-301. [DOI] [PubMed] [Google Scholar]
- 95.Lynch KL, et al. Synaptotagmin C2A loop 2 mediates Ca2+-dependent SNARE interactions essential for Ca2+-triggered vesicle exocytosis. Mol Biol Cell. 2007;18:4957–4968. doi: 10.1091/mbc.E07-04-0368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Martens S, Kozlov MM, McMahon HT. How synaptotagmin promotes membrane fusion. Science. 2007;316:1205–1208. doi: 10.1126/science.1142614. [DOI] [PubMed] [Google Scholar]
- 97.Tucker WC, Weber T, Chapman ER. Reconstitution of Ca2+-regulated membrane fusion by synaptotagmin and SNAREs. Science. 2004;304:435–438. doi: 10.1126/science.1097196. [DOI] [PubMed] [Google Scholar]
- 98.Stein A, Radhakrishnan A, Riedel D, Fasshauer D, Jahn R. Synaptotagmin activates membrane fusion through a Ca2+-dependent trans interaction with phospholipids. Nat Struct Mol Biol. 2007;14:904–911. doi: 10.1038/nsmb1305. [DOI] [PubMed] [Google Scholar]
- 99.Pabst S, et al. Selective interaction of complexin with the neuronal SNARE complex. Determination of the binding regions. J Biol Chem. 2000;275:19808–19818. doi: 10.1074/jbc.M002571200. [DOI] [PubMed] [Google Scholar]
- 100.McMahon HT, Missler M, Li C, Sudhof TC. Complexins: cytosolic proteins that regulate SNAP receptor function. Cell. 1995;83:111–119. doi: 10.1016/0092-8674(95)90239-2. [DOI] [PubMed] [Google Scholar]
- 101.Giraudo CG, Eng WS, Melia TJ, Rothman JE. A clamping mechanism involved in SNARE-dependent exocytosis. Science. 2006;313:676–680. doi: 10.1126/science.1129450. [DOI] [PubMed] [Google Scholar]
- 102.Schaub JR, Lu X, Doneske B, Shin YK, McNew JA. Hemifusion arrest by complexin is relieved by Ca2+–synaptotagmin I. Nat Struct Mol Biol. 2006;13:748–750. doi: 10.1038/nsmb1124. [DOI] [PubMed] [Google Scholar]
- 103.Tang J, et al. A complexin/synaptotagmin 1 switch controls fast synaptic vesicle exocytosis. Cell. 2006;126:1175–1187. doi: 10.1016/j.cell.2006.08.030. [DOI] [PubMed] [Google Scholar]
- 104.Huntwork S, Littleton JT. A complexin fusion clamp regulates spontaneous neurotransmitter release and synaptic growth. Nat Neurosci. 2007;10:1235–1237. doi: 10.1038/nn1980. [DOI] [PubMed] [Google Scholar]
- 105.Reim K, et al. Complexins regulate a late step in Ca2+-dependent neurotransmitter release. Cell. 2001;104:71–81. doi: 10.1016/s0092-8674(01)00192-1. [DOI] [PubMed] [Google Scholar]; Complexins I and II are shown to play a critical active role in the Ca2+-triggered step of neurotransmitter release.
- 106.Reim K, et al. Structurally and functionally unique complexins at retinal ribbon synapses. J Cell Biol. 2005;169:669–680. doi: 10.1083/jcb.200502115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Roggero CM, et al. Complexin/synaptotagmin interplay controls acrosomal exocytosis. J Biol Chem. 2007;282:26335–26343. doi: 10.1074/jbc.M700854200. [DOI] [PubMed] [Google Scholar]
- 108.Xue M, et al. Distinct domains of complexin I differentially regulate neurotransmitter release. Nat Struct Mol Biol. 2007;14:949–958. doi: 10.1038/nsmb1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Fuson KL, Montes M, Robert JJ, Sutton RB. Structure of human synaptotagmin 1 C2AB in the absence of Ca2+ reveals a novel domain association. Biochemistry. 2007;46:13041–13048. doi: 10.1021/bi701651k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Weimer RM, et al. UNC-13 and UNC-10/rim localize synaptic vesicles to specific membrane domains. J Neurosci. 2006;26:8040–8047. doi: 10.1523/JNEUROSCI.2350-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Voets T, et al. Munc18–1 promotes large dense-core vesicle docking. Neuron. 2001;31:581–591. doi: 10.1016/s0896-6273(01)00391-9. [DOI] [PubMed] [Google Scholar]
- 112.de Wit H, Cornelisse LN, Toonen RF, Verhage M. Docking of secretory vesicles is syntaxin dependent. PLoS ONE. 2006;1:e126. doi: 10.1371/journal.pone.0000126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hammarlund M, Palfreyman MT, Watanabe S, Olsen S, Jorgensen EM. Open syntaxin docks synaptic vesicles. PLoS Biol. 2007;5:e198. doi: 10.1371/journal.pbio.0050198. [DOI] [PMC free article] [PubMed] [Google Scholar]


