Abstract
Engineered protein channels have many potential applications in biosensing at the single molecule level. A future generation of biosensor could be an array of target-specific ion channels, where each protein pore acts as a sensor element. An important step toward this goal is to create a portable, durable, single protein channel-integrated chip device. Here we report a versatile, modular chip that contains a single ion channel for single molecular biosensing. The core of the device is a long-lived lipid membrane that has been sandwiched between two air-insulated agarose layers which gel in situ. A single protein pore embedded in the membrane serves as the sensor element. The modular device is highly portable, allowing a single ion channel to continuously function following detachment of the chip from the instrument and independent transportation of the device. The chip also exhibits high durability, which is evidenced from long-duration continuous observation of single channel dynamics. Once engineered protein pores are installed, the chip becomes a robust stochastic sensor for real-time targeting such as detection of the second messenger IP3. This pluggable biochip could be incorporated with many applicable devices, such as a micro-fluidic system, and be made into a micro-array for both biomedical detection and membrane protein research.
Keywords: Biosensor(s), Single molecule detection, Membrane, Ion channel(s), Biochip, Second Messenger
INTRODUCTION
Engineered ion channels have been demonstrated for many potential applications, from ultra-sensitive single molecule detections1-5 and potential single DNA sequencing6-11 to modulation of ion selectivity12, nano-electroosmotic transportation13 and nanochemistry14;15. It is reasonable to predict that a future generation of biosensor could be an array of target-specific ion channels1 where each protein pore acts as a sensor element in a long-lived, highly sealed (>10 GΩ) lipid membrane. This modular device should be portable for both independent storage and free transportation, while in real-time applications, it should act as an independent pluggable component in coupling with other systems.
However, the creation of such a robust, versatile device that works with single protein pores is very challenging, and has not been achieved so far. For example, lipid bilayers have been formed on micrometer-sized apertures fabricated on a thin substrate16-20, but their lifetimes have not been substantially prolonged. Bilayers tethered on solid surfaces21;22 suffer from low insulating seals and high resistance of electrolyte reservoir between the bilayer and the substrate, making it problematic in single molecule detection and long-term electrical measurement. Lipid membrane by painted method23 has also been formed on pre-cast gel slab, and then covered with another gel slab as double support24;25. But this membrane may not suit single ion channel measurement due to the reported low resistance. In an improvement, the UV-triggered hydrogel has been used in replacing agarose gel26, but this work still lacks convincing evidences for portability and durability required for device design. In addition, the applicability is also limited because UV light may potentially damage or alter molecular functionalities, such as engineered photo-sensitive protein pores.
Here we report for the first time a portable, long-lived, modular biochip that integrates single ion channels for both biomedical detection and membrane protein research. Particularly, the high portability and durability of the device has been directly evidenced from both continuous movie and long-time electrical recording.
EXPERIMENTAL SECTION
Reagents and Proteins
The following chemicals were purchased from Sigma-Aldrich: agarose Type-IX, Type-VII, inositol-1,4,5-triphosphate (IP3), adenosine triphosphate (ATP), cyclodextrins, potassium chloride, magnesium chloride. Solvent hexadecane and pentane were purchased from Sigma-Aldrich and Burdick & Jackson, respectively. The lipid diphytanoyl-phosphatidylcholine (DPhPC) was obtained from Avantilipids Ins. The protein of small potassium channel Kcv encoded by chlorella virus27;28 and the DNAs of wild-type and mutant α-hemolysin M113R/T145R (RL2 background)29 were provided by Stephen Cheley at Hagan Bayley's laboratory. α-hemolysin proteins were prepared following the protocol as described previously30. Briefly, the protein was synthesized in the coupled in vitro transcription and translation kit (IVTT, Promega). In a 25-μl reaction, 2 μl DNA template (0.4 μg/μl in stock solution) was incubated with IVTT components containing complete amino acid mixture (including [35S]methionine) for 1 h at 37°C, followed by mixing with rabbit blood cell membrane (rabbit blood was provided by Pel-Freez Biologicals Ins.) in order to assemble protein heptamer. The protein assembly was collected using electrophoresis on the SDS-polyacrylamide gel, and dissolved in 50 μl water as stock. The protein amount was 10−100 ng per 50 μl.
Pico-Ampere Electrical Recording and Data Analysis
Single channel currents were recorded at a specific holding potential (protein addition side was ground) by using a patch-clamp amplifier (Axopatch 200B, Molecular Device Inc.), then low-pass filtered with a built-in 4-pole Bessel filter at 1−5 kHz and sampled at 5−20 kHz with a Digidata 1332 A/D converter (Molecular Device Inc.). Current amplitudes and dwell times were obtained from histograms constructed using pClamp 9.0 software (Molecular Device Inc.). All measurements were given as mean±SD based on n separate experiments (n≥3).
Modular Ion Channel Chip—Composition and Fabrication
While compositions of the chip device may vary to suit individual applications, one exemplary chip prototype can be assembled by the following components (Fig.1A): partition, compartment blocks, lids, electrodes, agarose layers, lipid membrane, and protein ion channel.
Figure 1.
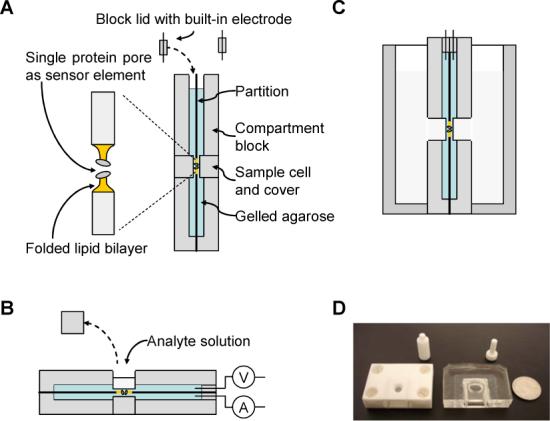
Composition, fabrication, usage and prototype of the modular ion channel chip. (A) The chip is composed of two flat compartments that are separated by a partition, with each compartment enclosing a thin layer of ultra-low gelling temperature agarose which has been premixed with salt at the desired concentration. The two agarose layers on both sides of the partition sandwich a lipid bilayer membrane over a 100 μm-aperture in the center of partition, where single protein pores are embedded as sensor elements; (B) Usage of the modular ion channel chip as an independent device. The analyte can be added from the sample cell on the back of either compartment and delivered to the sensor element through agarose between the sample cell and the membrane; (C) Usage of the chip as a pluggable device; (D) Pictures of prototype chips made from Teflon (left) and silicone elastomer polydimethylsiloxane (PDMS, middle). The partition is a 25-μm Teflon film. The quarter (right) is to compare the chip dimension.
The partition was a 25-μm thin Teflon film (Goodfellow Ltd.) with a tiny aperture (typically 100 μm) punctured in the center, over which the lipid bilayer membrane can be effectively formed. The two compartment blocks were made of solid and electrically insulating polymer material, such as Teflon and silicone elastomer polydimethylsiloxane (PDMS, SylgardR 184, Dow Corning). The Teflon blocks were fabricated at University Scientific Instrument Shop, while the PDMS blocks were self-made according to the product instruction book. There was a hole of 3-mm wide acting as the sample cell on the back of each block. The sample cell was initially closed with a cover, and was used for loading sample when the cover was removed. The assembly of the partition and compartment blocks forms two compartments (cis and trans) that are separated by the partition. In our design, the dimension of each compartment was 1.4-ml (2×2×0.38 cm). The interfaces between compartment blocks and partition were sealed with vacuum grease to prevent leaking.
The gel solution contained 1.5% (w/v) ultra-low gelling temperature agarose (Type-IX, Tg < 17 °C), 1 M KCl, and 10 mM Tris at pH7.2. The mixture was heated up to 55−65 °C to melt the agarose; then cooled down to room temperature, at which this type of agarose is capable of remaining in liquid form for days. To form the lipid bilayer membrane by the mono-layer folding process31, we initially placed the chip vertically, and add the melted agarose solution (at room temperature) to each compartment up to the level below the central hole of the Teflon partition. Then, we used a 10 μl micropipet to drop 5 μl of lipid solution (DPhPC was dissolved in pantane at 10 mg/ml) to the gel solution in each compartment. The lipid can immediately spread and form a molecular mono-layer on the solution surface. After one minute of evaporating the solvent, more gel solution was added until the liquid surface was raised over the aperture on the partition. Immediately, the lipid mono-layer on both sides of the partition automatically hybridized each other to form a bilayer membrane covering the partition aperture. A high sealing, good quality bilayer was identified according to the membrane resistance (>10 GΩ), capacitance (∼100 pF), and current noise (the value of IRMS on the amplifier panel was 1.2−1.5 pA).
Upon bilayer formation, we used a micropipetter to slowly release 0.5 μl of protein solution to the gel solution on one side of bilayer to the final concentration of 0.07−0.7 ng/ml, followed by gentle mixing to control the protein insertion into the membrane. Once single ion channels were identified, the compartment on the protein side was gently perfused with new gel solution to prevent additional protein incorporation. The single ion channel embedded in the membrane serves as the sensor element.
Once the insertion of the single protein pore was verified, both electrodes were plugged off from the solutions to disconnect the electrical monitoring. We found that this process does not break the membrane formed in agarose solution. The whole chip was then gently transferred to an environment below gelling temperature, for example 10 °C refrigerator. After gelling for 10 minutes, the gel solution on both sides was solidified. Thus, the membrane was sandwiched between two gel layers and stabilized.
Finally, the top openings of two compartments were sealed by two block lids, a very critical component in keeping the water content in the gel. The electrodes fixed in the middle of blocks can connect with gel, providing the membrane potential and receiving the pico-ampere current information.
RESULTS AND DISCUSSION
The chip fabricated following the above steps is highly durable and portable, and can be independently stored at room temperature without instrument connection. It can also be physically carried, manually rotated and transported (as shown below). It can either be placed vertically or be laid horizontally as a biosensor (Fig.1B). When the sample cell cover on the upper compartment wall is removed, the analyte solution can be added directly to the sample cell. Since agarose gel is permeable to analytes of various molecular weights (pore size of 2% agarose gel is 470 nm)32, the analyte in the solution can transport or diffuse through the gel and reach the protein pore in the membrane for single molecule detection. Since the chip is pluggable, it can also be incorporated into other devices. For example, it is possible to plug the chip into a recording chamber as shown in Fig. 1C in order to be accessible by different analytes from both sides of the chip.
Durability
The chip device that we have constructed is highly durable. The agarosesandwiched membrane in the chip can last 57±9 hours on average, whereas a qualified agarose-free membrane is only 6±3 hours. This is a dramatically improved property for both biosensing and research applications. To visualize this capability, we have performed a continuous observation of ion channel functionalities in the chip (Fig.2). The protein in this experiment is Kcv, the smallest potassium channel encoded by chlorella viruses27. So far, there has been no published study on single Kcv channels (in preparation)28. Fig.2A represents a 65-hour continuous current recording (+40 mV) of chip-based Kcv channels (screen-captured picture), which clearly demonstrates that the lipid membrane in the chip is of good quality and consistently retains high-sealing property (>10 GΩ in 1M KCl) throughout the recording.
Figure 2.
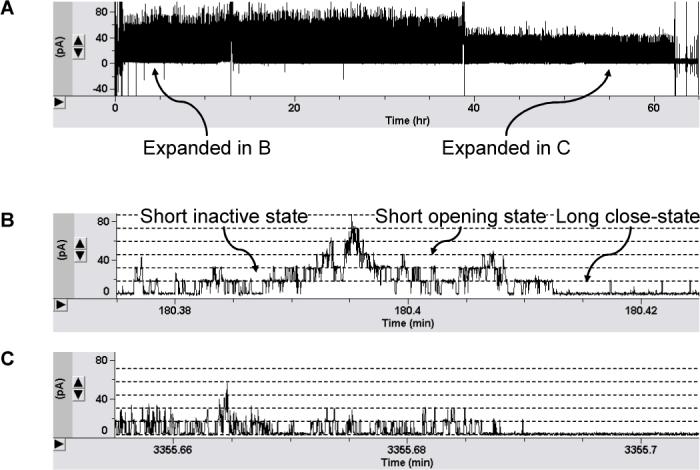
(A) A 65-hour continuous current recording of long-lived single Kcv channels in the chip. The trace is screen-captured. Agarose layers contain 1 M KCl, 10 mM Tris, and 1.5% ultra-low gelling temperature agarose (pH7.2). Detailed current recordings at the 3rd and 55th hour in (A) are expanded and screen-captured in (B) and (C) respectively.
We further compared the functions of Kcv in agarose-supported and un-supported membrane. In order to visualize the activity of Kcv channel in the chip, we extracted a current domain at the 3rd hour of the recording shown in Fig.2A and expanded it in Fig.2B. By single channel analysis, we found that the Kcv channel in the chip reversibly transits between the long closed (5±1 s, 0 pS), the short open (3.5±0.1 ms, 452±26 pS), and the short inactivated (9.3±3.5 ms, 0 pS) states in 1 M KCl and at +40 mV. These data are in consistence with the Kcv gating kinetics and channel conductance as studied in a traditional planar bilayer system (in preparation)28.
In summary, the results from these comparisons suggest that the chip containing agarose-supported membrane is a highly durable device, and the integrated ion channel is functionally identical to that in an un-supported membrane. In addition, we have also observed that the opening duration for all the Kcv channel in membrane becomes shortened (2.3±0.1 ms) after continuous voltage stimulation for long hours (Fig.2C), whereas a similar kinetic change has not been identified for Kcv channels in a short-lived agarose-free bilayer. At the current stage, the nature of this phenomenon is not known yet. Probably it is related to many factors such as protein-lipid interactions. However, we at least learn from this observation that the durable ion channel chip could be a potential tool for monitoring long-term protein dynamics in the membrane.
Portability
The ion channel chip is also highly portable, another important capability required for practical applications in biosensing and research. It can be repeatedly disconnected and reconnected to any device, such as an electrophysiology recording instrument; the disconnected chip is capable of independent storage, and can be transported from place to place, whereas a membrane without solid support does not have these capabilities.
To exhibit the portable chip as an independent modular device, we have produced a movie to continuously visualize activity of a live ion channel during the entire procedure for chip handling (Supplementary Movie S1). Several important observations in the movie have also been captured in the pictures in Fig.3. We believe this is an unprecedented real-time proof of a portable membrane protein device. In this experiment, we still use Kcv as the target channel protein because its gating current can be easily recognized. The movie starts with the current recording of a single Kcv channel in the chip (Fig.3A), followed by disconnection of the chip from the electrical recording instrument (zero conductance, as shown in the movie). Then the researcher removes the chip from the testing box (Fig.3B), carries it out of the laboratory, travels around the building (Fig.3C), and returns to the testing site. Upon re-connecting the chip to the recording device (Fig.3D), the original Kcv channel in the chip immediately appears on the screen as reflected by the gating current (Fig.3E). The success rate of the portability test is 90% (n>10). In addition to the safe transport capability, we can also rotate the chip to any angle and flip it over to suit different application platforms. In one example, the chip can be laid horizontally for stochastic sensing. By comparison, a membrane in solution without solid support (agarose-free configuration) could not survive such mechanical impact. It easily breaks down when the chamber is tilted at such an angle that causes solution to level down below the central aperture on the partition, leaving the bilayer exposed to the air.
Figure 3.
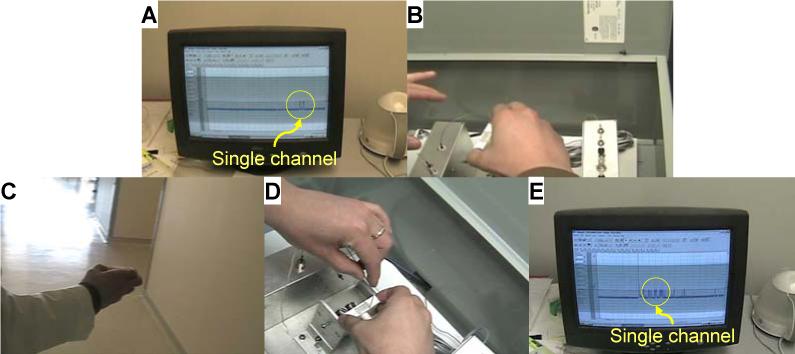
Pictures captured from a movie showing the portability of the chip (Movie S1 in Supplementary Materials). (A) Visualizing a single potassium channel Kcv in the chip. The gating profile is marked with a yellow circle; (B) Disconnecting the chip from the electrical recording devices and moving the chip out of the testing box; (C) Handling the chip and transporting it around the building; (D) Re-connecting the chip to the electrical system; (E) Recovery of single channel current from the chip.
In addition to durability and portability, we also compared the breakage voltage for different membrane configurations. Our test indicated both agarose-supported and agarose-free membrane function properly at a voltage above 200 mV, but the supported membrane in the chip can survive up to 326±72 mV, higher than 287±98 mV for an un-supported membrane.
Determinants
We have concluded several determinants in achieving high portability and durability for the chip. The core of the chip (Fig.1) is a series of treatments designed to form and retain an ion channel-integrated lipid membrane in temperature-triggered agarose which gels in situ. The lipid membrane encapsulated in the chip should be initially formed by the mono-layer folding technique31, rather than the painting method23, in melted agarose solution. The mono-layer folding process is a unique method of instant formation of solvent-free bilayer, which is not only consistently stable in properties such as capacity and high sealing, but also allows quick insertion of proteins in a controllable manner. At lower temperature, the agarose solutions on both sides of the membrane are solidified into agarose layers. When returning to the room temperature, the agarose should retain its solid format so that the lipid membrane is tightly “clipped” in between while the ion channel functionalities are retained. Such sandwiched membrane definitely possesses much greater mechanical resistance than that supported from single side only33. We first succeeded in fabricating the chip by using ultra-low gelling temperature agarose (Type IX-A, Tg = 17 °C at 1.5%), which retains liquid form at room temperature after melting. In an subsequent effort, with the help of new technique for achieving high temperature lipid membrane34, we further succeeded in using agarose with a higher gelling temperature (Type VII-A, Tg = 26 °C at 1.5%).
Another key determinant to successful chip fabrication is effective preservation of the water component in agarose for long-term storage of the chip. This requires the agarose enclosed in the chip to be isolated as much as possible to minimize the water loss. Furthermore, the successful use of agarose and diphytanoyl-phosphatidylcholine (DPhPC), rather than UV-cured polymer and specially synthesized lipid, greatly expands the chip applicability. This is because agarose is non-toxic and biocompatible, and particularly possesses much larger pore sizes for efficient diffusion of various analytes (the pore size of 2% agarose gel is ∼470 nm)32. Similarly, DPhPC is a commonly used lipid for reconstitution of various ion channels, from α-helical proteins such as Kcv, as studied above, to β-barrel species, such as α-hemolysin, as described in the next section. The modular chip can be made of any hard, electrically insulating polymer material such as silicone elastomer polydimethylsiloxane (PDMS) and Teflon (Fig.1D), and can be potentially minimized if micro- or nano-fabricating techniques are applied.
Applications in Biosensing
The unique portability and durability of the ion channel chip makes it an independent, pluggable, modular biosensor device. For real-time assay, we first lay the chip horizontally, as shown in Fig.1B, then gently remove the cover of the upper sample cell. After adding analyte solution into the sample cell, the analyte molecule can diffuse through the thin gel layer, then reach and block the ion channel sensor element in the membrane for single molecule detection. The analyte transportation can be driven by diffusion, voltage or the electroosmotic force depending on analyte dimension, molecular weight, and charge property.
To demonstrate the capability of the chip as a biosensor, we first incorporated pore-forming protein α-hemolysin (αHL) into the chip and measured the time-dependent pore block with β-cyclodextrin (βCD). The αHL-βCD system has been well established as the model for single-molecule stochastic sensing4. The goal of the time-dependent measurement is to evaluate the transportation time of βCD in the agarose. Specifically, the chip incorporating a single wild-type αHL pore was fabricated according to steps described above (Fig.1A). The gel layers enclosed in the chip contained 1 M KCl, 10 mM Tris, and 1.5% ultra-low gelling temperature agarose (pH7.2). 20 μl of cyclodextrin solution (40 μM) was added to the sample cell using a micropipetter. The pico-Ampere current from the chip was continuously monitored at +40 mV.
From the current traces shown in Fig.4A-left, we can clearly identify individual pore block events by single βCD molecules, which become more frequent as the time progresses. The single molecule block occurrence (f) was defined as 1/τon, where τon is the average duration between two adjacent block events. f should correspond to the local concentration of βCD around the pore. Therefore, analysis of a series of current blocks at different stages (Fig.4A-left) leads to the plot of block occurrence-time relationship (Fig.4A-right) where the block occurrence reaches the maximum in 40 minutes for a 1 mm-thick 1.5% agarose layer. We expect that this duration could be further shortened for a thinner agarose layer if micro-fabrication technology is applied, or for a charged analyte which can be driven by voltage.
Figure 4.
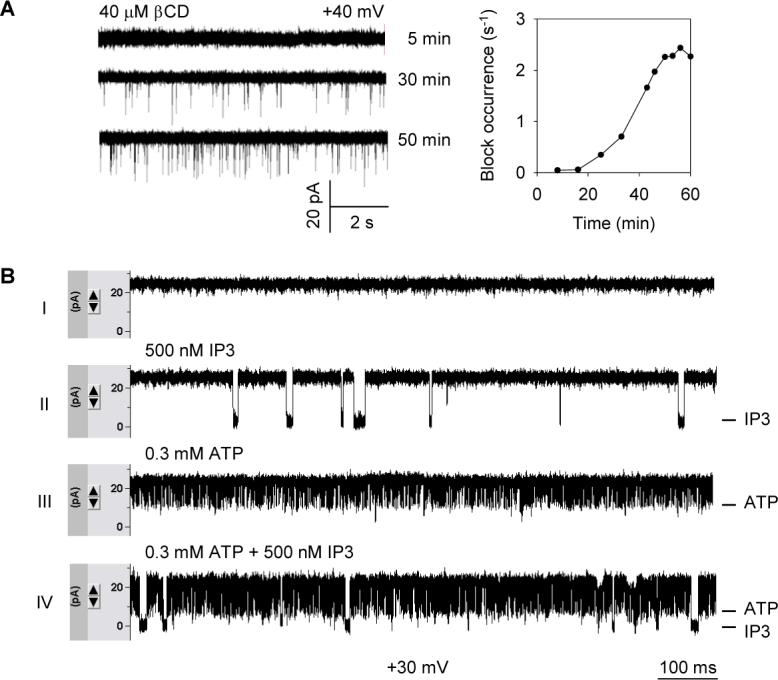
Stochastic sensing on the modular ion channel chip. (A) Time-dependent transportation of analyte from the sample cell to the sensor element in the chip. Single pores of wild-type αHL were incorporated into the sandwiched bilayer as the sensor element. 40 μM βCD was added to the chip from the sample cell (Fig.1B). Agarose layers contained 1 M KCl, 10 mM Tris, and 1.5% ultra-low gelling temperature agarose (pH7.2). Left panel shows current traces recorded after 5, 30 and 40 minutes after addition of βCD. Right panel is the time-dependent block occurrence with βCD; (B) Detection of second messenger IP3 on chips. Mutant αHL, M113R/T145R, was used as the single pore sensor element. Agarose layers contained the same components as in A, with the addition of 0.5 mM MgCl2. All traces were recorded at +30 mV. Trace (I), control test without analyte; Trace (II), 500 nM IP3 was loaded into the sample cell of the same chip; Trace (III) 0.3 mM ATP was added in a separate control test; and Trace (IV), mixture of 500 nM IP3 and 0.3 mM ATP was loaded in the chip. All traces were captured after the block occurrence reached the maximum.
Finally, we exemplified chip-based stochastic sensing by using engineered αHL as the sensor element to detect second messenger inositol-1,4,5-triphosphate (IP3) (Fig.4B). The sensor pore that is equipped with two arginine rings (14 arginines) in the most restrictive site in the lumen (M113R/T145R) has been made for high specific discrimination of various phosphate compounds29. This time, we employed the same gel composition as for the cyclodextrin measurement, plus 0.5 mM MgCl2, and the chip was monitored at +30 mV. First, in a control test without analyte (Fig.4B-I), we recorded a flat background conductance for the chip containing a single sensor pore. Then, loading 20 μl of 500 nM IP3 to the same chip produces characteristic long current blocks (5.7±0.7 ms, Fig.4B-II), which correspond to the binding of single IP3 molecules to the sensor pore. In comparison, in a separate control test the presence of 0.3 mM ATP in the chip produces distinctly faster blockade (0.28 ±0.06 ms, Fig.4B-III). As predicted, when 500 nM IP3 is presented in the mixture with 0.3 mM ATP, like it is in a living cell, we can clearly discriminate two binding patterns from a single current recording: the short and long blocks caused by ATP and IP3 respectively (Fig.4B-VI). Because the event occurrence for each component in mixture is corresponding to its concentration29, its quantity in the mixture could be readily determined by single-molecule stochastic analysis4.
CONCLUSIONS AND PERSPECTIVES
In summary, we construct a portable, long-lived, modular sensor chip that integrates single ion channels into sandwiched, therefore stabilized lipid membrane between temperature-triggered agarose layers. We for the first time use continuous movie and long-term electrical recording to demonstrate high portability and durability of the device, and exhibit the chip capability of single-molecule detection by sensing biological compounds, such as second messenger IP3.
The robust, versatile device is capable of being programmed because any membrane protein can be used to make the chip for various biomedical detections, such as screening of enzyme35 and detection of glucose36 or neural transmitters. For research purposes, the chip also has potential applications: examples are DNA and protein detection in genomics and proteomics, or dynamics of protein-membrane interaction. In addition to agarose, any polymer material, such as smart polymer, that solidifies with pH or salt concentration and is permeable to ions and molecules but does not damage bilayer membrane and protein, could be used as membrane support for the biosensor.
There are several advantages of the modular chip biosensor that contains a single protein nanopore for stochastic sensing. One is the simultaneous analysis of the identity and quantity of multi-analyte in mixture by a single sensor element. Another advantage is the ability to create digital output with a high Signal/Noise ratio, due to the single molecule binding. As a modular device, the chip may be hybridized with other applicable devices. For example, the chip can be coupled with a micro-fluidic system and perform electrical detection combined with optical technologies. Furthermore, the chip may provide a micro-array in future for high throughput screening , with each array element containing a single stochastic sensor. This speculation is reasonable because micro-patterned hydrogels have been created37;38. In addition, the chip features quick fabrication. The bilayer formation by a mono-layer folding process takes as little as 5 minutes. The single channel incorporation time is no more than 10 minutes. The gelling time of agarose at 10 °C is 15 minutes. Therefore the biosensor fabrication time is less than 1 hour.
Certainly, we also foresee many challenges for these technological hybridizations, such as significant miniaturization relative to our current device for interfacing to a microfluidic system, and methods for indexing each individual ion channel on the array. Hopefully, with the help of powerful micro-/nano-fabrication tools including bioMEMS techniques, new protein pore-based nano-systems could ultimately be developed. This possibility is further enhanced by two recent technological advances: the ability to form a protein pore-incorporated lipid membrane across a microfluidic channel39, and the ability to quickly transferring ion channels into membrane for single channel assay40.
Supplementary Material
Acknowledgements
We thank Dr. Stephen Cheley for the Kcv proteins and α-hemolysin DNAs, which have been started in Dr. Hagan Bayley's laboratory at Texas A&M University. This investigation was supported by a NSF Career Award and the University of Missouri Research Board. This investigation was conducted in a facility constructed with support from Research Facilities Improvement Program Grant Number C06-RR-016489−01 from the National Center for Research Resources, National Institutes of Health. In addition, we also thank Dr. Kevin Gillis for invaluable comments on manuscript.
Footnotes
Supporting Information Movie S1 (SingleChannelOnPortableChip.wmv): The movie shows the portability of the modular ion channel chip. At the beginning, we can visualize a gating single potassium channel Kcv in the chip by an electrophysiology recording instrument. Then the researcher disconnect the chip from the electrical recording devices and moves the chip out of the testing box, handles the chip and transports it around the building. When the chip is taken back to the research site, it is re-connected to the electrical system, and immediately the recovery of single channel current from the chip can be visualized from the computer.
Reference List
- 1.Bayley H, Cremer PS. Nature. 2001;413:226–30. doi: 10.1038/35093038. [DOI] [PubMed] [Google Scholar]
- 2.Bezrukov SM, Vodyanoy I, Parsegian VA. Nature. 1994;370:279–81. doi: 10.1038/370279a0. [DOI] [PubMed] [Google Scholar]
- 3.Braha O, Gu LQ, Zhou L, Lu XF, Cheley S, Bayley H. Nature Biotechnology. 2000;18:1005–07. doi: 10.1038/79275. [DOI] [PubMed] [Google Scholar]
- 4.Gu LQ, Braha O, Conlan S, Cheley S, Bayley H. Nature. 1999;398:686–90. doi: 10.1038/19491. [DOI] [PubMed] [Google Scholar]
- 5.Movileanu L, Howorka S, Braha O, Bayley H. Nature Biotechnology. 2000;18:1091–95. doi: 10.1038/80295. [DOI] [PubMed] [Google Scholar]
- 6.Kasianowicz JJ, Brandin E, Branton D, Deamer DW. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:13770–73. doi: 10.1073/pnas.93.24.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meller A, Nivon L, Brandin E, Golovchenko J, Branton D. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:1079–84. doi: 10.1073/pnas.97.3.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vercoutere W, Winters-Hilt S, Olsen H, Deamer D, Haussler D, Akeson M. Nature Biotechnology. 2001;19:248–52. doi: 10.1038/85696. [DOI] [PubMed] [Google Scholar]
- 9.Mathe J, Visram H, Viasnoff V, Rabin Y, Meller A. Biophysical Journal. 2004;87:3205–12. doi: 10.1529/biophysj.104.047274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sanchez-Quesada J, Saghatelian A, Cheley S, Bayley H, Ghadiri MR. Angewandte Chemie-International Edition. 2004;43:3063–67. doi: 10.1002/anie.200453907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Howorka S, Cheley S, Bayley H. Nature Biotechnology. 2001;19:636–39. doi: 10.1038/90236. [DOI] [PubMed] [Google Scholar]
- 12.Gu LQ, Dalla Serra M, Vincent JB, Vigh G, Cheley S, Braha O, Bayley H. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:3959–64. doi: 10.1073/pnas.97.8.3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gu LQ, Cheley S, Bayley H. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:15498–503. doi: 10.1073/pnas.2531778100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luchian T, Shin SH, Bayley H. Angewandte Chemie-International Edition. 2003;42:3766–71. doi: 10.1002/anie.200351313. [DOI] [PubMed] [Google Scholar]
- 15.Gu LQ, Cheley S, Bayley H. Science. 2001;291:636–40. doi: 10.1126/science.291.5504.636. [DOI] [PubMed] [Google Scholar]
- 16.Fertig N, Blick RH, Behrends JC. Biophysical Journal. 2002;82:3056–62. doi: 10.1016/S0006-3495(02)75646-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mayer M, Kriebel JK, Tosteson MT, Whitesides GM. Biophysical Journal. 2003;85:2684–95. doi: 10.1016/s0006-3495(03)74691-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Romer W, Steinem C. Biophysical Journal. 2004;86:955–65. doi: 10.1016/S0006-3495(04)74171-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sackmann E. Science. 1996;271:43–48. doi: 10.1126/science.271.5245.43. [DOI] [PubMed] [Google Scholar]
- 20.Schmidt C, Mayer M, Vogel H. Angewandte Chemie-International Edition. 2000;39:3137–40. doi: 10.1002/1521-3773(20000901)39:17<3137::aid-anie3137>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 21.Cornell BA, BraachMaksvytis VLB, King LG, Osman PDJ, Raguse B, Wieczorek L, Pace RJ. Nature. 1997;387:580–83. doi: 10.1038/42432. [DOI] [PubMed] [Google Scholar]
- 22.Knoll W, Frank CW, Heibel C, Naumann R, Offenhausser A, Ruhe J, Schmidt EK, Shen WW, Sinner A. J Biotechnol. 2000;74:137–58. doi: 10.1016/s1389-0352(00)00012-x. [DOI] [PubMed] [Google Scholar]
- 23.Mueller P, Rudin DO, Tien HT, Wescott WC. Nature. 1962;194:979–80. doi: 10.1038/194979a0. [DOI] [PubMed] [Google Scholar]
- 24.Costello RF, Peterson IP, Heptinstall J, Byrne NG, Miller LS. Advanced Materials for Optics and Electronics. 1998;8:47–52. [Google Scholar]
- 25.Costello RF, Peterson IR, Heptinstall J, Walton DJ. Biosensors & Bioelectronics. 1999;14:265–71. [Google Scholar]
- 26.Jeon TJ, Malmstadt N, Schmidt JJ. Journal of the American Chemical Society. 2006;128:42–43. doi: 10.1021/ja056901v. [DOI] [PubMed] [Google Scholar]
- 27.Plugge B, Gazzarrini S, Nelson M, Cerana R, Van Etten JL, Derst C, DiFrancesco D, Moroni A, Thiel G. Science. 2000;287:1641–44. doi: 10.1126/science.287.5458.1641. [DOI] [PubMed] [Google Scholar]
- 28.Shim JW, Cheley S, Yang M, Bayley H, Gu LQ. (unpublished work)
- 29.Cheley S, Gu LQ, Bayley H. Chemistry & Biology. 2002;9:829–38. doi: 10.1016/s1074-5521(02)00172-2. [DOI] [PubMed] [Google Scholar]
- 30.Cheley S, Braha G, Lu XF, Conlan S, Bayley H. Protein Science. 1999;8:1257–67. doi: 10.1110/ps.8.6.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Montal M, Mueller P. Proc Natl Acad Sci U S A. 1972;69:3561–6. doi: 10.1073/pnas.69.12.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pluen A, Netti PA, Jain RK, Berk DA. Biophysical Journal. 1999;77:542–52. doi: 10.1016/S0006-3495(99)76911-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ide T, Takeuchi Y, Aoki T, Yanagida T. Japanese Journal of Physiology. 2002;52:429–34. doi: 10.2170/jjphysiol.52.429. [DOI] [PubMed] [Google Scholar]
- 34.Kang XF, Gu LQ, Cheley S, Bayley H. Angewandte Chemie-International Edition. 2005;44:1495–99. doi: 10.1002/anie.200461885. [DOI] [PubMed] [Google Scholar]
- 35.Xie HZ, Braha O, Gu LQ, Cheley S, Bayley H. Chemistry & Biology. 2005;12:109–20. doi: 10.1016/j.chembiol.2004.11.013. [DOI] [PubMed] [Google Scholar]
- 36.Kullman L, Gurnev PA, Winterhalter M, Bezrukov SM. Physical Review Letters. 2006;96 doi: 10.1103/PhysRevLett.96.038101. [DOI] [PubMed] [Google Scholar]
- 37.Mayer M, Yang J, Gitlin I, Gracias DH, Whitesides GM. Proteomics. 2004;4:2366–76. doi: 10.1002/pmic.200300748. [DOI] [PubMed] [Google Scholar]
- 38.Klajn R, Fialkowski M, Bensemann IT, Bitner A, Campbell CJ, Bishop K, Smoukov S, Grzybowski BA. Nature Materials. 2004;3:729–35. doi: 10.1038/nmat1231. [DOI] [PubMed] [Google Scholar]
- 39.Malmstadt N, Nash MA, Purnell RF, Schmidt JJ. Nano Letters. 2006;6:1961–65. doi: 10.1021/nl0611034. [DOI] [PubMed] [Google Scholar]
- 40.Holden MA, Jayasinghe L, Daltrop O, Mason A, Bayley H. Nature Chemical Biology. 2006;2:314–18. doi: 10.1038/nchembio793. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


