Summary
Transcription complex components frequently show coupled folding and binding but the functional significance of this mode of molecular recognition is unclear. IκBα binds to and inhibits the transcriptional activity of NF-κB via its ankyrin repeat (AR) domain. The β-hairpins in ARs 5–6 in IκBα are weakly folded in the free protein, and their folding is coupled to NF-κB binding. Here we show that introduction of two stabilizing mutations in IκBα AR 6 causes ARs 5–6 to cooperatively fold to a conformation similar to that in NF-κB-bound IκBα. Free IκBα is degraded by a proteasome-dependent but ubiquitin-independent mechanism, and this process is slower for the pre-folded mutants both in vitro and in cells. Interestingly, the pre-folded mutants bind NF-κB more weakly as shown by both SPR and ITC in vitro and immunoprecipitation experiments from cells. One consequence of the weaker binding is that resting cells containing these mutants show incomplete inhibition of NF-κB activation; they have significant amounts of nuclear NF-κB. Additionally, the weaker binding combined with the slower degradation rate of the free protein results in reduced levels of nuclear NF-κB upon stimulation. These data clearly demonstrate that the coupled folding and binding of IκBα is critical for its precise control of NF-κB transcriptional activity.
Keywords: coupled folding and binding, transcription factor regulation, protein-protein interactions, ankyrin repeat, ubiquitin-independent proteasome degradation
Introduction
The nuclear factor κB (NF-κB) family of transcription factors play key roles in normal growth and development, in inflammatory and immune responses, and in numerous human diseases 1; 2. While the most abundant NF-κB is the p50/p65 heterodimer, the NF-κB family is composed of homo- and heterodimers formed from the combinatorial assembly of the p65 (RelA), RelB, c-Rel, p50, and p52 subunits 1. The inhibitor proteins IκBα, IκBβ, and IκBε tightly regulate the transcriptional activity of p65 and c-Rel containing NF-κB dimers 3. In resting cells, IκBα binds extremely tightly to NF-κB, preventing its nuclear accumulation and association with DNA 4; 5; 6. Upon stimulation, NF-κB-bound IκBα is specifically phosphorylated (by the IκB kinase, IKK), ubiquitinated, and degraded by the proteasome 7; 8; 9; 10; 11. NF-κB then enters the nucleus, binds DNA, and regulates transcription of its numerous target genes 12. NF-κB activates transcription of its own inhibitor, IκBα, resulting in a negative-feedback loop 13; 14; 15; 16. The newly synthesized IκBα enters the nucleus and is responsible for rapid post-induction repression of NF-κB transcriptional activity 17. IκB regulation of NF-κB transcriptional activity is so critical that misregulation results in many different diseases 2. In fact, constitutive activation of NF-κB is observed in many types of cancer, and improper IκBα function is observed in B-cell and Hodgkin's lymphomas 18.
Free IκBα has a low thermodynamic stability, and it is rapidly degraded by the proteasome in a process that does not require phosphorylation or ubiquitination 19; 20. The in vivo half-life of free IκBα is less than 10 min 20; 21; 22. However, NF-κB-bound IκBα is stable for hours, and its degradation requires phosphorylation and ubiquitination 20; 21. These distinct degradation pathways for free and NF-κB-bound IκBα appear to be critical for signal-responsive NF-κB activation. Decreases in NF-κB-bound IκBα phosphorylation reduce NF-κB activation upon stimulation in a mathematical model of NF-κB signaling 21. Furthermore, IκBα mutants with slower basal degradation rates result in slower activation of NF-κB upon stimulation with TNF-α 20. A recent study shows that rapid synthesis and degradation of IκBα provides a mechanism for resistance to metabolic stresses 23. Additionally, this study showed that both degradation pathways are critical for proper control of NF-κB activation in response to UV.
IκBα is composed of an N-terminal signal response region where phosphorylation and ubiquitination occur, an ankyrin repeat (AR) domain that binds to NF-κB (Figure 1A), and a C-terminal PEST sequence 24; 25. The PEST sequence is important for basal degradation of free IκBα 20. The AR, a structural motif of ~30–40 amino acids composed of a β-hairpin followed by two anti-parallel α-helices and a variable loop, is found in more than 3,000 different proteins with highly varied functions 26. AR domains function by mediating specific protein-protein interactions 27. AR consensus sequences based on statistical analyses were developed 28; 29. Many consensus designed AR proteins have been made, and they are generally more stable than naturally occurring AR proteins 28; 29; 30; 31. The GXTPLHLA motif (Figure 1B) is the most prevalent signature in the consensus sequence, and mutation of IκBα ARs 4 and 5 to these residues resulted in a stability increase of ~1.5 kcal/mol 32.
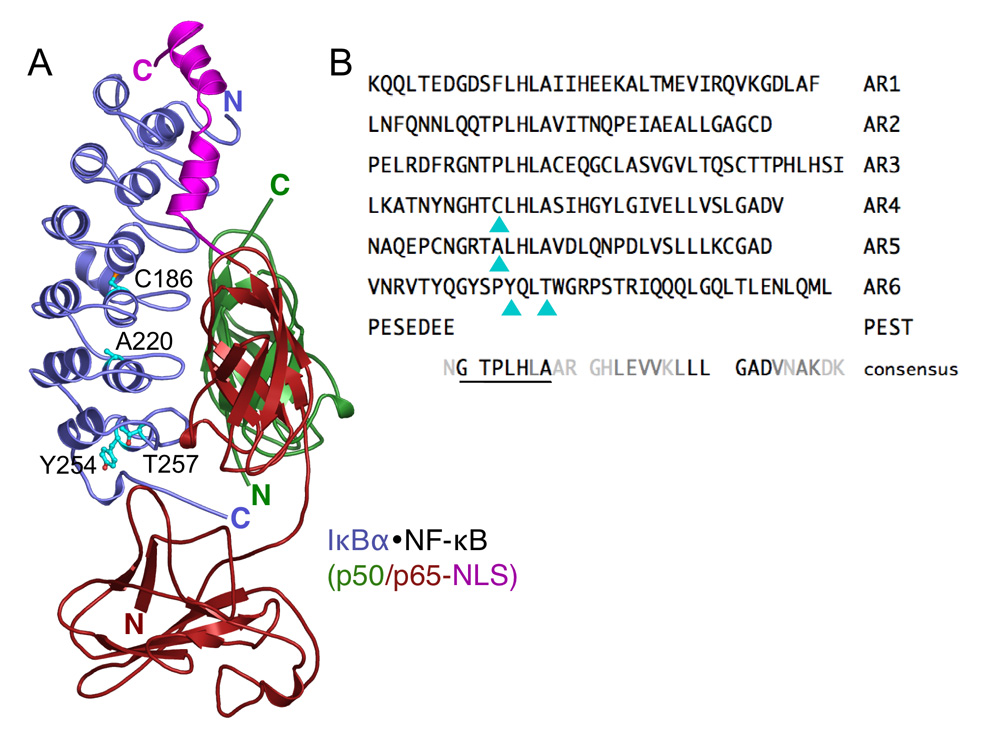
Figure 1.
(A) The crystal structure of IκBα (blue) bound to NF-κB (p50, green; p65, red; p65 nuclear localization sequence (NLS), magenta) 24. Residues mutated in this study, Y254, T257, C186, and A220, do not contact NF-κB, and they are depicted with ball-and-stick representation and colored cyan. The figure was prepared using PyMOL 74. (B) The sequences of the IκBα ankryin repeats (ARs) are aligned with the consensus sequence for a stable AR 29. Cyan triangles indicate residues mutated in this study. In the consensus sequence, black letters indicate highly conserved residues and gray letters indicate weaker conservation.
The IκBα•NF-κB interface buries more than 4,000 Å2 and all six IκBα ARs contact NF-κB 24; 25 (Figure 1A). In free IκBα, only ARs 1–4 of IκBα are compactly folded, whereas, ARs 5 and 6 are weakly folded and highly flexible 32; 33; 34. ARs 1–4 fold cooperatively 32 and show protection from amide H/2H exchange that is consistent with a compact structure 33. In fact, the extent of exchange in ARs 1–4 correlates with the solvent accessible surface area (SASA) calculated for IκBα from the IκBα•NF-κB crystal structure with NF-κB removed, suggesting that ARs 1–4 adopt the same conformation in free and NF-κB-bound IκBα 33. In contrast, ARs 5–6 do not fold cooperatively 32 and the β-hairpins in ARs 5–6 exchange nearly all of their amide protons and they exchange more than predicted by their SASA, suggesting that they are flexible 33. However, when IκBα is bound to NF-κB, the β-hairpins in ARs 5–6 show large decreases in amide H/2H exchange. The extent of exchange in NF-κB-bound IκBα correlates with the SASA calculated from the IκBα•NF-κB crystal structure, suggesting that ARs 5–6 are compactly folded when bound to NF-κB. Thus, free IκBα is partially folded, ARs 1–4 are compactly folded and ARs 5–6 are weakly folded, but ARs 5–6 adopt a fully folded conformation only when IκBα binds to NF-κB 33.
Dynamic structures that fold upon binding to their targets are observed in many transcription factors 35; 36; 37; 38 and cell-cycle regulators 39; 40. Many eukaryotic transcription factors are predicted to have extended regions of intrinsic disorder 41. Coupled folding and binding also appears to be important for the recruitment of coactivators for transcriptional activation. Recent NMR studies elucidated the mechanism of coupled folding and binding of CREB with CBP 42, and p160 coactivators and CBP/p300 show mutual synergistic folding 43.
Despite the wealth of biophysical characterization of coupled folding and binding, the biological consequences of this process remain unclear. Folding-on-binding of the cyclin-dependent kinase (Cdk) inhibitor, p27, was shown to confer binding specificity for only Cdks that regulate cell division 44. Additional possibilities, such as facilitating rapid degradation, ability to bind multiple targets, and rapid binding kinetics, have been proposed, but functional characterizations remain elusive. We proposed that the coupled folding and binding of ARs 5–6 in IκBα might modulate the binding affinity between IκBα and NF-κB 6, and might be the switch between the basal and stimulated degradation mechanisms 33.
In experiments presented here, we took advantage of the stable consensus sequence to rationally design IκBα mutants with pre-folded ARs 5–6. We demonstrate that mutation of as few as two amino acids in AR 6 causes pre-folding of the two C-terminal ARs of IκBα. Evolution apparently selected for weakly folded sequences in ARs 5–6 in IκBα and this region confers at least two functions that are critical for proper control of NF-κB signaling: high affinity binding to NF-κB and rapid degradation of free IκBα.
Results
Rational design of stable, folded IκBα mutants
To conform AR 6 of IκBα more closely to the consensus sequence for a stable AR 28; 29, we introduced the Y254L and T257A substitutions (Figure 1), both individually and in combination, into the AR domain of IκBα (residues 67–287). These two amino acids in AR 6 do not contact NF-κB in the IκBα•NF-κB crystal structure 24; 25. The Y254L/T257A (YL/TA) substitutions were also combined with two other mutations located in ARs 4 and 5, C186P and A220P, which were previously shown to increase the overall stability by ~1.5 kcal/mol 32. The far-UV circular dichroism (CD) spectra of WT IκBα and the mutants showed no significant differences (data not shown), indicating that the secondary structure of the mutants is unchanged. To confirm that the mutations did not appreciably change the binding interface, we calculated the structure similarity factor from the NMR chemical shifts of 15N-NF-κB resonances bound to wild-type (WT) or YL/TA IκBα 45. Structural similarity factors of less than 10 Hz are considered to be insignificant and the structural similarity factor (monitoring NF-κB) that we obtained was 2.3 Hz. Thus, no significant differences in the binding interface were introduced by the mutations.
WT IκBα folding by CD shows a cooperative transition involving ARs 1–4, and a non-cooperative transition involving ARs 5–6 32. The overall stability of the proteins (ΔGCDs) obtained from the CD measurements shows that the Y254L single mutation is sufficient to stabilize the IκBα AR domain, but the T257A mutation does not change the overall stability (Figure 2 and Table 1). Although the C186P/A220P (CP/AP) mutant is more stable than WT IκBα, combination of the CP/AP mutations with YL/TA does not result in additional stability (Table 1). Folding studies of other AR domains show that increasing the number of repeats in the cooperatively folded unit does not necessarily add to the overall stability 46; 47.
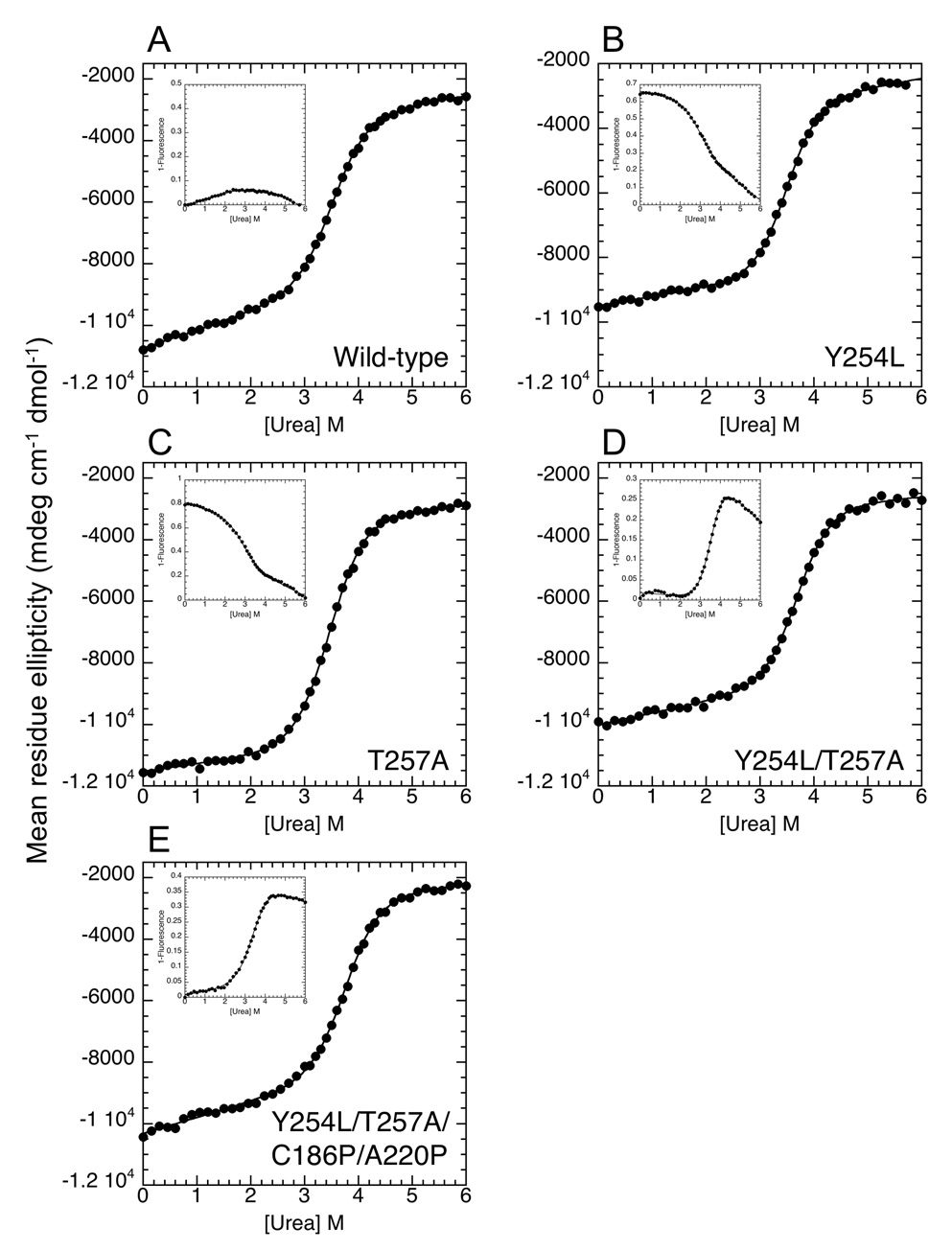
Figure 2.
Equilibrium unfolding of WT and mutant IκBα using urea as a denaturant. The CD signal and the fluorescence of the single tryptophan in IκBα, W258, located in AR 6 (insets) were recorded simultaneously for urea titrations of various IκBα proteins. The cooperative CD unfolding transition shows that the Y254L (B), Y254L/T257A (D), and Y254L/T257A/C186P/A220P (E) mutants are slightly more stable than WT (A) IκBα, but the T257A (C) mutant has the same thermodynamic stability. Only mutants containing both Y254L and T257A (D and E) show cooperative unfolding transitions in the fluorescence of W258, which is located in AR 6 (insets).
Table 1.
IκBα equilibrium folding by urea.
| Protein | ΔGCD (kcal/mol) | mCD (kcal/mol•M) | mprea (mdeg/cm•dmol•M) | ΔGFL (kcal/mol) | mFL (kcal/mol•M) |
|---|---|---|---|---|---|
| Wild-type | 6.5 ± 0.2 | 1.8 ± 0.1 | 590 ± 20 | N/Ab | N/A |
| Y254L | 7.1 ± 0.2 | 2.0 ± 0.1 | 280 ± 20 | N/A | N/A |
| T257A | 6.3 ± 0.2 | 1.8 ± 0.1 | 260 ± 30 | N/A | N/A |
| Y254L/T257A | 7.2 ± 0.3 | 2.0 ± 0.1 | 380 ± 30 | 6.7 ± 0.2 | 1.9 ± 0.1 |
| Y254L/T257A/C186P/A220P | 7.0 ± 0.3 | 1.9 ± 0.1 | 520 ± 30 | 5.0 ± 0.2 | 1.4 ± 0.1 |
| C186P/A220P c | 8.3 ± 0.8 | 2.2 ± 0.2 | 400 ± 30 | N/A | N/A |
mpre is the slope of the pre-transition baseline.
ΔGFL and mFL could not be calculated for non-cooperative folding transitions.
Taken from Ferreiro et al.32.
While CD monitors the entire AR domain, a single tryptophan, W258, in AR 6 monitors the folding transition of only the C-terminal part of the IκBα AR domain. Therefore, measuring the equilibrium folding of IκBα using CD and fluorescence signals simultaneously enables us to distinguish between the cooperative folding transition and the non-cooperative folding of ARs 5–6 (Figure 2). In WT IκBα, the CD and the W258 fluorescence both show a non-cooperative transition that can be assigned to ARs 5–6 32. Although the Y254L mutation was sufficient to increase the overall stability as measured by CD, it was not sufficient to pre-fold AR 6 as measured by a cooperative folding transition of W258. In contrast, both of the YL/TA-containing mutants did show a cooperative folding transition of W258, indicating that ARs 5–6 now fold cooperatively (Figure 2). The W258 fluorescence revealed that the C-terminal ARs in the YL/TA mutants are more stable (ΔGFLs) than those in the YL/TA/CP/AP mutant (Table 1).
IκBα mutants have compactly folded ARs 5–6
To further probe the "foldedness" of the YL/TA and YL/TA/CP/AP mutants, we measured their conformational flexibility using amide H/2H exchange, which probes the solvent accessibility of the amide protons in the protein. In non-globular proteins, regions of the protein that are compact exchange fewer amide protons, whereas regions that are weakly folded exchange more amide protons 48. The β-hairpins in free WT IκBα are compactly folded in ARs 1–4, and exchange only a few amide protons in these regions, but they are weakly folded in ARs 5–6, and exchange nearly all of their amide protons in these regions 33. All three IκBα proteins show similar exchange behavior in ARs 1–4. Remarkably, the YL/TA and YL/TA/CP/AP mutants both show much less exchange in the β-hairpins in ARs 5–6 compared to WT IκBα (Figure 3 & Supplementary Table 1).
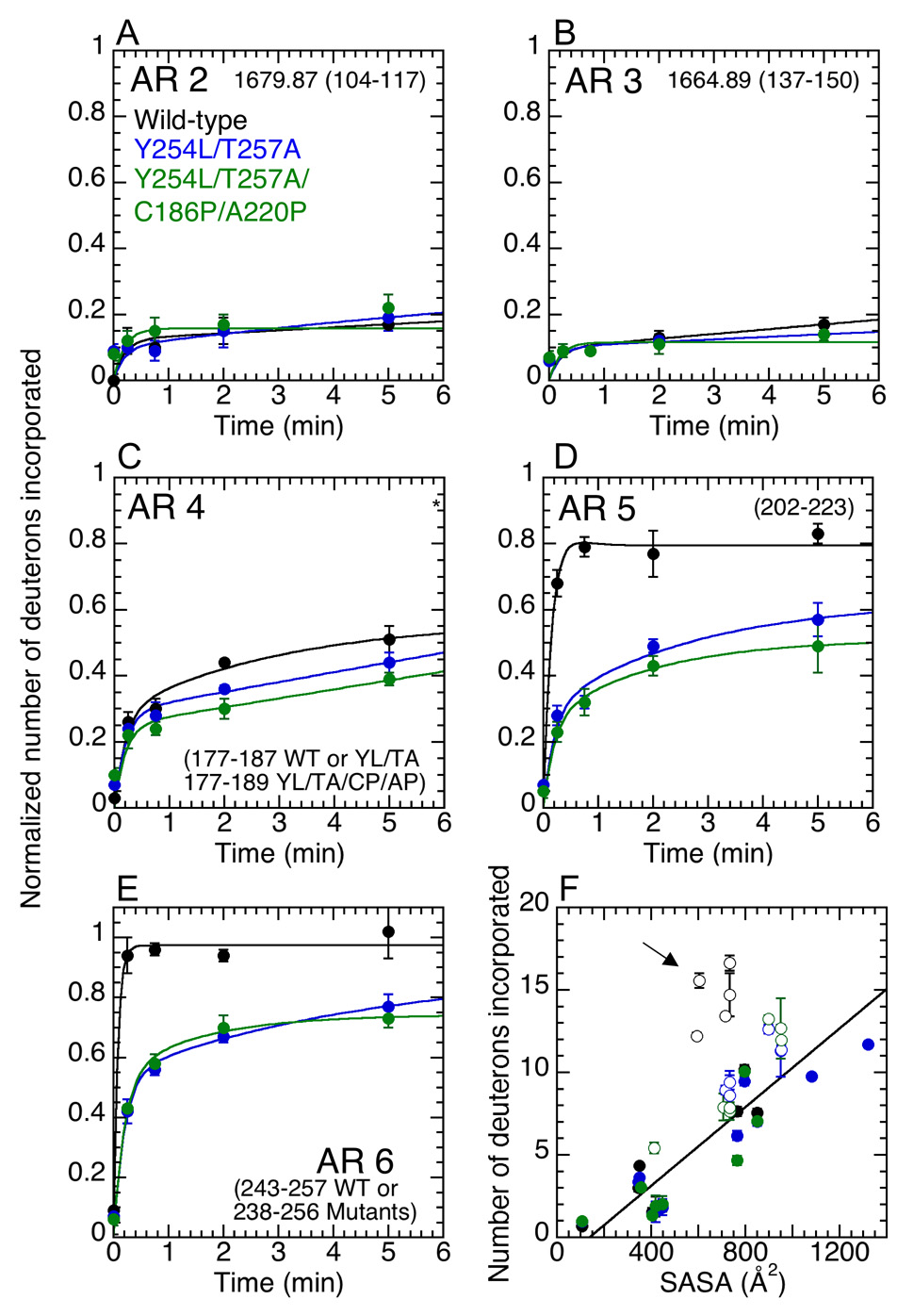
Figure 3.
Amide H/2H exchange in wild-type (black), Y254L/T257A (blue), and Y254L/T257A/C186P/A220P (green) IκBα. Deuterium incorporation in the β-hairpins in ARs 2 (A), 3 (B), and 4 (C) is similar in all three proteins; however, the β-hairpins in ARs 5 (D) and 6 (E) incorporate much less deuterium in the pre-folded mutants than in WT IκBα. The deuterium incorporation is normalized according to the number of backbone amides in the peptide. (F) The number of deuterons incorporated in each peptide in ARs 1–4 (closed circles) correlates extremely well with the calculated solvent accessible surface area (SASA) of the corresponding region of IκBα. The β-hairpins in ARs 5 and 6 (open circles) in free WT IκBα exchange to a much greater extent than predicted by their SASA (see cluster indicated by arrow), whereas the extent of exchange in these regions in the mutants are well correlated with their SASA. The average of three independent exchange experiments is reported, and the error bars represent the standard deviation of these experiments.
Calculations of the solvent accessible surface area (SASA) of regions in the protein can be used to account for the structural determinants of their exchange 48. If the extent of exchange correlates with the calculated SASA, then the structure of the region is the primary determinant of the exchange. However, exchange that is much greater than predicted by the SASA indicates conformational flexibility. SASA calculations using a model for the free IκBα structure from the IκBα•NF-κB crystal structure with NF-κB removed showed that the β-hairpins in WT IκBα ARs 5–6 exchange much more than predicted by their SASA, indicating that they are flexible in free IκBα 33. Similar analyses show that the exchange in all regions of the mutant IκBα proteins is well correlated with their SASA (Figure 3F), suggesting that the mutants adopt a folded structure similar to that of NF-κB-bound IκBα.
NMR 1H, 15N heteronuclear single quantum coherence (HSQC) spectra of WT IκBα showed only 169 of the 208 expected cross peaks, nearly all of which have been assigned to ARs 1–4 (Supplementary Figure 1). In contrast, the HSQC spectrum of the YL/TA mutant shows all of the expected cross peaks (Supplementary Figure 1). The cross peaks assigned to ARs 1–4 in WT IκBα show significant overlap with cross peaks in the YL/TA mutant spectrum. The structural similarity factor calculated for ARs 1–4 comparing WT and YL/TA IκBα of 4.1 Hz is less than 10 Hz, suggesting that the structures are similar in these regions 45. This high degree of similarity is especially striking since the presence of the folded ARs 5–6 is expected to perturb the chemical shifts of ARs 1–4 slightly. The presence of a large number of new cross peaks that likely correspond to ARs 5–6 provide additional evidence that these ARs are compactly folded in the YL/TA mutant.
Pre-folded IκBα mutants are degraded more slowly in vitro and in vivo
Robust NF-κB activation in response to extracellular signals depends on the basal degradation rate of free IκBα 20; 21. Degradation of free IκBα is independent of phosphorylation and ubiquitination and instead appears to be mediated by its C-terminal PEST sequence 20. Free IκBα degradation is ~5-times slower when the PEST sequence is deleted 20. IκBα is readily degraded in vitro with the 20S proteasome 20; 49. A common feature of ubiquitin-independent substrates of the 20S proteasome is that they require an unfolded region to initiate degradation 50. Since the C-terminal ARs are more compact in the YL/TA and YL/TA/CP/AP "pre-folded" mutants than in WT IκBα, we tested to see if the degradation of the free proteins was altered. In vitro degradation experiments utilized proteins that were purified by size-exclusion chromatography, since the presence of aggregates slows degradation (data not shown). Although WT IκBα was almost completely degraded within 30 minutes, the mutants persist for longer than 60 min (Figure 4A).
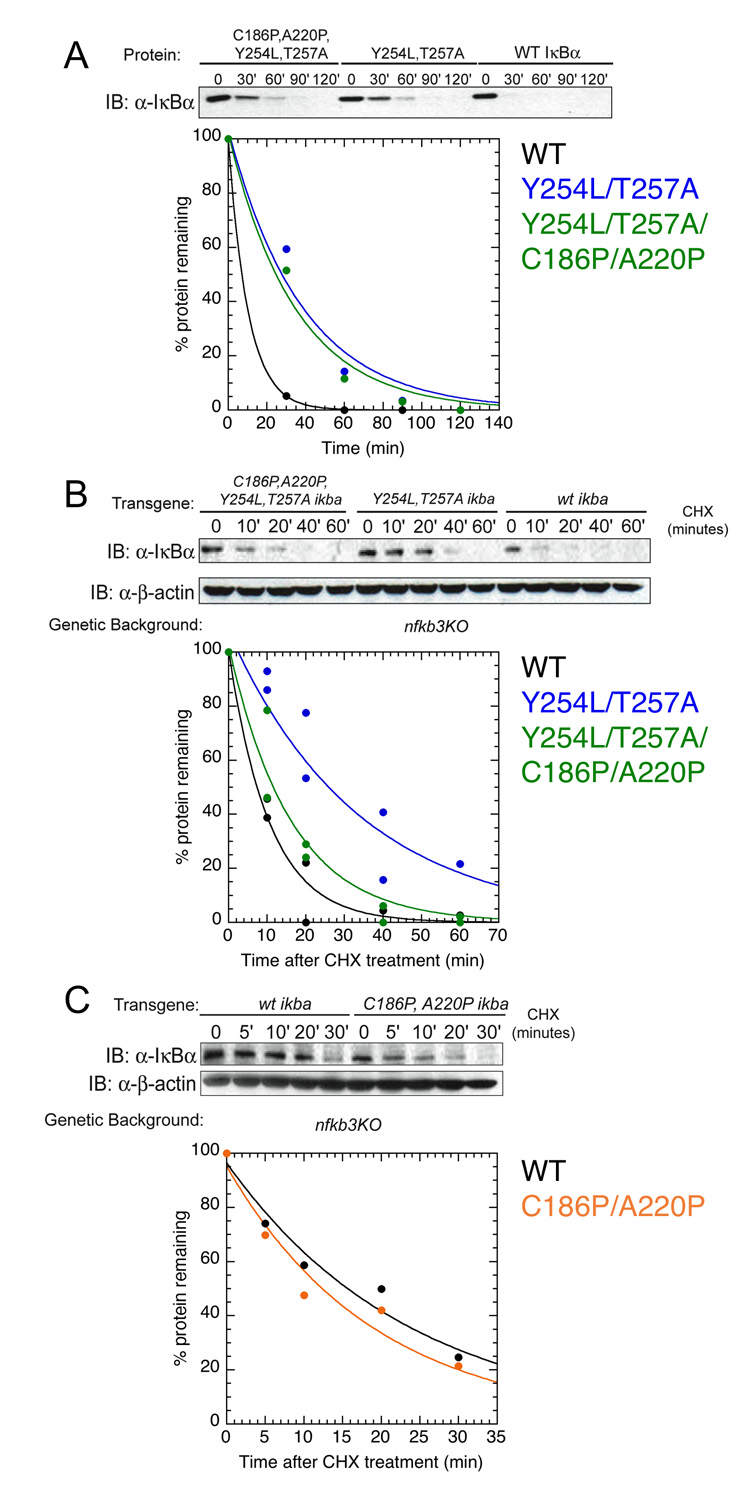
Figure 4.
Y254L/T257A (blue) and Y254L/T257A/C186P/A220P (green) are degraded more slowly than WT IκBα (black) in vitro and in vivo. (A) Purified 20S proteasome was incubated with WT and mutant IκBα and the amount of protein remaining was detected by western blot (top) and quantified by densitometry measurements (bottom). (B) Stable cell-lines containing IκBα transgenes were treated with cycloheximide (CHX) to stop translation and the amount of protein remaining over time was detected by western blot (top). Densitometry quantification of two independent experiments is shown (bottom) with a combined fit of the data. (C) The C186P/A220P mutant (orange) is degraded faster than WT IκBα (black). An α-β-actin western blot, shown in panels B and C, shows the equivalent loading of all samples.
To measure the in vivo half-lives of full-length WT, YL/TA, and YL/TA/CP/AP IκBα we introduced these transgenes into stable mouse embryonic fibroblast (MEF) cell lines deficient in the NF-κB proteins known to associate with IκBα 3 (nfkb3KO: nfkb1−/−rela−/−crel−/−), since NF-κB binding slows the degradation of IκBα 13; 20; 49. Transgenic free WT IκBα is degraded at the same rate as endogenous free IκBα 20. After treatment with cycloheximide (CHX) to stop translation, the amount of IκBα remaining was measured by western blot. WT IκBα was degraded with a half-life of ~7 min, whereas the YL/TA and YL/TA/CP/AP mutants were degraded more slowly, with half-lives of ~23 and ~11 min, respectively (Figure 4B). Importantly, the in vivo degradation rates inversely correlate with the stability of AR 6 (ΔGFL) in each protein (Figure 4B and Table 1).
Comparison of the results from the stabilized mutants allows us to show whether the proteasome degradation rate depends on overall IκBα stability or on the local stability of AR 6. The YL/TA and YL/TA/CP/AP mutants show increases in both their overall stability (ΔGCD) and the local stability of AR 6 (ΔGFL) (Figure 2 and Table 1). In contrast, the previously characterized CP/AP mutant has increased overall stability, but does not show cooperative folding of AR 6 by fluorescence 32. This mutant is degraded at approximately the same rate as WT IκBα, both by the 20S proteasome in vitro (data not shown) and in stable nfkb3KO cell lines treated with CHX in vivo (Figure 4C). This result clearly points to local stability in AR 6 as a key determinant of IκBα's susceptibility to proteasome degradation.
Pre-folded IκBα mutants bind NF-κB with reduced affinity
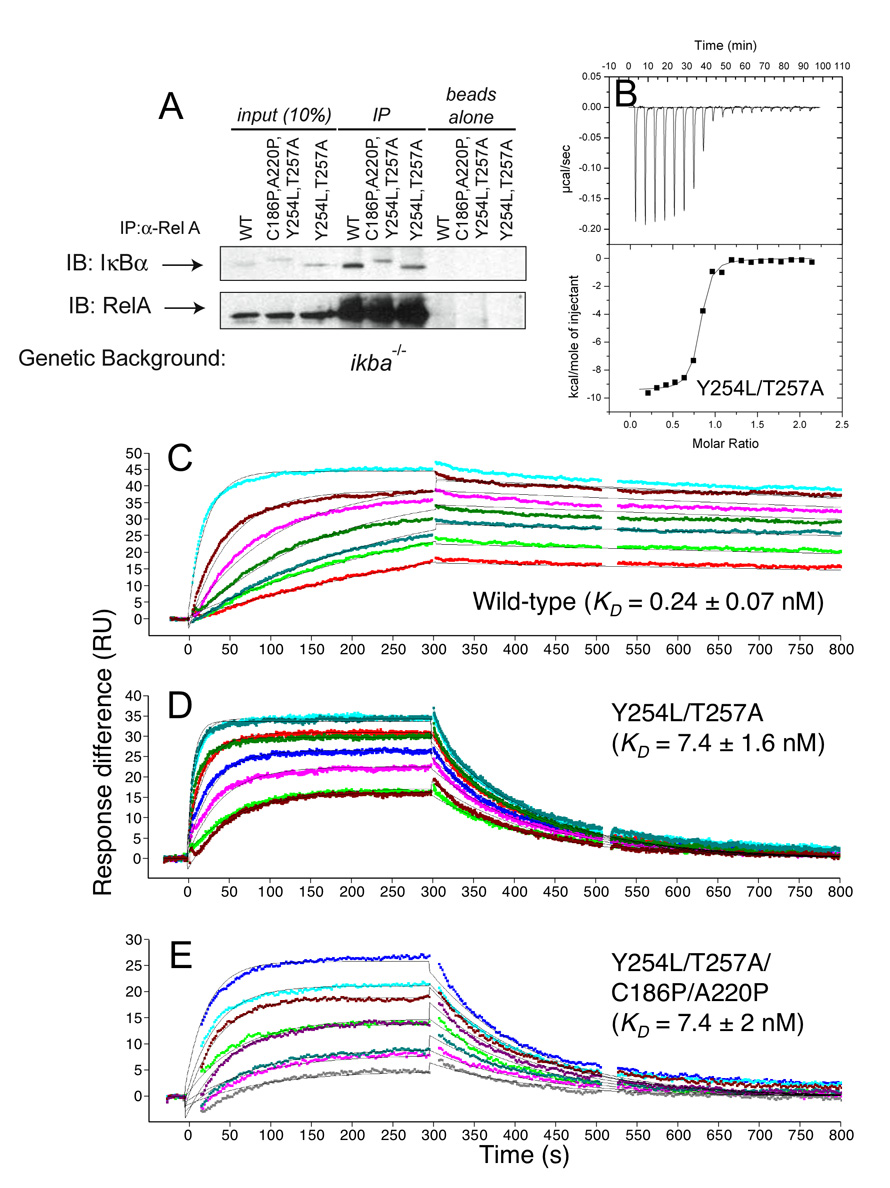
The transcriptional activity of NF-κB is highly regulated 51, in part, through the extremely tight-binding of IκBα to NF-κB 4; 5; 6. To determine whether pre-folding of IκBα alters its NF-κB binding affinity, we first introduced WT, YL/TA, and YL/TA/CP/AP IκBα into stable MEF cells lines deficient in endogenous IκBα (ikba−/−). We then immunoprecipitates RelA (p65) and measured the amount of NF-κB-bound IκBα by western blot. Consistent with their slower degradation rates (Figure 4A–B), the steady-state levels of IκBα are slightly higher for the mutants compared to WT IκBα (input samples, Figure 5A). In contrast, the levels of NF-κB-bound IκBα in the immunoprecipitated samples are much lower for the mutants than for WT IκBα (Figure 5A), indicating a significantly weaker binding affinity. Densitometric quantification of the amounts of total (input samples, Figure 5A, multiplied by 10) and NF-κB-bound IκBα (IP samples, Figure 5A) shows that the amount of free IκBα is twice as high for the pre-folded mutants.
Figure 5.
Y254L/T257A and Y254L/T257A/C186P/A220P bind more weakly than WT IκBα to NF-κB (p50248–350/p65190–321) in vitro and in vivo. (A) NF-κB (p50/p65 and p65/p65) was immunoprecipitated from lysates of stable cell-lines containing IκBα transgenes. Total IκBα (10% input samples) and NF-κB-bound IκBα (IP samples) levels were detected by western blot (top). The starting levels of IκBα are higher in the Y254L/T257A and Y254L/T257A/C186P/A220P mutants compared to WT IκBα, but much lower levels of NF-κB-bound IκBα are observed for both mutants compared to WT IκBα. The starting and immunoprecipitated levels of NF-κB (p65) are similar in all three cell-lines (bottom). There is no non-specific binding of IκBα or NF-κB to the protein G beads (beads alone samples). (B) ITC binding isotherm of NF-κB titrated into Y254L/T257A IκBα at 25 °C. Data were analyzed using a model for a single set of identical binding sites, and the observed KD is 23 nM. (C–E) Surface plasmon resonance (Biacore) was used to determine the binding kinetics of NF-κB (immobilized via an N-terminal biotin tag on the p65 subunit) with (C) wild type IκBα (at concentrations of 1.55–59.7 nM), (D) Y254L/T257A IκBα (at concentrations of 6.89–118 nM) and (E) Y254L/T257A/C186P/A220P IκBα (at concentrations of 1.40–106 nM). The pre-folded mutants both dissociate much faster than WT. Data were analyzed using a 1:1 Langmuir binding model.
We also measured the binding kinetics by surface plasmon resonance (SPR) using N-terminally biotinylated NF-κB (p50248–350/p65190–321 dimerization domains) immobilized on a streptavidin chip. All of the IκBα proteins associated with NF-κB at exactly the same rapid rate (ka = 1.1 ± 0.2 × 106 M−1s−1); however, the mutants dissociated from NF-κB much more rapidly than WT (Figure 5C–E). The YL/TA and YL/TA/CP/AP mutants dissociate 28-times faster than WT IκBα (Table 2A). The much faster dissociation of the pre-folded mutants results in reversible NF-κB binding, unlike the nearly irreversible binding seen for WT IκBα.
Table 2.
IκBα and NF-κB (p50191–321/p65248–350) binding kinetics and thermodynamics.
| A. SPR binding kinetics and affinities. | ||||
|---|---|---|---|---|
| Protein | ka (106 M−1s−1) | kd (10−3 s−1) | KD (nM) | χ2 |
| Wild-type | 1.1 ± 0.2 | 0.28 ± 0.05 | 0.24 ± 0.07 | 1.5 |
| Y254L/T257A | 1.1 ± 0.2 | 7.9 ± 0.1 | 7.4 ± 1.6 | 0.44 |
| Y254L/T257A/C186P/A220P | 1.1 ± 0.2 | 8.0 ± 0.3 | 7.4 ± 2 | 0.27 |
| B. ITC binding thermodynamics. | ||||
| Protein | KD,ITC | ΔH | −TΔS (kcal/mol) | −TΔS (kcal/mol) |
| (nM) | (kcal/mol) | (KD from ITC) | (KD from SPR) | |
| Wild-type | N/Aa | −15 | N/Aa | 1.9 |
| Y254L/T257A | 23 | −9.4 | −1.0 | −1.7 |
| Y254L/T257A/C186P/A220P | 21 | −9.8 | −0.7 | −1.5 |
The KD,ITC and corresponding −TΔS for WT IκBα binding to NF-κB could not be determined due to the high c value for the interaction, where c is defined by Wiseman et al.73.
The dissociation constants (KD) determined by isothermal titration calorimetry (ITC) for the YL/TA and YL/TA/CP/AP mutants were 23 and 21 nM, respectively (Figure 5B and Table 2B). As in previous studies 6, the affinities determined by ITC are ~3-times weaker than those determined in SPR experiments, which is within the expected range (Table 2). For both the YL/TA and YL/TA/CP/AP mutants, binding to NF-κB is mainly driven by favorable enthalpy at 298 K, but the entropy is also favorable (Table 2B). WT IκBα binding to NF-κB has a much larger favorable enthalpy; however, unlike the mutants, the entropy of binding is slightly unfavorable at 298 K (Table 2B).
NF-κB transcriptional activation
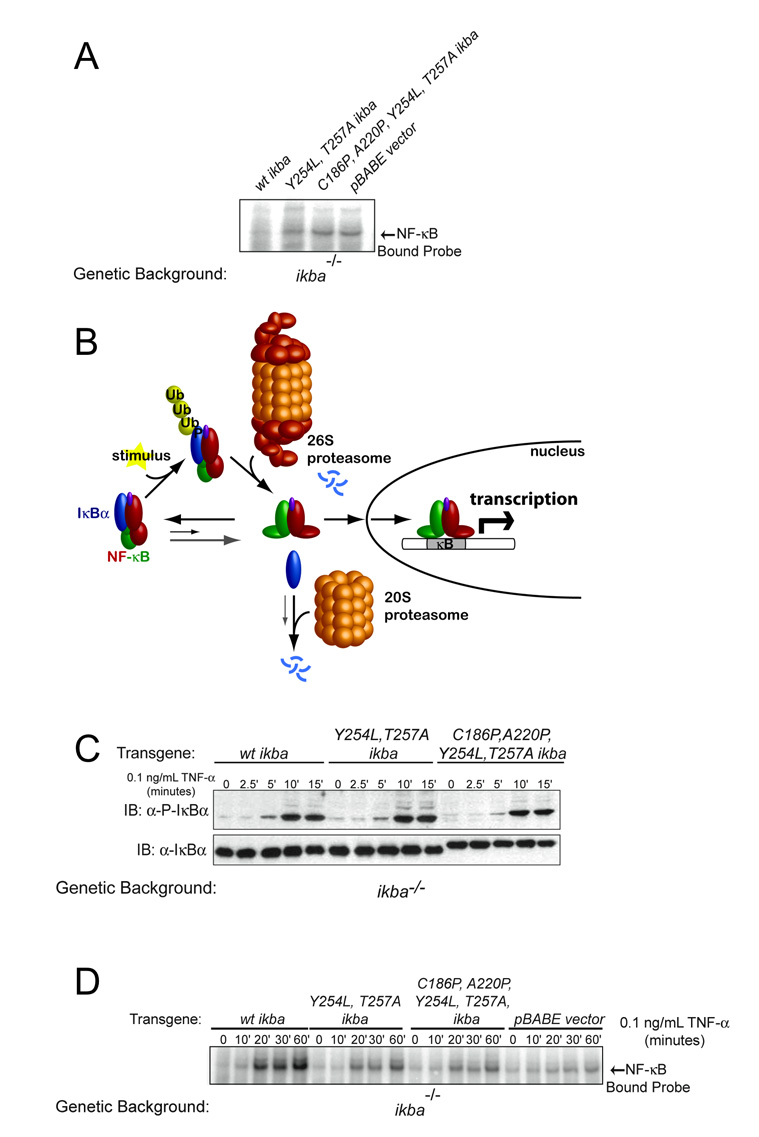
In resting cells, IκBα's extremely tight binding to NF-κB retains the transcription factor in the cytosol 4; 5; 6. We measured the levels of nuclear NF-κB in resting cells containing WT IκBα or the pre-folded mutants to determine what effects the weaker binding affinity and the slower basal degradation rates of the free pre-folded mutants have on basal NF-κB activation. NF-κB is almost completely inhibited in resting cells containing WT IκBα, as shown by the extremely small amount of nuclear NF-κB measured by electrophoretic mobility shift assays (EMSAs). In contrast, resting cells containing the pre-folded IκBα mutants have a significant amount of nuclear NF-κB, similar to that seen for cells completely lacking IκBα (pBABE vector) (Figure 6A). All of these cells still contain IκBβ and ε, which can partially, but not completely, compensate for IκBα deficiency 21; 52.
Figure 6.
Cells containing pre-folded mutants show altered amounts of nuclear NF-κB compared to WT IκBα. (A) Nuclear NF-κB levels in resting cells, measured by EMSA, show an extremely small amount of nuclear NF-κB in cells containing WT IκBα, whereas a significant amount of nuclear NF-κB is seen in cells containing the pre-folded mutants, which is equivalent to the amount seen in cells deficient in IκBα (pBABE vector). (B) Schematic outlining stimulus-induced activation of NF-κB. IκBα binds to NF-κB and, in resting cells, this prevents its nuclear localization. However, the faster dissociation rates for the pre-folded mutants (gray arrow) result in a significant amount of free IκBα and unbound NF-κB, which can translocate into the nucleus. Furthermore, free IκBα basal degradation is slower in cells containing the pre-folded mutants (gray arrow), resulting in a further increase in free IκBα levels. Upon stimulation, NF-κB-bound IκBα is phosphorylated, which initiates rapid ubiquitination and degradation by the 26S proteasome. This releases NF-κB, which can then translocate into the nucleus, bind DNA, and activate transcription. (C) Measurement of the amount of phosphorylated IκBα after stimulation with TNF-α shows that the pre-folded mutants are phosphorylated at the same rate as WT IκBα. Since phosphorylation initiates signal-dependent degradation of NF-κB-bound IκBα, we expect that the pre-folded mutants will be degraded at the same rate as WT in response to stimulus, in contrast to the slower basal degradation rates of the free pre-folded mutants. (D) Upon stimulation with TNF-α, cells containing WT IκBα show a robust increase in nuclear NF-κB, measured by EMSA. Cells containing the pre-folded mutants also show an increase in nuclear NF-κB upon stimulation; however, the response is reduced compared to cells containing WT IκBα, but higher than that observed in cells deficient in IκBα (pBABE vector).
NF-κB is activated when, in response to extracellular stimuli, the IκB that is bound to it is phosphorylated, ubiquitinated, and degraded by the proteasome (Figure 6B). We tested NF-κB activation in cells containing the pre-folded mutants, which have altered NF-κB binding affinities and basal degradation rates (for the free protein), to understand the effects of these parameters. We first sought to verify that the signal-dependent degradation rate was unaffected by the mutations. Since phosphorylation of IκBα initiates signal-dependent degradation, followed by rapid ubiquitination and proteasomal degradation of IκBα 8; 11; 53, we measured the phosphorylation rate of the different IκBα proteins in cells stimulated with 0.1 ng/mL TNF-α. Both of the pre-folded mutants are phosphorylated at the same rate as WT IκBα (Figure 6C). Therefore, any changes in the observed activation of NF-κB in cells containing the pre-folded mutants should reflect the alterations in their stability and binding affinities.
We measured the amount of nuclear NF-κB in cells containing the different IκBα proteins after stimulation with 0.1 ng/mL TNF-α. Cells containing WT IκBα show a robust increase in nuclear NF-κB levels upon stimulation, due to the signal-dependent degradation of IκBα that releases NF-κB, which translocates into the nucleus. Cells containing the pre-folded mutants also show an increase in nuclear NF-κB levels upon stimulation; however, these levels are reduced compared to those in cells containing WT IκBα (Figure 6D). This may be due to the fact that there are lower levels of NF-κB-bound IκBα in cells containing the pre-folded mutants. The nuclear NF-κB levels were higher upon stimulation in cells containing the pre-folded mutants than in cells lacking IκBα, suggesting that the IκBα mutants continue to play a role in NF-κB activation (Figure 6D).
Discussion
Like many other transcriptional activators and cell-cycle regulators that fold upon binding to their targets 54, the folding of IκBα is coupled to its binding to NF-κB 33. In most cases, the functional significance of coupled folding and binding remains a mystery. The simplicity of the ankyrin-repeat architecture of IκBα presents a unique opportunity to rationally perturb the "foldedness" of free IκBα, since many determinants of folding and stability in AR proteins are understood. In addition, the repeat architecture allows engineering of local changes in stability 55; 56. The wealth of prior characterization of the NF-κB signaling module provides the biological framework within which we should be able to interpret the functional consequences of our rational perturbations of IκBα.
Only two mutations are required to fold ARs 5–6 in IκBα
The GXTPLHLA motif is the strongest signature in the AR consensus sequence 28; 29. Mutations to this consensus stabilize AR domains, and mutations away from it destabilize them 32; 57; 58; 59; 60. IκBα deviates from this consensus signature in ARs 1, 2, 4, 5, and 6 (Figure 1B). Interestingly, these deviations are generally conserved among species. While some of these deviations are amino acids that contact NF-κB (F77, Q111, and Q255) 24; 25, many do not contact NF-κB and can be substituted without affecting NF-κB binding 32. We show here that mutation of only two residues, Y254 and T257, to their consensus counterparts causes a dramatic increase in the "foldedness" of ARs 5–6 (Figure 2 and Figure 3), which are weakly folded in free WT IκBα, but are compactly folded in NF-κB-bound IκBα 33. In similar experiments, the tetratricopeptide repeat (TPR) domain of protein phosphatase 5, which contains three repeats, required only one mutation to fold before binding 61. These results emphasize the utility of the non-globular architecture of repeat proteins to address questions of coupled folding and binding.
CD experiments show that the helical secondary structure is fully formed in free WT IκBα 34. Therefore, it seems that ARs 5–6 in free WT IκBα are poised to fold, but lack a few stabilizing interactions. While WT IκBα gains this additional "foldedness" upon interaction with NF-κB 33, two substitutions (Y254L/T257A) provide sufficient local stability for the pre-folded mutants to attain essentially the same folded structure as the NF-κB-bound form, but in the absence of NF-κB. Y254L and T257A do not contact NF-κB (Figure 1A), and the similarity of 15N-NF-κB chemical shifts when bound to WT or YL/TA IκBα suggests that the binding interface is unchanged.
While many consensus-designed AR proteins have been made 28; 29; 30; 31, it remains unclear exactly how these sequences stabilize the protein. Our data show that both the Y254L and T257A substitutions are required to pre-fold IκBα (Figure 2). The packing of residues in these conserved positions in IκBα ARs suggests that the Y254L substitution may be important for intrarepeat stabilization and the T257A substitution may be important for interrepeat stabilization.
IκBα foldedness controls its intracellular half-life
Free WT IκBα is rapidly degraded in cells, with an in vivo half-life of ~7 min (Figure 4B). In fact, it is degraded so quickly that phosphorylation and ubiquitination of free IκBα is unnecessary 20. Binding to NF-κB dramatically slows the degradation of IκBα, making phosphorylation and ubiquitination required for the degradation of NF-κB-bound IκBα. We found that "pre-folding" the C-terminal repeats of IκBα caused it to be degraded more slowly than WT IκBα (Figure 4A–B). The disordered PEST sequence C-terminal to the AR domain appears to mediate the basal degradation of free IκBα 20. Pre-folding the C-terminal ARs in the mutants may cause a conformational change in the PEST sequence or may play a direct role in influencing the degradation rate.
Comparison of the CP/AP mutant with the pre-folded mutants, which all have similar increases in overall protein stability, shows that the determining factor in susceptibility to degradation is not overall stability, but instead the local stability at the C-terminus. Local stability of specific regions in many proteins control their degradation rates 62, although in some cases the overall thermodynamic stability of the protein is also influential in the degradation process 63. The 20S proteasome core, without any regulatory subunits, degrades some proteins, including IκBα, in an ubiquitin-independent "default" degradation mechanism 64. A unifying characteristic of substrates of the 20S proteasome is the presence of unstructured regions 50. It is likely that local rather than global stability will be the predominant determinant of protein degradation for substrates of ubiquitin-independent proteasomal degradation.
IκBα foldedness controls NF-κB binding affinity
Wild-type IκBα binds extremely tightly to NF-κB, preventing its nuclear localization 4; 5; 6. Interestingly, in vitro binding kinetics and thermodynamics show that pre-folding substantially reduces the overall binding energy, which is consistent with the weaker binding that is observed for the pre-folded mutants in vivo (Figure 5). Remarkably, the overall affinities of the pre-folded mutants for NF-κB (7.4 nM) are weaker than the affinity of NF-κB for DNA (4.7 nM) 65. This result suggests that coupled folding and binding is necessary to achieve the high-affinity binding that is required for effective inhibition of NF-κB transcriptional activity.
Since folding events are generally accompanied by a large entropic penalty, our observation that the pre-folded mutants bind NF-κB with weaker affinity compared to WT IκBα may be somewhat unexpected. Indeed, a similar study of coupled folding and binding in a tetratricopeptide repeat (TPR) protein found that increases in the favorable folding enthalpy, due to the coupled folding reaction, were not realized in the binding affinity due to nearly equivalent entropic penalties 61. In our study, we find that NF-κB binding is accompanied by a much larger favorable change in enthalpy in WT IκBα compared to the pre-folded mutants (Table 2B). This additional enthalpy most likely arises from interactions within WT IκBα that are realized in the folding of ARs 5–6 that is coupled to NF-κB binding. As expected, WT IκBα binding to NF-κB is accompanied by an unfavorable change in entropy; however, the magnitude of this entropic penalty is quite small (Table 2B). This may be due to the fact that free IκBα is partially folded. Only the folding of ARs 5–6 is coupled to NF-κB binding, and ARs 5–6 in free IκBα are weakly folded, but not completely unfolded 32; 33; 34. Intriguingly, a human growth hormone (hGH) variant with a helix that is highly flexible in the unbound state, but is compactly folded in the unbound wild-type hGH, actually binds ~400-times more tightly to the cognate receptor 66. Similar to IκBα binding to NF-κB, hGH binding is enthalpically driven and the high-affinity variant shows a much larger favorable change in enthalpy for binding that is not fully compensated by its unfavorable change in entropy, resulting in the higher overall binding energy for the variant hGH. These data suggest that there may be a more complex thermodynamic balance in the binding of partially folded proteins that, in some cases, allows for an increase in binding affinity due to coupled folding and binding.
All three IκBα proteins bind to NF-κB with the same association rate, but the pre-folded mutants dissociate from NF-κB 28-times faster compared to WT IκBα (Figure 5C–E and Table 2A). Since dissociation would then require unfolding and disruption of the favorable intra-IκBα interactions, it is possible to understand how a favorable enthalpy for folding WT IκBα can result in a marked slowing of its dissociation from NF-κB. A previous investigation of the thermodynamics of two protein-protein interactions with different binding kinetics found that the dissociation of the complex was slow in the enthalpically driven interaction 67. Coupled folding and binding in IκBα appears to be optimized to slow dissociation through increased favorable enthalpy, which requires minimization of the associated entropic penalty to result in an increase in binding affinity.
IκBα foldedness controls NF-κB transcription activation
In resting cells, extremely tight binding to IκBα retains NF-κB in the cytosol, effectively eliminating NF-κB transcriptional activity 4; 5; 6. However, the weaker binding of the pre-folded IκBα mutants results in incomplete inhibition and a significant amount of nuclear NF-κB is present (Figure 6A). In fact, there is nearly as much nuclear NF-κB in cells containing the pre-folded mutants as there is in cells deficient in IκBα. This is a situation similar to that seen in Hodgkin's disease where altered forms of IκBα are unable to bind NF-κB, resulting in sustained NF-κB transcriptional activity 18.
Upon stimulation, phosphorylation, ubiquitination, and proteasomal degradation of the NF-κB-bound IκB releases NF-κB, which translocates into the nucleus 7; 8; 9; 10; 11 (Figure 6B). Accordingly, cells containing WT IκBα show a robust increase in nuclear NF-κB in response to stimulation (Figure 6D). Importantly, we found no difference in the stimulus-induced phosphorylation, which initiates signal-dependent degradation of IκBα, among any of the IκBα proteins (Figure 6C). Thus, any differences observed in the activation of NF-κB reflect their different in vivo stabilities and NF-κB binding properties. Cells containing the pre-folded mutants show an increase in nuclear NF-κB when stimulated; however, the response is reduced compared to cells containing WT IκBα (Figure 6D). This reduction is likely due to a number of factors. The amount of NF-κB-bound IκBα is lower in cells containing the pre-folded mutants (Figure 5A), due to their weaker NF-κB binding. While this leads to an increase in nuclear NF-κB levels in resting cells, IκBβ and ε probably bind some of the excess free NF-κB, since they can partially compensate for the lack of IκBα in ikba−/− cells 21; 52. Stimulus-induced degradation of IκBβ and ε also lead to NF-κB activation; however, the response is delayed compared to IκBα 21; 52. Since stimulation increases the amount of nuclear NF-κB in cells containing the pre-folded mutants compared to those containing only the empty pBABE vector, the pre-folded IκBα mutants appear to still play a role in NF-κB activation in response to stimulation (Figure 6D). IκBα mutants with truncations of their C-terminal PEST sequence show slower basal degradation rates (for the free protein), but they show no change in NF-κB binding 20. Stimulation of cells containing these degradation mutants also results in less nuclear NF-κB compared to cells containing WT IκBα, due to the higher levels of free IκBα resulting from their slower basal degradation rates 20. A similar phenomenon may be contributing to the observed weaker response to stimulation observed for the pre-folded mutants. Clearly, cells containing the pre-folded mutants show misregulation of NF-κB.
We have shown that the weakly folded C-terminal repeats of IκBα and their coupled folding and binding are required for full repression of NF-κB in resting cells and robust activation of NF-κB upon stimulation. Coupled folding and binding of WT IκBα to NF-κB, results in extremely high-affinity binding. The weakly folded C-terminal repeats of IκBα are also determinants of the rapid basal degradation rate. Both of these properties effectively eliminate free IκBα in the cell and facilitate a robust activation response upon stimulation. These results also demonstrate the diverse functional consequences of coupled folding and binding. This phenomenon allows a single protein to develop extremely different functional properties for its free and bound states, and provides a rapid mechanism to switch between these distinct functional states. Undoubtedly, coupled folding and binding will play a critical role in other highly regulated cellular signaling systems.
Materials and Methods
Protein expression and purification
Human IκBα67–287 was expressed and purified as described previously, except cultures were induced at 18 °C 6; 34. IκBα mutations were introduced using QuikChange mutagenesis 68. NF-κB p50248–350 and p65190–321, with an added N-terminal Cys, were expressed, purified, and quantified as described previously 6. IκBα protein concentrations were determined by spectrophotometry, using molar absorptivities of 12,950 M−1cm−1 for WT and T257A IκBα and 11,460 M−1cm−1 for Y254L, Y254L/T257A, and Y254L/T257A/C186P/A220P IκBα.
Cell line preparation
Full-length human IκBα (WT, YL/TA, and YL/TA/CP/AP) was introduced into immortalized 3T3 mouse embryonic fibroblasts using the pBabe-puro retroviral transgenic system 69. 293T cells (80% confluent) in Dulbecco's modified Eagle's medium (DMEM) were transiently transfected with 20 µL Lipofectamine 2000 (Invitrogen). Retroviral vector (8 µg) was co-transfected with 3 µg pCl-Eco (Imgenex). After 3 hours of growth, the media was changed and these cells were allowed to grow for 40–48 hours in DMEM supplemented with penicillin/streptomycin/gentamycin (Invitrogen) and 10% fetal bovine serum. The supernatant was then filtered and placed onto the target 40–50% confluent 3T3 mouse embryonic fibroblasts (ikba−/− or nfkb3KO: nfkb1−/−rela−/−crel−/−) along with 8 µg/mL polybrene (Sigma) in DMEM supplemented with penicillin/streptomycin/gentamycin (Invitrogen) and 10% bovine calf serum. These cells grew for another 48 hours before selection with 10 µg/mL puromycin (Calbiochem). Cells containing the transgenes were grown in DMEM supplemented with penicillin/streptomycin/gentamycin and 10% bovine calf serum. Protein concentrations of cellular extracts and nuclear extracts were determined by Bradford assays.
Equilibrium folding
Equilibrium folding curves were measured using circular dichroism and fluorescence simultaneously as described previously 32, except 2 µM IκBα was used in all cases. Additionally, 50 mL of 8 M urea was deionized by stirring for 1 hr with 2.5 g AG501X8 mixed bed resin (BioRad). After removal of the resin, buffer was added to a final concentration of 25 mM Tris, 50 mM NaCl, 0.5 mM EDTA, 1 mM DTT (pH 7.5). The urea concentration in the denatured samples (7.21–7.39 M) was determined by refractometry 70. Folding curves were fit to a two-state folding model, where the pre- and post-transition baselines were treated with a linear dependence on the concentration of denaturant, as described previously 32.
Amide H/2H exchange
Exchange reactions were performed as described previously, except the reactions proceeded for 0, 0.25, 0.75, 2, or 5 min 32. For WT IκBα, 14 peptides that cover 60% of the sequence were analyzed. For YL/TA IκBα, 17 peptides yielded 70% coverage. For YL/TA/CP/AP IκBα, 14 peptides yielded 63% coverage. The β-hairpin in AR 4 contains the C186, which is mutated to a proline in the YL/TA/CP/AP mutant, and it results in a peptide that covers two extra residues (188–189) of the protein. All peptides are listed in Supplementary Table 1. The solvent accessible surface area (SASA) of each peptide was calculated as described previously 48, using the structure from the IκBα•NF-κB crystal structure with NF-κB removed as a model 24. Similar correlations are observed whether or not mutated peptides are included, even though SASA was calculated from the WT protein model.
NMR spectroscopy
Free 15N-IκBα67–287 (0.1 mM) or 0.08 mM 2H-15N p50248–350/p65190–321 bound to 0.1 mM IκBα67–287 were prepared in 25 mM Tris, 50 mM NaCl, 0.5 mM EDTA, 2 mM DTT, and 2 mM NaN3 at pH 7.5 in 90% H2O/10% D2O. 1H-15N TROSY-HSQC NMR spectra were acquired at 20 °C on Bruker AVANCE 750 and Bruker AVANCE 800 spectrometers. Spectra were processed with NMRPipe 71 and analyzed with NMRView 72.
Proteasome degradation assay
In vitro experiments were performed with human 20S proteasome (a gift from Drs. Rechsteiner and Pratt, University of Utah). IκBα67–287 (1 µM), purified by size exclusion chromatography within 30 hours, was incubated with 20S proteasome (56 nM) for 0, 30, 60, 90, or 120 min at 25 °C in 20 mM Tris, 200 mM NaCl, 10 mM MgCl2, 1 mM DTT (pH 7.0). Degradation reactions were quenched by boiling with SDS-PAGE sample buffer. Intact IκBα was separated by 12.5% SDS-PAGE and visualized using western blots probed with sc-847 (Santa Cruz Biotechnologies) followed by anti-rabbit HRP conjugate. Reactions without proteasome did not degrade (data not shown). Densitometry measurements were performed using ImageQuant TL (GE Healthcare).
Full-length IκBα transgenes were introduced into mouse embryonic fibroblasts deficient in the NF-κB proteins known to associate with it (nfkb3KO: nfkb1−/−rela−/−crel−/−), since NF-κB binding slows the degradation of IκBα 13; 20; 49. Cells were grown to 70% confluency and treated with 10 µg/mL cycloheximide resuspended in 50% ethanol. Cells were washed twice with ice-cold phosphate buffered saline and lysed in 100 µL of 20 mM Tris (pH 7.5), 200 mM NaCl, 1% TritonX, 2 mM DTT, 5 mM p-nitrophenylphosphate, 2 mM sodium phosphate, 1 mM phenylmethanesulphonylfluoride, and protease inhibitor cocktail. Cell extract (50 µg) was separated using 12.5% SDS-PAGE and visualized using western blots probed with sc-371 (Santa Cruz Biotechnologies) followed by anti-rabbit HRP conjugate. Densitometry measurements were performed using ImageQuant TL (GE Healthcare), and IκBα measurements were normalized for loading using the α-β-actin control.
Immunoprecipitation
NF-κB was immunoprecipitated from mouse embryonic fibroblasts deficient in endogenous IκBα (ikba−/−) containing full-length IκBα transgenes grown to 95% confluency. Cell lysates were prepared as described above, and 500 µg total cellular protein was treated with sc-372-G (Santa Cruz Biotechnologies) overnight. Immunoprecipitates were captured with protein G agarose (Upstate), washed three times with 10 mM Tris (pH 7.5), 150 mM NaCl, and 1% TritonX, and analyzed by SDS-PAGE. IκBα and NF-κB were visualized by western blot, probed with sc-371 and sc-372 (Santa Cruz Biotechnologies) followed by anti-rabbit HRP conjugate. Densitometry measurements were performed using ImageQuant TL (GE Healthcare).
Surface plasmon resonance
Sensorgrams were recorded on a Biacore 3000 instrument using streptavidin chips as described previously 6. NF-κB was biotinylated and immobilized as described previously 6. 150, 250, and 350 RU of NF-κB (p50248–350/p65190–321) were immobilized. For NF-κB (p50/p65) binding, WT IκBα67–287 (1.55–59.7 nM) was injected for 5 min and dissociation was measured for 20 min at 25 °C at 50 µL/min. Regeneration was achieved by a 1 min pulse of 3 M urea in 0.5X running buffer, as determined by repeat injections. YL/TA IκBα67–287 (6.89–118 nM) and YL/TA/CP/AP IκBα67–287 (1.40–106 nM) were injected for 5 min, dissociation was measured for 15 min at 25 °C and 50 µL/min, and no regeneration was required.
Isothermal titration calorimetry
ITC experiments for IκBα67–287 binding to NF-κB (p50248–350/p65190–321) were performed as described previously 6. The KD,obs for WT IκBα binding to NF-κB could not be determined due to the high c value for the interaction, where c is defined by Wiseman et al. 73, therefore the value of −TΔS was calculated from the affinity obtained by SPR.
IκBα phosphorylation assay
Mouse embryonic fibroblasts deficient in endogenous IκBα (ikba−/−) containing full-length human IκBα transgenes were grown to 95% confluency. Cells were stimulated with 0.1 ng/mL TNF-α, and lysed as described above. Cell extract (50 µg) was separated using 12.5% SDS-PAGE and visualized using western blots probed with 5a5 (antibody for S32/36 phosphorylated IκBα from Cell signaling) and sc-371 (antibody for total IκBα from Santa Cruz Biotechnologies) followed by anti-mouse and anti-rabbit HRP conjugate, respectively. Densitometry measurements were performed using ImageQuant TL (GE Healthcare), and phosphorylated IκBα measurements were normalized according to the total IκBα level in each sample.
Electrophoretic mobility shift assay
EMSAs were performed as described previously 17, except cells were grown to confluency, stimulated with 0.1 ng/mL TNF-α and 6 µg of nuclear protein was used.
Supplementary Material
Acknowledgments
We thank A. Hoffmann, A. Derman, A. Shiau, M. Guttman, D. Ferreiro, M. Beach, and S. Bergqvist for many helpful discussions. Human 20S proteasome was a generous gift from Drs. Rechsteiner and Pratt (University of Utah). SMET was supported by the Irvington Institute Fellowship Program of the Cancer Research Institute. EM was supported by the Heme training grant T32DK007233. Research funding was provided by NIH grant GM071862.
Abbreviations
- AR
ankyrin repeat
- WT
wild-type
- YL/TA
Y254L/T257A
- CP/AP
C186P/A220P
- YL/TA/CP/AP
Y254L/T257A/C186P/A220P
- CD
circular dichroism
- SASA
solvent accessible surface area
- HSQC
heteronuclear single quantum coherence
- MEF
mouse embryonic fibroblast
- CHX
cycloheximide
- SPR
surface plasmon resonance
- ITC
isothermal titration calorimetry
- EMSA
electrophoretic mobility shift assay
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ghosh S, May MJ, Kopp EB. NF-kappa B and Rel proteins: evolutionarily conserved mediators of immune responses. Annu. Rev. Immunol. 1998;16:225–260. doi: 10.1146/annurev.immunol.16.1.225. [DOI] [PubMed] [Google Scholar]
- 2.Kumar A, Takada Y, Boriek AM, Aggarwal BB. Nuclear factor-kappaB: its role in health and disease. J. Mol. Med. 2004;82:434–448. doi: 10.1007/s00109-004-0555-y. [DOI] [PubMed] [Google Scholar]
- 3.Verma IM, Stevenson JK, Schwarz EM, Van Antwerp D, Miyamoto S. Rel/NF-kappa B/I kappa B family: intimate tales of association and dissociation. Genes Dev. 1995;9:2723–2735. doi: 10.1101/gad.9.22.2723. [DOI] [PubMed] [Google Scholar]
- 4.Baeuerle PA, Baltimore D. I kappa B: a specific inhibitor of the NF-kappa B transcription factor. Science. 1988;242:540–546. doi: 10.1126/science.3140380. [DOI] [PubMed] [Google Scholar]
- 5.Baldwin AS., Jr The NF-kappa B and I kappa B proteins: new discoveries and insights. Annu. Rev. Immunol. 1996;14:649–683. doi: 10.1146/annurev.immunol.14.1.649. [DOI] [PubMed] [Google Scholar]
- 6.Bergqvist S, Croy CH, Kjaergaard M, Huxford T, Ghosh G, Komives EA. Thermodynamics reveal that helix four in the NLS of NF-kappaB p65 anchors IkappaBalpha, forming a very stable complex. J. Mol. Biol. 2006;360:421–434. doi: 10.1016/j.jmb.2006.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Traenckner EB, Baeuerle PA. Appearance of apparently ubiquitin-conjugated I kappa B-alpha during its phosphorylation-induced degradation in intact cells. J. Cell Sci. Suppl. 1995;19:79–84. doi: 10.1242/jcs.1995.supplement_19.11. [DOI] [PubMed] [Google Scholar]
- 8.Traenckner EB, Pahl HL, Henkel T, Schmidt KN, Wilk S, Baeuerle PA. Phosphorylation of human I kappa B-alpha on serines 32 and 36 controls I kappa B-alpha proteolysis and NF-kappa B activation in response to diverse stimuli. EMBO J. 1995;14:2876–2883. doi: 10.1002/j.1460-2075.1995.tb07287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Traenckner EB, Wilk S, Baeuerle PA. A proteasome inhibitor prevents activation of NF-kappa B and stabilizes a newly phosphorylated form of I kappa B-alpha that is still bound to NF-kappa B. EMBO J. 1994;13:5433–5441. doi: 10.1002/j.1460-2075.1994.tb06878.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen ZJ, Parent L, Maniatis T. Site-specific phosphorylation of IkappaBalpha by a novel ubiquitination-dependent protein kinase activity. Cell. 1996;84:853–862. doi: 10.1016/s0092-8674(00)81064-8. [DOI] [PubMed] [Google Scholar]
- 11.Brown K, Franzoso G, Baldi L, Carlson L, Mills L, Lin YC, Gerstberger S, Siebenlist U. The signal response of IkappaB alpha is regulated by transferable N- and C-terminal domains. Mol. Cell. Biol. 1997;17:3021–3027. doi: 10.1128/mcb.17.6.3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pahl HL. Activators and target genes of Rel/NF-kappaB transcription factors. Oncogene. 1999;18:6853–6866. doi: 10.1038/sj.onc.1203239. [DOI] [PubMed] [Google Scholar]
- 13.Scott ML, Fujita T, Liou HC, Nolan GP, Baltimore D. The p65 subunit of NF-kappa B regulates I kappa B by two distinct mechanisms. Genes Dev. 1993;7:1266–1276. doi: 10.1101/gad.7.7a.1266. [DOI] [PubMed] [Google Scholar]
- 14.Brown K, Park S, Kanno T, Franzoso G, Siebenlist U. Mutual regulation of the transcriptional activator NF-kappa B and its inhibitor, I kappa B-alpha. Proc. Natl. Acad. Sci. U. S. A. 1993;90:2532–2536. doi: 10.1073/pnas.90.6.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun SC, Ganchi PA, Ballard DW, Greene WC. NF-kappa B controls expression of inhibitor I kappa B alpha: evidence for an inducible autoregulatory pathway. Science. 1993;259:1912–1915. doi: 10.1126/science.8096091. [DOI] [PubMed] [Google Scholar]
- 16.de Martin R, Vanhove B, Cheng Q, Hofer E, Csizmadia V, Winkler H, Bach FH. Cytokine-inducible expression in endothelial cells of an I kappa B alpha-like gene is regulated by NF kappa B. EMBO J. 1993;12:2773–2779. doi: 10.1002/j.1460-2075.1993.tb05938.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoffmann A, Levchenko A, Scott ML, Baltimore D. The IkappaB-NF-kappaB signaling module: temporal control and selective gene activation. Science. 2002;298:1241–1245. doi: 10.1126/science.1071914. [DOI] [PubMed] [Google Scholar]
- 18.Lee CH, Jeon YT, Kim SH, Song YS. NF-kappaB as a potential molecular target for cancer therapy. Biofactors. 2007;29:19–35. doi: 10.1002/biof.5520290103. [DOI] [PubMed] [Google Scholar]
- 19.Krappmann D, Wulczyn FG, Scheidereit C. Different mechanisms control signal-induced degradation and basal turnover of the NF-kappaB inhibitor IkappaB alpha in vivo. EMBO J. 1996;15:6716–6726. [PMC free article] [PubMed] [Google Scholar]
- 20.Mathes E, O'Dea E, Hoffmann A, Ghosh G. NF-kappaB dictates the degradation pathway of IkappaBalpha. EMBO J. doi: 10.1038/emboj.2008.73. Manuscript in revision. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O'Dea EL, Barken D, Peralta RQ, Tran KT, Werner SL, Kearns JD, Levchenko A, Hoffmann A. A homeostatic model of IkappaB metabolism to control constitutive NF-kappaB activity. Mol. Syst. Biol. 2007;3:111. doi: 10.1038/msb4100148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rice NR, Ernst MK. In vivo control of NF-kappa B activation by I kappa B alpha. EMBO J. 1993;12:4685–4695. doi: 10.1002/j.1460-2075.1993.tb06157.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O'Dea EL, Kearns JD, Hoffmann A. UV as an amplifier rather than inducer of NF-kappaB activity. Mol. Cell. doi: 10.1016/j.molcel.2008.03.017. Manuscript in revision. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jacobs MD, Harrison SC. Structure of an IkappaBalpha/NF-kappaB complex. Cell. 1998;95:749–758. doi: 10.1016/s0092-8674(00)81698-0. [DOI] [PubMed] [Google Scholar]
- 25.Huxford T, Huang DB, Malek S, Ghosh G. The crystal structure of the IkappaBalpha/NF-kappaB complex reveals mechanisms of NF-kappaB inactivation. Cell. 1998;95:759–770. doi: 10.1016/s0092-8674(00)81699-2. [DOI] [PubMed] [Google Scholar]
- 26.Mosavi LK, Cammett TJ, Desrosiers DC, Peng ZY. The ankyrin repeat as molecular architecture for protein recognition. Protein Sci. 2004;13:1435–1448. doi: 10.1110/ps.03554604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li J, Mahajan A, Tsai MD. Ankyrin repeat: a unique motif mediating protein-protein interactions. Biochemistry. 2006;45:15168–15178. doi: 10.1021/bi062188q. [DOI] [PubMed] [Google Scholar]
- 28.Kohl A, Binz HK, Forrer P, Stumpp MT, Pluckthun A, Grutter MG. Designed to be stable: crystal structure of a consensus ankyrin repeat protein. Proc. Natl. Acad. Sci. U. S. A. 2003;100:1700–1705. doi: 10.1073/pnas.0337680100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mosavi LK, Minor DL, Jr, Peng ZY. Consensus-derived structural determinants of the ankyrin repeat motif. Proc. Natl. Acad. Sci. U. S. A. 2002;99:16029–16034. doi: 10.1073/pnas.252537899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Binz HK, Amstutz P, Kohl A, Stumpp MT, Briand C, Forrer P, Grutter MG, Pluckthun A. High-affinity binders selected from designed ankyrin repeat protein libraries. Nat. Biotechnol. 2004;22:575–582. doi: 10.1038/nbt962. [DOI] [PubMed] [Google Scholar]
- 31.Mosavi LK, Peng ZY. Structure-based substitutions for increased solubility of a designed protein. Protein Eng. 2003;16:739–745. doi: 10.1093/protein/gzg098. [DOI] [PubMed] [Google Scholar]
- 32.Ferreiro DU, Cervantes CF, Truhlar SM, Cho SS, Wolynes PG, Komives EA. Stabilizing IkappaBalpha by "consensus" design. J. Mol. Biol. 2007;365:1201–1216. doi: 10.1016/j.jmb.2006.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Truhlar SM, Torpey JW, Komives EA. Regions of IkappaBalpha that are critical for its inhibition of NF-kappaB.DNA interaction fold upon binding to NF-kappaB. Proc. Natl. Acad. Sci. U. S. A. 2006;103:18951–18956. doi: 10.1073/pnas.0605794103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Croy CH, Bergqvist S, Huxford T, Ghosh G, Komives EA. Biophysical characterization of the free IkappaBalpha ankyrin repeat domain in solution. Protein Sci. 2004;13:1767–1777. doi: 10.1110/ps.04731004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Love JJ, Li X, Chung J, Dyson HJ, Wright PE. The LEF-1 high-mobility group domain undergoes a disorder-to-order transition upon formation of a complex with cognate DNA. Biochemistry. 2004;43:8725–8734. doi: 10.1021/bi049591m. [DOI] [PubMed] [Google Scholar]
- 36.Kumar R, Betney R, Li J, Thompson EB, McEwan IJ. Induced alpha-helix structure in AF1 of the androgen receptor upon binding transcription factor TFIIF. Biochemistry. 2004;43:3008–3013. doi: 10.1021/bi035934p. [DOI] [PubMed] [Google Scholar]
- 37.Lee BM, Xu J, Clarkson BK, Martinez-Yamout MA, Dyson HJ, Case DA, Gottesfeld JM, Wright PE. Induced fit and "lock and key" recognition of 5S RNA by zinc fingers of transcription factor IIIA. J. Mol. Biol. 2006;357:275–291. doi: 10.1016/j.jmb.2005.12.010. [DOI] [PubMed] [Google Scholar]
- 38.Radhakrishnan I, Perez-Alvarado GC, Parker D, Dyson HJ, Montminy MR, Wright PE. Solution structure of the KIX domain of CBP bound to the transactivation domain of CREB: a model for activator:coactivator interactions. Cell. 1997;91:741–752. doi: 10.1016/s0092-8674(00)80463-8. [DOI] [PubMed] [Google Scholar]
- 39.Lacy ER, Filippov I, Lewis WS, Otieno S, Xiao L, Weiss S, Hengst L, Kriwacki RW. p27 binds cyclin-CDK complexes through a sequential mechanism involving binding-induced protein folding. Nat. Struct. Mol. Biol. 2004;11:358–364. doi: 10.1038/nsmb746. [DOI] [PubMed] [Google Scholar]
- 40.Kriwacki RW, Hengst L, Tennant L, Reed SI, Wright PE. Structural studies of p21 Waf1/Cip1/Sdi1 in the free and Cdk2-bound state: conformational disorder mediates binding diversity. Proc. Natl. Acad. Sci. U. S. A. 1996;93:11504–11509. doi: 10.1073/pnas.93.21.11504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu J, Perumal NB, Oldfield CJ, Su EW, Uversky VN, Dunker AK. Intrinsic disorder in transcription factors. Biochemistry. 2006;45:6873–6888. doi: 10.1021/bi0602718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sugase K, Dyson HJ, Wright PE. Mechanism of coupled folding and binding of an intrinsically disordered protein. Nature. 2007;447:1021–1025. doi: 10.1038/nature05858. [DOI] [PubMed] [Google Scholar]
- 43.Demarest SJ, Martinez-Yamout M, Chung J, Chen H, Xu W, Dyson HJ, Evans RM, Wright PE. Mutual synergistic folding in recruitment of CBP/p300 by p160 nuclear receptor coactivators. Nature. 2002;415:549–553. doi: 10.1038/415549a. [DOI] [PubMed] [Google Scholar]
- 44.Lacy ER, Wang Y, Post J, Nourse A, Webb W, Mapelli M, Musacchio A, Siuzdak G, Kriwacki RW. Molecular basis for the specificity of p27 toward cyclin-dependent kinases that regulate cell division. J. Mol. Biol. 2005;349:764–773. doi: 10.1016/j.jmb.2005.04.019. [DOI] [PubMed] [Google Scholar]
- 45.Camarero JA, Shekhtman A, Campbell EA, Chlenov M, Gruber TM, Bryant DA, Darst SA, Cowburn D, Muir TW. Autoregulation of a bacterial sigma factor explored by using segmental isotopic labeling and NMR. Proc. Natl. Acad. Sci. U. S. A. 2002;99:8536–8541. doi: 10.1073/pnas.132033899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tripp KW, Barrick D. The tolerance of a modular protein to duplication and deletion of internal repeats. J. Mol. Biol. 2004;344:169–178. doi: 10.1016/j.jmb.2004.09.038. [DOI] [PubMed] [Google Scholar]
- 47.Ferreiro DU, Cho SS, Komives EA, Wolynes PG. The energy landscape of modular repeat proteins: topology determines folding mechanism in the ankyrin family. J. Mol. Biol. 2005;354:679–692. doi: 10.1016/j.jmb.2005.09.078. [DOI] [PubMed] [Google Scholar]
- 48.Truhlar SME, Croy CH, Torpey JW, Koeppe JR, Komives EA. Solvent accessibility of protein surfaces by amide H/2H exchange MALDI-TOF mass spectrometry. J. Am. Soc. Mass Spectrom. 2006;17:1490–1497. doi: 10.1016/j.jasms.2006.07.023. [DOI] [PubMed] [Google Scholar]
- 49.Alvarez-Castelao B, Castano JG. Mechanism of direct degradation of IkappaBalpha by 20S proteasome. FEBS Lett. 2005;579:4797–4802. doi: 10.1016/j.febslet.2005.07.060. [DOI] [PubMed] [Google Scholar]
- 50.Liu CW, Corboy MJ, DeMartino GN, Thomas PJ. Endoproteolytic activity of the proteasome. Science. 2003;299:408–411. doi: 10.1126/science.1079293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tergaonkar V, Correa RG, Ikawa M, Verma IM. Distinct roles of IkappaB proteins in regulating constitutive NF-kappaB activity. Nat. Cell Biol. 2005;7:921–923. doi: 10.1038/ncb1296. [DOI] [PubMed] [Google Scholar]
- 52.Kearns JD, Basak S, Werner SL, Huang CS, Hoffmann A. IkappaBepsilon provides negative feedback to control NF-kappaB oscillations, signaling dynamics, and inflammatory gene expression. J. Cell Biol. 2006;173:659–664. doi: 10.1083/jcb.200510155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brown K, Gerstberger S, Carlson L, Franzoso G, Siebenlist U. Control of I kappa B-alpha proteolysis by site-specific, signal-induced phosphorylation. Science. 1995;267:1485–1488. doi: 10.1126/science.7878466. [DOI] [PubMed] [Google Scholar]
- 54.Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 55.Kloss E, Courtemanche N, Barrick D. Repeat protein folding: New insights into origins of cooperativity, stability, and topology. Arch. Biochem. Biophys. 2008;469:83–99. doi: 10.1016/j.abb.2007.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barrick D, Ferreiro DU, Komives EA. Folding landscapes of ankyrin repeat proteins: experiments meet theory. Curr. Opin. Struct. Biol. 2008 doi: 10.1016/j.sbi.2007.12.004. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zweifel ME, Leahy DJ, Hughson FM, Barrick D. Structure and stability of the ankyrin domain of the Drosophila Notch receptor. Protein Sci. 2003;12:2622–2632. doi: 10.1110/ps.03279003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lowe AR, Itzhaki LS. Rational redesign of the folding pathway of a modular protein. Proc. Natl. Acad. Sci. U. S. A. 2007;104:2679–2684. doi: 10.1073/pnas.0604653104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tang KS, Guralnick BJ, Wang WK, Fersht AR, Itzhaki LS. Stability and folding of the tumour suppressor protein p16. J. Mol. Biol. 1999;285:1869–1886. doi: 10.1006/jmbi.1998.2420. [DOI] [PubMed] [Google Scholar]
- 60.Tripp KW, Barrick D. Enhancing the stability and folding rate of a repeat protein through the addition of consensus repeats. J. Mol. Biol. 2007;365:1187–1200. doi: 10.1016/j.jmb.2006.09.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cliff MJ, Williams MA, Brooke-Smith J, Barford D, Ladbury JE. Molecular recognition via coupled folding and binding in a TPR domain. J. Mol. Biol. 2005;346:717–732. doi: 10.1016/j.jmb.2004.12.017. [DOI] [PubMed] [Google Scholar]
- 62.Penrose KJ, Garcia-Alai M, de Prat-Gay G, McBride AA. Casein Kinase II phosphorylation-induced conformational switch triggers degradation of the papillomavirus E2 protein. J. Biol. Chem. 2004;279:22430–22439. doi: 10.1074/jbc.M314340200. [DOI] [PubMed] [Google Scholar]
- 63.Parsell DA, Sauer RT. The structural stability of a protein is an important determinant of its proteolytic susceptibility in Escherichia coli. J. Biol. Chem. 1989;264:7590–7595. [PubMed] [Google Scholar]
- 64.Asher G, Reuven N, Shaul Y. 20S proteasomes and protein degradation "by default". Bioessays. 2006;28:844–849. doi: 10.1002/bies.20447. [DOI] [PubMed] [Google Scholar]
- 65.Malek S, Huxford T, Ghosh G. Ikappa Balpha functions through direct contacts with the nuclear localization signals and the DNA binding sequences of NF-kappaB. J. Biol. Chem. 1998;273:25427–25435. doi: 10.1074/jbc.273.39.25427. [DOI] [PubMed] [Google Scholar]
- 66.Horn JR, Kraybill B, Petro EJ, Coales SJ, Morrow JA, Hamuro Y, Kossiakoff AA. The role of protein dynamics in increasing binding affinity for an engineered protein-protein interaction established by H/D exchange mass spectrometry. Biochemistry. 2006;45:8488–8498. doi: 10.1021/bi0604328. [DOI] [PubMed] [Google Scholar]
- 67.Baerga-Ortiz A, Bergqvist S, Mandell JG, Komives EA. Two different proteins that compete for binding to thrombin have opposite kinetic and thermodynamic profiles. Protein Sci. 2004;13:166–176. doi: 10.1110/ps.03120604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Papworth C, Bauer JC, Braman J, Wright DA. Site-directed mutagenesis in one day with >80% efficiency. Strategies. 1996;8:3–4. [Google Scholar]
- 69.Morgenstern JP, Land H. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pace CN. Determination and analysis of urea and guanidine hydrochloride denaturation curves. Methods Enzymol. 1986;131:266–280. doi: 10.1016/0076-6879(86)31045-0. [DOI] [PubMed] [Google Scholar]
- 71.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 72.Johnson BA, Blevins RA. Nmr View - a Computer-Program for the Visualization and Analysis of Nmr Data. J. Biomol. NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 73.Wiseman T, Williston S, Brandts JF, Lin LN. Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal. Biochem. 1989;179:131–137. doi: 10.1016/0003-2697(89)90213-3. [DOI] [PubMed] [Google Scholar]
- 74.DeLano WL. The PyMOL Molecular Graphics System. San Carlos, CA, USA: DeLano Scientific; 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.