Abstract
The lipooligosaccharide (LOS) biosynthesis region is one of the more variable genomic regions between strains of Campylobacter jejuni. Indeed, eight classes of LOS biosynthesis loci have been established previously based on gene content and organization. In this study, we characterize additional classes of LOS biosynthesis loci and analyze various mechanisms that result in changes to LOS structures. To gain further insights into the genomic diversity of C. jejuni LOS biosynthesis region, we sequenced the LOS biosynthesis loci of 15 strains that possessed gene content that was distinct from the eight classes. This analysis identified 11 new classes of LOS loci that exhibited examples of deletions and insertions of genes and cassettes of genes found in other LOS classes or capsular biosynthesis loci leading to mosaic LOS loci. The sequence analysis also revealed both missense mutations leading to “allelic” glycosyltransferases and phase-variable and non-phase-variable gene inactivation by the deletion or insertion of bases. Specifically, we demonstrated that gene inactivation is an important mechanism for altering the LOS structures of strains possessing the same class of LOS biosynthesis locus. Together, these observations suggest that LOS biosynthesis region is a hotspot for genetic exchange and variability, often leading to changes in the LOS produced.
Campylobacter jejuni is a major cause of acute bacterial gastroenteritis worldwide. The main routes of infection are the consumption of contaminated meat (particularly poultry), contaminated water, or unpasteurized milk products (27). In most cases the infections are self-limiting; however, in rare cases, some patients with C. jejuni enteritis later develop neurological conditions such as Guillain-Barré or Fisher syndrome (34). Most strains of C. jejuni isolated from patients with various neurological conditions synthesize lipooligosaccharides (LOS) exhibiting molecular mimicry with the carbohydrate moieties of gangliosides enriched in peripheral nerves (20). Ganglioside mimicry by C. jejuni LOS, therefore, could induce the production of autoantibodies against gangliosides and the development of neurological disease (36). Indeed, the sera of many patients with C. jejuni-associated Guillain-Barré or Fisher syndrome possess high titers of antibodies to gangliosides and cross-react with C. jejuni LOS (2). However, many C. jejuni strains isolated from clinical samples do not synthesize ganglioside mimics. Therefore, it is not completely understood what other roles the C. jejuni LOS structures may play in bacterium-host interactions.
The LOS is a surface-exposed molecule, and variability of the C. jejuni LOS may have arisen as a result of selection for antigenic diversity to evade the immune system of its various hosts. Variation in LOS structures is due to the diversity of monosaccharide components and the linkages between them and the derivatization of the monosaccharides with noncarbohydrate moieties. The formation of these linkages is determined by genes encoding glycosyltransferases and other transferases for the addition of various moieties located in the LOS biosynthesis locus. Also, the monosaccharides available are often determined by genes in the LOS biosynthesis locus that are involved in the synthesis of sugar intermediates such as CMP-N-acetylneuraminic acid (7, 25).
The identification of the LOS biosynthesis locus in NCTC 11168 facilitated the cloning and characterization of LOS biosynthesis genes from other C. jejuni strains involved in the transfer of galactose, N-acetylgalactosamine, and sialic acid to the LOS outer core (7, 12, 13). The LOS biosynthesis region was identified as a hypervariable region within C. jejuni strains by whole-genome microarray analyses (5, 23, 24, 29, 30, 33), and sequencing of the LOS biosynthesis loci from several C. jejuni strains revealed differences in gene content and organization (7, 10, 12, 13, 28). In addition, comparisons of LOS structures and the corresponding DNA sequences of the LOS biosynthesis loci demonstrated that the structural diversity can be the consequence of either major or minor genetic differences at the LOS biosynthesis loci of the strains (9). Eight LOS biosynthesis locus classes were defined previously based on major genetic differences, gene content, and organization. Three of these LOS locus classes—A, B, and C—encode genes responsible for the production of sialylated LOS that are ganglioside mimics (10), while five other loci (D to H) lack a cst gene that encodes a sialic acid transferase (6, 9, 28). Sequence analysis also revealed minor genetic variation between C. jejuni strains that resulted in major LOS structural differences between strains that possessed the same LOS locus classes (8, 10, 11, 13). These minor genetic variations included (i) phase-variable homopolymeric tracts, (ii) gene inactivation by the deletion or insertion of a single base (without phase variation), (iii) missense or nonsense mutations leading to the inactivation of a glycosyltransferase, and (iv) single or multiple missense mutations leading to “allelic” glycosyltransferases (9-11, 28).
Moreover by using a PCR-based screening of LOS gene content, ca. 80% of the LOS loci from more than 100 C. jejuni strains were assigned into these eight distinct locus classes (A through H) (28). In the present study, we report the sequencing and characterization of 20 additional C. jejuni LOS outer core biosynthesis loci between waaM (Cj1134 in NCTC 11168) and waaV (Cj1146c in NCTC 11168), including 15 with an unknown LOS locus class. Based on gene content and organization, we identified 11 novel LOS locus classes, and each of these newly characterized LOS locus classes possessed regions with some similarity in gene content and organization, as observed in previously described classes (A through H), suggesting high levels of recombination within the locus (28). In addition, we observed in these new classes many of the genetic mechanisms of variation previously observed with other C. jejuni LOS classes, including frameshift mutations and missense mutations. The importance of these findings with regard to the evolution of both recognition specificity and different LOS structures is also discussed.
MATERIALS AND METHODS
Preparation of C. jejuni genomic DNA.
The bacterial strains used in the present study are listed in Table 1. The C. jejuni strains were cultured and genomic DNA was isolated from C. jejuni cells as described previously (28).
TABLE 1.
C. jejuni strains used in this study
| Strain | Descriptiona | LOS class | GenBank accession no. |
|---|---|---|---|
| RM1163 | CjT11 (no. 244); Lior 11 | J | EU414554 |
| RM1503 | HS:43, Penner serotype reference strain | M | EF140720 |
| RM1508 | LCDC 17385; Lior 11, HS:53 | J | EU404104 |
| RM1552 | NADC5591, ATCC 43434, HS:6 | E | EU404105 |
| RM1553 | NADC5592, ATCC 43435, HS:7 | H | EU404106 |
| RM1850 | D983 | I | EU404107 |
| RM1861 | L19; Lior 19, HS:42,15 | K | EU410350 |
| RM2095 | D2722, subsp. doylei | N | AY816330 |
| RM2227 | HS:15; NARMS 72522 | K | EF143353 |
| RM2229 | NARMS 72737 | K | EF143354 |
| RM3415 | HS:11; Penner serotype reference strain | F | EU404108 |
| RM3418 | HS:17; Penner serotype reference strain | D | EU404109 |
| RM3419 | HS:18; Penner serotype reference strain | S | EU404110 |
| RM3423 | HS:27; Penner serotype reference strain | O | EF143352 |
| RM3435 | HS:53; Penner serotype reference strain | L | EU404111 |
| RM3437 | HS:56; Penner serotype reference strain | Q | EU404112 |
| GB4 | HS:37, Guillain-Barré | P | AY943308 |
| GB15 | HS:5,34, Guillain-Barré | F | AY423554 |
| GB24 | HS:31, Guillain-Barré | K | AY573819 |
| GC149 | HS:31, Guillain-Barré | R | AY962325 |
That is, the strain name, Penner type (HS).
XL-PCR.
Long PCR (XL-PCR) reagents were supplied by Epicenter (Madison, WI). Each XL-PCR consisted of 1× MasterAmp Extra-Long PreMix 5, LOSXL primers (LOSXL1, 5′-AAGCGTCCTATTATCTTCACAACTGCACACTATGG; LOSXL2, 5′-ATGCCACAACTTTCTATCATAATCCCGCTT) at 0.2 μM each, 2.5 U of MasterAmp Extra-Long DNA polymerase, and 250 ng of genomic DNA (final reaction volume, 50 μl). The cycling conditions for XL PCR products were as follows: 25 cycles of 30 s at 94°C, 45 s at 52°C, and 15 min at 68°C, followed by a final extension at 68°C for 15 min. Thermal cycling was performed with a Tetrad Thermocycler (MJ Research, Waltham, MA). All PCR products were analyzed by agarose gel electrophoresis. Positive samples were identified based on the presence of bands of anticipated sizes. The primers were purchased from Operon Technologies (Alameda, CA).
DNA sequencing, assembly, and analysis.
The sequencing reactions were performed on a Tetrad Thermocycler using the ABI Prism BigDye terminator cycle sequencing kit (version 3.0; Applied Biosystems, Foster City, CA) and standard protocols as recommended by the manufacturer and then analyzed. All labeled products were purified on DyeEx spin columns (Qiagen, Valencia, CA). DNA sequencing was performed on an ABI Prism 3100 genetic analyzer (Applied Biosystems) using the POP-6 polymer and ABI Prism genetic analyzer data collection and sequencing analysis software. The DNA primers used for PCR or sequencing were designed by using Primer Premier 5.0 (Premier Biosoft International). PCR sequencing primers were purchased from either Qiagen or MWG-Biotech, Inc. (High Point, NC). Sequencing reads were trimmed and assembled by using Lasergene Seqman II (version 6.0; DNAstar, Madison, WI).
Nucleotide sequences were compared against the sequences of bacterial origin of the nonredundant DNA sequence NCBI database using the Basic Local Alignment Search Tool (BLAST) programs BLASTN and BLASTP analysis (1, 32) through the NCBI website (http://www.ncbi.nlm.nih.gov/BLAST/). Conserved domain searches were also performed during BLASTP analysis (26). For comparison of the different C. jejuni LOS loci, BLAST 2 (35) was used at the NCBI website (http://www.ncbi.nlm.nih.gov/BLAST/).
Examination of LOS.
LOS samples were prepared from bacteria that were subjected to complete digestion with proteinase K as described by Hitchcock and Brown (14). LOS samples were separated on 16% Tricine gels (Invitrogen, Carlsbad, CA) and then silver stained (Bio-Rad, Hercules, CA). An O-deacylated LOS sample of C. jejuni GC175 was prepared as described previously (21) and analyzed by capillary electrophoresis-electrospray ionization-mass spectrometry.
RESULTS
To increase our understanding of the diversity of the C. jejuni LOS, the LOS biosynthesis loci between the waaM (Cj1134 in NCTC 11168) and the waaV (Cj1146c in NCTC 11168) genes from 15 C. jejuni strains with unknown LOS classes (Table 1) were sequenced as described previously (28), identifying 11 novel LOS locus classes—I, J, K, L, M, N, O, P, Q, R, and S—based on gene content and organization. A new locus designation was used each time there was a gene content difference compared to previously identified LOS loci. Each new LOS locus was chronologically assigned a consecutive letter identification. Interestingly, each of these newly characterized LOS locus classes contained regions of gene content and organization that suggested these new classes related to previously described C. jejuni LOS classes.
Mosaic LOS classes related to LOS classes D and F.
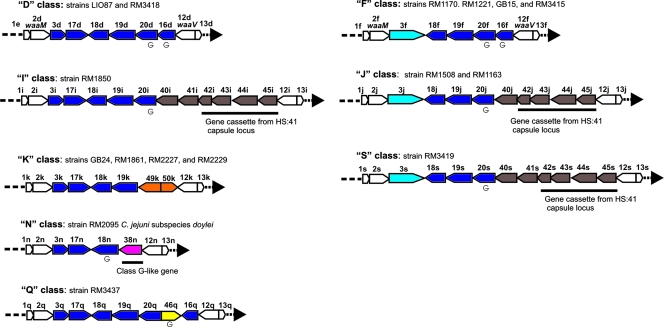
The majority of newly described LOS loci have gene content and organization similarities to classes D and F (Fig. 1). Indeed, classes D and F are quite similar to each other possessing four glycosyltransferase genes in common (orf18, orf19, orf20, and orf16). The classes D and F loci differ in that class D contains an orf3 encoding a one-domain glucosyltransferase, and this is followed by a glycosyltransferase gene (orf17). In contrast, class F possesses a larger orf3 that encodes a two-domain glucosyltransferase similar to Cj1135 from strain NCTC 11168 based on BLAST analysis. Previous structural analysis identified the two-domain product of orf3 as a glucosyltransferase that substitutes a glucose on both the LOS core heptoses, while the presence of the one-domain glucosyltransferase in class D and related classes suggests that only HepI is substituted with glucose (9).
FIG. 1.
LOS classes related to class D and F. Arrows represent ORFs. Genes colored white are common to all LOS classes. Genes colored blue are present in class D. The light blue-colored gene is present in class F. A “G” beneath a gene indicates the presence of an HGT. Gene size is not drawn to scale.
Based on the presence of orf3 that encodes a one-domain glucosyltransferase and at least the following two glycosyltransferase genes (orf17 and orf18), LOS classes I, K, N, and Q are in some way related to class D (Fig. 1). Classes I (strain RM1850) and Q (strain RM3437) LOS loci possess five of the class D open reading frames (ORFs; orf3, orf17, orf18, orf19, and orf20), and the 4,939 nucleotides (nt) containing these ORFs in class D (strain LIO87, GenBank accession no. AF400669, nt 1222 to 6172) shows 98.4% identity to class I by pairwise alignment and 92% identity to class Q. The class K LOS locus (strain RM2227) possesses four of these class D ORFs (orf3, orf17, orf18, and orf19), and the 3,865 nt containing these ORFs show 95% identity to class D strain LIO87 (nt 1222 to 5078). The class N LOS locus (strain RM2095) contains three of these ORFs (orf3, orf17, and orf18), and the 2,679 nt show 95% identity to class D strain LIO87 (nt 1222 to 3898).
The LOS classes J (strains RM1163 and RM1508) and S (strain RM3419) appear to be related to class F due to the presence of the first four ORFs (orf3, orf18, orf19, and orf20) adjacent to waaM of class F (Fig. 1) (strain RM1170, GenBank accession no. AY434498). Indeed, the 4,676 nt containing these four ORFs in class F (strainRM1170, nt 1222 to 5898) show 99% identity to class J and show 96% identity to class S by pairwise alignment.
In the regions that differ from classes D and F, classes I, J, and S are structurally similar to each other in terms of gene order and gene content and provide evidence for a set of common insertion or deletion events. These classes diverge from classes D and F in that orf16 is deleted and replaced by five (class J) or six genes (classes I and S). Classes I and S contain six additional ORFs, and five of these (orf40, orf42, orf43, orf44, and orf45) show similarity to capsular biosynthesis genes from the HS:41 C. jejuni strain 176.83 (GenBank accession no. BX545857). The class J LOS locus also contains these five capsular biosynthesis ORFs. The orf40 encoding a glycosyltransferase has a putative length of 1,053 nt and shows 92% identity over a 643-nt span to HS41.29 from the HS:41 C. jejuni strain 176.83. The four additional HS:41-like ORFs (orf42, orf43, orf44, and orf45) encode a nucleotidyl-sugar pyranose mutase, a sugar epimerase, putative UDP-glucose 6-dehydrogenase, and nucleotidyl-sugar pyranose mutase, respectively. These genes are in the same order as the ORFs 41.24, 41.25, 41.26, and 41.27 from the Penner HS:41 capsular biosynthesis region of C. jejuni strain 176.83 (GenBank accession no. BX545857) (18). Indeed, the 4,094-nt region spanning these four ORFs in classes I, J, and S shows 95% identity to the capsular region from the HS:41 strain 176.83, suggesting that the whole gene cassette (orf42, orf43, orf44, and orf45) was transferred together. Other than the HS:41 genes, the class I and S LOS loci possess a sixth ORF, orf41, that shows similarity at the amino acid level to a number of putative group 1 glycosyltransferases from other bacteria (pfam00535). Considering that the 5′ 279 nt of orf40j and orf41i(s) are 98% identical, it is likely that orf41 recombined into this region of orf40. Thus, it appears that an additional insertion event (orf41) gave rise to class S from class J. This six-gene cassette could then homologously recombine from class S to class D, giving rise to class I.
The remaining three LOS classes (class K, N, and Q) that have similarity to the class D LOS locus exhibit distinct insertion and deletion events. The class K LOS locus contains two additional ORFs that replace the class D orf20 and orf16. The first, orf49k, has 46% similarity to the capsular gene Cj1431c at the amino acid level (GenBank accession no. CAB73855), and orf50k has 73% similarity to orf30h from LOS class H at the amino acid level (GenBank accession no. AAW79071). However, this region shows little identity to any bacterial sequences by BLASTN analysis. It should be noted that the 3,473-nt region (nt 4294 to 7766 of EF143353, strain RM2227) containing these two genes and 100 nt on each side is more than 82% A+T-rich (data not shown), and this nucleotide content may be a factor in the recombination event. The class N LOS locus contains ORF (orf38n) that replaces orf19, orf20, and orf16. This ORF shows similarity to the RfaJ family of glycosyltransferases (COG1442) and has 86% similarity to orf38g from LOS class G at the amino acid level (GenBank accession no. AAR98510). The deletion of orf19 and orf20 can be attributed to a recombination event between orf18 and orf20, which have 93% nt identity over their first 300 nt. Indeed, evidence of such recombination is the presence of a homopolymeric G-tract (HGT) within this 300-nt region of orf18n that is generally found in orf20 of other LOS loci. Finally, the class Q LOS locus (strain RM3437) possesses all ORFs present in class D, with the addition of orf46q inserted between orf20q and orf16q. This ORF shows the conserved domain of group 1 glycosyltransferases (pfam00534) and the RfaG family (COG0438). The insertion altered the 3′-terminal end of orf16q and the 5′-terminal end of orf20q compared to orf16 and orf20 from classes D and F. The HGTs that generally are present in orf20 and orf16 genes are absent from these genes in the class Q LOS locus with the orf46q gene possessing an HGT.
Identification of two new LOS loci containing the cstII gene.
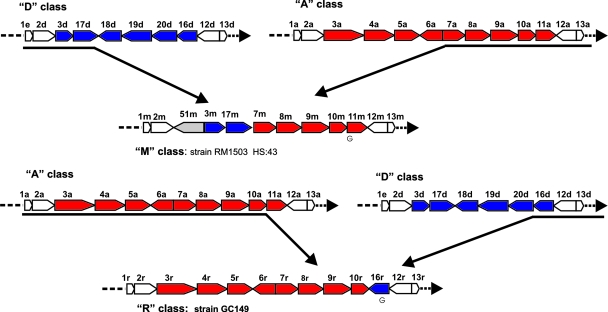
The class A locus has the potential to synthesize sialylated LOS that are ganglioside mimics, and two LOS classes were identified that possessed portions of class A. The class M locus appears to be a mosaic of LOS classes A and D genes (Fig. 2). This locus resembles class A in the regions adjacent to waaV, containing a cassette of five genes. This cassette with genes encoding a sialic acid transferase (cstII; orf7) and enzymes involved in CMP-N-acetyl neuraminic acid biosynthesis (neuBCA: orf8, orf9, and orf10) (7, 25) could also allow the synthesis of a sialylated LOS. The organization of genes adjacent to waaM (orf2m) is different from all of the C. jejuni LOS loci previously examined. Inserted between waaM and the class D gene cassette of orf3 and orf17 is orf51, with 74% similarity to orf19 of class D and class F. This insertion appears to have caused a 9-nt truncation of the waaM gene, while the orf3 gene is not disrupted. It is not clear what recombination mechanism may be responsible for the insertion of orf51, but the regions where the insertion is likely to have occurred at the 3′ end of waaM (nt 474 to 578) and 5′ end before orf3m (nt 1752 to 1815) in RM1503 are ca. 90% A+T-rich (data not shown).
FIG. 2.
LOS classes M and R contain cstII. Arrows represent ORFs. Genes colored white are common to all LOS classes. Genes colored red are genes present in classes M and R that are similar to class A. Genes colored blue are genes present in classes M and R that are similar to class D. A “G” beneath a gene indicates the presence of an HGT.
Another locus that contains a large portion the of LOS class A is the class R locus (strain GC149). The first 9,700 nt show 97% sequence identity to LOS class A, including an orf3 that encodes a two-domain glucosyltransferase, orf4, orf5 (cgtB), orf6 (cgtA), orf7 (cstII), orf8 (neuB), orf9 (neuC), and orf10 (neuA). The recently defined sialate O-acetyltransferase encoded by orf11 (16) has been removed during a recombination that introduced orf16 that is similar to that found in class D and class F LOS loci (Fig. 2). The insertion event appears to have altered the terminal 18 nt of the neuA gene from strain GC149 compared to the neuA genes from classes A, B, and M. Also altered are the adjacent, 125-nt terminal end of orf16r compared to orf16 from classes D and F. Moreover, this neuA-orf16r region is more than 83% A+T-rich.
LOS class related to LOS class G.
The class L LOS locus in strain RM3435 exhibits regions of similarity to several other LOS classes. Class L possesses three ORFs (orf35l, orf36l, and orf37l) in the middle of the locus with 93% identity across the 2,790 nt to class G (GenBank accession no. AAR98510) (Fig. 3). Also, like the class G locus, the class L LOS locus contains an orf3 that encodes a one-domain glucosyltransferase, but this is more similar to orf3 from classes E, H, and O loci with 97% nt identity. There are two additional ORFs: orf47l and orf48l. Orf47 shows the conserved domain of group 2 glycosyltransferases (pfam00535) and the WcaA family (COG0463) and has 81% identity to the orf19 glycosyltransferase gene from LOS class K. Orf48 also shows the conserved domain of group 2 glycosyltransferases (pfam00535) and the WcaA family (COG0463). This ORF is formed by a fusion between the 5′ terminus of orf16 and a capsule-related gene. Indeed, the first 705 nt of orf48 is 90% identical to the orf16 from the class F locus, and another 504 nt are 96% identical to the capsular biosynthesis gene HS19.08 from the HS:19 C. jejuni strain CJ12517 (GenBank accession no. BX545860). Elsewhere in class L, the region adjacent to orf3 is approximately 1.000 nt and appears to contain fragments of several genes. including a 390-nt span that is 88% similar to orf5/10 (Cj1143 in strain NCTC 11168) from class C; however, the origin of the remainder of this region is unknown.
FIG. 3.
Mosaic LOS class related to class G. Arrows represent ORFs. Genes colored white are common to all LOS classes. Genes colored pink are genes present in class L that are similar to class G. A “G” beneath a gene indicates the presence of an HGT.
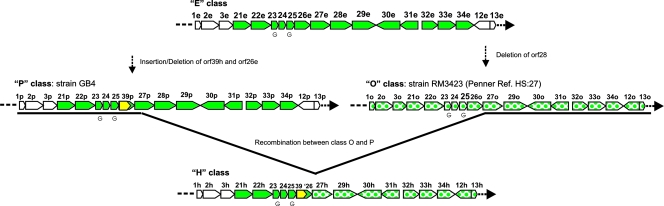
Both LOS classes O and P could be the intermediate between classes E and H.
The next pair of LOS classes, O (strain RM3423) and P (strain GB4), share gene content and organization very similar to LOS classes E and H; they both possess distinct differences (Fig. 4). The class O LOS locus has a deletion of orf28, while in the class P LOS locus, an insertion of a putative butyryltransferase gene (orf39) with the concomitant deletion of the 5′ region of orf26 has occurred. Both of these changes appear in LOS class H, and we previously hypothesized that an intermediate LOS locus between classes E and H, similar to class O or P, would exist (28). Indeed, the slightly higher sequence identity between class P (GB4) and class H (RM1047) from waaM to orf39 and slightly higher sequence identity between class O (RM3423) and class H (RM1047) from orf27 to the waaV LOS region suggests that class H arose from a recombination between class O and class P (Fig. 4). Finally, all of these classes (E, H, O, and P) contain HGTs in two genes, orf23 and orf25.
FIG. 4.
Intermediate LOS classes between class E and class H. Genes colored white are common to all LOS classes. The solid-green and spotted-green genes are merely used to distinguish the genes in class H that are more similar to class O or P. The orange colored gene is present in classes P and H. A “G” beneath a gene indicates the presence of an HGT.
Evidence for other genetic mechanisms to obtain LOS structural diversity.
Besides the deletions and insertions of genes and gene cassettes in the newly described LOS loci, sequence analysis has identified mutations affecting particular ORFs that can affect structural diversity. In the C. jejuni LOS loci, there are several occurrences of frameshift mutations that are phase variable due to HGT of eight or more consecutive Gs, as well as, non-phase-variable out-of-frame mutations (OOF) due to deletions or insertions (9, 11). Table 2 details the HGT and OOF mutations of orf3, orf17, orf18, orf19, orf20, and orf16 from the LOS locus classes that possess two or more of these ORFs. Of these ORFs, HGTs were common to orf20 and orf16. Indeed, differences in the phase variation at an HGT of orf20 between strains possessing the class F locus resulted in LOS differences, where an OOF mutation led to a truncated LOS and a variable HGT led to two LOS forms, as observed by gel migration (Fig. 5A). It should also be noted that HGTs were observed in all but one class of LOS, class K. Interestingly, we observed that a non-phase-variable OOF mutation in orf18 in strain RM1861 (class K) due to a 13-nt deletion resulted in a truncated LOS compared to RM2227 another class K strain (Fig. 5B).
TABLE 2.
Variants of the glycosyltransferases related to class D and class Fa
| Strain | LOS class | orf3 | orf17 | orf18 | orf19 | orf20 | orf16 |
|---|---|---|---|---|---|---|---|
| LIO87 | D | One domain | IF | IF | IF | HGT | HGT |
| RM3418 | D | One domain | IF | IF | IF | HGT | HGT |
| RM1170 | F | Two domain | NA | IF | OOF | HGT, OOF | HGT |
| RM1221 | F | Two domain | NA | IF | IF | HGT | HGT |
| GB15 | F | Two domain | NA | IF | IF | HGT, OOF | HGT |
| RM3415 | F | Two domain | NA | IF | IF | IF | HGT |
| RM1850 | I | One domain | OOF | IF | IF | OOF, HGT | NA |
| RM1508 | J | Two domain | NA | IF | IF | OOF, HGT | NA |
| RM1861 | K | One domain | IF | OOF | IF | NA | NA |
| RM2227 | K | One domain | IF | IF | IF | NA | NA |
| RM2229 | K | One domain | IF | IF | IF | NA | NA |
| GB24 | K | One domain | IF | IF | IF | NA | NA |
| RM1503 | M | One domain | IF | N/A | IF | NA | NA |
| RM2095 | N | One domain | IF | HGT | NA | NA | NA |
| RM3437 | Q | One domain | IF | IF | IF | OOF | IF |
| RM3419 | S | One domain | NA | IF | IF | OOF, HGT | NA |
Abbreviations: IF, in-frame; NA, not applicable.
FIG. 5.
Effect of mutations on LOS structure. (A) Differences in HGT in orf20 leads to in-frame (IF) (strain RM3415), out-of-frame (OOF) (variant of strain RM1221), and variable (IF/OOF) (strain RM1221) states in class F. (B) A 13-nt deletion disrupts orf18 in class K strain RM1861 compared to strain RM2227 with an intact orf18.
Previously, we reported two class A strains (OH4382 and OH4384) that differed only by the deletion of a single base pair in cgtA (orf5), which encodes the β-1,4-N-acetylgalactosaminyltransferase (7). These two strains were isolated from Japanese siblings (37), and both had LOS outer cores with di-sialic acid, but OH4382 expressed a truncated LOS outer core (Fig. 6A). We have now characterized another pair of Japanese strains (CF90-26 and GC175) that also differ only by the deletion of a single base pair in cgtA but have LOS outer cores with mono-sialic acid rather di-sialic acid (Fig. 6A). These four strains are all Penner serotype HS:19 and provide an interesting example of variation of LOS outer core structures among closely related strains. Each pair (OH4382/OH4384 and CF90-26/GC175) can vary between a truncated and full-length LOS outer core based on the deletion or insertion of an A. The deletion or insertion is relatively stable since we did not observe phase variation. The interconversion of the “mono-sialic acid pair” (CF90-26/GC175) and the “di-sialic acid” pair (OH4382/OH4384) could happen by the mutation of residue 51 with Thr51 giving a monofunctional CstII and Asn giving a bifunctional CstII (10). Since there are 24 base differences in cstII between CF90-26/GC175 and OH4382/OH4384, we also have to consider the possibility of lateral gene transfer of the cstII gene in this case. However, it is possible that a Thr51Asn substitution occurred in an ancestor of these strains and that other mutations have accumulated. Strains OH4382 and OH4384 are epidemiologically linked since they were isolated from siblings, but there is no evidence for epidemiological linkage between OH4382/OH4384 and CF90-26/GC175 or between the later two strains. Together, these four strains show that high conservation in gene content can result in various structures and the possibility to vary between these structures by deletion or insertion of a single base or substitution of a single amino acid (Fig. 6B).
FIG. 6.
LOS outer core structures of four class A C. jejuni (all Penner type HS:19) from Japanese patients. (A) The LOS outer core structures of OH4382, OH4384, and CF90-26 were reported previously (3, 22). The LOS outer structure of GC175 is proposed based on the mass spectrometry data presented in Table S1 in the supplemental material. (B) Comparison of the DNA sequences of the five glycosyltransferases involved in the biosynthesis of the LOS outer core. The identities are indicated between strains for each gene. The only difference between GC175 and CF90-26 is the deletion of an A in cgtA. This deletion of an A in cgtA is also the only difference between OH4382 and OH4384.
Single or multiple missense mutations leading to “allelic” glycosyltransferases have been observed for several C. jejuni LOS genes (9-11, 28). Specifically, different enzymatic specificities were previously observed for Orf7 (CstII) from classes A and B. There were alleles that were bifunctional (both α-2,3- and α-2,8-sialyltransferase activities) and monofunctional alleles (α-2,3-sialyltransferase activity). The alignment of the protein sequences showed alleles with 92% identity, and site-directed mutagenesis showed that residue Asn51 was critical for the bifunctional activity (10). However, a CstII variant with Asn51 and that has only α-2,3-sialyltransferase activity has recently been characterized (11). This CstII variant has diverged significantly from the other CstII sequences, and it is possible that one (or several) amino acid substitution(s) have inactivated the α-2,8-sialyltransferase activity in that variant. We examined the diversity of the glycosyltransferases encoded by orf18 and orf19 that are present in multiple LOS classes. From the alignment of the Orf18 glycosyltransferase amino acid sequences among 16 C. jejuni strains that possess the full-length ORF, 12 protein variants ranging from 84 to 99% identity were observed (see Fig. S1 in the supplemental material). It should be noted that a particular LOS class did not necessarily possess a particular variant. For example, although Orf18 from both class D strains had identical amino acid sequences, Orf18 in the four class F variants were distinct. Moreover, the Orf18 from RM1221 (class F) was identical to the Orf18 from strains RM1163 and RM1508 (class J). There was similar diversity among the 10 variants of the 14 Orf19 glycosyltransferases ranging from 87 to 99% identity (see Fig. S2 in the supplemental material).
DISCUSSION
Many bacterial pathogens exhibit variable cell surface polysaccharides as a result of differences in the LOS, lipopolysaccharide, capsules, and/or glycoproteins that they present. These variable glycoconjugates likely play important roles in the interaction of the bacteria with the various hosts and their immune systems. To achieve such structural variability, bacteria use several genetic mechanisms, including differences in the gene content, as seen for many of the 90 capsular polysaccharides of Streptococcus pneumoniae (4), and sequence variation that leads to changes in gene function and expression that has been observed in Haemophilus influenzae (15) and Neisseria species (17). Indeed, strains of C. jejuni exhibit variation in their capsule which is the basis of the Penner serotyping scheme, their LOS, and also their O-linked flagellar glycosylation (19).
In the present study, we compared the sequences of the 19 distinct classes of LOS biosynthetic loci of C. jejuni, 11 of which were newly described here. The sequence analysis highlights genetic mechanisms that are utilized by C. jejuni to generate LOS diversity. The most obvious mechanism is the variation in gene content resulting in the 19 different LOS biosynthesis loci. Based on shared gene content, several of these loci are related genetically and appear to have arisen by the insertion and/or deletion of genes or gene cassettes. There is a group of four LOS biosynthesis loci that possess the necessary genes to synthesize sialylated LOS but differ by gene insertions or gene duplication LOS (classes A, B, M, and R; Fig. 3). In particular, each class possesses genes encoding a sialyltransferase (cstII), a sialic acid synthase (neuB), an N-acetylglucosamine-6-phosphate 2-epimerase (neuC), and a CMP-Neu5Ac synthetase (neuA). The class C locus also possesses genes to synthesize sialylated LOS, but it appears more distantly related to these others classes based on sequence comparison (9). There were three other groups of LOS biosynthesis loci that did not possess the genes necessary to synthesize sialylated LOS. Six newly identified LOS biosynthesis loci (I, J, K, N, Q, and S) possessed genes found in classes D and F (Fig. 1), and three of these loci possessed a gene cassette of four HS:41 capsular genes. Furthermore, LOS biosynthesis classes G and L possess orf3 and orf16 and could be distantly related to the class D locus. However, these two classes exhibit multiple insertion and deletion events that make parsimonious descriptions difficult. On the other hand, the organizations of classes E, H, O, and P LOS biosynthesis loci do allow parsimonious recombination event predictions that explain derivation of class H from the others in this class (Fig. 4). Together, each group of LOS biosynthetic loci demonstrates the mosaic nature of gene and gene cassette insertion and deletion that can result in LOS structure variation.
It has been observed recently that the introduction of a complete LOS biosynthesis locus class can occur between strains by horizontal transfer (8, 31). Presumably, these exchanges involve recombination between homologous regions that flank the LOS biosynthesis locus. Our sequence characterization of new classes of LOS loci demonstrates that recombination events often occur within the LOS locus, where no obvious regions of homology exist. Thus, the exact mechanism for the production of new LOS loci described would potentially involve recombination between the large number of A and T polynucleotides or specific A+T-rich sequences that may be common between these loci. Indeed, the G+C content of the LOS loci (22 to 28%) is slightly lower than the G+C content for the rest of the genome (30%) and is often even lower than 20% in regions where insertions have occurred.
It is also not clear whether all of the newly described mosaic LOS loci create functional biosynthetic gene clusters. It is quite possible that the newly imported glycosyltransferases or modification enzymes may not recognize the existing LOS structures. Also, it is possible that during recombination only portions of genes could be transferred or that resident genes could be disrupted. Certainly, there is evidence of gene disruption in LOS classes H and P where orf39 has disrupted orf26. Also, the event that inserted orf16 in strain GC149 altered the 3′ terminal ends of the adjacent neuA gene and the incoming orf16 gene. In addition, it appears that the class L locus acquired only a portion of the orf5/10.
Aside from the differences in gene content, the sequence analysis also highlights the subtle genetic differences that are utilized by C. jejuni to generate LOS diversity. These include phase-variable HGTs, gene inactivation by the deletion of a single base or multiple bases (without phase variation), and missense mutations leading to “allelic” glycosyltransferases. Indeed, we demonstrated that OOF mutations in orf18, orf19, and orf20 (phase variable and non-phase variable) affected LOS structures (Fig. 5 and 6). Moreover, the importance of phase variation is highlighted by the fact that 18 of the 19 LOS biosynthetic loci examined thus far possess at least one HGT. As for the allelic glycosyltransferases, it is still to be determined whether any of the protein variants encoded by orf18 and orf19 exhibit different enzymatic specificities until the LOS structures for all of these strains are determined. Previously, different enzymatic specificities were observed for Orf7 (CstII) from classes A and B (10). There were alleles that were bifunctional (both α-2,3- and α-2,8-sialyltransferase activities) and alleles that were monofunctional (α-2,3-sialyltransferase activity). The alignment of the protein sequences showed that most of the bifunctional CstII alleles possessed the residue Asn51 (10, 11).
Finally, despite the ability to form a variety of LOS biosynthesis loci, we previously observed that over 60% of LOS biosynthesis loci from more than 100 clinical and environmental strains of C. jejuni belonged to classes A, B, or C (28). This suggests that possessing these particular loci that have the potential to synthesize a sialylated LOS may be advantageous in host interactions. It is also noteworthy that variation in other C. jejuni glycan structures (capsule and O-linked flagellar glycosylation) occurs by similar mechanisms (18, 19). Together, it points to the fact that despite our knowledge of locus sequences and glycan structural data, we still require a greater understanding of the significance of the C. jejuni glycan variability.
Supplementary Material
Acknowledgments
We are indebted to Sharon Horn, Anna Bates, Felicidad Bautista, and Marie-France Karwaksi for technical assistance in this study. We thank Jianjun Li for the mass spectrometry analysis of the LOS of C. jejuni GC175. We thank Beatriz Quiñones for critical reading of the manuscript.
Part of this study was supported by the U.S. Department of Agriculture, Agricultural Research Service CRIS project 5325-42000-045 (to C.T.P. and R.E.M.) and by Human Frontier Science Program grant RGP 38/2003 (to M.G., N.Y., and H.P.E.).
Footnotes
Published ahead of print on 13 June 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 253389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ang, C. W., J. D. Laman, H. J. Willison, E. R. Wagner, H. P. Endtz, M. A. De Klerk, A. P. Tio-Gillen, N. van den Braak, B. C. Jacobs, and P. A. van Doorn. 2002. Structure of Campylobacter jejuni lipopolysaccharides determines antiganglioside specificity and clinical features of Guillain-Barré and Miller Fisher patients. Infect. Immun. 701202-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aspinall, G. O., A. G. McDonald, H. Pang, L. A. Kurjanczyk, and J. L. Penner. 1994. Lipopolysaccharides of Campylobacter jejuni serotype O:19: structures of core oligosaccharide regions from the serostrain and two bacterial isolates from patients with the Guillain-Barré syndrome. Biochemistry 33241-249. [DOI] [PubMed] [Google Scholar]
- 4.Bentley, S. D., D. M. Aanensen, A. Mavroidi, D. Saunders, E. Rabbinowitsch, M. Collins, K. Donohoe, D. Harris, L. Murphy, M. A. Quail, G. Samuel, I. C. Skovsted, M. S. Kaltoft, B. Barrell, P. R. Reeves, J. Parkhill, and B. G. Spratt. 2006. Genetic analysis of the capsular biosynthetic locus from all 90 pneumococcal serotypes. PLoS Genet. 2e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dorrell, N., J. A. Mangan, K. G. Laing, J. Hinds, D. Linton, H. Al-Ghusein, B. G. Barrell, J. Parkhill, N. G. Stoker, A. V. Karlyshev, P. D. Butcher, and B. W. Wren. 2001. Whole genome comparison of Campylobacter jejuni human isolates using a low-cost microarray reveals extensive genetic diversity. Genome Res. 111706-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gilbert, M., J. Bellefeuille, M. F. Karwaski, A. M. Cunningham, and W. W. Wakarchuk. 2001. Partial sequence of the lipooligosaccharide biosynthesis locus of Campylobacter jejuni O:3. Institute for Biological Sciences, National Research Council of Canada, Montreal, Canada.
- 7.Gilbert, M., J. R. Brisson, M. F. Karwaski, J. Michniewicz, A. M. Cunningham, Y. Wu, N. M. Young, and W. W. Wakarchuk. 2000. Biosynthesis of ganglioside mimics in Campylobacter jejuni OH4384: identification of the glycosyltransferase genes, enzymatic synthesis of model compounds, and characterization of nanomole amounts by 600-mhz 1H and 13C NMR analysis. J. Biol. Chem. 2753896-3906. [DOI] [PubMed] [Google Scholar]
- 8.Gilbert, M., P. C. Godschalk, M. F. Karwaski, C. W. Ang, A. van Belkum, J. Li, W. W. Wakarchuk, and H. P. Endtz. 2004. Evidence for acquisition of the lipooligosaccharide biosynthesis locus in Campylobacter jejuni GB11, a strain isolated from a patient with Guillain-Barré syndrome, by horizontal exchange. Infect. Immun. 721162-1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gilbert, M., P. C. R. Godschalk, C. T. Parker, H. P. Endtz, and W. W. Wakarchuk. 2005. Genetic bases for the variation in the lipooligosaccharide outer core of Campylobacter jejuni and possible association of glycosyltransferase genes with post-infectious neuropathies, p. 219-248. In J. M. Ketley and M. E. Konkel (ed.), Campylobacter: molecular and cellular biology. Horizon Bioscience, Norwich, United Kingdom.
- 10.Gilbert, M., M. F. Karwaski, S. Bernatchez, N. M. Young, E. Taboada, J. Michniewicz, A. M. Cunningham, and W. W. Wakarchuk. 2002. The genetic bases for the variation in the lipo-oligosaccharide of the mucosal pathogen, Campylobacter jejuni: biosynthesis of sialylated ganglioside mimics in the core oligosaccharide. J. Biol. Chem. 277327-337. [DOI] [PubMed] [Google Scholar]
- 11.Godschalk, P. C., M. L. Kuijf, J. Li, F. St Michael, C. W. Ang, B. C. Jacobs, M. F. Karwaski, D. Brochu, A. Moterassed, H. P. Endtz, A. van Belkum, and M. Gilbert. 2007. Structural characterization of Campylobacter jejuni lipooligosaccharide outer cores associated with Guillain-Barré and Miller Fisher syndromes. Infect. Immun. 751245-1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guerry, P., C. P. Ewing, T. E. Hickey, M. M. Prendergast, and A. P. Moran. 2000. Sialylation of lipooligosaccharide cores affects immunogenicity and serum resistance of Campylobacter jejuni. Infect. Immun. 686656-6662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guerry, P., C. M. Szymanski, M. M. Prendergast, T. E. Hickey, C. P. Ewing, D. L. Pattarini, and A. P. Moran. 2002. Phase variation of Campylobacter jejuni 81-176 lipooligosaccharide affects ganglioside mimicry and invasiveness in vitro. Infect. Immun. 70787-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hitchcock, P. J., and T. M. Brown. 1983. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J. Bacteriol. 154269-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hood, D. W., M. E. Deadman, M. P. Jennings, M. Bisercic, R. D. Fleischmann, J. C. Venter, and E. R. Moxon. 1996. DNA repeats identify novel virulence genes in Haemophilus influenzae. Proc. Natl. Acad. Sci. USA 9311121-11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Houliston, R. S., H. P. Endtz, N. Yuki, J. Li, H. C. Jarrell, M. Koga, A. van Belkum, M. F. Karwaski, W. W. Wakarchuk, and M. Gilbert. 2006. Identification of a sialate O-acetyltransferase from Campylobacter jejuni: demonstration of direct transfer to the C-9 position of terminal α-2, 8-linked sialic acid. J. Biol. Chem. 28111480-11486. [DOI] [PubMed] [Google Scholar]
- 17.Kahler, C. M., and D. S. Stephens. 1998. Genetic basis for biosynthesis, structure, and function of meningococcal lipooligosaccharide (endotoxin). Crit. Rev. Microbiol. 24281-334. [DOI] [PubMed] [Google Scholar]
- 18.Karlyshev, A. V., O. L. Champion, C. Churcher, J. R. Brisson, H. C. Jarrell, M. Gilbert, D. Brochu, F. St Michael, J. Li, W. W. Wakarchuk, I. Goodhead, M. Sanders, K. Stevens, B. White, J. Parkhill, B. W. Wren, and C. M. Szymanski. 2005. Analysis of Campylobacter jejuni capsular loci reveals multiple mechanisms for the generation of structural diversity and the ability to form complex heptoses. Mol. Microbiol. 5590-103. [DOI] [PubMed] [Google Scholar]
- 19.Karlyshev, A. V., J. M. Ketley, and B. W. Wren. 2005. The Campylobacter jejuni glycome. FEMS Microbiol. Rev. 29377-390. [DOI] [PubMed] [Google Scholar]
- 20.Kimoto, K., M. Koga, M. Odaka, K. Hirata, M. Takahashi, J. Li, M. Gilbert, and N. Yuki. 2006. Relationship of bacterial strains to clinical syndromes of Campylobacter-associated neuropathies. Neurology 671837-1843. [DOI] [PubMed] [Google Scholar]
- 21.Koga, M., M. Gilbert, J. Li, S. Koike, M. Takahashi, K. Furukawa, K. Hirata, and N. Yuki. 2005. Antecedent infections in Fisher syndrome: a common pathogenesis of molecular mimicry. Neurology 641605-1611. [DOI] [PubMed] [Google Scholar]
- 22.Koga, M., M. Gilbert, M. Takahashi, J. Li, S. Koike, K. Hirata, and N. Yuki. 2006. Comprehensive analysis of bacterial risk factors for the development of Guillain-Barré syndrome after Campylobacter jejuni enteritis. J. Infect. Dis. 193547-555. [DOI] [PubMed] [Google Scholar]
- 23.Leonard, E. E., II, T. Takata, M. J. Blaser, S. Falkow, L. S. Tompkins, and E. C. Gaynor. 2003. Use of an open reading frame-specific Campylobacter jejuni DNA microarray as a new genotyping tool for studying epidemiologically related isolates. J. Infect. Dis. 187691-694. [DOI] [PubMed] [Google Scholar]
- 24.Leonard, E. E., II, L. S. Tompkins, S. Falkow, and I. Nachamkin. 2004. Comparison of Campylobacter jejuni isolates implicated in Guillain-Barré syndrome and strains that cause enteritis by a DNA microarray. Infect. Immun. 721199-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Linton, D., A. V. Karlyshev, P. G. Hitchen, H. R. Morris, A. Dell, N. A. Gregson, and B. W. Wren. 2000. Multiple N-acetyl neuraminic acid synthetase (neuB) genes in Campylobacter jejuni: identification and characterization of the gene involved in sialylation of lipo-oligosaccharide. Mol. Microbiol. 351120-1134. [DOI] [PubMed] [Google Scholar]
- 26.Marchler-Bauer, A., and S. H. Bryant. 2004. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 32W327-W231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller, W. G., and R. E. Mandrell. 2005. Prevalence of Campylobacter in the food and water supply: incidence, outbreaks, isolation and detection, p. 101-163. In M. E. Konkel and J. M. Ketley (ed.), Campylobacter: molecular and cellular biology. Horizon Scientific Press, Norwich, United Kingdom.
- 28.Parker, C. T., S. T. Horn, M. Gilbert, W. G. Miller, D. L. Woodward, and R. E. Mandrell. 2005. Comparison of Campylobacter jejuni lipooligosaccharide biosynthesis loci from a variety of sources. J. Clin. Microbiol. 432771-2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parker, C. T., B. Quiñones, W. G. Miller, S. T. Horn, and R. E. Mandrell. 2006. Comparative genomic analysis of Campylobacter jejuni strains reveals diversity due to genomic elements similar to those present in C. jejuni strain RM1221. J. Clin. Microbiol. 444125-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pearson, B. M., C. Pin, J. Wright, K. I'Anson, T. Humphrey, and J. M. Wells. 2003. Comparative genome analysis of Campylobacter jejuni using whole genome DNA microarrays. FEBS Lett. 554224-230. [DOI] [PubMed] [Google Scholar]
- 31.Phongsisay, V., V. N. Perera, and B. N. Fry. 2006. Exchange of lipooligosaccharide synthesis genes creates potential Guillain-Barré syndrome-inducible strains of Campylobacter jejuni. Infect. Immun. 741368-1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schäffer, A. A., L. Aravind, T. L. Madden, S. Shavirin, J. L. Spouge, Y. I. Wolf, E. V. Koonin, and S. F. Altschul. 2001. Improving the accuracy of PSI-BLAST protein database searches with composition-based statistics and other refinements. Nucleic Acids Res. 292994-3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taboada, E. N., R. R. Acedillo, C. D. Carrillo, W. A. Findlay, D. T. Medeiros, O. L. Mykytczuk, M. J. Roberts, C. A. Valencia, J. M. Farber, and J. H. Nash. 2004. Large-scale comparative genomics meta-analysis of Campylobacter jejuni isolates reveals low level of genome plasticity. J. Clin. Microbiol. 424566-4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takahashi, M., M. Koga, K. Yokoyama, and N. Yuki. 2005. Epidemiology of Campylobacter jejuni isolated from patients with Guillain-Barré and Fisher syndromes in Japan. J. Clin. Microbiol. 43335-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tatusova, T. A., and T. L. Madden. 1999. BLAST 2 Sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol. Lett. 174247-250. [DOI] [PubMed] [Google Scholar]
- 36.Yuki, N., K. Susuki, M. Koga, Y. Nishimoto, M. Odaka, K. Hirata, K. Taguchi, T. Miyatake, K. Furukawa, T. Kobata, and M. Yamada. 2004. Carbohydrate mimicry between human ganglioside GM1 and Campylobacter jejuni lipooligosaccharide causes Guillain-Barré syndrome. Proc. Natl. Acad. Sci. USA 10111404-11409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yuki, N., and Y. Tsujino. 1995. Familial Guillain-Barré syndrome subsequent to Campylobacter jejuni enteritis. J. Pediatr. 126162. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.