Abstract
Splicing of human immunodeficiency virus type 1 (HIV-1) exon 6D is regulated by the presence of a complex splicing regulatory element (SRE) sequence that interacts with the splicing factors hnRNP H and SC35. In this work, we show that, in the context of the wild-type viral sequence, hnRNP H acts as a repressor of exon 6D inclusion independent of its binding to the SRE. However, hnRNP H binding to the SRE acts as an enhancer of exon 6D inclusion in the presence of a critical T-to-C mutation. These seemingly contrasting functional properties of hnRNP H appear to be caused by a change in the RNA secondary structure induced by the T-to-C mutation that affects the spatial location of bound hnRNP H with respect to the exon 6D splicing determinants. We propose a new regulatory mechanism mediated by RNA folding that may also explain the dual properties of hnRNP H in splicing regulation.
Replication of human immunodeficiency virus type 1 (HIV-1) is dependent on the balanced expression of several viral genes. Multiple cellular factors interact with viral sequences to properly process the single viral primary transcript into more than 40 mRNAs (32). These messages fall into three classes; a ∼2-kb class of multiple spliced mRNAs coding for the viral proteins Tat, Rev, and Nef, a ∼4 -kb class of partially unspliced mRNAs coding for the viral proteins Env, Vpu, Vif, and Vpr, and the 9.2-kb unspliced transcript coding for the viral proteins Gag and Pol and which is also packaged into new virions as the viral genome (Fig. 1A). The HIV-1 genome is characterized by a high mutation rate that generates a heterogeneous population of viral isolates. This can lead to the appearance of noncanonical splice sites and unusual patterns of splicing. An example of this unusual splicing was described for the 6D exon, located within the env gene. Its inclusion results in the production of a series of aberrant mRNAs yielding aberrant Tat and Rev proteins (2, 33) (Fig. 1A). Although the splicing machinery rarely recognizes this exon, a naturally arising point mutation (T to C) within the env gene that activates its inclusion into most viral messages was identified. This splicing inclusion of exon 6D leads to downregulation of the production of all viral genes and leads to inefficient viral replication (39). Sequence analysis of viral isolates indicates that the sequences regulating exon 6D splicing are highly conserved at the RNA level but do not correlate with conservation of protein sequence (7).
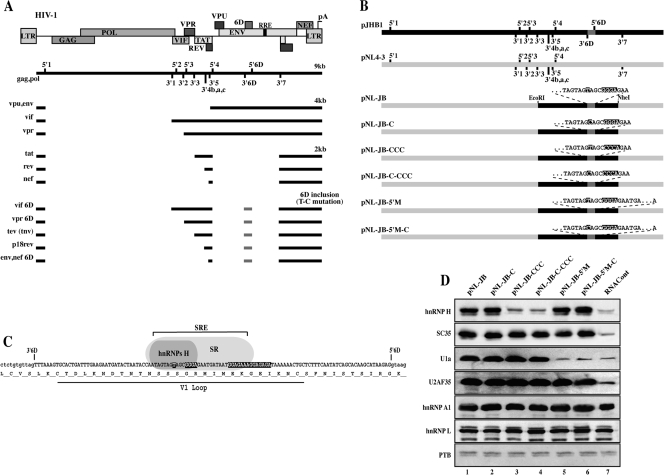
FIG. 1.
A) Schematic representation of the main HIV-1 RNA species. The relative position of the viral genes is indicated on the map on top. The viral genomic (∼9-kb unspliced) mRNA shows the location of 5′ss and 3′ss (numbered). The main mRNAs coding for the viral genes are grouped in three categories of ∼9 kb, ∼4 kb, and ∼2 kb. A class of aberrant transcript is created by the inclusion of the cryptic exon 6D (6D inclusion). The solid line represents the exons in each mRNA. B) Schematic map of the proviral clones utilized in this study. The proviral clone pJHB1 is not infective due to a deletion in the gag/pol region and carries the exon 6D specific splice sites. The proviral clone pNL4-3 does not carry the exon 6D splice sites. For cloning purposes the EcoRI-NheI fragment from pHB1 was inserted within pNL4-3 to generate the proviral clone pNL-JB. Mutations of the T or GGG motifs are indicated. In clones pNL-JB-5′M and pNL-JB-5′M-C the silent mutation GT to GC disrupts the 6D 5′ss. C) Exon 6D nucleotide and protein sequence for the HXB2 isolate. Location of the SRE that regulates the assembly of hnRNP H and SR proteins is indicated. The T, GGG, and the polypurine tract within the SRE are indicated by dark boxes. Mutation of the T to C activates exon 6D splicing. Location of the hypervariable V1 loop is indicated. D) Assembly of hnRNPs H, A1, L, and PTB, SR protein SC35, and the spliceosomal component U1a and the U2AF35 factor onto exon 6D was assayed by RAC. RNA baits comprehensive of the exon 6D were generated from the proviral clones shown in panel B. The control RNAcont bait was generated from the antisense strand of the pNL-JB clone. RNA baits were incubated in HeLa Nuclear extracts, and the bound proteins were eluted and analyzed by immunoblotting.
We have previously shown that a multiprotein complex, which includes members of the hnRNP H and serine/arginine-rich (SR) protein families, specifically binds to a tripartite splicing regulatory element (SRE) within exon 6D (7) (Fig. 1C). Members of the SR protein family have been shown to regulate splicing by binding purine-rich sequences and directing the assembly of prespliceosomal and spliceosomal factors to 5′ and 3′ splice sites (17). In agreement with these observations, our previous data indicated that several SR proteins bind to a purine-rich region in the central portion of exon 6D and promote in vitro splicing of a heterologous splicing substrate containing the central portion of exon 6D (7). Previous work also confirmed that mutation of the purine-rich region decreased exon 6D splicing in viruses carrying the T-to-C mutation, thus increasing the levels of gag/pol and env transcripts and enhancing viral replication (39).
Little is known on the precise role played in splicing regulation by the members of the hnRNP H protein family. hnRNPs H, H′, F, 2H9, and GRSF-1 constitute a family of highly homologous, ubiquitously expressed proteins that have been implicated in splicing, polyadenylation, capping, export, and translation of cellular and viral mRNAs. Members of this protein family have been shown to activate splicing in several systems (7, 10, 15, 19, 20, 27, 29, 34) while acting as splicing repressors in others (9, 14). It is not understood whether the basal splicing machinery requires such proteins to function (25), although we have recently reported that they can enhance splicing of a viral substrate by promoting the formation of ATP-dependent spliceosomal complexes (34). While the molecular mechanism by which hnRNP H family proteins regulate mRNA processing remains largely unknown, their ability to bind RNA sequences containing the DGGGD (D is A, G, or T) motif has been well-characterized (34).
As the mechanism by which the T-to-C mutation and hnRNP H proteins regulate splicing of exon 6D is still unclear, we set out to determine the role of these cis- and trans-regulatory factors in the context of a complete proviral clone. Since a growing number of reports have indicated that RNA secondary structure can play an important role in splicing control in cellular and viral systems (5, 8, 11, 16, 21), we investigated the potential role for RNA structure in exon 6D splicing regulation. We report that hnRNP H regulates viral splicing and infectivity by binding to a conserved sequence within exon 6D. By reproducing the regulation of exon 6D splicing in a heterologous minigene system, we have identified a structural change in the SRE region of the viral mutant that activates splicing of exon 6D. Structural compensatory mutations were able to rescue the wild-type splicing pattern and infectivity from the mutant virus. Surprisingly, hnRNP H activity was differentially modulated by the structural change in the viral transcript, suggesting a novel regulatory mechanism by which structural changes in the pre-mRNA can modify the activity of a splicing factor without altering its binding to the substrate.
MATERIALS AND METHODS
Plasmid construction.
pNL-JB proviral construct was obtained by subcloning a fragment from the proviral construct pJHB1 (a gift from David Rekosh and Marie-Louise Hammarskjold, University of Virginia) into pBluescript II KS (Stratagene) to generate pBK-JB. An EcoRI-NheI fragment from pBK-JB was then substituted for the EcoRI-Nhe fragment in the proviral vector pNL4-3 to generate pNL-JB. pNL4-3 and pNL-JB differ for the recognition site of several restriction enzymes within the EcoRI-NheI fragment, thus facilitating the screening for the introduction of mutation within the exon 6D region. All the pNL-JB mutants described in this work were obtained by site-directed PCR mutagenesis, subcloning of the mutated PCR fragment into pBK-JB, and cloning of the mutated EcoRI-NheI fragment into pNL4-3. The plasmids utilized for the transcription of the RNA substrates utilized for the RNA affinity chromatography (RAC) assays and the enzymatic probing of the RNA structures were generated by cloning into pBluescript II KS (Stratagene) a fragment amplified from pNL-JB and the various mutant proviruses utilizing primers 6D5′ ins (CTAGTCTAGAGTTTATGGGATCAAAGCCTAAAG) and 6D3′ins (CTAGCTCGAGATTATCTATTGGTATTATATCAAG) to obtain pBK6D and related mutants. pCbcl2-6D and other mutants were obtained by amplifying a viral fragment containing exon 6D and neighboring sequences from pNL-JB and mutants primers MC33.51 (AGCTGAATTCGGATCAAAGCCTAAAGCCATGTG) and MC42.1 (AGCTACCTTCAGAAGGAAAATGCATATTCTTTCTGCACCTTA) and inserting them within the intron of the pCbcl2 construct.
Cell transfections and RT-PCR analysis.
HEK-293T cells were maintained at below 80% confluence in Dulbecco's modified Eagle's medium (Gibco BRL) supplemented with 5% fetal calf serum and gentamicin (0.5 mg/ml). Cells were seeded in six-well plates 2 h before transfection at 30% confluence in fresh medium containing 5% fetal calf serum and no antibiotics. A mixture containing 3 μl Lipofectamine 2000 (Invitrogen) and 2 μg of expression plasmids was added to the cells. In parallel, 0.3 μg of plasmid pEGFP-N1 (Clontech) was added to each transfection mixture as a normalizer for transfection, RNA extraction, and reverse transcription (RT) efficiency. Cells were harvested after transfection as described for each experiment. Total RNA was extracted with the Stratagene RNA extraction kit and DNase treated twice with Turbo DNase (Ambion). Total mRNA was reverse transcribed utilizing a random pd(N)6 primer. PCRs for the pbcl2-6D constructs were performed utilizing primers pCbcl5 (AGAGCGTCAACCGGGAGATG) and pCbcl3 (GCCCTCTAGATGCATGCTCG). PCR products were separated on an agarose gel and quantified utilizing the Kodak 1D software. Quantitative PCR analysis of the viral transcripts was obtained with the following primer pairs: for all viral mRNAs, Ex8_5a (TTGCTCAATGCCACAGCCAT) and Ex8_3a (TTTGACCACTTGCCACCCAT); for gag/pol mRNas, 5′gag/pol_a (TTCTTCAGAGCAGACCAGAGC) and 3′gag/pol_a (GCTGCCAAAGAGTGATCTGA); for env mRNA, env(f) (GGCGGCGACTGGAAGAAGC) and Env(r) (CTATGATTACTATGGACCACAC); for tat, rev and nef mRNAs, 20.1_Spl (TCTATCAAAGCAACCCACCT) and 20.2_Spl (CGTCCCAGATAAGTGCTAAG); for 6D mRNAS, MC20.1_6D (CACAAGCATAAGAG ACCCAC) and MC20.3_6D (TCTCAAGCGGTGGTAGCTGA). Each sample was normalized by the relative content in enhanced green fluorescent protein (EGFP) transcript detected with the primers E5a (ACCACATGAAGCAGCACGACTTCT) and E5b (TCACCTTGATGCCGTTCTTCTGCT). quantitative PCR (qPCR) was performed utilizing a Stratagene Mx3005P real-time PCR system and analyzed with MxPro V3.0 software. Each assay was carried out with a minimum of three independent transfections while qPCR assays were carried out in duplicate.
siRNA assay.
HEK-293T cells were seeded in six-well plates 2 h before transfection at 30% confluence in fresh medium containing 5% fetal calf serum and no antibiotics. Cells were cotransfected with a mixture containing 3 μl of Lipofectamine 2000 (Invitrogen) and 80 pmol small interfering RNA (siRNA). Qiagen HP-validated siRNAs for hnRNP H (SI02654799) and SC35 (SI00301777) and negative control siRNA (1022076) were utilized in the assays. Cells were harvested and RNA analyzed 72 h after siRNA treatment.
Infection assay.
TZM-bl cells (obtained from the NIH AIDS Research and Reference Reagent Program) were seeded 6 h before infection in 96-well plates at 40% confluence in 200 μl of Dulbecco's modified Eagle's medium supplemented with 5% fetal calf serum and gentamicin (0.5 mg/ml). Next, we added 20 ml of the supernatant recovered from the HEK-293T cells 72 h after the transfection carried out with the proviral constructs. Finally, 48 h after infection cells were lysed and luciferase activity was assayed and quantified utilizing a Molecular Devices Spetromex-M5 and SoftMax Pro 5.2 software. Each infection was carried out in six replicates.
RAC.
Substrate RNAs for RAC assays were synthesized utilizing T7 RNA polymerase. RNAs were covalently linked to adipic acid dihydrazide agarose beads as previously described (34). The beads containing immobilized RNA were incubated in a reaction mixture containing 250 μl of HeLa cell nuclear extract in a final volume of 400 μl (20 mM HEPES-KOH pH 7.9, 5% glycerol, 0.1 M KCl, 0.2 mM EDTA, 0.5 mM dithiothreitol, 4 mM ATP, 4 mM MgCl2). Beads were then washed and the proteins specifically bound to the immobilized RNA eluted in 2% sodium dodecyl sulfate (SDS), separated on polyacrylamide-SDS gels, electroblotted, and probed with antibodies against SC35 (provided by J. Stevenin, INSERM, Strasbourg, France), hnRNP H/H1 (provided by D. L. Black, University of California, Los Angeles), hnRNP L, A1, and PTB (provided by G. Dreyfuss, University of Pennsylvania), U1a (provided by I. Mattaj, EMBL, Heidelbergh, Germany), and U2AF35 (provided by T. Maniatis, Harvard University).
Enzymatic analysis of RNA secondary structure.
RNA secondary structure determination with the use of limited V1 RNase (Ambion), T1 RNase (Ambion), and S1 nuclease (Fermentas) digestion has been described in detail elsewhere (6). Substrate RNAs were synthesized utilizing T7 RNA polymerase, and 1-μg aliquots of RNA were digested in a 100-μl final volume with 0.006 U of RNase V1, 0.06 U of RNase T1, and 0.9 U of S1 nuclease for 10 min at 30°C. An enzyme-free aliquot was processed together and used as a control. RNA substrates did not undergo a denaturation-renaturation step prior to the digestion. The cleaved RNAs were retrotranscribed according to standard protocols using the following antisense primer end labeled with 32P: 5′-ATTATCTATTGGTATTATATCAAGTTTA-3′. In silico secondary structure predictions were performed using the mFold program (42).
RESULTS
Construction of a proviral system to measure exon 6D splicing in vivo.
To facilitate the introduction of mutations within a proviral construct, we created a cloning cassette that is capable of splicing the exon 6D. To do this we substituted exon 6D and its flanking intronic sequences into the proviral construct pNL 4-3 (Fig. 1B). The pNL 4-3 provirus does not carry the exon 6D specific splice sites, while the cloning cassette derived from the viral isolate HXB2 does. The new provirus was named pNL-JB, and throughout the paper we refer to this construct and the sequences derived from it as wild type. Next, we introduced the T-to-C mutation activating exon 6D splicing (pNL-JB-C) and mutated the sequence that characterizes the hnRNP H binding site both in the wild-type sequence to obtain pNL-JB-CCC and in the T-to-C mutation context to obtain pNL-JB-C-CCC. To prove that the effects on viral replication and infectivity observed in the mutants were dependent on splicing of the exon 6D, we also mutated the exon 6D 5′ splice site in the plasmids carrying either the wild-type or the T-to-C mutated sequence (pNL-JB-5′M and pNL-JB-5′M-C).
Characterization of the protein binding profile of the HIV-1 SRE.
To confirm that hnRNP H and the SR protein SC35 assembled onto the exon 6D sequence within the proviral clones utilized in this study, we performed RAC assays. RNA bait substrates, containing exon 6D and portions of the flanking intron sequences, were generated from each proviral clone and incubated with HeLa nuclear extracts; proteins specifically bound to the bait RNAs were then eluted and analyzed. The control bait RNA (RNAcont) was generated by transcribing the antisense strand of the amplicon generated from the pNL-JB substrate. Mutation of the hnRNP H binding site within exon 6D strongly reduces binding of this protein (Fig. 1D, lanes 3 and 4). The SR protein SC35, which we had previously shown to activate splicing of a short in vitro splicing substrate containing the exon 6D polypurinic element (7), binds with similar efficiency to all the substrates, with the exception of the control RNA (Fig. 1D, lane 7). Mutation of the exon 6D splice site did not influence the recruitment of hnRNP H or SC35 (Fig. 1D, lanes 5 and 6).
The same approach allowed us to determine whether these mutations were capable of altering the assembly of prespliceosomal components to exon 6D splice sites. We had previously shown that mutations within the SRE did not alter binding of U2AF65/35 to the 3′ splice site, while mutation of the hnRNP H binding site appeared to downregulate the assembly of the U1snRNP to the 5′ splice site (7). Antibodies against U2AF35 and the U1a component of U1snRNP were used to detect the presence of U2AF65/35 and U1snRNP on the substrates. As shown in Fig. 1D, lane 2, the T-to-C mutation, which strongly activates exon 6D splicing, does not alter assembly of U2AF65/35 and U1snRNP to the splice sites, suggesting that the SRE regulates a later step in spliceosome assembly. The only inconsistency between these data and those previously obtained with the partial exon 6D substrates (7) is that the mutation of the hnRNP H binding site did not induce a decrease in the recruitment of U1snRNP (Fig. 1D, lane 3). We believe that this can be attributed to the longer substrate, containing both the 5′ and 3′ splice sites, that we have utilized in this assay.
Characterization of the mRNA transcripts profiles generated by each proviral clone.
Next, we analyzed the viral mRNA transcripts generated by each proviral clone following transient transfection of HEK-293T cells. The EGFP expression vector pEGFP-N1 was cotransfected with each proviral clone and utilized as a normalizing control for efficiency of transfection, RNA extraction, and reverse transcription. Total RNA preparations were treated extensively with DNase to eliminate traces of contaminating plasmids; transcripts were than quantified by RT-qPCR analysis. Three primer sets spanning different splice junctions were designed to specifically detect the env (4-kb) mRNA, the 2-kb messages (coding for tat, rev, and nef), and the transcripts that include exon 6D. A fourth set of primers was designed to anneal to a region downstream of the final 3′ splice site (Fig. 1A, 3′ss 7), common to all viral mRNAs; this primer set detects all the viral mRNA species and thus can be used to measure overall viral transcription. A fifth set of primers was designed to anneal immediately downstream of the major HIV 5′ splice site (Fig. 1A, 5′ss 1) and is specific for the gag/pol messengers.
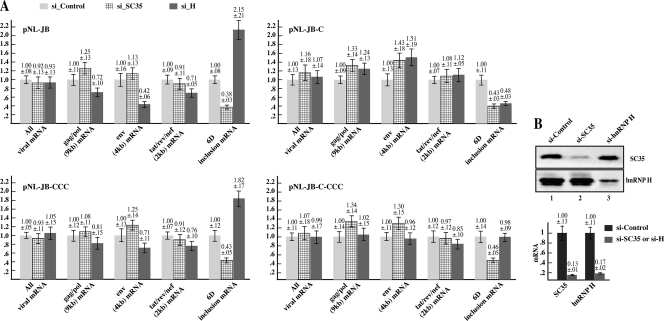
FIG. 3.
Downregulation of SC35 and hnRNP H by siRNA alters exon 6D expression. A) HEK-293T cells were transfected with the indicated proviral constructs or pEGFP-N1 as normalizing control and treated with either hnRNP H, SC35, or control siRNAs. mRNA was extracted 72 h after the siRNA treatment and analyzed by qPCR as described for Fig. 2. The product obtained from each reaction was normalized for the content in the pEGFP-N1 gene product and plotted versus the amount expressed by the si-Control-treated cells. Each graph shows the analysis of the viral mRNA species expressed by the indicated proviral clone. B) The graph indicates the quantification by qPCR of the SC35 and hnRNPH mRNA extracted from the respective siRNA-treated cells in comparison with the si-Control treated cells. The right panel shows the amounts of SC35 and hnRNP H proteins present in the siRNA-treated cells in comparison with the si-Control-treated cells.
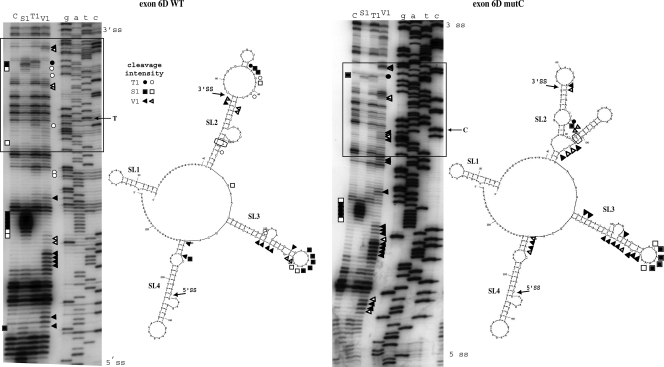
FIG. 5.
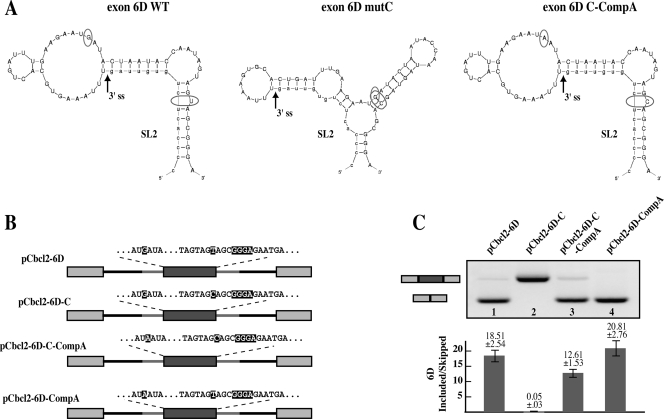
Structural analysis of exons 6D wild-type (WT) and 6DmutC. In vitro-transcribed 6D WT and 6DmutC RNAs were enzymatically digested with S1 nuclease (squares), T1 (circles), and V1 RNases (triangles). No enzyme was added to the RNA in a control reaction mixture (lane C). Black and white symbols indicate high and low cleavage intensity, respectively. The RNA substrate on which this analysis was performed consisted of the entire RNA transcript from pBK6D-WT and pBK6D-C plasmids (approximately 230 nucleotides), which included exon 6D and flanking intronic sequences. The cleaved fragments were detected by performing an RT reaction using a labeled 32P-end-labeled antisense oligo and separating them in a denaturing 6% polyacrylamide gel. A sequencing reaction performed with the same RT primer was run in parallel to the cleavages in order to determine the cleavage sites (lanes G, A, T, and C). The box indicates the region containing the T-to-C substitution and the main changes in the enzymatic digestions. On the right of each gel a schematic diagram representing the best-fitting mFold prediction for these two mRNAs is shown (the observed cleavages are superimposed on the different stem-loop structures SL1 to SL4). Also in these diagrams the positions of the splice sites are indicated by arrows while the T-to-C mutation is circled.
The parent proviral clone pNL4-3 expresses roughly double the amount of total viral transcripts as pNL-JB (Fig. 2, all viral mRNAs). Since pNL-JB differs from pNL4-3 for a central portion of roughly 1.4 kb, it is likely that sequences regulating viral mRNA stability contained within this region alter the overall amount of viral mRNA present (40). Nevertheless, all the proviral vectors derived from pNL-JB express similar amounts of viral transcripts, showing that the mutations within exon 6D do not interfere with viral transcription or stability. We also analyzed the total amount of viral mRNA present in the nuclear and cytoplasmic RNA fractions and observed no difference in the ratio of nuclear versus cytoplasmic mRNAs generated by the different constructs. This indicates that RNA export was not affected by the mutations introduced to create a provirus capable of splicing exon 6D (data not shown).
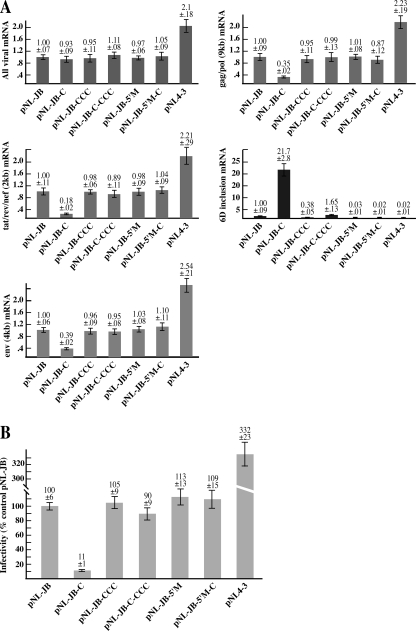
FIG. 2.
A) qPCR analysis of the viral mRNA transcripts transcribed by each proviral clone. HEK-293T cells were transfected with the indicated proviral clone and pEGFP-N1 (normalizing control). mRNA was extracted 72 h after transfection and analyzed by qPCR. Five primer sets were designed to detect the following viral mRNA species: all viral transcripts, env (4kb) mRNA, gag/pol (9kb) mRNA, the 2-kb transcripts (tat, rev and nef), and all the transcripts that include exon 6D. Each graph summarizes the quantification of the indicated mRNA species. The product obtained by each reaction was normalized for the sample content in the pEGFP-N1 gene product and plotted versus the amount expressed by the cells transfected with the pNL-JB provirus. The amount of each mRNA species generated by pNL-JB was utilized as a reference point and assigned a value of 1. B) Infectivity of the proviral mutants. Supernatant from HEK-293T cells transfected with the proviral clones was used to infect cultures of TZM-bl cells. At 48 h after infection, virus entry was measured by analyzing the indicator cells for luciferase activity. Virus infectivity was calculated as the percentage of the luciferase activity value for each proviral clone versus the mean value of the control provirus pNL-JB.
Analysis of the levels of the various viral mRNA species shows that the T-to-C mutation increased exon 6D splicing 21-fold (Fig. 2A, 6D inclusion mRNA). The same mutation decreased the relative amount of env, gag/pol threefold and the 2-kb mRNAs fivefold (Fig. 2A, gag/pol, env and tat/rev/nef mRNAs). Mutation of the hnRNP H binding site in combination with the T-to-C mutation (clone pNL-JB-C-CCC) restored the level of mRNAs including exon 6D to the wild-type level, increasing the amount of env, gag-pol, and 2-kb mRNAs. Thus, the single T-to-C mutation dramatically alters the splicing of the provirus in vivo, and this point mutation-induced radical change in splicing is strongly dependent on the downstream hnRNP H binding site. Mutation of the 6D 5′ splice site abolished inclusion of exon 6D and increasing the unspliced (gag/pol) and normally spliced (env, tat, rev, and nef) viral mRNAs in both the wild type and T-to-C mutant construct.
FIG. 6.
Exon 6D splicing and viral infectivity are reestablished by a structural compensatory mutation. A) SL2 mFold structures for the wild-type (6D WT), 6D T-to-C mutant (6D mutC), and predicted structure for the stem-loop carrying the 6D T-to-C mutation combined with the −20G-to-A compensatory mutation (6D C-CompA). Locations of the T to C and the compensatory −20G-to-A mutations are indicated by circles. B) Schematic representation of the heterologous pCbcl2-exon6D constructs and sequence of the region carrying the T to C and the compensatory −20G to A mutations. C) Alternative splicing ratios of the pCbcl2-6D minigenes. HEK-293T cells were transiently transfected with the pCbcl2 minigenes. mRNA was extracted 24 h after transfection and analyzed by RT-PCR. The splicing products representing exon 6D exclusion (lower band) and inclusion (upper band) were quantified and their ratio is indicated in the graph at the bottom of the figure.
We next sought to correlate the altered expression of the viral mRNA species with viral infectivity. To this end, supernatant collected from the HEK-293T cells transfected with the different proviral clones was used to infect TZM-bl cells (38), which contain an integrated copy of the luciferase gene under control of the HIV-1 long terminal repeat promoter. TZM-bl cells are easily infected with HIV-1, and once integrated into the cellular genome the virus begins expressing the viral protein Tat, which activates the viral promoter and luciferase expression. Thus, luciferase expression in these infected cells is a reliable reporter for viral infectivity. The T-to-C mutation decreases viral infectivity by 90%, and disruption of the hnRNP H binding site reestablishes viral infectivity to a similar level as wild type (Fig. 2B). Both the mutation of the hnRNP H binding site within the wild-type sequence background and also the mutation of the exon 6D 5′ splice site caused a 5 to 10% increase in viral infectivity. This strongly correlates with a decrease in the rate of exon 6D inclusion observed in both clones. The infectivity of the pNL4-3 clone was threefold higher than that of pNL-JB. This was not unexpected, given the higher level of viral mRNA expression of the pNL4-3 clone.
hnRNP H and the SR protein SC35 regulate exon 6D splicing in vivo.
In order to test whether hnRNP H or SC35 regulate the inclusion of exon 6D in vivo as they do in vitro, HEK-293T cells were treated with either hnRNP H, SC35, or control siRNAs (Fig. 3B) and transfected with the proviral constructs (Fig. 3A). The EGFP expression plasmid pEGFP-N1 was cotransfected as a normalizing control. Downregulation of SC35 decreased the exon 6D-containing mRNAs in all constructs (Fig. 3A, cf. si_Control with si_SC35). These data are in agreement with previous work that showed that SC35 activates the exon 6D polypurine ESE (7) and that disruption of the ESE correlates with a decrease in exon 6D splicing and higher viral replication (39). The 2-kb tat, rev, and nef mRNAs did not vary significantly in the SC35 siRNA-treated proviral clones. It should be noted that SC35 has been shown to upregulate splicing of tat mRNAs in vivo (22) and in vitro (18, 41). Therefore, its downregulation would have likely acted to reduce the 2-kb mRNAs by directly decreasing splicing to the tat, rev, and nef specific splice sites. However, the same mRNAs are upregulated by reducing the amount of exon 6D-containing mRNA species, possibly explaining why little change could be observed in the expression of 2-kb mRNAs.
Expression of the mRNAs that efficiently splice exon 6D from the wild-type pNL-JB vector more than doubled after treatment with hnRNP H siRNA (Fig. 3A, pNL-JB). This indicates that in the context of the wild-type viral sequence, hnRNP H acts as a repressor of efficient splicing of exon 6D. Surprisingly, siRNA treatment of the proviral clone pNL-JB-CCC also induced a substantial increase in exon 6D mRNAs (Fig. 3A, pNL-JB-CCC). This indicates that the splicing repressor activity exerted by hnRNP H is independent of the hnRNP H binding site present within exon 6D. Sequence analysis of the proviral constructs reveals more than 50 putative hnRNP H binding sites within the viral transcript, 7 of which are contained within the exon 6D upstream intron and 11 within the downstream one. Thus, it is conceivable that binding of hnRNP H to sites other than the characterized one within the exon may regulate exon 6D splicing. Interestingly, hnRNP H siRNA treatment reduced by 50% the exon 6D-containing mRNAs expressed from the pNL-JB-C vector, while causing little change in the expression of exon 6D-containing viral mRNAs generated by pNL-JB-C-CCC (Fig. 3A). Therefore, the T-to-C mutation abolishes the hnRNP H splicing repressor activity that is independent of the exon 6D GGG binding site. Altogether, these data suggest that in the presence of the T-to-C mutation hnRNP H promotes splicing of exon 6D, while in the absence of the T-to-C mutation hnRNP H represses the splicing of this exon.
The regulation of exon 6D splicing can be reproduced in a heterologous system.
Given the complexity of the mechanisms regulating HIV-1 splicing and the possibility of long-range interactions, we wanted to determine if the cis regulatory elements required for the regulation of exon 6D splicing were confined to the exon and the neighboring intronic sequences. To this end, we inserted exon 6D, together with the flanking intronic sequences, into the intron of the pCbcl2 minigene (Fig. 4A). Alternative splicing ratios of the pCbcl2-6D minigenes were determined by RT-PCR following transient transfection of HEK-293T cells (Fig. 4B).
FIG. 4.
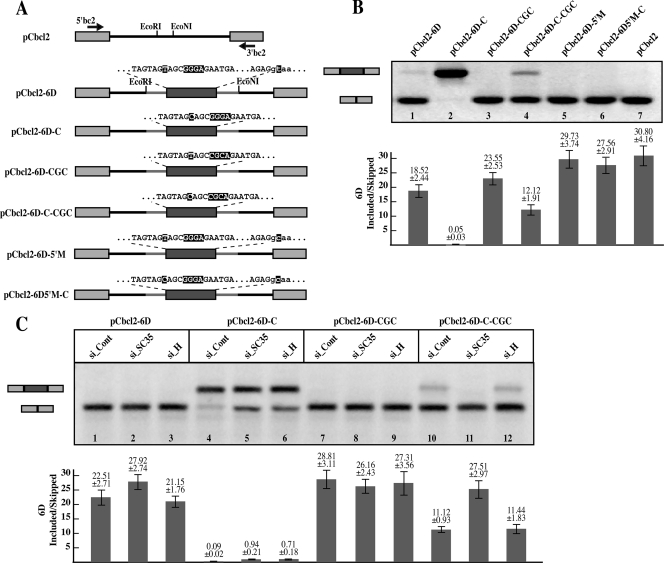
Exon 6D splicing is reproducible within a heterologous system. A) Exon 6D sequence inserted within the intron of the pCbcl2 minigene is comprehensive of the full exon, the first 30 nucleotides of the downstream intron, and 50 nucleotides of the upstream intron. Exon 6D mutations carried out in each minigene are indicated. B) Alternative splicing ratios of the pCbcl2 minigenes. HEK-293T cells were transfected with the pCbcl2 minigenes, and mRNA was extracted 48 h after transfection and analyzed by RT-PCR with the indicated primers (5′bc2 and 3′bc2). The endogenous bcl2 transcripts do not interfere with amplification of the pCbcl2 minigene since in the endogenous gene the size of the intervening intron is over 200 kb. The splicing products representing exon 6D exclusion (lower band) and inclusion (upper band) were quantified and their ratio is indicated in the graph at the bottom of the figure. C) Downregulation of SC35 and hnRNP H by siRNA alters exon 6D expression in the pCbcl2 minigene. HEK-293T cells were transfected with the constructs indicated on top of the figure and treated with either hnRNP H, SC35, or control siRNAs. mRNA was extracted 72 h after the siRNA treatment.
In this context, the wild-type exon 6D is nearly completely skipped (Fig. 4B, lane 1). Nonetheless, the T-to-C mutation caused exon 6D to be constitutively included in the messages (Fig. 4B, lane 2). Mutation of the GGG hnRNP H binding site in combination with the T-to-C mutation restored exclusion of the viral exon, similar to the effect observed in the proviral clones (Fig. 4B, lane 4). Exon 6D exclusion was also restored by mutation of the 6D 5′ splice site (Fig. 4B, lane 6). This indicates that both the T-to-C mutation and the hnRNP H binding site are required for efficient splicing of exon 6D in the heterologous construct, similarly to what was observed in the full-length viral transcripts.
To show that hnRNP H and SC35 were regulating exon 6D expression within the heterologous construct, HEK-293T cells were then treated with silencing RNAs directed against either hnRNP H, SC35, or a control and transfected with the constructs indicated in Fig. 4C. Similar to the results obtained with the full proviral constructs, both SC35 and hnRNP H were required for efficient splicing of exon 6D in the minigene carrying the T-to-C mutation (Fig. 4C, lanes 5 and 6). The requirement of hnRNP H for inclusion of exon 6D was lost in the construct carrying both the T-to-C and hnRNP H binding site mutations (Fig. 4C, lane 12).
Little difference was observed after siRNA treatments of the constructs pCbcl2-6D and pCbcl2-6D-CCC carrying the wild-type sequence or only the mutation of the hnRNP H binding site, respectively (Fig. 4C, lanes 1 to 3 and 7 to 9). However, both constructs generate mRNAs that show a nearly complete absence of exon 6D, thus it is unlikely that an effect further enhancing its exclusion might be observed. Furthermore, the absence of an increase in exon 6D inclusion after hnRNP H siRNA treatment of the cells transfected with the minigene carrying the wild-type exon 6D sequence, as opposed to the effect seen with the pNL-JB proviral vector (Fig. 3A), might also be attributable to the absence of hnRNP H binding sites outside exon 6D in the heterologous construct.
The T-to-C mutation induces a specific structural change in a stem-loop region of the viral transcript.
Extensive analysis of the cellular factors that bind to the wild type (T) and mutant (T-to-C) exon 6D sequences carried out by RNA affinity chromatography coupled with fractionation by SDS-polyacrylamide gel electrophoresis followed by mass spectrometry analysis and immunostaining for known splicing factors failed to identify additional cellular or viral factors whose recruitment was modified by the mutation (Fig. 1D, hnRNP A1, L, and PTB, and data not shown). Although we cannot completely rule out the involvement of other trans-acting factors whose recruitment changes upon the T-to-C mutation, all the evidence we have gathered up to now points to the conclusion that only members of the hnRNP H and SR protein families are regulating exon 6D splicing.
We set out to investigate whether the T-to-C mutation was inducing a structural change in the viral mRNA secondary structure. In silico analysis of the region containing the 6D wild-type construct, including the upstream and downstream intronic sequences, performed utilizing the mFold program, suggests that this sequence could fold upon itself to form several stem-loop elements that we named SL1 through SL4 (Fig. 5). Their existence was then tested experimentally by partial RNA digestion using specific RNases, including V1 (which cleaves double-stranded RNA and stacked RNA regions), T1 (which cleaves single-stranded guanosines), and S1 nuclease (which cleaves single-stranded RNA without sequence specificity). As shown in Fig. 5A, the observed cleavages are entirely consistent with the folding prediction for the SL1, SL2, and SL3 stem-loop structures. SL2 and SL3 are well-defined in terms of double-stranded (V1) and single-stranded (T1 and S1) cleavages, especially in their apical loop elements. Following the introduction of the T-to-C mutation, very few changes were observed in the cleavage pattern of SL3 and SL4 in the wild-type and mutant RNAs, suggesting that these structures are not affected by the mutation. The cleavage pattern undergoes an extensive change in the stem loop 2 (SL2) where the predominant presence of single-stranded cleavages observed in the wild-type structure that correspond to the apical SL2 loop element (Fig. 5A) is replaced by the predominance of double-stranded cleavages (V1) in the mutant SL2 structure (Fig. 5B). These results suggest that a striking change has occurred in the folding of this element. This is also entirely consistent with the folding predictions obtained with mFold when we introduced the T-to-C substitution in the exon 6D RNA sequence. The reason for this change was consistent with the prediction that the T-to-C mutation could induce a base pairing of the newly inserted C with a G positioned 20 nucleotides upstream (−20G) of the T-to-C mutation. Most importantly, this structural change remains localized in SL2, as can be observed by the fact that cleavage specificities and intensities in the SL3 loop remain almost unchanged in both 6D and 6D mutC structures (Fig. 5A and B, lower halves of the panels).
It has been observed in both viral and mammalian systems (3, 13, 23, 26, 36, 37) that mRNA secondary structure can affect the recognition of 5′ or 3′ splice sites by modifying their availability to the splicing machinery. This does not appear to be the case in exon 6D, since in both the wild-type and the 6D mutC structures, the 6D 3′ss remains located in a short stem just upstream of a loop formed by the first exonic nucleotides. This conclusion is also confirmed by the RAC data in Fig. 1, where no change was observed in the binding of U2AF or U1snRNP factors in both the wild-type and mutant 6D RNAs. The fact that the accessibility of niether the 6D 5′ss nor the 3′ss seems to be affected by the structural changes induced by the T-to-C mutation suggests that these structural changes affect splicing in ways other than by improving splice site accessibility.
A compensatory mutation restores splicing and infectivity in the T-to-C mutant provirus.
Since the T-to-C mutation induced a conformational change in stem-loop 2 (SL2) we wanted to investigate the possibility that this was the cause for the striking differences in exon 6D splicing efficiency observed in both the viral and heterologous constructs. Accordingly, mFold analysis of SL2 predicted that mutation of the −20 G to A would restore the wild-type SL2 structure (Fig. 6A, exon 6D C-CompA). To validate this prediction we inserted the compensatory −20G-to-A mutation in the pCbcl2-C construct, generating pCbcl2-C-CompA. This second compensatory mutation dramatically restored the wild-type splicing pattern (Fig. 6B and C, lanes 2 and 3). Insertion of the compensatory point mutation alone in the wild-type background did not modify the splicing pattern of the heterologous construct (Fig. 6B and C, lane 4, pCbcl2-CompA).
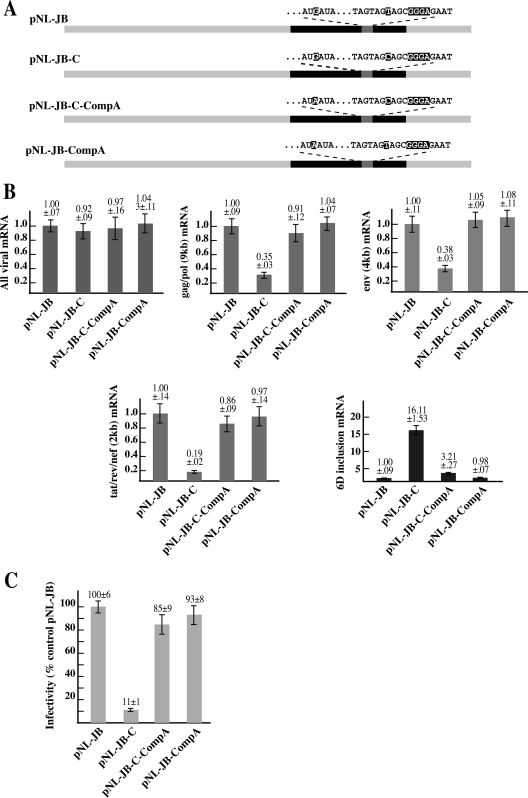
Next, we set to investigate if the compensatory −20G-to-A mutation could also restore the wild-type splicing pattern and infectivity in the pNL-JB-C provirus. qPCR analysis of the viral mRNA species generated by the pNL-JB-C-CompA provirus (Fig. 7A and B) showed that the compensatory mutation induced a fivefold reduction of exon 6D, including mRNAs and an increase of the gag/pol, env, and 2-kb mRNAs, thus reestablishing the splicing patterns of the wild-type pNL-JB proviral construct. Finally, we tested the infectivity of the proviruses generated by the clones containing the compensatory mutation by infecting TZM-bl cells and assaying for luciferase expression of the infected cells. The −20G-to-A mutation restored viral infectivity to the pNL-JB-C mutant to 85% of the wild-type level. No differences in the relative amount of mRNA species or infectivity were detected with the control pNL-JB-CompA construct.
FIG. 7.
A) Schematic map of the proviral clones. The compensatory −20G-to-A mutation was inserted in the proviral clone pNL-JB to generate pNL-JB-CompA and into pNL-JB-C to generate pNL-JB-C-CompA. B) qPCR analysis of the viral mRNAs transcribed by the proviral clones carrying the regulatory and compensatory mutations. HEK-293T cells were transfected with the indicated proviral clone and pEGFP-N1 (normalizing control). mRNA was extracted 72 h after transfection and analyzed by qPCR. Each graph summarizes the quantification of the indicated mRNA species. The product obtained by each reaction was normalized for the sample content in pEGFP-N1 gene product and plotted versus the amount expressed by the cells transfected with the pNL-JB provirus. C) Relative infectivity of the proviral mutants. Supernatant from the HEK-293T cells transfected with the proviral clones was used to infect cultures of TZM-bl cells. At 48 h after infection, virus entry was measured by analyzing the indicator cells for luciferase activity. Virus infectivity was calculated by dividing the luciferase activity value for each proviral clone by the mean value of the control provirus pNL-JB.
DISCUSSION
During the process of transcription, the emerging nascent transcript is quickly assembled into large heterogeneous ribonucleoprotein particles and may be allowed only a very short time span to fold into stable secondary and tertiary structures (12). However, an increasing number of reports suggest the presence of specific secondary structure along the transcript, which might influence the splicing machinery (5). Indeed, the binding of several positive and negative splicing factors to the substrate RNA has been shown to depend on the RNA secondary structure as well as on the target nucleotide sequence (4, 5, 11, 30, 35), suggesting a straightforward way for RNA structure to influence splicing. Another mechanism by which RNA secondary structure might influence splicing is the regulation of the distance between splicing regulatory elements, as demonstrated for the stem-loop element in the FGFR-2 transcript (1). Recent evidence in the Drosophila melanogaster Dscam gene has shown that alternative regulatory mechanisms mediated by RNA structure may also work through different and still-to-be-defined mechanisms (16, 31).
The results presented in this work demonstrate that a structural change in the HIV-1 transcript can modulate the activity of a complex SRE within exon 6D whose functions require members of the hnRNP H and SR protein families (Fig. 8). Intriguingly, downregulation of hnRNP H within the context of the wild-type virus induces an increase in exon 6D inclusion, and this effect is not dependent on the hnRNP H binding site present within the exon. On the contrary, downregulation of hnRNP H in the context of the T-to-C mutant virus reduces inclusion of the exon, and the effect is dependent on the hnRNP H binding site within exon 6D. In parallel, however, we have demonstrated that the T-to-C mutation can induce a conformational change within a stem-loop element that contains the SRE sequence (SL2). This conformational change does not alter hnRNP H binding to the SRE but might affect hnRNP H spatial location with respect to other splicing factors and the splice sites. The finding that hnRNP H activity could be modulated by the secondary structure of the RNA in proximity to its binding site may prove to be key in understanding the ability of the hnRNP H family members to act as both splicing repressors or enhancers, in addition to other peculiar functional features of this protein. Previous work has indicated that hnRNPs of the H family may promote intron removal by binding multiple intronic sequences and possibly by interacting with other splicing regulators, such as hnRNP A1 (19, 28). Nevertheless, not all hnRNP H binding sites appear to be equally functional. The DGGGD consensus binding sequences for the members of the hnRNP H family can be found more than 50 times within the HIV-1 viral transcript, but not all those binding sites appear to be functional splicing enhancers or silencers, although they all appear to recruit hnRNPs of the H family. In previous work we showed that out of a stretch of eight putative hnRNP H binding sites within a region of the gag/pol gene, only one was functional in regulating splicing of a viral substrate (34). Why are only a few hnRNP H binding sites functional? The data presented in this work suggest that a proper conformation of the substrate mRNA in the proximity of the hnRNP H binding site modulates the activity of this protein. An interesting parallel can be made with SMN2 exon 7, where the splicing repressor activity of hnRNPA1 binding to different ESS is dependent on the nucleotide context in which the splicing silencers are localized (24). Taken together, these results demonstrate that protein binding sites and functional activities must be assessed while taking into account the context in which they are localized within the RNA molecule.
FIG. 8.
Model for exon 6D splicing regulation. A) A specific structural conformation is required for efficient splicing of exon 6D. Presence of the T nucleotide induces a secondary structure that prevents efficient utilization of the exon 6D splice sites. hnRNP H binding to GGG sequences outside exon 6D inhibits efficient splicing of the exon while SR proteins promote it. In this scenario, specific binding of hnRNP H within exon 6D does not affect splicing. The yellow circles represent the spliceosome, while red bars represent putative hnRNP H binding sites. B) The T-to-C mutation causes a conformational change in the structure of stem-loop 2. This structural change activates splicing to the exon 6D splice sites. In this case, however, hnRNP H bound to the exon 6D GGG enhances the exon splicing efficiency.
Finally, it is still unclear if the change in RNA secondary structure alters the positioning of the bound hnRNP H with respect to other spliceosomal/nonspliceosomal factors and other regulatory sequences or whether it might disrupt some other mechanism leading to inhibition of splicing of the 6D exon. The available data suggest that a later step in spliceosome assembly might be differentially regulated by the presence of hnRNP H in the T-to-C mutant. In agreement with this view, our previous work showed that members of the hnRNP H family could affect splicing by promoting the formation of ATP-dependent spliceosomal complexes (34). Furthermore, Martinez-Contreras and coworkers showed that regulation of splicing via hnRNP H interaction with sequences different from the 5′ splice site does not alter binding of the U1 snRNP (28). We propose that the presence of the T nucleotide induces a secondary structure that prevents efficient utilization of the exon 6D splice sites by the spliceosome. hnRNP H binding to GGG sequences outside exon 6D are likely to inhibit splicing of the exon by upregulating the splicing of other viral mRNA species. While in the wild-type virus binding of hnRNP H within exon 6D does not affect splicing, the T-to-C mutation induces a conformational change in the structure of stem-loop 2 that results in the activation of exon 6D splicing mediated by the bound hnRNP H (Fig. 8).
Properly balanced splicing of the single HIV-1 pre-mRNA is essential for the viral life cycle and is dependent on the host cell splicing machinery. In this work, we have shown that mutations in any of the elements constituting the SRE or in the cellular factors interacting with it can alter the amount of exon 6D-containing 2-kb mRNAs. Furthermore, we have shown how a structural compensatory mutation can reconstitute infectivity to the virus, mimicking a process that, thanks to the low fidelity of the reverse transcriptase, the virus can carry out during the course of the infection. More importantly, we have also shown how the secondary structure of the viral transcript can modify the activity of key splicing factors, in particular hnRNP H, by changing their spatial localization within the exon 6D structure.
Acknowledgments
We thank Francisco Baralle and Alan Zahler for helpful comments.
This work was supported by NIH NIAID grants R01AI052820 to M.C. E.B. and C.S. are supported by Telethon Onlus Foundation (Italy) (GGP06147), FIRB (RBNE01W9PM), and by a European community grant (EURASNET-LSHG-CT-2005-518238).
Footnotes
Published ahead of print on 11 June 2008.
REFERENCES
- 1.Baraniak, A. P., E. L. Lasda, E. J. Wagner, and M. A. Garcia-Blanco. 2003. A stem structure in fibroblast growth factor receptor 2 transcripts mediates cell-type-specific splicing by approximating intronic control elements. Mol. Cell. Biol. 23:9327-9337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benko, D. M., S. Schwartz, G. N. Pavlakis, and B. K. Felber. 1990. A novel human immunodeficiency virus type 1 protein, tev, shares sequences with tat, env, and rev proteins. J. Virol. 64:2505-2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blanchette, M., and B. Chabot. 1997. A highly stable duplex structure sequesters the 5′ splice site region of hnRNP A1 alternative exon 7B. RNA 3:405-419. [PMC free article] [PubMed] [Google Scholar]
- 4.Buckanovich, R. J., and R. B. Darnell. 1997. The neuronal RNA binding protein Nova-1 recognizes specific RNA targets in vitro and in vivo. Mol. Cell. Biol. 17:3194-3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buratti, E., and F. E. Baralle. 2004. Influence of RNA secondary structure on the pre-mRNA splicing process. Mol. Cell. Biol. 24:10505-10514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buratti, E., M. Baralle, L. De Conti, D. Baralle, M. Romano, Y. M. Ayala, and F. E. Baralle. 2004. hnRNP H binding at the 5′ splice site correlates with the pathological effect of two intronic mutations in the NF-1 and TSHβ genes. Nucleic Acids Res. 32:4224-4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caputi, M., and A. M. Zahler. 2002. SR proteins and hnRNP H regulate the splicing of the HIV-1 tev-specific exon 6D. EMBO J. 21:845-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheah, M. T., A. Wachter, N. Sudarsan, and R. R. Breaker. 2007. Control of alternative RNA splicing and gene expression by eukaryotic riboswitches. Nature 447:497-500. [DOI] [PubMed] [Google Scholar]
- 9.Chen, C. D., R. Kobayashi, and D. M. Helfman. 1999. Binding of hnRNP H to an exonic splicing silencer is involved in the regulation of alternative splicing of the rat beta-tropomyosin gene. Genes Dev. 13:593-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chou, M. Y., N. Rooke, C. W. Turck, and D. L. Black. 1999. hnRNP H is a component of a splicing enhancer complex that activates a c-src alternative exon in neuronal cells. Mol. Cell. Biol. 19:69-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Damgaard, C. K., T. O. Tange, and J. Kjems. 2002. hnRNP A1 controls HIV-1 mRNA splicing through cooperative binding to intron and exon splicing silencers in the context of a conserved secondary structure. RNA 8:1401-1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eperon, L. P., I. R. Graham, A. D. Griffiths, and I. C. Eperon. 1988. Effects of RNA secondary structure on alternative splicing of pre-mRNA: is folding limited to a region behind the transcribing RNA polymerase? Cell 54:393-401. [DOI] [PubMed] [Google Scholar]
- 13.Estes, P. A., N. E. Cooke, and S. A. Liebhaber. 1992. A native RNA secondary structure controls alternative splice-site selection and generates two human growth hormone isoforms. J. Biol. Chem. 267:14902-14908. [PubMed] [Google Scholar]
- 14.Fogel, B. L., and M. T. McNally. 2000. A cellular protein, hnRNP H, binds to the negative regulator of splicing element from Rous sarcoma virus. J. Biol. Chem. 275:32371-32378. [DOI] [PubMed] [Google Scholar]
- 15.Garneau, D., T. Revil, J. F. Fisette, and B. Chabot. 2005. Heterogeneous nuclear ribonucleoprotein F/H proteins modulate the alternative splicing of the apoptotic mediator Bcl-x. J. Biol. Chem. 280:22641-22650. [DOI] [PubMed] [Google Scholar]
- 16.Graveley, B. R. 2005. Mutually exclusive splicing of the insect Dscam pre-mRNA directed by competing intronic RNA secondary structures. Cell 123:65-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Graveley, B. R. 2000. Sorting out the complexity of SR protein functions. RNA 6:1197-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hallay, H., N. Locker, L. Ayadi, D. Ropers, E. Guittet, and C. Branlant. 2006. Biochemical and NMR study on the competition between proteins SC35, SRp40, and heterogeneous nuclear ribonucleoprotein A1 at the HIV-1 Tat exon 2 splicing site. J. Biol. Chem. 281:37159-37174. [DOI] [PubMed] [Google Scholar]
- 19.Han, K., G. Yeo, P. An, C. B. Burge, and P. J. Grabowski. 2005. A combinatorial code for splicing silencing: UAGG and GGGG motifs. PLoS Biol. 3:e158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hastings, M. L., C. M. Wilson, and S. H. Munroe. 2001. A purine-rich intronic element enhances alternative splicing of thyroid hormone receptor mRNA. RNA 7:859-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hiller, M., Z. Zhang, R. Backofen, and S. Stamm. 2007. Pre-mRNA secondary structures influence exon recognition. PLoS Genet. 3:e204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jacquenet, S., D. Decimo, D. Muriaux, and J. L. Darlix. 2005. Dual effect of the SR proteins ASF/SF2, SC35 and 9G8 on HIV-1 RNA splicing and virion production. Retrovirology 2:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jacquenet, S., D. Ropers, P. S. Bilodeau, L. Damier, A. Mougin, C. M. Stoltzfus, and C. Branlant. 2001. Conserved stem-loop structures in the HIV-1 RNA region containing the A3 3′ splice site and its cis-regulatory element: possible involvement in RNA splicing. Nucleic Acids Res. 29:464-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kashima, T., N. Rao, C. J. David, and J. L. Manley. 2007. hnRNP A1 functions with specificity in repression of SMN2 exon 7 splicing. Hum. Mol. Genet. 16:3149-3159. [DOI] [PubMed] [Google Scholar]
- 25.Krecic, A. M., and M. S. Swanson. 1999. hnRNP complexes: composition, structure, and function. Curr. Opin. Cell Biol. 11:363-371. [DOI] [PubMed] [Google Scholar]
- 26.Loeb, D. D., A. A. Mack, and R. Tian. 2002. A secondary structure that contains the 5′ and 3′ splice sites suppresses splicing of duck hepatitis B virus pregenomic RNA. J. Virol. 76:10195-10202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marcucci, R., F. E. Baralle, and M. Romano. 2006. Complex splicing control of the human thrombopoietin gene by intronic G runs. Nucleic Acids Res. 35:132-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martinez-Contreras, R., J. F. Fisette, F. U. Nasim, R. Madden, M. Cordeau, and B. Chabot. 2006. Intronic binding sites for hnRNP A/B and hnRNP F/H proteins stimulate pre-mRNA splicing. PLoS Biol. 4:e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Min, H., R. C. Chan, and D. L. Black. 1995. The generally expressed hnRNP F is involved in a neural-specific pre-mRNA splicing event. Genes Dev. 9:2659-2671. [DOI] [PubMed] [Google Scholar]
- 30.Nagel, R. J., A. M. Lancaster, and A. M. Zahler. 1998. Specific binding of an exonic splicing enhancer by the pre-mRNA splicing factor SRp55. RNA 4:11-23. [PMC free article] [PubMed] [Google Scholar]
- 31.Olson, S., M. Blanchette, J. Park, Y. Savva, G. W. Yeo, J. M. Yeakley, D. C. Rio, and B. R. Graveley. 2 December 2007, posting date. A regulator of Dscam mutually exclusive splicing fidelity. Nat. Struct. Mol. Biol. [Epub ahead of print.] doi: 10.1038/nsmb1339. [DOI] [PMC free article] [PubMed]
- 32.Purcell, D. F. J., and M. A. Martin. 1993. Alternative splicing of human immunodeficiency virus type 1 mRNA modulates viral protein expression, replication, and infectivity. J. Virol. 67:6365-6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salfeld, J., H. G. Gottlinger, R. A. Sia, R. E. Park, J. G. Sodroski, and W. A. Haseltine. 1990. A tripartite HIV-1 tat-env-rev fusion protein. EMBO J. 9:965-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schaub, M. C., S. R. Lopez, and M. Caputi. 2007. Members of the heterogeneous nuclear ribonucleoprotein (hnRNP) H family activate splicing of an HIV-1 splicing substrate by promoting formation of ATP-dependent spliceosomal complexes. J. Biol. Chem. 282:13617-13626. [DOI] [PubMed] [Google Scholar]
- 35.Shi, H., B. E. Hoffman, and J. T. Lis. 1997. A specific RNA hairpin loop structure binds the RNA recognition motifs of the Drosophila SR protein B52. Mol. Cell. Biol. 17:2649-2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tu, M., W. Tong, R. Perkins, and C. R. Valentine. 2000. Predicted changes in pre-mRNA secondary structure vary in their association with exon skipping for mutations in exons 2, 4, and 8 of the Hprt gene and exon 51 of the fibrillin gene. Mutat. Res. 432:15-32. [DOI] [PubMed] [Google Scholar]
- 37.Varani, L., M. G. Spillantini, M. Goedert, and G. Varani. 2000. Structural basis for recognition of the RNA major groove in the tau exon 10 splicing regulatory element by aminoglycoside antibiotics. Nucleic Acids Res. 28:710-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wei, X., J. M. Decker, H. Liu, Z. Zhang, R. B. Arani, J. M. Kilby, M. S. Saag, X. Wu, G. M. Shaw, and J. C. Kappes. 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 46:1896-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wentz, P. M., B. E. Moore, M. V. Cloyd, S. M. Berget, and L. A. Donehower. 1997. A naturally arising mutation of a potential silencer of exon splicing in human immunodeficiency virus type I induces dominant aberrant splicing and arrests virus production. J. Virol. 71:8542-8551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wolff, H., R. Brack-Werner, M. Neumann, T. Werner, and R. Schneider. 2003. Integrated functional and bioinformatics approach for the identification and experimental verification of RNA signals: application to HIV-1 INS. Nucleic Acids Res. 31:2839-2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zahler, A. M., C. K. Damgaard, J. Kjems, and M. Caputi. 2004. SC35 and heterogeneous nuclear ribonucleoprotein A/B proteins bind to a juxtaposed exonic splicing enhancer/exonic splicing silencer element to regulate HIV-1 tat exon 2 splicing. J. Biol. Chem. 279:10077-10084. [DOI] [PubMed] [Google Scholar]
- 42.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]