SUMMARY
Natural killer (NK) cells eliminate virally infected and tumor cells. Among the receptors regulating NK cell function is 2B4 (CD244), a member of the signaling lymphocyte-activation molecule (SLAM) family that binds CD48. 2B4 is the only heterophilic receptor of the SLAM family, whose other members, e.g., NK-T-B-antigen (NTB-A), are self-ligands. We determined the structure of the complex between the N-terminal domains of mouse 2B4 and CD48, as well as the structures of unbound 2B4 and CD48. The complex displayed an association mode related to, yet distinct from, that of the NTB-A dimer. Binding was accompanied by the rigidification of flexible 2B4 regions containing most of the polymorphic residues across different species and receptor isoforms. We propose a model for 2B4-CD48 interactions that permits the intermixing of SLAM receptors with major histocompatibility complex-specific receptors in the NK cell immune synapse. This analysis revealed the basis for heterophilic recognition within the SLAM family.
INTRODUCTION
Natural killer (NK) cells are a fundamental component of innate immunity against tumors and virally infected cells. The cytolytic activity of NK cells is regulated by a dynamic interplay between activating and inhibitory signals transmitted by distinct classes of receptors found on their surface (Plougastel and Yokoyama, 2003; Lanier, 2005). The dominant signal received by an NK cell through its interaction with normal numbers of major histocompatibility complex (MHC) class I molecules on target cells is inhibitory. If the number of MHC class I molecules is reduced through infectious or tumorigenic processes, this inhibitory signal is attenuated and the NK cell is activated. Inhibitory receptors specific for MHC class I include the killer immunoglobulin-like receptors (KIRs), members of the Ly49 family, and CD94-natural killer group 2A (CD94-NKG2A) (Natarajan et al., 2002; Deng and Mariuzza, 2006). Activating receptors include NK-T-B-antigen (NTB-A), natural killer receptor protein 1 (NKR-P1), DNAX accessory molecule 1 (DNAM-1), NKp30, and NKp46, whose ligands are non-MHC molecules (e.g., C-type lectin-related molecule 1 [Clr1] for NKR-P1 and CD155 for DNAM-1) (Moretta et al., 2006). In addition, the NK receptor 2B4 (CD244), which binds CD48, has been shown to mediate both activating and inhibitory functions (see below). The balance between positive-signaling receptors (resulting in target-cell lysis) and negative-signaling receptors (preventing lysis) ultimately determines the outcome of NK cell-target cell encounters (Plougastel and Yokoyama, 2003; Lanier, 2005). During such encounters, highly organized complexes of receptors and other molecules, termed NK cell immune synapses, are formed at the NK cell-target cell interface. Moreover, these synapses appear to differ in cytolytic and noncytolytic interactions (Davis et al., 1999; Vyas et al., 2001; Vyas et al., 2002; Orange et al., 2003).
2B4 and NTB-A belong to the signaling lymphocyte-activation molecule (SLAM) family of receptors, which also includes SLAM (CD150), CD84, lymphocyte antigen 9 (Ly9; CD229), and CD2-like receptor-activating cytotoxic cells (CRACC; CD319) (Veillette, 2006; Ma et al., 2007). The SLAM family is a subset of the greater CD2 family, whose members comprise a ligand-binding immunoglobin V (IgV) domain, distal from the cell membrane, and a membrane-proximal IgC2 domain. SLAM family receptors are expressed by a wide range of hematopoietic cells and regulate both innate and adaptive immunity (Veillette, 2006; Ma et al., 2007). With the sole exception of 2B4, which is present on all NK cells, in addition to subsets of γδ and CD8+ T cells, all SLAM family receptors are homophilic because they are self-ligands (Assarsson et al., 2005; Veillette, 2006). By contrast, 2B4 participates in heterophilic interactions with CD48, which is broadly expressed on immune cells (Brown et al., 1998; Latchman et al., 1998). As demonstrated with gene-deficient mice, 2B4-CD48 interactions are essential for interleukin-2 (IL-2)-driven expansion and activation of NK cells, as well as for efficient NK-mediated cytotoxicity and tumor clearance (Lee et al., 2006).
In humans, NK cell activation through 2B4 is accompanied by the phosphorylation of immunoreceptor tyrosine-based switch motifs (ITSMs) in its cytoplasmic tail and the recruitment of SLAM-associated protein (SAP), an SH2 domain-containing adaptor (Chen et al., 2004; Eissmann et al., 2005). This association is crucial for 2B4 to deliver activating signals to NK cells (Tangye et al., 2000; Nakajima et al., 2000; Parolini et al., 2000; Bloch-Queyrat et al., 2005). SAP is mutated in human X-linked lymphoproliferative disease, a severe immunodeficiency characterized by an inability to control Epstein-Barr virus (EBV) infection (Veillette, 2006), such that it cannot associate with 2B4. In these patients, 2B4 fails to activate NK-mediated lysis of EBV-infected cells upon binding CD48.
Surprisingly, 2B4 has also been found to act as an inhibitory NK receptor, at least in mice (Assarsson et al., 2005; Veillette, 2006). Thus, several studies of 2B4-deficient mice have shown that that 2B4 inhibits, rather than increases, NK-mediated killing of CD48-expressing target cells (Lee et al., 2004; Vaidya et al., 2005; McNerney et al., 2005). In this way, 2B4 could participate in conferring self-tolerance to immature NK cells that lack inhibitory receptors for self-MHC class I molecules (Fernandez et al., 2005; Kumar and McNerney, 2005). Although the factors influencing whether 2B4 is stimulatory or inhibitory are unknown, this heterophilic SLAM family receptor appears to be more multifunctional than previously appreciated.
Despite the importance of 2B4-CD48 interactions for the regulation of NK cell activity, structural studies have been confined to the individual proteins: a solution nuclear magnetic resonance (NMR) structure of mouse 2B4 (Ames et al., 2005) and an X-ray crystal structure of rat CD48 (Evans et al., 2006). To understand 2B4-CD48 recognition, we determined the crystal structures of mouse 2B4 and CD48 in both bound and free forms and identified key interactions through the mutational analysis of 2B4. The comparison of the 2B4-CD48 structure with those of the NTB-A and CD84 homophilic dimers (Cao et al., 2006; Yan et al., 2007), and of the CD2-CD58 complex (Wang et al., 1999), revealed the basis for hetero- versus homophilic recognition within the SLAM family of receptors. We further propose a model for 2B4-CD48 interactions at the NK cell immune synapse that would allow the localization of SLAM family receptors with NK receptors that monitor MHC or MHC-like molecules on target cells.
RESULTS
Structure Determination
A soluble version of 2B4, consisting of the ligand-binding IgV domain alone, was expressed in bacterial inclusion bodies and folded in vitro as described previously (Ames et al., 2005). However, initial attempts to produce the IgV portion of CD48 by this approach were unsuccessful, possibly because of the instability of the isolated domain. Accordingly, we used in vitro-directed evolution (yeast display) to isolate stable variants of CD48. It has been demonstrated for several proteins that the efficiency of yeast surface display can serve as a proxy screening variable for the enhanced thermal stability and folding efficiency of mutant proteins (Kowalski et al., 1998; Kieke et al., 1999; Esteban and Zhao, 2004). Accordingly, a library of randomly mutated CD48 molecules was displayed on the surface of yeast by fusion to the agglutinin receptor. We sorted the library by flow cytometry for binding to a fluorescent-labeled CD48 antibody and isolated variants with increased reactivity. For further study, we chose a clone that differed from the wild-type at two positions (Thr-Tyr12 and Lys-Arg30). As revealed subsequently by the 2B4-CD48 structure, neither position is involved in interactions with 2B4. In marked contrast to the wild-type, the mutant CD48 IgV domain could be efficiently folded in vitro after expression in bacterial inclusion bodies; this mutant was used for all subsequent experiments.
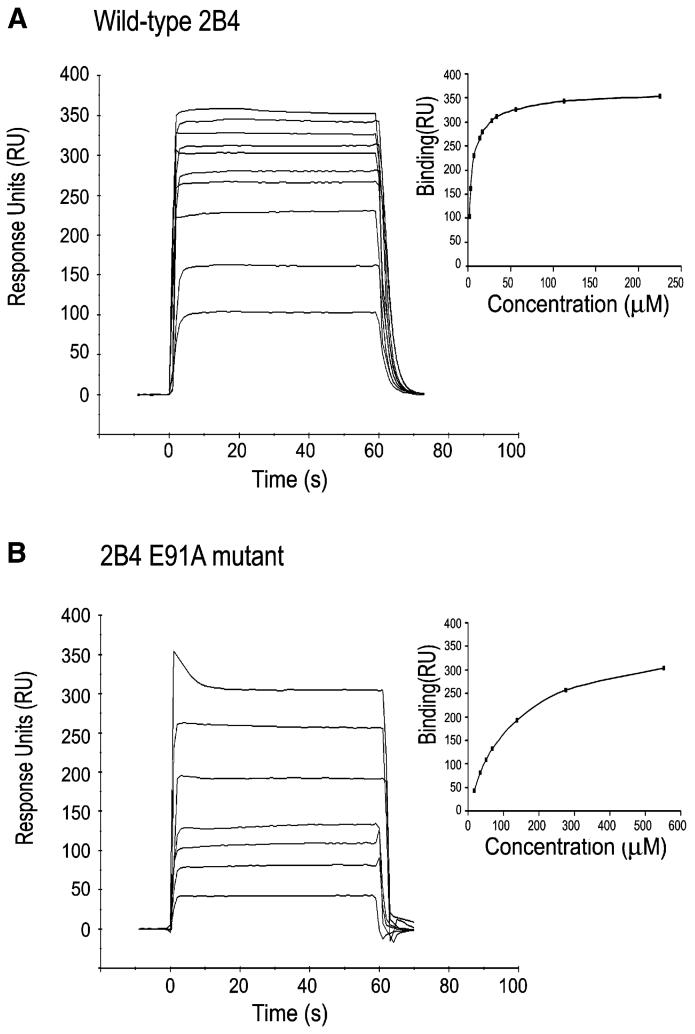
As measured by surface plasmon resonance (SPR), soluble CD48 bound to immobilized 2B4 with an equilibrium dissociation constant (KD) of 4 μM, with very fast on and off rates (Figure 1A). This affinity is similar to that previously reported for recombinant mouse 2B4 and CD48 produced in eukaryotic cells (KD = 16 μM) (Brown et al., 1998), demonstrating that the bacterially expressed proteins are correctly folded and fully active.
Figure 1. Equilibrium Binding of Wild-Type and Mutant 2B4 to CD48.
SPR sensograms for the interaction of wild-type 2B4 (225, 112, 56, 33.8, 28.2, 16.9, 14.1, 7.0, 3.5, 1.8 μM) (A) and 2B4 E91A mutant (552, 276, 138, 69, 55.2, 34.4, 27.6 μM) (B) with immobilized CD48 (700 RU) after correction for nonspecific binding. Inset plots show non-linear steady-state affinity analysis of the equilibrium binding data. KD = 4 μM for wild-type 2B4; KD = 130 μM for 2B4 E91A.
The crystal structure of the 2B4-CD48 complex was determined by molecular replacement to a resolution of 1.6 Å (Table 1). The final model was refined to Rwork and Rfree values of 19.0% and 24.3%, respectively. All residues were in unambiguous electron density, except a few at the N and C termini of both proteins. In addition, we determined the structures of 2B4 and CD48 in free form to resolutions of 2.4 Å and 1.7 Å, respectively (see below).
Table 1. X-ray Crystallographic Statistics.
| 2B4-CD48 | 2B4 | CD48 | |
|---|---|---|---|
| Data-Processing Statistics | |||
| Resolution limit (Å)a | 1.63 (1.69-1.63) | 2.38 (2.44-2.38) | 1.66 (1.72-1.66) |
| Space group | C2 | P21212 | I41 |
| Cell dimensions (Å, °) | a = 67.7 b = 37.0 c = 94.6 β = 108.7 | a = 79.0 b = 183.3 c = 59.7 | a = 78.8 c = 31.3 |
| Unique reflectionsa | 28061 (2733) | 34221 (2963) | 11309 (1041) |
| Completeness (%)a | 99.1 (97.4) | 97.0 (85.7) | 97.2 (81.5) |
| Rmerge (%)a,b | 5.5 (18.0) | 4.5 (28.8) | 4.7 (20.0) |
| I/σI a | 53.6 (8.0) | 33.4 (4.2) | 66.7 (8.4) |
| Refinement Statistics | |||
| Rwork (%)c | 19.0 | 22.4 | 20.2 |
| Rfree (%)c | 24.3 | 26.9 | 26.1 |
| Number of atoms: protein/H2O/SO42- | 1704/213/5 | 3307/128 | 797/81 |
| Mean B Values (Å2) | |||
| Main chains | 18.3 | 47.1 | 22.6 |
| Side chains | 22.0 | 47.5 | 26.0 |
| H2O | 31.2 | 47.5 | 34.6 |
| RMSDs from Ideality | |||
| Bond lengths (Å) | 0.019 | 0.014 | 0.018 |
| Bond angles (°) | 1.78 | 1.55 | 1.76 |
| Ramachandran-Plot Statistics | |||
| Most favored (%) | 89.7 | 88.3 | 91.6 |
| Additionally allowed (%) | 9.2 | 10.9 | 7.2 |
| Generally allowed (%) | 1.1 | 0.8 | 1.2 |
| Disallowed (%) | 0 | 0 | 0 |
Values in the parentheses are statistics of the highest resolution shell.
, where Ij is the intensity of an individual reflection and < I > is the average intensity of that reflection.
, where Fc is the calculated structure factor. Rfree is as for Rwork but calculated for a randomly selected 5.0% of reflections not included in the refinement.
Overall Structure
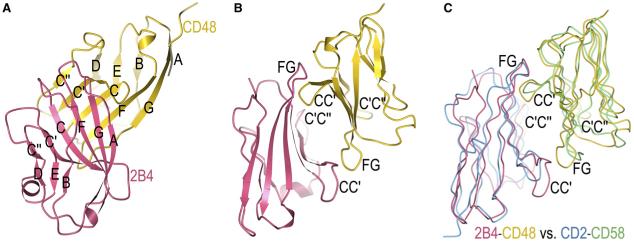
The 2B4 and CD48 N-terminal domains each contain nine β strands assembled into two antiparallel β sheets of strands designated AGFCC’C” and DEB (Figure 2A). Like other CD2 family members, the 2B4 and CD48 ligand-binding domains lack the canonical disulfide bond that links the B and F strands in other IgV domains. However, 2B4 has a noncanonical disulfide bridge between Cys3 and Cys100 that stabilizes an unusually long parallel β structure between strands A and G, imparting a convex curvature to the outer surface of the six-stranded β sheet (AGFCC’C”). The mouse CD48 IgV domain is similar in overall topology to the corresponding domains of CD2 and CD58 (room mean square [r.m.s.] differences of 1.1, 1.1, and 1.3 Å for 74, 74, and 82 equivalent Cα atoms for rat CD2, human CD2, and human CD58, respectively). 2B4 and CD48 differ from each other (and from other CD2 family members) in several important respects, involving regions implicated in ligand recognition. Most prominently, the CC’ loop of 2B4 is four residues longer than the corresponding loop of CD48 (two and five residues longer when compared to CD2 and CD58, respectively).
Figure 2. Structure of the 2B4-CD48 Complex.
(A) Ribbon diagram of the 2B4-CD48 complex showing the face-to-face interaction between the AGFCC’C” β sheets of the two IgV domains. 2B4 is rose and CD48 is gold; β strands are labeled.
(B) The complex rotated 90° clockwise about the vertical axis with respect to the view in (A). Loops that contribute to the binding interface are labeled.
(C) Superposition of the 2B4-CD48 and CD2-CD58 complexes (tube diagram). Equivalent Cα atoms of 2B4 were superposed onto CD2, and equivalent Cα atoms of CD48 were superposed onto CD58, with the program Superpose Molecules (Collaborative Computational Project, Number 4, 1994). 2B4 is rose, CD48 is gold, CD2 is cyan, and CD58 is green. The complexes are oriented as in (B).
In the 2B4-CD48 complex, the AGFCC’C” β sheet of 2B4 interacts face to face with the corresponding β sheet of CD48, such that the two sheets contact each other at an ∼75° angle, defined as the amount 2B4 must be rotated to align its AGFCC’C” β sheet with that of CD48 (Figure 2A). The contact area is centered on the C and C’ strands for both 2B4 and CD48. This association mode shows overall similarities to that observed in the CD2-CD58 heterophilic complex (Wang et al., 1999), but it differs in the detailed architecture and physicochemical properties of the interface (see below). In the NTB-A and CD84 homophilic complexes, whose interfaces are also formed by juxtaposed AGFCC’C” sheets from the IgV domains (Cao et al., 2006; Yan et al., 2007), the β strands associate at a markedly different angle (∼115°).
The 2B4-CD48 Heterophilic Interface
The interface between 2B4 and CD48 (Figure 2B) is considerably more symmetrical than that between CD2 and CD58, but it is less so than the NTB-A (or CD84) homodimer interface, which displays approximate 2-fold symmetry. In the 2B4-CD48 complex, the FG loop of each IgV domain contacts the CC’ loop of its partner (Figure 2B). At one corner of the interface, the FG loop of 2B4 makes six hydrogen bonds with the CC’ loop of CD48, all involving main chain atoms of 2B4 (Table 2). At the opposite corner, the FG loop of CD48 engages the CC’ loop of 2B4 through a combination of two salt bridges and five hydrogen bonds, as well as numerous main chain-main chain contacts. By contrast, in the less symmetrical CD2-CD58 interface, only the FG loop of CD2 makes main chain-main chain interactions with the CC’ loop of CD58, a distinction best appreciated by the superimpositon the two heterophilic complexes (Figure 2C). In addition to the FG and CC’ loops, β strands C, C’, F and G, and loop C’C” of both 2B4 and CD48 contribute to the binding interface, as does loop C”D of CD48 (Figures 3A and 3B). The only contacting residue outside of this region is Asp5 of 2B4, located immediately before β strand A, which forms two side chain-side chain hydrogen bonds with Asn39 of CD48 (Table 2). However, Asp5 is replaced by glycine in rat and human 2B4, implying that these interactions are not requisite for complex stabilization.
Table 2. Interactions between 2B4 and CD48.
| CD48 |
CD48 |
|
|---|---|---|
| 2B4 | Hydrogen Bonds and Salt Bridges (Underlined) | Van der Waals Contacts |
| D5 | D5(Oδ1)-N39(Nδ2), D5(Oδ2)-N39(Nδ2) | K38, N39 |
| Q33 | Q33(Oε1)-R31(Nη2) | R31 |
| K35 | K35(Nζ)-E93(Oε1) | E93 |
| E38 | T92 | |
| R43 | R43(Nε)-T92(Oγ1), R43(Nη2)-E91(Oε2), R43(Nη2)-T92(O), R43(Nη2)-T92(Oγ1) | E91, T92, E93, N94 |
| K44 | K44(N)-E93(O), K44(O)-E93(N) | T92, E93 |
| E46 | E46(Oε2)-R31(Nη1), E46(Oε2)-R31(Nη2) | R31 |
| N49 | N49(Oδ1)-R31(Nη1) | R31 |
| Y51 | R31, N46 | |
| E91 | E91(Oε2)-R87(Nη1), E91(Oε1)-R87(Nη1) | R87 |
| T93 | T93(Oγ1)-K41(Nζ) | K41, F54 |
| N94 | F54 | |
| T95 | T95(O)-E55(N) | F54, E55 |
| G96 | G96(O)-Q40(Nε2) | Q40, F54, E55, S56 |
| G97 | G97(O)-Q40(Nε2), G97(O)-K41(N) | N39, Q40, K41, F54 |
| K98 | N39, Q40 | |
| V99 | V99(N)-N39(O), V99(O)-N39(Nδ2) | K38, N39, K41 |
| N101 | N101(Oδ1)-K38(O), N101(Oδ1)-N39(Nδ2) | K38 |
Figure 3. Comparison of Hetero- and Homophilic Interactions by SLAM Family Receptors.
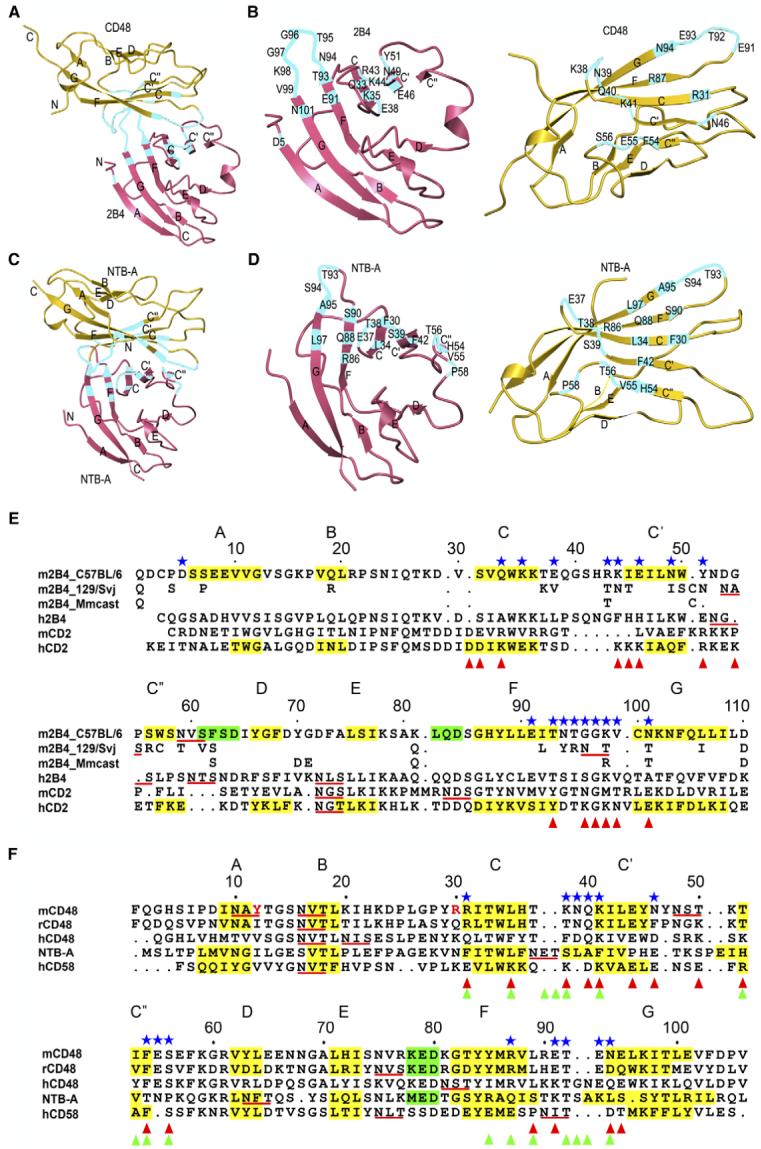
(A and C) Overall arrangement of the 2B4-CD48 heterophilic and NTB-A homophilic dimers. CD48 and the upper NTB-A monomer are in the same orientation. Residues contributing to the binding interfaces are highlighted in cyan. The N and C termini are indicated.
(B and D) Detailed view of the interfaces. CD48 and the upper NTB-A monomer are each rotated ∼90° about the horizontal axis relative to (A) and (C), respectively. Contacting residues are labeled.
(E and F) Structure-based alignment of CD2 family IgV domains. The β strands are shaded yellow and labeled A through G; α helices are shaded green. Potential N-linked glycosylation sites are underlined red. Contacting residues in the 2B4-CD48 heterophilic dimer are denoted with blue stars. Residues involved in homophilic interactions in the NTB-A dimer are marked with green triangles (Cao et al., 2006). Residues contributing to the CD2-CD58 heterophilic interface are denoted with red triangles (Wang et al., 1999). In (E), residues of 2B4 isoforms 129/Svj (UniProtKB-TrEMBL: Q3TDI8) and Mmcast (Mus musculus castaneus, UniProtKB-TrEMBL: Q3S4B1) that are identical to 2B4 isoform C57BL/6 (UniProtKB-Swiss-Prot: Q07763) residues have been omitted. In (F), residues of yeast-displayed mouse CD48 that differ from the wild-type sequence (Thr12, Lys30) are indicated in red.
Whereas most (20 of 32) contacting residues in the 2B4-CD48 heterophilic interface are located in loops (Figures 3B, 3E, and 3F), the NTB-A homophilic interface is formed mainly by strand-to-strand contacts involving strands C, C’, C”, and F (Figures 3D and 3F). This distinction results from different dimerization geometries in the hetero- versus homophilic complexes (Figures 3A and 3C), as evident from superimpositon CD48 onto one NTB-A monomer and comparing the placement of 2B4 and the second NTB-A monomer, which are related by a translation of ∼8 Å and rotation of ∼30°.
The 2B4-CD48 complex buries a total solvent-accessible surface of 1564 Å2, of which 803 Å2 is contributed by 2B4 and 761 Å2 by CD48. This is similar to the total surface buried in the NTB-A complex (1583 Å2) but substantially greater than that in the CD2-CD58 complex (1161 Å2). The structural features that account for the difference between the two heterophilic complexes include the relatively long CC’ loop and C’ β strand of 2B4. In addition, CD48 uses not only the FG loop but also β strands F and G to engage 2B4, whereas CD58 contacts CD2 only through the FG loop (Figure 3F).
The 2B4-CD48 interface is predominantly hydrophilic and includes 32 bound water molecules, of which nine are completely inaccessible to bulk solvent (Figure 4A). Ten of the interfacial waters mediate 28 hydrogen bonds between 2B4 and CD48, which form an extensive interlinking network. By contrast, no interfacial waters were reported for the CD2-CD58 and NTB-A complexes, possibly because of the lower resolution of these structures (∼3 Å). Indeed, interfacial waters were found in the CD84 homodimer (2 Å resolution), whose interface, like that of the 2B4-CD48 heterodimer, is mainly hydrophilic (Yan et al., 2007). The 2B4-CD48 interface is characterized by moderate shape complementarity, based on a shape correlation statistic (Sc) of 0.69 (Sc = 1.0 for perfect geometric fits) (Lawrence and Colman, 1993), which is comparable to the Sc value of the NTB-A dimer (0.68) but greater than that of the CD2-CD58 interface (0.56). Despite these differences, and those in buried surface areas indicated above, all three complexes display surprisingly similar affinities (KD ∼5 μM) (van der Merwe et al., 1994; Cao et al., 2006).
Figure 4. The 2B4-CD48 Binding Interface.
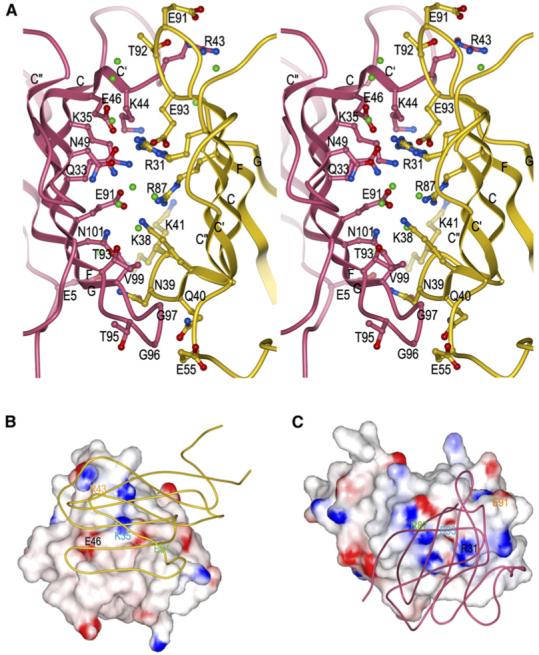
(A) Stereo view of residues that participate in hydrogen bond or salt bridge interactions at the 2B4/CD48 interface (Table 2). 2B4 is rose and CD48 is gold. The side chains of interacting residues are drawn in ball-and-stick representation, with carbon atoms in rose (2B4) or gold (CD48), nitrogen atoms in blue, and oxygen atoms in red. Bound waters mediating hydrogen bonds between 2B4 and CD48 are shown as green balls.
(B) Electrostatic potential surface of 2B4 viewed from the perspective of CD48 (tube diagram). The view of the complex in (B) is rotated ∼90° counterclockwise about the vertical axis compared to the view in (A). Solvent-accessible surfaces of 2B4 are colored according to electrostatic potential, with positively charged regions in blue and negatively charged regions in red. Electrostatic surface potentials were calculated with GRASP (Nicholls et al., 1991).
(C) Electrostatic potential surface of CD48 from the perspective of 2B4 (tube diagram). The view of the 2B4-CD48 complex in (C) is rotated 180° about the vertical axis with respect to the view in (B). Four selected pairs of charged residues (each pair labeled with a different color) that form salt bridges across the 2B4-CD48 interface are indicated on the surfaces, illustrating their matched charged potentials and relative positions on 2B4 (B) or CD48 (C).
In the complex, 18 2B4 residues interact with 14 CD48 residues through a combination of 18 direct hydrogen bonds and six salt bridges (Table 2; Figure 4A). The interface shows clear electrostatic complementarity, based on mapping pairs of interacting charged residues onto the electrostatic potential surfaces of the two proteins (Figures 4B and 4C). Although the 2B4-CD48 interface, like that of CD2-CD58, is dominated by polar interactions, the CD2-CD58 interface contains nearly twice the number of salt bridges (ten versus six) and considerably fewer hydrogen bonds (five versus 18). Both heterophilic interfaces are less hydrophobic than the NTB-A homophilic interface, which includes a tight cluster of symmetry-related aromatic residues (Cao et al., 2006). In the 2B4-CD48 complex, contacting residues 2B4 Gly97 and CD48 Phe54 contribute the greatest number of hydrophobic interactions.
Both 2B4 and CD48 exist as highly glycosylated proteins on the cell surface. (Schatzle et al., 1999; Killeen et al., 1988). However, all predicted N-linked glycosylation sites in the 2B4 and CD48 IgV domains lie outside of the binding interface (Figures 3E and 3F) or are not conserved between different isoforms, suggesting that oligosaccharides do not directly influence recognition. For example, although 2B4 from 129/Svj mice possesses a potential N-linked glycosylation site in the FG loop, which contacts CD48 in the structure, this site is not conserved across mouse strains, including C57BL/6.
Basis for Binding Specificity
Of the 18 residues of mouse 2B4 that contact CD48 in the complex, only five (Lys35, Glu91, Thr93, Gly97, and Val99) are conserved in human and rat 2B4 and in different isoforms of the mouse receptor (Figure 3E). By comparison, a higher proportion of contacting residues of mouse CD48 (8 of 14) are conserved in the rat and human proteins. We have identified several key interactions by mutational analysis. Among the conserved contacting residues of 2B4, Lys35 is located in strand C, Glu91 and Thr93 in strand F, and Gly97 and Val99 in loop FG. In the complex, Lys35 and Glu91 each form salt bridges to conserved residues of CD48 (Asp93 and Arg87, respectively), and these salt bridges could serve as key anchor points for 2B4 across different species (Table 2). Consistent with this idea, the mutation of 2B4 Glu91 to alanine reduced binding to CD48 by 97% (KD = 130 μM versus 4 μM for wild-type 2B4), as measured by SPR (Figure 1B; Table 3). Similarly, alanine substitution of 2B4 Lys35 decreased the affinity for CD48 by 98%. A less pronounced (91%), though still significant, effect on binding resulted from mutation of 2B4 Thr93, which forms a side chain-side chain hydrogen bond with conserved CD48 residue Lys41. The remaining two conserved contacting residues of 2B4, Gly97, and Val99, also interact with CD48 at conserved (Gln40, Lys41), or conservatively substituted (Asn or Asp39) positions (Figure 3F). By contrast, alanine substitution of 2B4 Thr95, which is not conserved in human 2B4 and certain mouse isoforms (Figure 3E), has no effect on affinity (Table 3). In a mutational study of human 2B4, residues Lys68 and Glu70, which correspond to mouse 2B4 Asn49 and Tyr51 in the numbering system used here, were shown to be critical for CD48 binding (Mathew et al., 2005). Although not conserved in human 2B4, both residues contact CD48 in the mouse complex (Figure 3E).
Table 3. Affinity of 2B4 and 2B4 Mutants for CD48.
Wild-type 2B4 from mouse strain C57BL/6.
Wild-type 2B4 from mouse strain 129/Svj.
The relatively low sequence conservation on the 2B4 side of the interface (Figure 3E) can be understood in terms of the complex structure, in conjunction with examination of 2B4 isoforms from mouse strains BALB/c, A.CA, 129/Svj, and NOD (the isoform studied here is from C57BL/6). Amino acid differences among 2B4 receptors from different strains are mainly restricted to loop regions, including CD48-contacting loops CC’, C’C”, and FG (Figure 3E). To assess whether different 2B4 isoforms likely bind CD48 with similar affinities, we measured the affinity of 129/Svj 2B4 for CD48. This isoform bound CD48 with KD = 10 μM, which is comparable to the KD of C57BL/6 2B4 (4 μM) (Table 3), despite sequence differences at 10 of 18 contacting positions, mostly in loops CC’ and FG (Figure 3E). The ability of loop FG to tolerate substitutions is explained, in part, by the finding that all six hydrogen bonds to CD48 in the structure involve main chain, rather than side chain, atoms of 2B4 (Table 2). Similarly, in loop CC’, both hydrogen bonds between Lys44 (Asn in 129/Svj 2B4) and CD48 Glu93 are main chain-main chain. Other interacting residues that differ between C57BL/6 and 129/Svj 2B4, for example Glu/Val38 and Tyr/Asn51, make only a few van der Waals contacts with CD48. More difficult to explain is the apparent lack of an effect on binding from substitution of 2B4 Arg43 by threonine (in 129/Svj 2B4) or alanine (Table 3), even though this residue makes multiple interactions with CD48 through its side chain (Table 2). However, 2B4 Arg43 is located at the periphery of the interface with CD48, where solvent rearrangements could compensate for the loss of direct protein-protein interactions at the mutation site, as observed in antigen-antibody complexes (Li et al., 2003).
In addition to 2B4 and CD2, rat CD48 has been shown to bind a third ligand, designated 2B4R (Kumaresan et al., 2000) with an affinity comparable to 2B4 (Evans et al., 2006). Unlike 2B4, 2B4R does not contain ITSMs in its 65 amino acid cytoplasmic domain, implying a different role in NK cell function. Although rat 2B4R and 2B4 are closely related (89% sequence identity), many (12 of 30) of the residues that distinguish the IgV domains of these proteins correspond to ones forming the AGFCC’C” ligand-binding face of mouse 2B4, such that rat 2B4R and 2B4 differ at 11 of 18 predicted CD48-contacting positions (data not shown). However, as in the case of mouse 2B4 isoforms, the conserved residues include Lys35, Glu91, and Thr93, which are located in β strands and are hot spots for CD48 recognition by mouse 2B4 (Table 3), whereas the differences are mostly confined to loops CC’ and FG.
Conformational Changes upon 2B4 Binding to CD48
To identify possible conformational changes in 2B4 or CD48 associated with complex formation, we determined the structures of the unbound proteins to resolutions of 2.4 Å and 1.7 Å, respectively (Table 1). The crystals contained one (CD48) or four (2B4) molecules in the asymmetric unit. Superposition of the four unbound 2B4 molecules gave r.m.s. differences in the range of 0.4 to 0.5 Å for 99 α-carbon atoms, indicating close similarity. However, the CC’ loop (residues 38-43) of 2B4 is disordered in two of the molecules in the asymmetric unit. In the other two molecules, the CC’ loop is well defined, but two different conformations are evident, with an average displacement of 4.0 Å in the position of their in α-carbon atoms (Figure 5A). Notably, one of these loop conformations is close to that found in liganded 2B4 (r.m.s. difference of 1.5 Å in α-carbon positions), whereas the other would create steric clashes with CD48. Thus, the CC’ loop of 2B4 appears to be intrinsically flexible, a property that might allow it to sample a wide range of conformations to rapidly select an optimum one for recognizing the FG loop of CD48. Conceivably, this flexibility might also enable 2B4 to recognize still-unidentified ligands besides CD48. By contrast, the FG loop of 2B4, comprising residues 94-99, is ordered in all four molecules in the asymmetric unit and adopts nearly the same conformation in each case, without involving any crystal contacts. Moreover, this conformation closely resembles that present in the 2B4-CD48 complex (Figure 5A). Surprisingly, the opposite was observed in the NMR structure of 2B4: Whereas the CC’ loop was accurately defined, the FG loop appeared to be dynamically disordered. This discrepancy might be due in part to the different methods of structure determination. The C’C” loop (residues 51-55) of CD48-bound 2B4 displays a different conformation from that in free 2B4 (average displacement of 2.8 Å in α-carbon positions) (Figure 5A). However, it is unclear whether this difference is attributable to ligand binding because only one C’C” loop residue (Tyr51) contacts CD48 (Table 2).
Figure 5. Conformational Changes in 2B4 and CD48 upon Complex Formation.
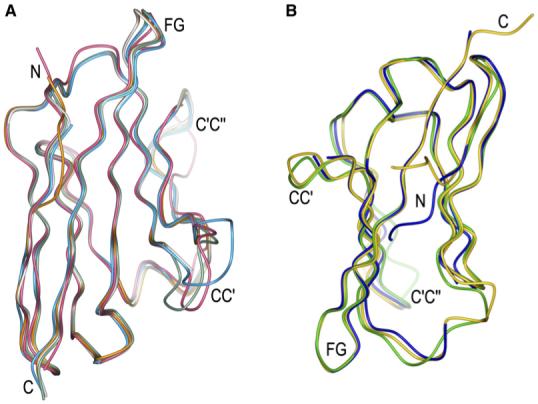
(A) Superposition of bound (rose) and unbound (dark green, cyan, gray, and orange, representing the four molecules in the asymmetric unit) 2B4.
(B) Superposition of bound (gold) and unbound (blue) mouse CD48 and of unbound rat CD48 (light green) (Evans et al., 2006).
Superposition of free CD48 onto CD48 in the complex with 2B4 gave an r.m.s. difference in α-carbon positions of 0.6 Å, indicating no substantial conformational changes upon binding (Figure 5B). However, as in the case of 2B4, the CC’ loop of CD48 appears to be flexible because the tip of this loop (residues 38-39) is disordered in the unbound structure. In the 2B4-CD48 complex, by contrast, Lys38 and Asn39 are well defined and form several hydrogen bonds with 2B4 (Table 2). In the structure of rat CD48 (Evans et al., 2006), which is very similar overall to that of mouse CD48 (r.m.s. difference of 0.9 Å for 87 α-carbon atoms), the entire CC’ loop is ordered. Moreover, its conformation differs significantly from that of the CC’ loop of 2B4-bound mouse CD48, with displacements in α-carbon positions up to 4.6 Å (Figure 5B). Hence, it remains to be determined whether the CC’ loop of rat CD48 adopts a conformation resembling that of mouse CD48 upon binding 2B4. Similar considerations apply to the C’C” loop of rat CD48 (Figure 5B).
DISCUSSION
The SLAM family of receptors is a subset of the greater CD2 family, which is believed to be the product of successive gene-duplication events. The overall structural similarity of the 2B4-CD48 (or CD2-CD58) heterophilic and NTB-A homophilic complexes, with interfaces formed by juxtaposed AGFCC’C” β sheets of the IgV domains, further suggests that these molecules evolved from common ancestors having homophilic association properties. However, because of differences in dimerization geometry, most of the contacting residues in the 2B4-CD48 heterophilic interface are located in loops, whereas the NTB-A homophilic interface is composed mainly of strand-to-strand contacts. In both cases, however, the key binding interactions are mediated by β strand (rather than loop) residues, as indicated by mutational analysis of 2B4 and by the fact that amino acid differences among 2B4 isoforms are largely confined to loop regions, in particular CD48-contacting loops CC’ and FG. Indeed, the ability of the 2B4-CD48 interface to tolerate multiple mutations while maintaining affinity and specificity is demonstrated by our finding that 2B4 alleles from C57BL/6 and 129/Svj mice, whose sequences differ at most (10 of 18) contacting positions, nevertheless bind CD48 with comparable affinities (KD ∼5 μM). Accordingly, 2B4 function is probably conserved across mouse strains.
It is unknown why 2B4 acts mainly as an activating receptor in humans but as an inhibitory receptor in mice because it seems likely from the present analysis that human and mouse 2B4 engage CD48 with similar docking topologies. Moreover, the human and mouse 2B4-CD48 interactions have comparable affinities (Brown et al., 1998). On the other hand, there exist two isoforms of mouse 2B4 that differ in their cytoplasmic domains and signaling capacities, such that the relative quantities of these isoforms could determine the specific response of NK cells to 2B4 ligation (Ma et al., 2007).
In rodents, CD48 binds CD2 in addition to 2B4, albeit with ∼90% lower affinity (KD = 60 μM for the rat CD2-CD48 pair) (Davis et al., 2003). In human, the interaction between CD48 and CD2 is much weaker (KD > 0.5 mM) and probably not biologically relevant (Brown et al., 1998). Because sequence and structural homology between CD2 and 2B4 are lower than those between CD58 and CD48 (Figures 3E and 3F), especially at the binding sites, the CD2-CD48 complex is likely to more closely resemble the CD2-CD58 complex (Wang et al., 1999) than the 2B4-CD48 complex. This conclusion is also consistent with available mutagenesis data. For example, mutation of rat CD48 Glu44 abolished binding to CD2 (Davis et al., 1998), as did mutation of the corresponding residue of human CD58 (Glu37) (Wang et al., 1999). In the CD2-CD58 structure, CD58 Glu37 forms a salt bridge with CD2 Arg48, which is conserved in a model of the CD2-CD48 complex constructed by least-squares superposition of CD2 and CD48 onto their counterparts in the CD2-CD58 complex (data not shown). However, in the 2B4-CD48 structure, CD48 Glu44 makes no contacts with 2B4 (Table 2) and probably contributes little to binding.
Based on comparisons of free and bound structures, 2B4 appears to be more intrinsically flexible than CD48, most notably in loops CC’ and FG. In addition, CD48 exhibits considerably less polymorphism than 2B4 across different species and mouse strains. These differences might reflect structural constraints on CD48 imposed by the requirement that it recognize both 2B4 and CD2, whereby variations in CD48 that seriously affect binding to either partner are not tolerated.
Upon NK cell-target cell interaction, a highly organized complex of molecules, termed the NK cell immune synapse, is formed at the contact area of both cells (Davis et al., 1999; Vyas et al., 2001; Vyas et al., 2002). These molecules include NK receptors that monitor MHC class I expression on target cells (KIRs and Ly49s in human and mouse, respectively), cytoskeletal proteins (F-actin, tailin), integrins (CD11a, CD11b), and signaling molecules (Lck, SHP-1) (Davis et al., 1999; Vyas et al., 2002; Orange et al., 2003). Additionally, 2B4 and SAP are recruited to the NK cell immune synapse during recognition of EBV-virus-infected B cells by NK cells (Roda-Navarro et al., 2004).
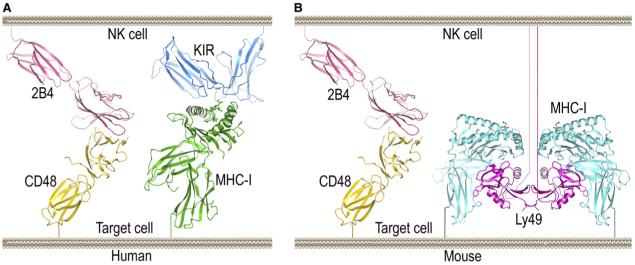
It has been proposed that the unique compartmentalization of molecules observed at immune synapses is regulated by a sorting mechanism based on molecular dimensions (van der Merwe et al., 2000; Bromley et al., 2001). To compare the dimensions of 2B4-CD48 and KIR-MHC complexes, both of which accumulate at the NK cell immune synapse (Davis et al., 1999; Roda-Navarro et al., 2004), we constructed a model of the interaction between the entire extracellular regions of 2B4 and CD48 (IgV and IgC2 domains), based on the current structure of the complex between their respective IgV domains and the structure of full-length rat CD2 (Jones et al., 1992) (Figure 6). The overall rod-like architecture of full-length CD2 is the consequence of specific interactions between the IgV and IgC2 domains, several of which appear to be conserved in 2B4 and CD48. These include hydrophobic interactions between 2B4 Val11 and Arg182 (corresponding to CD2 Tyr7 and Arg162), and between Val13 and Ala183 (CD2 Ala9 and Val163). In addition, 2B4 Leu109 (CD2 Leu98) is predicted to contact several residues from the IgC2 domain that are conserved in CD2. Similarly, the interdomain interface of CD48 might include a salt bridge between Lys78 and Asp104, corresponding to that between Arg70 and Glu99 in CD2.
Figure 6. Hypothetical Models of Interactions at the NK Cell Immune Synapse.
(A) Human NK cell immune synapse. The distance spanned by the modeled 2B4-CD48 complex is comparable to that spanned by the KIR2DL2-HLA-Cw3 complex (PDB accession code 1EFX). The model of the extracellular region of 2B4 is based on the present 2B4-CD48 complex and the structure of full-length CD2 (1HNG). The model of full-length CD48 is based on the chimeric rat CD48-CD2 structure (2DRU).
(B) Mouse NK cell immune synapse. The structure of the Ly49C-H-2Kb complex (1P4L) does not include the 70-residue stalk regions that connect the Ly49C homodimer to the NK cell membrane. The stalks are drawn arbitrarily here.
In the modeled complex between full-length 2B4 and CD48, the distance spanned by the molecules is ∼115 Å, which is approximately the distance bridged by human KIR-HLA-C complexes (Boyington et al., 2000; Fan et al., 2001) (Figure 6A). In the case of mouse Ly49-MHC complexes (Dam et al., 2003; Deng and Mariuzza, 2006), the C-type lectin-like domains of the receptor are tethered to the NK cell membrane by exceptionally long (70 residues), and most likely flexible, stalk regions that should allow the recruitment of Ly49-MHC and 2B4-CD48 complexes to the immune synapse (Figure 6B).
On the basis of the structure of the NTB-A homophilic dimer, which spans an end-to-end distance of ∼100 Å, it was proposed that all homophilic receptor pairs of the SLAM family, with the exception of Ly9, have similar linear dimensions (Cao et al., 2006). Because the 2B4-CD48 heterophilic dimer also measures ∼100 Å, homo- and heterophilic receptor pairs could potentially colocalize at the NK cell immune synapse with a common set of SAP-related adaptors (SAP, EAT2, ERT) that recruit specific protein tyrosine kinases or phosphatases to the synapse. This intermixing of signaling molecules would permit signal modulation by SLAM family receptors in response to different stimuli, resulting in stimulation or inhibition of interferon-γ secretion and NK cell-mediated killing.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
The ligand-binding domain of 2B4 (residues 1-112) from C57BL/6 mice was expressed by in vitro folding from Escherichia coli inclusion bodies as described (Ames et al., 2005). Mutants of 2B4 (K35A, R43A, E91A, T93A, and T95A) were produced and purified similarly to the wild-type protein.
Initial attempts to express the ligand-binding domain of CD48 from C57BL/6 mice by in vitro folding were unsuccessful. Accordingly, we used yeast display to select stable variants (Kieke et al., 1999). The IgV and IgC2 domains (residues 1-189) of CD48, minus the leader sequence and glycosylphosphatidylinositol anchor signal, were cloned into a modified yeast display vector containing a tobacco etch virus (TEV) cleavage site and c-myc C-terminal tag (pYD-TEV). We performed error-prone polymerase chain reaction (PCR) on CD48 by adjusting the ratio of Mg2+ to Mn2+ to 6.67 to 1 to achieve an error rate of approximately 1%. PCR products were ligated in vivo into pYD-TEV by homologous recombination in yeast strain EBY-100 (Boder and Wittrup, 2000). A library of approximately 1 × 107 independent transformants was stained with anti-CD48-PE (BD PharMingen) and sorted four times with a BD FACSAria high-speed cell sorter with progressively lower concentrations of antibody at each successive sort. Populations from the third and fourth sorts were plated on synthetic dextrose-casamino acids (SD-CAA) plates so that individual clones could be isolated.
Several CD48-expressing clones were selected for higher expression and heat stability on the yeast surface. All these clones contained a truncation at the beginning of the IgC2 domain; however, they bound both CD48 antibody and soluble monomeric 2B4 at high levels. Of these, a clone from the fourth sort was chosen for further study because it contained the fewest mutations, T12Y and K30R, both of which were predicted to be outside the putative 2B4-binding site. The mutant DNA sequence, encoding CD48 residues 1-106, was cloned into pT7-7 (Novagen). Bacteria were grown at 37°C to an absorbance of 0.6-0.8 at 600 nm, and 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) was added. After incubation for 3 hr, the bacteria were harvested by centrifugation and resuspended in 50 mM Tris-HCl (pH 8.0) containing 0.1 M NaCl and 2 mM ethylenediamine tetraacetic acid (EDTA); cells were disrupted by sonication. Inclusion bodies were washed with 50 mM Tris-HCl (pH 8.0), 0.1 M NaCl, and 0.5% (v/v) Triton X-100 and then solubilized in 6 M guanidine-HCl and 100 mM Tris-HCl (pH 8.0).
For in vitro folding, solubilized CD48 was diluted to a final concentration of 10 μg/ml into 0.8 M arginine, 100 mM Tris-HCl (pH 8.0), 2 mM EDTA, and 1 mM dithiothreitol. After 2 days at 4°C, the folding mixture was concentrated, dialyzed against phosphate-buffered saline (pH 7.4), and applied to a Superdex 75 HR column (Amersham Biosciences). Further purification was carried out with a MonoS column in 20 mM MES (pH 5.5).
Crystallization and Data Collection
Crystallization trials of purified proteins at 10 mg/ml in 20 mM Tris-HCl (pH 7.5) were carried out at 4°C in hanging drops. Crystals of unbound 2B4 grew in 0.1 mM Tris-HCl (pH 8.5) and 2.0 M ammonium dihydrogen phosphate. Free CD48 crystals were obtained from 30% (w/v) PEG 8000, 100 mM sodium imidazole (pH 7.8), and 0.2 M NaCl. Crystals of the 2B4-CD48 complex, purified with a Superdex 75 HR column, grew in 1.6 M ammonium sulfate, 0.1 mM MES (pH 6.5), and 10% dioxane.
For data collection, crystals of unbound 2B4 and CD48 were cryo-protected in 20% (v/v) glycerol. Ammonium sulfate (3.5 M) was used for the flash freezing of crystals of the 2B4-CD48 complex. X-ray diffraction data for 2B4, CD48, and 2B4-CD48 were collected to resolutions of 2.38 Å, 1.66 Å, and 1.63 Å, respectively, at beamline X29 of the Brookhaven National Synchrotron Light Source with an ADSC Quantum-315 CCD detector. Data were indexed, integrated, and scaled with HKL2000 (Otwinowski and Minor, 1997) (Table 1).
Structure Determination and Refinement
Diffraction data for the 2B4-CD48 complex were phased with coordinates of a truncated model of rat CD2 (Protein Data Bank [PDB] accession code 1HNG) (Jones et al., 1992) with the program Phaser (Storoni et al., 2004). All of the residues of the model were replaced by polyalanine before refinement. The program RefMac 5.0 (Murshudov et al., 1997) implemented in the CCP4 program suite (Collaborative Computational Project, Number 4, 1994) was used for initial rigid-body refinement and then iterative maximum-likelihood refinement. Between refinement rounds, σA-weighted 2Fo - Fc and Fo - Fc maps were calculated and used for the identification and building of the side chains, truncated loops, and solvent molecules with the program XtalView (McRee, 1999), and these were included in subsequent refinement cycles. The final refinement statistics are shown in Table 1.
Orientational and positional parameters of unbound 2B4 and CD48 molecules in their respective unit cells were obtained by molecular replacement with Phaser (Storoni et al., 2004). The truncated structures of bound 2B4 and CD48 in the 2B4-CD48 complex were used as search models. The resulting models were refined with RefMac 5.0 (Murshudov et al., 1997). Model building and identification of water molecules were done with XtalView (McRee, 1999). The refinement statistics for 2B4 and CD48 are given in Table 1.
Stereochemical parameters of all structures were evaluated with PROCHECK (Laskowski et al., 1993). Interchain contact residues were identified with the program CONTACT (Collaborative Computational Project, Number 4, 1994) and were defined as residues containing an atom ≤ 4.0 Å from a residue of the partner chain. Buried surface areas were calculated with SURFACE (Collaborative Computational Project, Number 4, 1994) with a 1.4 Å probe radius. BobScript (Esnouf, 1999), Raster 3D (Bacon and Anderson, 1988), and GRASP (Nicholls et al., 1991) were used for the preparation of the figures.
SPR Analysis
All binding experiments were performed at 25°C with a BIAcore 1000 biosensor in phosphate-buffered saline (pH 7.4) running buffer. CD48 or 2B4 (100 μg/ml) were covalently coupled to a CM5 sensor chip via primary amine groups with an Amine Coupling Kit (BIAcore). Various concentrations of 2B4 or CD48 were then serially injected over the CD48 or 2B4 surface, respectively. Equilibrium affinity measurements were performed at different flow rates (5-60 μl/min). Specific binding data were fitted with a 1:1 Langmuir binding model so that dissociation constants (KDs) could be calculated.
ACKNOWLEDGMENTS
We thank H. Robinson (Brookhaven National Synchrotron Light Source) for X-ray data collection, R. Guan for initial studies, and N. Wais for technical assistance. We also thank the flow cytometry facility at the University of Chicago for cell sorting of yeast libraries and use of analysis instruments. This work was supported by National Institutes of Health Grant AI47990 to R.A.M. L.D. is a Cancer Research Institute Postdoctoral Fellow.
Footnotes
Accession Numbers
Atomic coordinates and structure factors for 2B4, CD48, and the 2B4-CD48 complex have been deposited in the Protein Data Bank under accession codes 2PTU, 2PTV, and 2PTT, respectively.
REFERENCES
- Ames JB, Vyas V, Lusin JD, Mariuzza R. NMR structure of the natural killer cell receptor 2B4 (CD244): Implications for ligand recognition. Biochemistry. 2005;44:6416–6423. doi: 10.1021/bi050139s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assarsson E, Kambayashi T, Persson CM, Chambers BJ, Ljunggren HG. 2B4/CD48-mediated regulation of lymphocyte activation and function. J. Immunol. 2005;175:2045–2049. doi: 10.4049/jimmunol.175.4.2045. [DOI] [PubMed] [Google Scholar]
- Bacon DJ, Anderson WF. A fast algorithm for rendering space-filling molecule pictures. J. Mol. Graph. 1988;6:219–220. [Google Scholar]
- Bloch-Queyrat C, Fondaneche MC, Chen R, Yin L, Relouzat F, Veillette A, Fischer A, Latour S. Regulation of natural cytotoxicity by the adaptor SAP and the Src-related kinase Fyn. J. Exp. Med. 2005;202:181–192. doi: 10.1084/jem.20050449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boder ET, Wittrup KD. Yeast surface display for directed evolution of protein expression, affinity, and stability. Methods Enzymol. 2000;328:430–444. doi: 10.1016/s0076-6879(00)28410-3. [DOI] [PubMed] [Google Scholar]
- Boyington JC, Motyka SA, Schuck P, Brooks AG, Sun PD. Crystal structure of an NK cell immunoglobulin-like receptor in complex with its MHC class I ligand. Nature. 2000;405:537–543. doi: 10.1038/35014520. [DOI] [PubMed] [Google Scholar]
- Bromley SK, Burack WR, Johnson KG, Somersalo K, Sims TN, Sumen C, Davis MM, Shaw AS, Allen PM, Dustin ML. The immunological synapse. Annu. Rev. Immunol. 2001;19:375–396. doi: 10.1146/annurev.immunol.19.1.375. [DOI] [PubMed] [Google Scholar]
- Brown MH, Boles K, van der Merwe PA, Kumar V, Mathew PA, Barclay AN. 2B4, the natural killer and T cell immunoglobulin superfamily surface protein, is a ligand for CD48. J. Exp. Med. 1998;188:2083–2090. doi: 10.1084/jem.188.11.2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao E, Ramagopal UA, Fedorov A, Fedorov E, Yan Q, Lary JW, Cole JL, Nathenson SG, Almo SC. NTB-A receptor crystal structure: insights into homophilic interactions in the signaling lymphocytic activation molecule receptor family. Immunity. 2006;25:559–570. doi: 10.1016/j.immuni.2006.06.020. [DOI] [PubMed] [Google Scholar]
- Chen R, Relouzar F, Roncagalli R, Aoukaty A, Tan R, Latour S, Veillette A. Molecular dissection of 2B4 signaling: Implications for signal transduction by SLAM-related receptors. Mol. Cell. Biol. 2004;24:5144–5156. doi: 10.1128/MCB.24.12.5144-5156.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Dam J, Guan R, Natarajan K, Dimasi N, Chlewicki LK, Kranz DM, Schuck P, Margulies DH, Mariuzza RA. Variable MHC class I engagement by Ly49 NK cell receptors revealed by the crystal structure of Ly49C bound to H-2Kb. Nat. Immunol. 2003;4:1213–1222. doi: 10.1038/ni1006. [DOI] [PubMed] [Google Scholar]
- Davis SJ, Davies EA, Tucknott MG, Jones EY, van der Merwe PA. The role of charged residues mediating low affinity protein-protein recognition at the cell surface by CD2. Proc. Natl. Acad. Sci. USA. 1998;95:5490–5494. doi: 10.1073/pnas.95.10.5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis DM, Chiu I, Fassett M, Cohen GB, Mandelboim O, Strominger JL. The human natural killer cell immune synapse. Proc. Natl. Acad. Sci. USA. 1999;96:15062–15067. doi: 10.1073/pnas.96.26.15062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis SJ, Ikemizu S, Evans EJ, Fugger L, Bakker TR, van der Merwe PA. The nature of molecular recognition by T cells. Nat. Immunol. 2003;4:217–224. doi: 10.1038/ni0303-217. [DOI] [PubMed] [Google Scholar]
- Deng L, Mariuzza RA. Structural basis for recognition of MHC and MHC-like ligands by natural killer cell receptors. Semin. Immunol. 2006;18:159–166. doi: 10.1016/j.smim.2006.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissmann P, Beauchamp L, Wooters J, Tilton JC, Long EO, Watzl C. Molecular basis for positive and negative signaling by natural killer cell receptor 2B4 (CD244) Blood. 2005;105:4722–4729. doi: 10.1182/blood-2004-09-3796. [DOI] [PubMed] [Google Scholar]
- Esnouf RM. Further additions to MolScript version 1.4, including reading and contouring of electron-density maps. Acta Crystallogr. D Biol. Crystallogr. 1999;55:938–940. doi: 10.1107/s0907444998017363. [DOI] [PubMed] [Google Scholar]
- Esteban O, Zhao H. Directed evolution of soluble single-chain human class II MHC molecules. J. Mol. Biol. 2004;340:81–95. doi: 10.1016/j.jmb.2004.04.054. [DOI] [PubMed] [Google Scholar]
- Evans EJ, Castro MAA, O’Brien R, Kearney A, Walsh H, Sparks LM, Tucknott MG, Davies EA, Carmo AM, van der Merwe PA, et al. Crystal structure and binding properties of the CD2 and CD244 (2B4) binding protein CD48. J. Biol. Chem. 2006;281:29309–29320. doi: 10.1074/jbc.M601314200. [DOI] [PubMed] [Google Scholar]
- Fan QR, Long EO, Wiley DC. Crystal structure of the human natural killer inhibitory receptor KIR2DL1-HLA-Cw4 complex. Nat. Immunol. 2001;2:452–460. doi: 10.1038/87766. [DOI] [PubMed] [Google Scholar]
- Fernandez NC, Treiner E, Vance RE, Jamieson AM, Lemieux S, Raulet DH. A subset of natural killer cells achieves self-tolerance without expressing inhibitory receptors specific for self MHC molecules. Blood. 2005;105:4416–4423. doi: 10.1182/blood-2004-08-3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones EY, Davis SJ, Williams AF, Harlos K, Stuart DI. Crystal structure at 2.8 Å resolution of a soluble form of the cell adhesion molecule CD2. Nature. 1992;360:232–239. doi: 10.1038/360232a0. [DOI] [PubMed] [Google Scholar]
- Kieke MC, Shusta EV, Boder ET, Teyton L, Wittrup KD, Kranz DM. Selection of functional T cell receptor mutants from a yeast surface-display library. Proc. Natl. Acad. Sci. USA. 1999;96:5651–5656. doi: 10.1073/pnas.96.10.5651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killeen N, Moessner R, Arvieux J, Willis A, Williams AF. The MRC OX-45 antigen of rat leukocytes and endothelium is in a subset of the immunoglobulin superfamily with CD2, LFA-3 and carcinoembryonic antigens. EMBO J. 1988;7:3087–3091. doi: 10.1002/j.1460-2075.1988.tb03174.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalski JM, Parekh RN, Mao J, Wittrup KD. Protein folding stability can determine the efficiency of escape from endoplasmic reticulum quality control. J. Biol. Chem. 1998;73:19453–19458. doi: 10.1074/jbc.273.31.19453. [DOI] [PubMed] [Google Scholar]
- Kumar V, McNerney ME. A new self: MHC class I-independent natural killer cell self-tolerance. Nat. Rev. Immunol. 2005;5:363–374. doi: 10.1038/nri1603. [DOI] [PubMed] [Google Scholar]
- Kumaresan PR, Stepp SE, Bennett M, Kumar V, Mathew PA. Molecular cloning of transmembrane and soluble forms of a novel rat natural killer cell receptor related to 2B4. Immunogenetics. 2000;51:306–313. doi: 10.1007/s002510050624. [DOI] [PubMed] [Google Scholar]
- Lanier LL. NK cell recognition. Annu. Rev. Immunol. 2005;23:225–274. doi: 10.1146/annurev.immunol.23.021704.115526. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: A program to check the stereo chemical quality of protein structures. J. Appl. Cryst. 1993;26:283–291. [Google Scholar]
- Latchman Y, McKay PF, Reiser H. Identification of the 2B4 molecule as a counter-receptor for CD48. J. Immunol. 1998;161:5809–5812. [PubMed] [Google Scholar]
- Lawrence MC, Colman PM. Shape complementarity at protein-protein interfaces. J. Mol. Biol. 1993;234:946–950. doi: 10.1006/jmbi.1993.1648. [DOI] [PubMed] [Google Scholar]
- Lee KM, McNerney ME, Stepp SE, Mathew PA, Schatzle JD, Bennett M, Kumar V. 2B4 acts as a non-major histocompatibility complex binding inhibitory receptor on mouse natural killer cells. J. Exp. Med. 2004;199:1245–1254. doi: 10.1084/jem.20031989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KM, Forman JP, McNerney ME, Stepp S, Kuppireddi S, Guzior D, Latchman YE, Sayegh MH, Yagita H, Park CK, et al. Requirement of homotypic NK-cell interactions through 2B4(CD244)/CD48 in the generation of NK effector functions. Blood. 2006;107:3181–3188. doi: 10.1182/blood-2005-01-0185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Urrutia M, Smith-Gill SJ, Mariuzza RA. Dissection of binding interactions in the complex between the anti-lysozyme antibody HyHEL-63 and its antigen. Biochemistry. 2003;42:11–22. doi: 10.1021/bi020589+. [DOI] [PubMed] [Google Scholar]
- Ma CS, Nichols KE, Tangye SG. Regulation of cellular and humoral responses by the SLAM and SAP families of molecules. Annu. Rev. Immunol. 2007;25:337–379. doi: 10.1146/annurev.immunol.25.022106.141651. [DOI] [PubMed] [Google Scholar]
- Mathew SO, Kumaresan PR, Lee JK, Huynh VT, Mathew PA. Mutational analysis of the human 2B4 (CD244)/CD48 interaction: Lys68 and Glu70 in the V domain of 2B4 are critical for CD48 binding and functional activation of NK cells. J. Immunol. 2005;175:1005–1013. doi: 10.4049/jimmunol.175.2.1005. [DOI] [PubMed] [Google Scholar]
- McNerney ME, Guzior D, Kumar V. 2B4 (CD244)-CD48 interactions provide a novel MHC class I-independent system for NK cell self-tolerance in mice. Blood. 2005;106:1337–1340. doi: 10.1182/blood-2005-01-0357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McRee DE. XtalView/Xfit-A versatile program for manipulating atomic coordinates and electron density. J. Struct. Biol. 1999;125:156–165. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- Moretta L, Bottino C, Pende D, Castriconi R, Mingari MC, Moretta A. Surface NK receptors and their ligands on tumor cells. Semin. Immunol. 2006;18:151–158. doi: 10.1016/j.smim.2006.03.002. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Nakajima H, Cella M, Bouchon A, Grierson HL, Lewis J, Duckett CS, Cohen JI, Colonna M. Patients with X-linked lymphoproliferative disease have a defect in 2B4 receptor-mediated NK cell cytotoxicity. Eur. J. Immunol. 2000;30:3309–3318. doi: 10.1002/1521-4141(200011)30:11<3309::AID-IMMU3309>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Natarajan K, Dimasi N, Wang J, Mariuzza RA, Margulies DH. Structure and function of natural killer cell receptors: Multiple molecular solutions to self, nonself discrimination. Annu. Rev. Immunol. 2002;20:853–885. doi: 10.1146/annurev.immunol.20.100301.064812. [DOI] [PubMed] [Google Scholar]
- Nicholls A, Sharp KA, Honig B. Protein folding and association: Insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- Orange JS, Harris KE, Andzelm MM, Valter MM, Geha RS, Strominger JL. The mature activating natural killer cell immunologic synapse is formed in distinct stages. Proc. Natl. Acad. Sci. USA. 2003;100:14151–14156. doi: 10.1073/pnas.1835830100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Parolini S, Bottino C, Falco M, Augugliaro R, Giliani S, Franceschini R, Ochs HD, Wolf H, Bonnefoy JY, Biassoni R, et al. X-linked lymphoproliferative disease. 2B4 molecules displaying inhibitory rather than activating function are responsible for the inability of natural killer cells to kill Epstein-Barr virus-infected cells. J. Exp. Med. 2000;192:337–346. doi: 10.1084/jem.192.3.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plougastel BFM, Yokoyama WM. Immune functions encoded by the natural killer gene complex. Nat. Rev. Immunol. 2003;3:304–316. doi: 10.1038/nri1055. [DOI] [PubMed] [Google Scholar]
- Roda-Navarro P, Mittelbrun M, Ortega M, Howie D, Terhorst C, Sanchez-Madrid F, Fernandez-Ruiz E. Dynamic redistribution of the activating 2B4/SAP complex at the cytotoxic NK cell immune synapse. J. Immunol. 2004;173:3640–3646. doi: 10.4049/jimmunol.173.6.3640. [DOI] [PubMed] [Google Scholar]
- Schatzle JD, Sheu S, Stepp SE, Mathew PA, Bennett M, Kumar V. Characterization of inhibitory and stimulatory forms of the murine natural killer cell receptor 2B4. Proc. Natl. Acad. Sci. USA. 1999;96:3870–3875. doi: 10.1073/pnas.96.7.3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storoni LC, McCoy AJ, Read RJ. Likelihood-enhanced fast rotation functions. Acta Crystallogr. D Biol. Crystallogr. 2004;60:432–438. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
- Tangye SG, Phillips JH, Lanier LL, Nichols KE. Functional requirement for SAP in 2B4-mediated activation of human natural killer cells as revealed by the X-linked lymphoproliferative syndrome. J. Immunol. 2000;165:2932–2936. doi: 10.4049/jimmunol.165.6.2932. [DOI] [PubMed] [Google Scholar]
- Vaidya SV, Stepp SE, McNerney ME, Lee JK, Lee K, Bennett M, Lee KM, Stewart CL, Kumar V, Mathew PA. Targeted disruption of the 2B4 gene in mice reveals an in vivo role of 2B4 (CD244) in the rejection of B16 melanoma cells. J. Immunol. 2005;174:800–807. doi: 10.4049/jimmunol.174.2.800. [DOI] [PubMed] [Google Scholar]
- van der Merwe PA, Barclay AN, Mason DW, Davies EA, Morgan BP, Tome M, Krishnam AK, Ianelli C, Davis SJ. Human cell-adhesion molecule CD2 binds CD58 (LFA-3) with a very low affinity and an extremely fast dissociation rate but does not bind CD48 or CD59. Biochemistry. 1994;33:10149–10160. doi: 10.1021/bi00199a043. [DOI] [PubMed] [Google Scholar]
- van der Merwe PA, Davis SJ, Shaw AS, Dustin ML. Cytoskeletal polarization and redistribution of cell-surface molecules during T cell antigen recognition. Semin. Immunol. 2000;12:5–21. doi: 10.1006/smim.2000.0203. [DOI] [PubMed] [Google Scholar]
- Veillette A. Immune regulation by SLAM family receptors and SAP-related proteins. Nat. Rev. Immunol. 2006;6:56–66. doi: 10.1038/nri1761. [DOI] [PubMed] [Google Scholar]
- Vyas YM, Mehta KM, Morgan M, Maniar H, Butros L, Jung S, Burkhardt JK, Dupont B. Spatial organization of signal transduction molecules in the NK cell immune synapses during MHC class I-regulated noncytolytic and cytolytic interactions. J. Immunol. 2001;167:4358–4367. doi: 10.4049/jimmunol.167.8.4358. [DOI] [PubMed] [Google Scholar]
- Vyas YM, Maniar H, Dupont B. Visualization of signaling pathways and cortical cytoskeleton in cytolytic and noncytolytic natural killer cell immune synapses. Immunol. Rev. 2002;189:161–178. doi: 10.1034/j.1600-065x.2002.18914.x. [DOI] [PubMed] [Google Scholar]
- Wang JH, Smolyar A, Tan K, Liu JH, Kim M, Sun ZY, Wagner G, Reinherz EL. Structure of a heterophilic adhesion complex between the human CD2 and CD58 (LFA-3) counterreceptors. Cell. 1999;97:791–803. doi: 10.1016/s0092-8674(00)80790-4. [DOI] [PubMed] [Google Scholar]
- Yan Q, Malashkevich VN, Federov A, Federov E, Cao E, Lary JW, Cole JL, Nathenson SG, Almo SC. Structure of CD84 provides insights into SLAM family function. Proc. Natl. Acad. Sci. USA. 2007;104:10538–10588. doi: 10.1073/pnas.0703893104. [DOI] [PMC free article] [PubMed] [Google Scholar]