Abstract
Constitutively active tyrosine kinases promote leukemogenesis by increasing cell proliferation and inhibiting apoptosis. However, mechanisms underlying apoptotic inhibition have not been fully elucidated. In many settings, apoptosis occurs by mitochondrial cytochrome c release, which nucleates the Apaf-1/caspase-9 apoptosome. Here we report that the leukemogenic kinases, Bcr-Abl, FLT3/D835Y, and Tel-PDGFRβ, all can inhibit apoptosome function. In cells expressing these kinases, the previously reported apoptosome inhibitor, Hsp90β, bound strongly to Apaf-1, preventing cytochrome c-induced Apaf-1 oligomerization and caspase-9 recruitment. Hsp90β interacted weakly with the apoptosome in untransformed cells. While Hsp90β was phosphorylated at Ser 226/Ser 255 in untransformed cells, phosphorylation was absent in leukemic cells. Expression of mutant Hsp90β (S226A/S255A), which mimics the hypophosphorylated form in leukemic cells, conferred resistance to cytochrome c-induced apoptosome activation in normal cells, reflecting enhanced binding of nonphosphorylatable Hsp90β to Apaf-1. In Bcr-Abl-positive mouse bone marrow cells, nonphosphorylatable Hsp90β expression conferred imatinib (Gleevec) resistance. These data provide an explanation for apoptosome inhibition by activated leukemogenic tyrosine kinases and suggest that alterations in Hsp90β-apoptosome interactions may contribute to chemoresistance in leukemias.
Apoptosis is a cellular suicide program critical for development and tissue homeostasis (11). Excess apoptosis is associated with degenerative disorders, while a failure of apoptosis contributes to oncogenesis (17, 55). In many cells, chemotherapeutic agents that cause genotoxic stress promote the release of cytochrome c from the mitochondria to the cytoplasm (26). Once cytoplasmic, cytochrome c induces formation of a caspase-activating complex called the apoptosome, consisting of cytochrome c, the adaptor protein Apaf-1, dATP/ATP, and zymogenic caspase-9 (25, 58). Apaf-1 is an adaptor protein containing a nucleotide-binding domain that binds to dATP/ATP, flanked by an N-terminal caspase recruitment domain (CARD) and C-terminal WD-40 repeats (58, 59). Binding of caspase-9 to Apaf-1 is CARD mediated (25, 34). The binding of cytochrome c and dATP/ATP to the WD-40 and nucleotide-binding domain, respectively, induces conformational changes of Apaf-1 to expose its CARD, thereby recruiting caspase-9 into the apoptosome (7, 21, 46, 59). Activated caspase-9 can then cleave and activate effector caspases to dismantle the dying cell. Such apoptotic pathways can be regulated either through modulation of cytochrome c release or by altering apoptosome formation.
There is a growing list of apoptosome inhibitors and activators (40). In most cases, how these apoptosome regulators modify apoptosome activity is unknown. Several chaperones, including Hsp90 and Hsp70, have been reported to inhibit the apoptosome (4). In the case of Hsp90, its binding to Apaf-1 was reported to block Apaf-1 oligomerization and caspase-9 recruitment (35). However, it is unclear how this abundant cellular protein might be regulated to allow it to alter apoptosome function in a controlled manner. This is of particular interest as Hsp90 is often upregulated in cancer cells (22, 52). Although one consequence of this upregulation is protection of oncogenic proteins from proteasomal degradation (3), it may be that apoptosome regulation is an important secondary effect, enhancing resistance to apoptosis and potentially contributing to chemoresistance.
Constitutively active leukemogenic tyrosine kinases increase cellular proliferation and inhibit apoptosis. For example, potent apoptotic inhibitors p190Bcr-Abl and p210Bcr-Abl are found in approximately 25% of adult patients with acute lymphocytic leukemia (ALL) and more than 95% of patients with chronic myeloid leukemia (CML). Bcr-Abl inhibits mitochondrial cytochrome c release by promoting the inhibitory phosphorylation of the proapoptotic Bcl-2 family protein Bad through the phosphatidylinositol 3-kinase (PI 3-kinase)/Akt pathway (31, 45). Additionally, Bcr-Abl augments expression of antiapoptotic Bcl-2 family members through the transcription factor STAT5 (2, 20, 39, 43). We have previously demonstrated that Bcr-Abl also prevents apoptosis downstream of mitochondrial cytochrome c release by perturbing caspase-9 recruitment to Apaf-1 (12). When purified wild-type Bcr-Abl was added to cytosolic extracts, cytochrome c-induced caspase activation was prevented. Furthermore, Bcr-Abl-expressing cells exhibited remarkable resistance to apoptotic death induced by cytochrome c microinjection. Since Bcr-Abl did not perturb the interaction of endogenous caspase-9 with the isolated recombinant Apaf-1 CARD, our data suggested that it might be Apaf-1 whose function was altered by Bcr-Abl. Although inhibitory phosphorylations of caspase-9 by Akt (8) and c-Abl (38) have been reported, caspase-9 was not phosphorylated in Bcr-Abl-expressing cells (12).
We report here that Tel-PDGFRβ (a fusion protein of the N terminus of Tel with the transmembrane and cytoplasmic domains of the platelet-derived growth factor receptor β [14]) and the activated FLT3 kinase mutants (FLT3/D835Y and FLT3-ITD [1, 30, 54]), prevalent in CML and acute myeloid leukemia (AML), respectively, also trigger resistance to cytochrome c-induced apoptosome formation. In investigating the mechanism of inhibition, we discovered that Hsp90β, a previously reported apoptosome inhibitor, was hypophosphorylated in cells expressing leukemogenic tyrosine kinases. Moreover, hypophosphorylation promoted increased binding of Hsp90β to Apaf-1. In untransformed cells, Hsp90β was phosphorylated on Ser 226 and Ser 255, while these sites were unphosphorylated in leukemic cells. Mutation of these residues to nonphosphorylatable forms resulted in stronger binding of Hsp90β to Apaf-1 and increased cytochrome c resistance in untransformed cells. Furthermore, expression of the nonphosphorylatable mutant conferred imatinib resistance in Bcr-Abl-positive mouse bone marrow cells. Our data suggest that modulation of Hsp90β-directed kinases/phosphatases underlies resistance to cytochrome c-induced apoptosome activation in leukemias expressing activated tyrosine kinases. Moreover, they point to a possible role of Hsp90β in modulating sensitivity of leukemias to chemotherapeutic agents.
MATERIALS AND METHODS
Antibodies and reagents.
Anti-caspase-9 (Neomarkers, Fremont, CA), anti-caspase-9 (mouse specific; Cell Signaling, Danvers, MA), anti-cleaved caspase-3, anti-caspase-3 (mouse specific), anti-cytochrome c, anti-Abl (BD Biosciences, San Diego, CA), anti-Apaf-1 (Anaspec, San Jose, CA; Alexis Biochemicals, San Diego, CA), anti-Hsp70 (Affinity Bioreagents, Golden, CO), anti-Hsp90 (Stressgen Bioreagents, Ann Arbor, MI; Upstate Biotechnology, Lake Placid, NY; Santa Cruz Biotechnology, Santa Cruz, CA), anti-PDGFRβ (Upstate Biotechnology), anti-FLT3, antiactin (Santa Cruz Biotechnology), goat anti-rabbit Alexa Fluor 647 (Molecular Probes, Eugene, OR), and anti-FLAG (Sigma, St. Louis, MO) were used for immunoblotting. Anti-Hsp90β antibody (D-19; Santa Cruz Biotechnology), anti-FLT3 antibody (S-18; Santa Cruz Biotechnology), and anti-Apaf-1 antibody (13F11; Alexis) were used to immunoprecipitate the respective proteins from Ba/F3 cell lysates. Immunoblots were imaged using the Li-Cor Odyssey IR Imaging System. Imatinib mesylate and PKC412 were purchased from LC Laboratories (Woburn, MA). Recombinant human FLT3 ligand was purchased from R&D Systems (Minneapolis, MN). Recombinant casein kinase 2 (CK2) and lambda phosphatase were purchased from New England Biolabs (Ipswich, MA).
Cell culture.
Control and Ba/F3 cells expressing p210Bcr-Abl, FLT3/D835Y, or Tel-PDGFRβ were generous gifts from D. G. Gilliland (Harvard Medical School). Ba/F3 cells expressing wild-type or T315I p210Bcr-Abl were kind gifts from G. Q. Daley (Harvard Medical School) (19). Control Ba/F3 cells were maintained in RPMI medium supplemented with 10% fetal bovine serum (FBS) and 1 ng/ml interleukin-3 (IL-3), whereas Ba/F3 cells expressing p210Bcr-Abl, FLT3/D835Y, or Tel-PDGFRβ were maintained without IL-3. THP-1, U-937, and Ku812 cells were cultured in RPMI medium supplemented with 10% FBS. MV4-11 and SUP-B15 cells were grown in Iscove's modified Dulbecco's medium with 20% FBS. TF-1 cells were cultured in RPMI medium supplemented with 10% FBS and 2 ng/ml recombinant human granulocyte-macrophage colony-stimulating factor (Sigma).
For stable expression, Ba/F3 cells were transduced with a murine stem cell virus (MSCV) retroviral vector bicistronically expressing green fluorescent protein (GFP) with either Hsp90β (S226A/S255A) or Hsp90β (S226E/S255E). GFP-positive cells were selected by fluorescence-activated cell sorting (FACS). For IL-3 withdrawal, cells were washed with phosphate-buffered saline and then cultured in RPMI lacking IL-3 for 20 h.
Cell extracts.
Cell lysates were prepared as described previously (26). Cells were harvested, washed with cold buffer A (20 mM HEPES [pH 7.4], 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 5 μg/ml leupeptin, and 5 μg/ml aprotinin), and pelleted. Pellets were resuspended in twice the pellet volume of hypotonic lysis buffer (buffer A containing 250 mM sucrose) and incubated on ice for 30 min. Cells were then Dounce homogenized and centrifuged at 14,000 rpm for 20 min at 4°C. Supernatants were assayed for protein concentrations with the Bradford assay (Bio-Rad, Hercules, CA) and used as the cell-free lysate. In vitro caspase assays were performed as previously described (12).
Plasmid construction and transfection.
Human Apaf-1 and human Hsp90β in pFastBac were gifts from X. Wang (University of Texas Southwestern) and T. Takenawa (University of Tokyo), respectively. myc-Bax in pCMV-tag3A vector was a gift from C. W. Distelhorst (Case Western Reserve University). Caspase-9 (C287A) in pET-23b was obtained from Addgene (Cambridge, MA) (47). Caspase-9 (C287S) with the N-terminal FLAG tag was generated from human caspase-9 and cloned into pcDNA3 (Invitrogen, Carlsbad, CA). Apaf-1 and Hsp90β deletion mutants were cloned into pGEX-KG for production of glutathione S-transferase (GST) fusion proteins. Hsp90β and p210Bcr-Abl (from A. M. Pendergast, Duke University Medical Center) were also cloned into the MSCV vector which bicistronically expresses GFP and yellow fluorescent protein (YFP), respectively (MSCV-Hsp90β-IRES-GFP and MSCV-p210Bcr-Abl-IRES-YFP). Hsp90 mutants with the N-terminal FLAG tag were cloned into pcDNA3. All point mutations were generated with the QuikChange mutagenesis kit (Stratagene, La Jolla, CA).
Transfection was performed using the Amaxa electroporation system (Nucleofection kit V, program X-01; Amaxa Biosystems, Gaithersburg, MD). Ten micrograms of empty vector or myc-Bax (or 2 μg of FLAG-tagged Hsp90β mutants) was transfected to 4 × 106 cells. Cells were fixed by 4% formaldehyde 8 h posttransfection and membrane permeabilized by 90% methanol. The cells were stained with cleaved caspase-3 antibody and with goat anti-rabbit Alexa Fluor-647 and subjected to FACS analysis.
siRNA transfection.
All small interfering RNAs (siRNAs) were designed and synthesized by Dharmacon RNA Technologies (Lafayette, CO); four siRNAs targeting mouse Hsp90β (or four nontargeting siRNAs) were mixed into one pool and used as single reactions. The target sequences for mouse Hsp90β (catalog no. L-050742-00) were 5′-GGA CAA GAU UCG AUA UGA G-3′, 5′-UGG AAG AGG UGG AUU AAA G-3′, 5′-GAU CAA AGA GAA GUA CAU U-3′, and 5′-GGU GUU AUG UAU UGU GGU U-3′. Nontargeting siRNA pool 1 (catalog no. D-001206-13) was used as a control. The siRNAs were prepared according to the manufacturer's instructions. RNA interference was carried out by electroporation using the Amaxa Cell Nucleofection kit V and program X-05 (10 μl of 20 μM siRNA stock to 2.5 × 106 Ba/F3 cells per reaction). Forty-eight hours after the treatment, the cells were subjected to lysis, resulting in cell-free lysates.
Gel filtration.
Ba/F3 cell lysate (5 μg/μl) was incubated in the presence or absence of 1 mM dATP and 2.5 ng/μl cytochrome c at 37°C for 30 min in a volume of 250 μl. In vitro reconstitution of apoptosome formation was performed by incubating 0.4 μM Apaf-1 and 0.8 μM caspase-9 (C287A) at 30°C for 30 min in the presence or absence of 1 mM dATP and cytochrome c (0.01 or 0.4 μM) in a final volume of 250 μl of buffer A with 100 mM NaCl. In certain experiments, Apaf-1 and caspase-9 (C287A) were preincubated with 1 μM Hsp90β at 30°C for 30 min before addition of dATP and cytochrome c. After incubation, the reaction mixture was loaded onto a Superdex 200 column at a flow rate of 0.3 ml/min.
Colony-forming assay.
Mouse bone marrow cells were isolated, enriched with c-kit beads, and stained with c-kit-APC, Sca1-PECy5, and lineage markers conjugated with phycoerythrin as described previously (56). KLS cells were sorted and cultured overnight in 10% Dulbecco modified Eagle medium with 50 ng/ml of stem cell factor and 10 ng/ml of IL-3 and IL-6 (R&D Systems). Cells were infected with MSCV-p210Bcr-Abl-IRES-YFP together with MSCV-Hsp90 (S226A/S255A)-IRES-GFP or Hsp90 (S226E/S255E)-IRES-GFP. Two days later, YFP and GFP double-positive cells were selected by FACS and plated in methylcellulose medium (M3434; Stem Cell Technologies, Vancouver, BC, Canada). Colonies were counted 7 days after plating.
RESULTS
Leukemogenic tyrosine kinases inhibit recruitment of caspase-9 to Apaf-1.
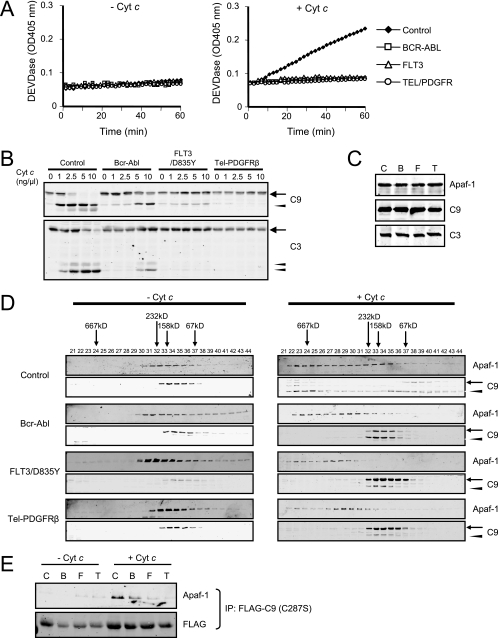
We reported previously that Bcr-Abl could impede apoptosis after mitochondrial cytochrome c release by perturbing caspase-9 recruitment to Apaf-1 (12). As shown in Fig. 1A, addition of cytochrome c and dATP to cell lysates prepared from untransformed Ba/F3 cells resulted in robust caspase-3 activation, as measured by DEVDase activity, while this activity was inhibited in lysates from Ba/F3 cells expressing Bcr-Abl. We wondered whether this property of Bcr-Abl might be shared by other leukemogenic tyrosine kinases and so repeated these experiments in lysates from Ba/F3 cells expressing FLT3/D835Y or Tel-PDGFRβ. In both cases, cytochrome c-induced caspase activation was markedly dampened (Fig. 1A). Consistent with these observations, immunoblotting to detect caspase-9 and caspase-3 cleavage in response to cytochrome c was significantly reduced in lysates from cells expressing the leukemogenic tyrosine kinases (Fig. 1B). Importantly, total protein levels of Apaf-1, caspase-9, and caspase-3 were unaltered by tyrosine kinase expression (Fig. 1C), suggesting that the inhibition might be at the level of Apaf-1 oligomerization or caspase-9 recruitment.
FIG. 1.
Leukemogenic tyrosine kinases inhibit recruitment of caspase-9 to Apaf-1. (A) Cell lysates were prepared from control Ba/F3 cells or Ba/F3 cells expressing Bcr-Abl, FLT3/D835Y, or Tel-PDGFRβ and incubated with 0 or 2.5 ng/μl cytochrome c (Cyt c) and 1 mM dATP. Caspase-3 activity was assayed by measuring cleavage of DEVD-pNA. (B) Lysates were incubated with 1 mM dATP and various concentrations of cytochrome c, and immunoblotting was performed for caspase-9 (C9) and caspase-3 (C3). Procaspases and cleaved caspases are indicated by arrows and arrowheads, respectively. (C) Total cell lysates from control Ba/F3 cells (C) or Ba/F3 cells expressing Bcr-Abl (B), FLT3/D835Y (F), or Tel-PDGFRβ (T) were immunoblotted with anti-Apaf-1, anti-caspase-9 (C9), and anti-caspase-3 (C3) antibodies. (D) Cell lysates were separated on a Superdex 200 column before and after incubation with 2.5 ng/μl cytochrome c and 1 mM dATP for 30 min. Immunoblotting was performed for Apaf-1 and caspase-9. Procaspase-9 and cleaved caspase-9 are indicated by arrows and arrowheads, respectively. (E) Ba/F3 cells were transfected with FLAG-tagged caspase-9 (C287S), and lysates were prepared. Immunoprecipitation (IP) was performed with or without addition of cytochrome c (2.5 ng/μl) and dATP (1 mM). Pellets were analyzed by immunoblotting with anti-Apaf-1 and anti-FLAG antibodies.
To identify the step(s) negatively affected by the leukemogenic kinases, we performed gel filtration chromatography with the untransformed and transformed cell-free lysates and immunoblotted each fraction for Apaf-1 and caspase-9. When cytochrome c and dATP were added to Ba/F3 lysates, Apaf-1 shifted from lower-molecular-mass (monomeric) to higher-molecular-mass (apoptosomal) fractions exceeding 667 kDa (Fig. 1D). Caspase-9 was also in the apoptosomal fractions, and a substantial fraction of the protein was cleaved to its smaller active form (Fig. 1D). When cytochrome c was added to the lysates from Ba/F3 cells expressing Bcr-Abl, FLT3/D835Y, or Tel-PDGFRβ, Apaf-1 oligomerization was impaired (Fig. 1D). Strikingly, caspase-9 recruitment to the apoptosomal fractions and its subsequent cleavage were significantly diminished in the kinase-containing lysates (Fig. 1D). To confirm that this failure of apoptosome assembly was due to inhibition of caspase-9 recruitment to Apaf-1 in FLT3 cells and Tel-PDGFRβ, as in Bcr-Abl-expressing cells (12), we attempted to coimmunoprecipitate Apaf-1 with a FLAG-tagged catalytically inactive caspase-9 mutant (C287S). Indeed, significantly less Apaf-1 coprecipitated with FLAG-caspase-9 (C287S) in lysates prepared from Ba/F3 cells expressing leukemogenic tyrosine kinases than in control Ba/F3 lysates (Fig. 1E), indicating that FLT3/D835Y and Tel-PDGFRβ share with Bcr-Abl the ability to block caspase-9 recruitment to Apaf-1.
Apaf-1 binds to Hsp90β in cells expressing leukemogenic tyrosine kinases.
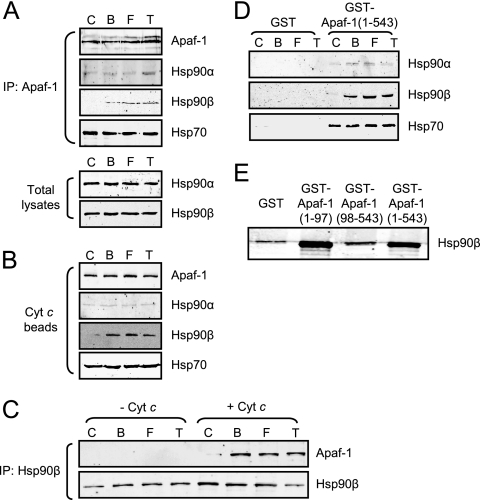
To determine how leukemogenic tyrosine kinases might inhibit apoptosome function, we examined a panel of known apoptosome inhibitors for interaction with Apaf-1 in normal and transformed cells. In most cases, we found no difference in association of these factors in the presence and absence of tyrosine kinase expression (for example, see Hsp70 in Fig. 2A, B, and D). In contrast, Hsp90β specifically coimmunoprecipitated with Apaf-1 from Ba/F3 cells expressing the leukemogenic tyrosine kinases, but not from control cells, though total Hsp90β protein levels were equivalent (Fig. 2A). Similarly, using cytochrome c-Sepharose to retrieve endogenous Apaf-1 from cell lysates, we found that Hsp90β copurified with Apaf-1 from the kinase-expressing Ba/F3 cells but not from control Ba/F3 cells (Fig. 2B). These data raised the possibility that leukemogenic tyrosine kinases could trigger the interaction of Hsp90β with Apaf-1 to block apoptosome assembly.
FIG. 2.
Apaf-1 binds to Hsp90β in cells expressing leukemogenic tyrosine kinases. (A) Control Ba/F3 cells (C) and those expressing Bcr-Abl (B), FLT3/D835Y (F), and Tel-PDGFRβ (T) were subjected to immunoprecipitation (IP) with anti-Apaf-1 antibody. The IP pellets were analyzed for Hsp90α, Hsp90β, and Hsp70 and Apaf-1 (top). Total Ba/F3 cell lysates were immunoblotted with anti-Hsp90α and -β antibodies (bottom). (B) Lysates were incubated with cytochrome c (Cyt c) beads, and pellets were subjected to immunoblotting with anti-Apaf-1, anti-Hsp90α and -β, and anti-Hsp70 antibodies. (C) IP with anti-Hsp90β antibody was carried out for the Ba/F3 cell lysates before and after cytochrome c (2.5 ng/μl) and dATP (1 mM) addition. The pellets were analyzed by immunoblotting with anti-Apaf-1 antibody. (D) Cell lysates were incubated with GST-Apaf-1 (1-543) or GST alone. Protein complexes were retrieved by using glutathione beads, and immunoblotting was performed for Hsp90α and -β or Hsp70. (E) Ba/F3 lysates expressing Tel-PDGFRβ were incubated with GST, GST-Apaf-1 (1-97), GST-Apaf-1 (98-543), or GST-Apaf-1 (1-543). Protein complexes were retrieved by using glutathione beads, and immunoblotting was performed for Hsp90β.
To further characterize the Apaf-1-Hsp90β interaction, Hsp90β was immunoprecipitated from untransformed and transformed Ba/F3 lysates before and after cytochrome c addition. Apaf-1 coimmunoprecipitated with Hsp90β only in kinase-expressing lysates (Fig. 2C). Moreover, Apaf-1 interaction with Hsp90β was seen only after addition of cytochrome c, suggesting that Apaf-1 conformational changes were necessary for Hsp90β binding (Fig. 2C). Indeed, recombinant Apaf-1 bearing a deletion of the WD-40 region [GST-Apaf-1 (1-543)] was able to retrieve Hsp90β from kinase-expressing Ba/F3 lysates in the absence of cytochrome c (Fig. 2D). Furthermore, this interaction was greatly diminished by deletion of the Apaf-1 CARD [GST-Apaf-1 (98-543) in Fig. 2E], and the Apaf-1 CARD was sufficient to bind Hsp90β [GST-Apaf-1 (1-97) in Fig. 2E]. These data suggest that in the presence of the leukemogenic tyrosine kinases, Hsp90β binds to the Apaf-1 CARD and that this binding depends on the cytochrome c-induced conformational change of Apaf-1. Although both the α and β isoforms of Hsp90 have been reported as inhibitors of apoptosome formation (35), we found the enhanced binding of Hsp90 to Apaf-1 in the presence of leukemogenic tyrosine kinases to be restricted to the β isoform (Fig. 2A, B, and D).
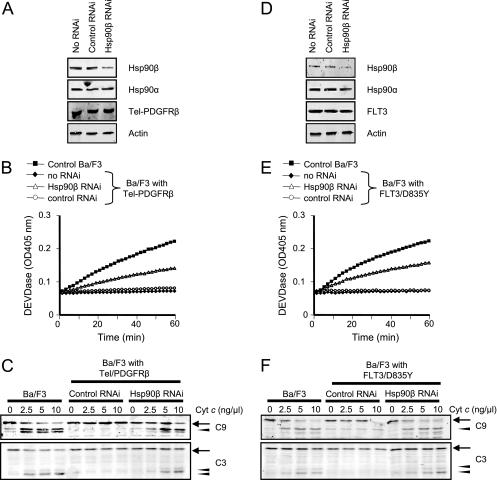
If the post-cytochrome c protection seen in Ba/F3 cells expressing the leukemogenic tyrosine kinases could be attributed to Hsp90β-Apaf-1 binding, then reduction in Hsp90β protein levels would be expected to abrogate the observed protection from cytochrome c-induced caspase activation. To test this, we attempted to silence Hsp90β expression using siRNA in Ba/F3 cells expressing Tel-PDGFRβ (Fig. 3A to C) or FLT3/D835Y (Fig. 3D to F), both of which exhibited stronger protection from cytochrome c than Bcr-Abl (Fig. 1B). Hsp90β knockdown (∼70% reduction; Fig. 3A and D) markedly restored sensitivity to cytochrome c; control siRNA had no such effect (Fig. 3B and E). Consistent with these data, at various doses of cytochrome c, caspase-3 and caspase-9 cleavages could be observed in Hsp90β but not control siRNA-treated tyrosine kinase-expressing cells (Fig. 3C and F).
FIG. 3.
Hsp90β knockdown partially restores sensitivity to cytochrome c in Ba/F3 cells expressing Tel-PDGFRβ (A to C) or FLT3/D835Y (D to F). (A and D) Ba/F3 cells expressing Tel-PDGFRβ (A) or FLT3/D835Y (D) were treated with Hsp90β-specific siRNA or control siRNA. Total cell lysates were immunoblotted for Hsp90β, Hsp90α, Tel-PDGFRβ, FLT3, and actin. (B and E) Caspase activity was assayed by measuring cleavage of DEVD-pNA following incubation of the cell lysates with 5 ng/μl cytochrome c and 1 mM dATP. (C and F) Immunoblotting was performed for caspase-9 and caspase-3 upon addition of various amounts of exogenous cytochrome c (Cyt c) to the lysates. Procaspase-9/procaspase-3 and cleaved caspase-9/caspase-3 are indicated by arrows and arrowheads, respectively. RNAi, RNA interference.
Hypophosphorylation of Hsp90β at Ser 226 and Ser 255 in cells expressing leukemogenic tyrosine kinases.
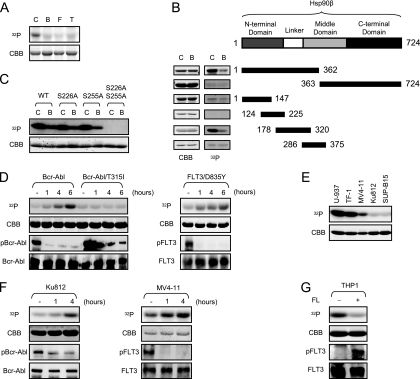
Although the findings above strongly suggested that Hsp90β binding to Apaf-1 could confer protection from apoptosis in leukemic cells, it remained unclear how these kinases might control Hsp90β binding to Apaf-1. In analyzing the effects of tyrosine kinase expression on Hsp90β, we found that recombinant Hsp90β protein was phosphorylated when incubated with control Ba/F3 cell lysates, whereas this phosphorylation was significantly suppressed when incubation was with Ba/F3 cell lysates expressing Bcr-Abl, FLT3/D835Y, or Tel-PDGFRβ (Fig. 4A). A literature search revealed that this hypophosphorylation of Hsp90β had been observed previously in Bcr-Abl (though not Tel-PDGFRβ or FLT3)-expressing cells (48). To identify phosphorylated sites on Hsp90β in control but not kinase-expressing cells, we generated a panel of GST-Hsp90β deletion mutants and incubated them with either control or Bcr-Abl-expressing Ba/F3 cell lysates (Fig. 4B). In vitro kinase assays revealed that differential phosphorylation occurred on GST fusion proteins containing the “linker” region of Hsp90β, connecting the N-terminal ATPase domain and the middle domain of the protein (Fig. 4B). Using GST-Hsp90β (178-300), we narrowed down the sites of differential Hsp90β phosphorylation to Ser 226 and Ser 255; mutation of these sites entirely abrogated phosphorylation (Fig. 4C).
FIG. 4.
Phosphorylation of Hsp90β at Ser 226/255 is suppressed in cells expressing the tyrosine kinases. (A) Recombinant His-tagged Hsp90β protein on nickel beads was incubated with the Ba/F3 cell lysates in the presence of [γ-32P]ATP; control Ba/F3 cells (C); and those expressing Bcr-Abl (B), FLT3/D835Y (F), and Tel-PDGFRβ (T). (B) Various deletion mutants of Hsp90β were made as GST fusion proteins and incubated with control Ba/F3 lysates (C) or lysates expressing Bcr-Abl (B) in the presence of [γ-32P]ATP. (C) Two point mutations (S226A and S255A) were introduced into GST-Hsp90β (178-300). The GST fusion proteins were incubated with control Ba/F3 lysates in the presence of [γ-32P]ATP. WT, wild type. (D) Ba/F3 cells expressing either wild-type Bcr-Abl or Bcr-Abl carrying the T315I mutation were treated with the Abl kinase inhibitor imatinib (1 μM) for the time indicated (left panel). Ba/F3 cells expressing FLT3/D835Y were treated with the FLT3 kinase inhibitor PKC412 (20 nM) over time (right panel). After each treatment, cell lysates were prepared and incubated with GST-Hsp90β (178-300) in the presence of [γ-32P]ATP. (E) GST-Hsp90β (178-300) was incubated with lysates from U-937, TF-1, MV4-11, Ku812, and SUP-B15 cells in the presence of [γ-32P]ATP. (F) Ku812 and MV4-11 cells were treated with imatinib (1 μM) and PKC412 (20 nM), respectively, for the indicated time. The cell lysates were incubated with GST-Hsp90β (178-300) in the presence of [γ-32P]ATP. (G) THP-1 cells were treated with or without recombinant human FLT3 ligand (FL; 50 ng/ml) for 2 h. The cell lysates were incubated with GST-Hsp90β (178-300) in the presence of [γ-32P]ATP. 32P incorporation and Coomassie blue staining (CBB) are shown. FLT3 was immunoprecipitated from whole-cell lysates with anti-FLT3 antibody. Western blotting was performed for Bcr-Abl, FLT3, and phosphotyrosine.
Importantly, this suppression of Hsp90β phosphorylation was greatly affected by an inhibitor of the leukemogenic kinases. Treatment of Ba/F3 cells expressing Bcr-Abl or FLT3/D835Y with the specific Abl kinase inhibitor imatinib (1 μM) or the FLT3 inhibitor PKC412 (20 nM), respectively, promoted in vitro phosphorylation of Hsp90β using cell lysates prepared from the treated cells (Fig. 4D). In contrast, the same treatment did not affect Hsp90β phosphorylation in lysates from Ba/F3 cells expressing Bcr-Abl/T315I, a known imatinib-resistant mutant of Bcr-Abl (Fig. 4D). These results suggest that the suppression of Hsp90β phosphorylation is, directly or indirectly, controlled by the activity of the leukemogenic tyrosine kinases. In support of this idea, lysates from two Bcr-Abl-positive cell lines, Ku812 (derived from a CML patient) and SUP-B15 (derived from an ALL patient), minimally phosphorylated the linker region of Hsp90β in vitro, while lysates from Bcr-Abl-negative lymphoid and myeloid cell lines, TF-1 and U-937, respectively, robustly phosphorylated this region (Fig. 4E). Likewise, although a lysate from MV4-11, a human AML cell line carrying the FLT3-ITD mutation, exhibited low-level in vitro phosphorylation of Hsp90β, this phosphorylation was markedly reduced compared to that in the control TF-1 and U-937 cells (Fig. 4E). When Ku812 and MV4-11 cells were treated with imatinib and PKC412, respectively, the resulting lysates gained the ability to phosphorylate the Hsp90β linker region over time (Fig. 4F), further confirming the results obtained from Ba/F3 cells. Conversely, addition of the FLT3 ligand (FL) induced tyrosine phosphorylation of FLT3 in THP-1 cells, an AML-M5 cell line expressing wild-type FLT3, and an in vitro kinase assay showed that phosphorylation of the Hsp90β linker region was attenuated in these lysates after FL stimulation (Fig. 4G). Together, these results indicate that phosphorylation of the Hsp90β linker region (Ser 226 and Ser 255) is greatly suppressed in the presence of leukemogenic tyrosine kinases.
Hypophosphorylation of Hsp90β at Ser 226 and Ser 255 promotes apoptosome inhibition.
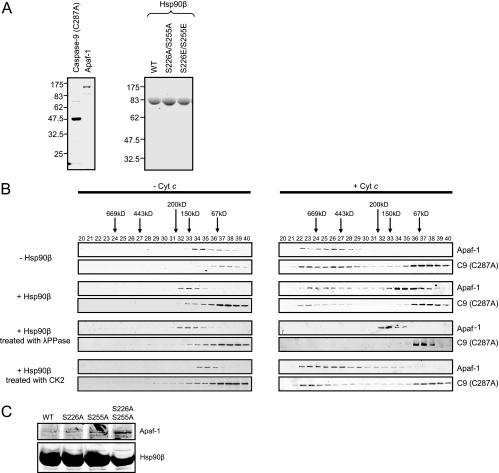
To assess the effects of phosphorylation on Hsp90β's ability to inhibit apoptosome formation, we examined apoptosome assembly by gel filtration using purified apoptosome components and recombinant Hsp90β (Fig. 5A). Recombinant human Hsp90β and Apaf-1 proteins were expressed in Sf9 cells and purified as described previously (59). Human caspase-9 was produced and purified from Escherichia coli BL21(DE3) cells; to simplify the detection of caspase-9 recruitment to the apoptosomal fractions, catalytically inactive caspase-9 (C287A) was used to eliminate caspase-9 cleavage products. Upon addition of cytochrome c and dATP (1 mM), Apaf-1 oligomerized, moving from monomeric to apoptosomal fractions (∼669 kDa) (Fig. 5B). The migration of caspase-9 (C287A) to the apoptosomal fractions was also observed (Fig. 5B). In the presence of Hsp90β, the migration of both Apaf-1 and caspase-9 (C287A) was shifted significantly to their monomeric forms, as previously reported (35). Importantly, in the presence of Hsp90β pretreated with lambda phosphatase, Apaf-1 oligomerization and caspase-9 recruitment were completely inhibited (Fig. 5B). Although the kinase responsible for Hsp90β phosphorylation in vivo is not known, it has been reported that CK2 can phosphorylate these sites in vitro (24). Thus, we prephosphorylated recombinant Hsp90β with CK2 before addition to the reaction. Remarkably, Hsp90β phosphorylation greatly reduced its ability to inhibit Apaf-1 oligomerization and caspase-9 recruitment (Fig. 5B). These results indicate that the phosphorylation status of Hsp90β significantly impacts its ability to inhibit apoptosome formation.
FIG. 5.
The phosphorylation of Hsp90β controls its inhibitory effect on Apaf-1 oligomerization. (A) Recombinant human caspase-9 (C287A) and human Apaf-1 proteins were produced and purified from BL21(DE3) and Sf9 cells, respectively, as described in Materials and Methods (left). Likewise, recombinant human Hsp90β proteins (wild type [WT] and mutants) were generated and purified from Sf9 cells (right). Shown is a sodium dodecyl sulfate-polyacrylamide gel stained with Coomassie blue; 5 μg of caspase-9 (C287A), 3 μg of Apaf-1, and 20 μg of Hsp90β were loaded per lane. Molecular masses (kDa) are indicated on the left side. (B) Purified recombinant Apaf-1 (0.4 μM) was mixed with catalytically inactive caspase-9 (C287A) (0.8 μM). After incubation with or without 1 mM dATP and 0.4 μM cytochrome c, the samples were loaded onto a Superdex 200 column, and each column fraction was analyzed for Apaf-1 and caspase-9 (C287A) by immunoblotting (top panel). The same experiment was performed in the presence of recombinant Hsp90β (1 μM) that was untreated (second panel) or pretreated with lambda phosphatase (λPPase; third panel) or CK2 (bottom panel). (C) His-Hsp90β wild type (WT), Hsp90β (S226A), Hsp90β (S255A), or Hsp90β (S226/255A) was incubated with control Ba/F3 cell lysates in the presence of 5 ng/μl cytochrome c. His-tagged proteins were retrieved on nickel beads, and the resultant pellets were analyzed for the presence of Apaf-1.
To determine whether Ser 226/255 are the primary phosphorylation sites regulating the binding of Hsp90β to Apaf-1, an Apaf-1 binding assay was performed using wild-type and mutant recombinant Hsp90β proteins. When His-tagged wild-type Hsp90β immobilized on nickel beads was incubated with Ba/F3 cell lysate in the presence of cytochrome c and dATP, only trace amounts of Apaf-1 bound (Fig. 5C). In contrast, Apaf-1 binding to nonphosphorylatable Hsp90β proteins (S226A, S255A, and S226A/S255A) was readily detectable (Fig. 5C), consistent with the idea that suppression of Ser 226/255 phosphorylation increases Hsp90β binding to Apaf-1. Similar results were obtained by overexpressing FLAG-tagged Hsp90β (wild type, S226A, S255A, and S226A/S255A) in 293T cells; transfected FLAG-Hsp90β showed increased binding of endogenous Apaf-1 to mutant forms of Hsp90β compared to the wild-type protein (data not shown). Although all of the mutant Hsp90β proteins exhibited enhanced binding to Apaf-1, the S226A/S255A mutant exhibited the strongest binding (Fig. 5C). These data indicate that Hsp90β binding to Apaf-1 is inhibited by phosphorylation at Ser 226/255 and that hypophosphorylation enhances binding to Apaf-1 following expression of leukemogenic tyrosine kinases.
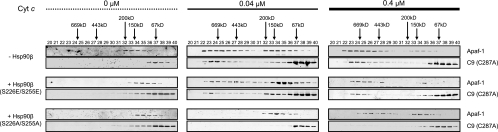
To extend the Hsp90-Apaf-1 binding data, apoptosome formation was reconstituted in vitro with recombinant proteins. When cytochrome c was added to Apaf-1 and caspase-9 (C287A) in the absence of Hsp90β, the apoptosome assembled in a dose-dependent manner (Fig. 6). Importantly, addition of Hsp90β (S226E/S255E), which carries mutations mimicking phosphorylation at both sites, had minimal effects on apoptosome assembly. In contrast, Apaf-1 oligomerization and caspase-9 recruitment were significantly delayed in the presence of Hsp90β (S226A/S255A) (Fig. 6). These results strongly suggest that the phosphorylation status of Ser 226/255 is linked to the ability of Hsp90β to bind Apaf-1 and that suppression of the phosphorylation leads to inhibition of apoptosome formation.
FIG. 6.
Hsp90β (S226A/S255A) inhibits Apaf-1 oligomerization and caspase-9 recruitment. Recombinant Apaf-1 (0.4 μM) was mixed with catalytically inactive caspase-9 [C9 (C287A)] (0.8 μM) and recombinant Hsp90β (1 μM; S226E/S255E and S226A/S255A) and then incubated with dATP (1 mM) and 0.1 μM or 0.4 μM of cytochrome c. After incubation, each sample was loaded onto a Superdex 200 column, and each column fraction was analyzed for Apaf-1 and C9 (C287A) by immunoblotting.
Hsp90β (S226A/S255A) renders normal Ba/F3 cells resistant to cytochrome c.
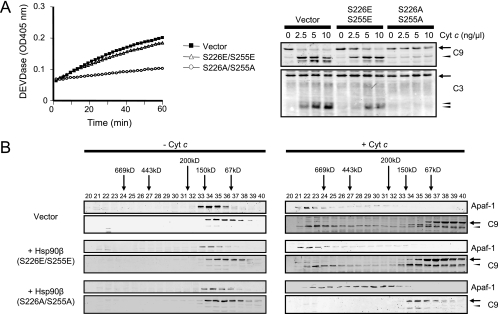
If Hsp90β hypophosphorylation were sufficient to inhibit apoptosome formation, then we might expect overexpression of Hsp90β (S226A/S255A) to promote apoptosome inhibition in Ba/F3 cells even without expression of leukemogenic tyrosine kinases. To test this, we generated normal Ba/F3 cells stably expressing Hsp90β (S226E/S255E) or Hsp90β (S226A/S255A) by using a bicistronic retroviral vector, MSCV-IRES-GFP, and monitored cytochrome c-induced caspase activation in cell-free lysates. Compared to the lysates from Ba/F3 cells expressing Hsp90β (S226E/S255E) or empty vector, lysates from Hsp90β (S226A/S255A)-expressing Ba/F3 cells were highly refractory to cytochrome c (Fig. 7A). Gel filtration showed that Apaf-1 oligomerization and caspase-9 recruitment were significantly delayed in lysates expressing Hsp90β (S226A/S255A), phenocopying cells expressing the leukemogenic tyrosine kinases (Fig. 7B). Apoptosome assembly was comparable in lysates from Hsp90β (S226E/S255E)-expressing and vector-infected cells (Fig. 7B). These data demonstrate that Hsp90β hypophosphorylation in normal cells is sufficient to confer resistance to cytochrome c-induced caspase activation.
FIG. 7.
Hsp90β (S226A/S255A) causes post-cytochrome c protection in normal Ba/F3 cell lysates. (A) Control Ba/F3 cells were infected with a retroviral vector encoding Hsp90β (S226E/S255E) or Hsp90β (S226A/S255A) or empty vector. GFP-positive cells were sorted by FACS. Cell lysates were prepared and incubated with 5 ng/μl cytochrome c and 1 mM dATP. Caspase-3 activity was assayed by measuring the cleavage of DEVD-pNA over time (left). Likewise, the lysates were incubated with 1 mM dATP and various concentrations of cytochrome c (Cyt c), and immunoblotting was performed for caspase-9 (C9) and caspase-3 (C3) (right). Procaspase-9/procaspase-3 and cleaved caspase-9/caspase-3 are indicated by arrows and arrowheads, respectively. (B) Ba/F3 cell lysates expressing the empty vector, Hsp90β (S226E/S255E), or Hsp90β (S226A/S255A) were incubated with or without cytochrome c (5 ng/μl) and dATP (1 mM) and loaded onto a Superdex 200 column. Each column fraction was analyzed for Apaf-1 and caspase-9 by immunoblotting.
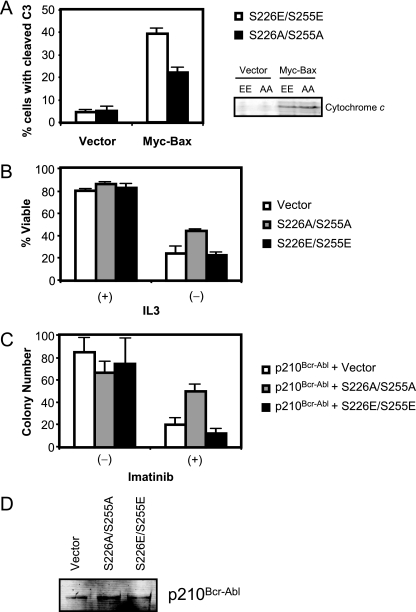
To evaluate the role of Hsp90β (S226A/S255A) in preventing cytochrome c-induced apoptosis, we transfected Bax into Ba/F3 cells stably expressing Hsp90β (S226E/S255E) or Hsp90β (S226A/S255A) and quantitated cells expressing active caspase-3. Transfection of Bax into cells expressing Hsp90β (S226E/S255E) produced large-scale cell death as manifested by active caspase-3 staining; this response was greatly dampened in cells expressing Hsp90β (S226A/S255A) despite equivalent amounts of cytoplasmic cytochrome c in the two cell lines (Fig. 8A). To further extend this finding to a more physiological setting, we monitored cell permeability to propidium iodide (PI) upon IL-3 withdrawal. Ba/F3 cells transfected with FLAG-Hsp90β (S226E/S255E) or Hsp90β (S226A/S255A) were cultured in IL-3-free medium, and then the live cell population was monitored for PI exclusion. As shown in Fig. 8B, Ba/F3 cells expressing Hsp90β (S226A/S255A) were markedly resistant to IL-3 withdrawal, compared to cells expressing Hsp90β (S226E/S255E). These data confirm the results from cell lysates demonstrating that hypophosphorylated Hsp90β is a strong inhibitor of apoptosome formation.
FIG. 8.
Expression of Hsp90β (S226A/S255A) renders cells resistant to proapoptotic stimuli. (A) Ba/F3 cells stably expressing Hsp90β (S226E/S255E) or Hsp90β (S226A/S255A) were transfected with myc-Bax. Five hours after transfection, the cells were fixed and stained with cleaved caspase-3 (C3) antibody and an Alexa 647-conjugated secondary antibody. The population of the cleaved caspase-3-positive cells was analyzed by FACS. (B) Ba/F3 cells were transfected with 2 μg of FLAG-tagged Hsp90β (S226E/S255E) or Hsp90β (S226A/S255A). Twenty-four hours after transfection, cells were transferred to IL-3-free medium (or IL-3-containing medium as a control) and further cultured for 20 h. The percentage of viable cells was analyzed by PI exclusion with FACS. (C) Mouse bone marrow KLS cells were coinfected with retroviral vectors encoding p210Bcr-Abl and Hsp90β (S226E/S255E) or Hsp90β (S226A/S255A). Coinfected cells were further selected by FACS and plated in methylcellulose medium. The colony numbers were counted 7 days after plating. Averages with standard errors of the means are shown. (D) Ba/F3 cells stably expressing p210Bcr-Abl were infected with a retroviral vector encoding Hsp90β (S226E/S255E) or Hsp90β (S226A/S255A) or empty vector. GFP-positive cells were selected by FACS and further cultured for 2 weeks. Cells were harvested and analyzed for expression of p210Bcr-Abl.
Hsp90β (S226A/S255A) confers imatinib resistance on Bcr-Abl-positive mouse bone marrow cells.
Tyrosine kinase inhibitors are an important clinical tool in treating leukemias expressing activated tyrosine kinases (51). These inhibitors block kinase activity, typically resulting in mitochondrion-dependent apoptotic death (20). Despite the clinical utility of these inhibitors, a majority of patients retain residual leukemic cells after treatment (5, 27). Additionally, many patients eventually develop drug resistance (15). Although resistance is generally due to either point mutations in or amplification of leukemogenic tyrosine kinases (10, 15, 27), failure to find such alterations in some resistant patients suggests that other mechanisms of resistance may exist. Given our data, it is possible that post-cytochrome c protection from apoptosis caused by suppression of Hsp90β phosphorylation may contribute to resistance of the kinase-expressing cells to chemotherapeutics (both kinase inhibitors and conventional agents). As a proof of principle, we performed a colony-forming assay using mouse bone marrow cells that were retrovirally cotransduced with Bcr-Abl and mutant Hsp90β proteins. Mouse bone marrow KLS cells were cotransduced with p210Bcr-Abl (bicistronically expressed with YFP) and Hsp90β (S226A/S255A) or Hsp90β (S226E/S255E; bicistronically expressed with GFP). Thereafter, GFP-YFP double-positive cells were selected by FACS and plated in the presence or absence of imatinib (1 μM), and colony numbers were counted 7 days later. As shown in Fig. 8C, there was no significant difference in the number of colonies formed in the absence of imatinib. Remarkably, when imatinib was added, Hsp90β (S226A/S255A)-expressing cells formed significantly more colonies than did Hsp90β (S226E/S255E)-expressing cells. Of note, stable expression of the Hsp90β mutants did not alter expression levels of Bcr-Abl in Ba/F3 cells (Fig. 8D). These data suggest that Hsp90β (S226A/S255A) expression by itself can confer imatinib resistance on Bcr-Abl-positive bone marrow cells, raising the interesting possibility that Hsp90 mutations could contribute to imatinib resistance in leukemic patients.
DISCUSSION
Chromosomal translocations or mutations leading to tyrosine kinase activation can result in leukemogenesis by promoting cellular proliferation and inhibiting cell death. Given the antiapoptotic potency of activated kinases, it is likely that multiple apoptotic pathways are inhibited through phosphorylation of a range of cellular substrates. Indeed, many molecular targets have been reported to contribute to the apoptotic resistance observed in leukemic cells (36). We have shown here that three leukemogenic tyrosine kinases, Bcr-Abl, FLT3/D835Y, and Tel-PDGFRβ, share the ability to suppress constitutive phosphorylation of the molecular chaperone Hsp90β. Although the detailed molecular pathways linking activated tyrosine kinases to Hsp90β are not yet known, Hsp90β hypophosphorylation impairs Apaf-1 oligomerization and subsequent caspase-9 recruitment, thereby inhibiting cytochrome c-induced caspase activation.
Regulation of Hsp90β phosphorylation.
We have shown that phosphorylation of Ser 226/255 of Hsp90β is negatively regulated by leukemogenic tyrosine kinases. Phosphorylation of these sites on Hsp90β in HeLa cells was previously reported (24). Furthermore, mass spectrometric analysis of endogenous Hsp90β proteins in porcine brain lysates revealed diphosphorylation (13), consistent with the idea that Ser 226 and Ser 255 are constitutively phosphorylated in some cell types and that such phosphorylations are suppressed by leukemogenic tyrosine kinases. It remains to be determined how phosphorylation/dephosphorylation on Ser 226/255 is regulated in normal and leukemic cells. In particular, it will be interesting to determine whether leukemogenic tyrosine kinases upregulate an Hsp90-directed phosphatase(s) or downregulate a kinase(s) targeting these sites. In an earlier study, CK2 was reported capable of phosphorylating both sites in vitro (24). However, it is not known if CK2 is the relevant kinase in vivo. Moreover, it is controversial whether Bcr-Abl promotes or inhibits the activity of CK2 in CML (18, 29). The serine/threonine protein phosphatase 5 (PP5) and its yeast homologue Ppt1 are known to associate with Hsp90 and modulate its function (9, 50). PP5 interacts with the C terminus of Hsp90 through its tetratricopeptide repeat domain (44). In the present study, however, we demonstrate differential phosphorylation of Hsp90β by control and tyrosine kinase-expressing cell lysates even using Hsp90β fragments [e.g., Hsp90β (178-300)] lacking the common tetratricopeptide repeat interaction region, an MEEVD motif at the C-terminal end of Hsp90 (Fig. 4). Thus, if PP5 is the sole phosphatase responsible for Ser 226/255 dephosphorylation, the activity of a Ser 226/255-directed kinase must be downregulated in leukemic cells.
Leukemogenic tyrosine kinases often activate the survival kinase Akt to drive tumorigenesis (6, 45). Thus, we initially hypothesized that Akt might modulate the activity of a Ser 226/255-directed kinase and/or phosphatase downstream of the leukemogenic tyrosine kinases. However, suppression of Akt activity in Bcr-Abl-expressing Ba/F3 cells by the PI 3-kinase inhibitor LY294002 or overexpression of constitutively active Akt (myr-Akt) in FL5.12 cells (37) had no effect on the phosphorylation status of Hsp90β (M. Kurokawa and S. Kornbluth, unpublished data). Moreover, LY294002 treatment or myr-Akt overexpression changed neither cytochrome c sensitivity nor the interaction of endogenous Hsp90β with GST-Apaf-1 (1-543) (M. Kurokawa and S. Kornbluth, unpublished data). Therefore, we believe that the suppression of phosphorylation on Ser 226/255 is not mediated through the PI 3-kinase/Akt pathway.
Phosphorylation controls Hsp90β-mediated apoptosome inhibition.
It is not yet clear how phosphorylation/dephosphorylation of Hsp90β controls its interaction with Apaf-1. Both phosphoserines are located in the linker region of Hsp90, which connects the N-terminal ATPase domain and the client protein-binding domain. It was shown that the presence of the linker region per se increases the binding affinity of yeast Hsp90 to client proteins, though the structural mechanism remains unclear (41). Therefore, it is possible that the phosphorylation/dephosphorylation of the linker region modulates conformational changes in adjacent domains, thereby regulating the binding affinity for client proteins.
Hsp90 chaperone activity is coupled to the binding and hydrolysis of ATP, which are regulated by specific cochaperone proteins (52). However, recent studies have raised the possibility that posttranslational modifications of Hsp90, such as acetylation (42) and phosphorylation (28, 33, 57), may also regulate Hsp90 function. It is noteworthy that we observed little binding of the Hsp90α isoform to Apaf-1 (Fig. 2). Although it is generally believed that the α and β isoforms are functionally redundant, there is some evidence to suggest that the two isoforms may have distinct chaperone activities (23, 49). Therefore, it will be interesting to determine what causes the differential binding of Hsp90α and Hsp90β to Apaf-1. In addition, it will be of interest to know whether phosphorylation of the two conserved serines, Ser 231/263, of Hsp90α (corresponding to Ser 226/255 of the β isoform) is also suppressed by the leukemogenic tyrosine kinases. If so, it may be that critical targets of Hsp90α, distinct from those of the β isoform, are affected by the leukemogenic tyrosine kinases.
Apaf-1 modulation by Hsp90β.
We demonstrated that the CARD is the primary binding site of Hsp90β on Apaf-1. It has been shown that surface hydrophobicity of client proteins determines binding to Hsp90 (53). In contrast, the caspase-9 CARD interacts with α-helices, α2 and α3, of the Apaf-1 CARD (34), which is comprised largely of hydrophilic amino acids, suggesting that Hsp90β may not directly compete with caspase-9 for the same Apaf-1 binding site. Interestingly, the caspase-9 binding site is located at the N terminus of the Apaf-1 CARD (amino acids 22 to 32 and amino acids 37 to 44 [34]), whereas there is a hydrophobic cluster located near the C terminus of the CARD (83LAALLHDGIPVV94). It would be interesting to determine whether a mutation within the hydrophobic region can alter or even override the inhibitory interaction of Hsp90β with Apaf-1.
Hsp90 binding does not appear to prevent cytochrome c-induced exposure of the Apaf-1 CARD, as binding does not occur in the absence of cytochrome c. Hsp90β binding to Apaf-1 CARD may prevent conformational changes in Apaf-1 that are necessary for proper Apaf-1 oligomerization/caspase-9 recruitment. In this regard, we note that Apaf-1 appears to partially oligomerize even in the presence of the leukemogenic tyrosine kinases (Fig. 1D), but it may be that this oligomerization is aberrant, preventing caspase-9 recruitment.
Hsp90 regulation and chemoresistance.
The α and β isoforms of Hsp90 comprise 1 to 2% of total cellular protein, even in an unstressed, untransformed cell. Importantly, an increase in Hsp90 over basal levels is believed to contribute to tumorigenesis in many types of cancers. Indeed, small-molecule Hsp90 inhibitors (e.g., geldanamycin and its derivatives) selectively kill certain types of cancer cells by promoting apoptosis (22). Hsp90 interacts with a large number of signaling proteins including oncogenic kinases, transcription factors, and hormone receptors (52). This diversity of partners has made it complicated to dissect its precise role in regulating apoptosis. Our data point to a direct role for Hsp90 in preventing apoptosome activation in leukemias.
Imatinib (Gleevec) has demonstrated remarkable success in the treatment of CML. However, many CML patients treated with the inhibitor eventually develop resistance, retaining Bcr-Abl-positive cells which are extremely difficult to eliminate. Moreover, the inhibitor is less effective at later stages of the disease. We demonstrated here that the nonphosphorylatable mutant (S226A/S255A) of Hsp90β, but not the phosphomimetic mutant (S226E/S255E), conferred imatinib resistance on Bcr-Abl-positive mouse bone marrow cells in a colony-forming assay, though these mutants did not appear to work through stabilization of Bcr-Abl, as reported for the Hsp90 inhibitor geldanamycin (3). These data suggest that suppression of Hsp90β phosphorylation triggers some degree of chemoresistance in the leukemic cells, potentially because of apoptosome inhibition. We note, however, that expression of Hsp90β (S226A/S255A) per se is not sufficient to transform normal Ba/F3 cells or primary mouse hematopoietic cells (M. Kurokawa, C. Zhao, T. Reya, and S. Kornbluth, unpublished data), suggesting that additional oncogenic signaling is necessary to drive tumorigenesis. It will be of great interest to investigate the relationship of Ser 226/255 phosphorylation to malignancy, chemoresistance, and prognosis of leukemias. In addition, it may be possible to extend these observations to other leukemias and, potentially, to solid tumors that are also driven by aberrant expression of constitutively active tyrosine kinases. Lastly, our results suggest kinases and phosphatases regulating Hsp90β phosphorylation as potential therapeutic targets. Histone deacetylase inhibitors and geldanamycin derivatives, both of which impair Hsp90 function, have shown promise for the treatment of leukemias in combination with tyrosine kinase inhibitors such as imatinib (16, 32). These agents appear to act, in part, by promoting degradation of the activated kinases. Since the hypophosphorylated Hsp90β did not alter Bcr-Abl levels in our experiments (Fig. 8D), it may be that agents affecting Hsp90 phosphorylation status would offer a distinct and possibly beneficial avenue to complement Hsp90 inhibitors.
Acknowledgments
We thank D. G. Gilliland, G. Q. Daley, A. M. Pendergast, T. Takenawa, X. Wang, L. Neckers, G. Salvesen, C. W. Distelhorst, M. Z. Wang, and J. C. Rathmell for kindly providing reagents and suggestions. We thank B. Harvat and M. Cook for FACS analysis and D. Ribar for technical assistance. We also thank L. K. Nutt, Z. T. Schafer, P. B. Deming, A. Parrish, and C. Johnson for valuable discussion and comments on the manuscript.
This work was supported by NIH 5R01 CA102707 to S.K. and an Irvington Institute Fellowship of the Cancer Research Institute to M.K.
Footnotes
Published ahead of print on 30 June 2008.
REFERENCES
- 1.Abu-Duhier, F. M., A. C. Goodeve, G. A. Wilson, R. S. Care, I. R. Peake, and J. T. Reilly. 2001. Identification of novel FLT-3 Asp835 mutations in adult acute myeloid leukaemia. Br. J. Haematol. 113983-988. [DOI] [PubMed] [Google Scholar]
- 2.Amarante-Mendes, G. P., A. J. McGahon, W. K. Nishioka, D. E. Afar, O. N. Witte, and D. R. Green. 1998. Bcl-2-independent Bcr-Abl-mediated resistance to apoptosis: protection is correlated with up regulation of Bcl-xL. Oncogene 161383-1390. [DOI] [PubMed] [Google Scholar]
- 3.An, W. G., T. W. Schulte, and L. M. Neckers. 2000. The heat shock protein 90 antagonist geldanamycin alters chaperone association with p210bcr-abl and v-src proteins before their degradation by the proteasome. Cell Growth Differ. 11355-360. [PubMed] [Google Scholar]
- 4.Beere, H. M. 2005. Death versus survival: functional interaction between the apoptotic and stress-inducible heat shock protein pathways. J. Clin. Investig. 1152633-2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhatia, R., M. Holtz, N. Niu, R. Gray, D. S. Snyder, C. L. Sawyers, D. A. Arber, M. L. Slovak, and S. J. Forman. 2003. Persistence of malignant hematopoietic progenitors in chronic myelogenous leukemia patients in complete cytogenetic remission following imatinib mesylate treatment. Blood 1014701-4707. [DOI] [PubMed] [Google Scholar]
- 6.Brandts, C. H., B. Sargin, M. Rode, C. Biermann, B. Lindtner, J. Schwäble, H. Buerger, C. Müller-Tidow, C. Choudhary, M. McMahon, W. E. Berdel, and H. Serve. 2005. Constitutive activation of Akt by Flt3 internal tandem duplications is necessary for increased survival, proliferation, and myeloid transformation. Cancer Res. 659643-9650. [DOI] [PubMed] [Google Scholar]
- 7.Cain, K., D. G. Brown, C. Langlais, and G. M. Cohen. 1999. Caspase activation involves the formation of the aposome, a large (∼700 kDa) caspase-activating complex. J. Biol. Chem. 27422686-22692. [DOI] [PubMed] [Google Scholar]
- 8.Cardone, M. H., N. Roy, H. R. Stennicke, G. S. Salvesen, T. F. Franke, E. Stanbridge, S. Frisch, and J. C. Reed. 1998. Regulation of cell death protease caspase-9 by phosphorylation. Science 2821318-1321. [DOI] [PubMed] [Google Scholar]
- 9.Chen, M. S., A. M. Silverstein, W. B. Pratt, and M. Chinkers. 1996. The tetratricopeptide repeat domain of protein phosphatase 5 mediates binding to glucocorticoid receptor heterocomplexes and acts as a dominant negative mutant. J. Biol. Chem. 27132315-32320. [DOI] [PubMed] [Google Scholar]
- 10.Clark, J. J., J. Cools, D. P. Curley, J. C. Yu, N. A. Lokker, N. A. Giese, and D. G. Gilliland. 2004. Variable sensitivity of FLT3 activation loop mutations to the small molecule tyrosine kinase inhibitor MLN518. Blood 1042867-2872. [DOI] [PubMed] [Google Scholar]
- 11.Danial, N. N., and S. J. Korsmeyer. 2004. Cell death: critical control points. Cell 116205-219. [DOI] [PubMed] [Google Scholar]
- 12.Deming, P. B., Z. T. Schafer, J. S. Tashker, M. B. Potts, M. Deshmukh, and S. Kornbluth. 2004. Bcr-Abl-mediated protection from apoptosis downstream of mitochondrial cytochrome c release. Mol. Cell. Biol. 2410289-10299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garnier, C., D. Lafitte, T. J. Jorgensen, O. N. Jensen, C. Briand, and V. Peyrot. 2001. Phosphorylation and oligomerization states of native pig brain HSP90 studied by mass spectrometry. Eur. J. Biochem. 2682402-2407. [DOI] [PubMed] [Google Scholar]
- 14.Golub, T. R., G. F. Barker, M. Lovett, and D. G. Gilliland. 1994. Fusion of PDGF receptor beta to a novel ets-like gene, tel, in chronic myelomonocytic leukemia with t(5;12) chromosomal translocation. Cell 77307-316. [DOI] [PubMed] [Google Scholar]
- 15.Gorre, M. E., M. Mohammed, K. Ellwood, N. Hsu, R. Paquette, P. N. Rao, and C. L. Sawyers. 2001. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science 293876-880. [DOI] [PubMed] [Google Scholar]
- 16.Gorre, M. E., K. Ellwood-Yen, G. Chiosis, N. Rosen, and C. L. Sawyers. 2002. BCR-ABL point mutants isolated from patients with imatinib mesylate-resistant chronic myeloid leukemia remain sensitive to inhibitors of the BCR-ABL chaperone heat shock protein 90. Blood 1003041-3044. [DOI] [PubMed] [Google Scholar]
- 17.Hanahan, D., and R. A. Weinberg. 2000. The hallmarks of cancer. Cell 10057-70. [DOI] [PubMed] [Google Scholar]
- 18.Hériché, J. K., and E. M. Chambaz. 1998. Protein kinase CK2α is a target for the Abl and Bcr-Abl tyrosine kinases. Oncogene 1713-18. [DOI] [PubMed] [Google Scholar]
- 19.Hoover, R. R., F. X. Mahon, J. V. Melo, and G. Q. Daley. 2002. Overcoming STI571 resistance with the farnesyl transferase inhibitor SCH66336. Blood 1001068-1071. [DOI] [PubMed] [Google Scholar]
- 20.Horita, M., E. J. Andreu, A. Benito, C. Arbona, C. Sanz, I. Benet, F. Prosper, and J. L. Fernandez-Luna. 2000. Blockade of the Bcr-Abl kinase activity induces apoptosis of chronic myelogenous leukemia cells by suppressing signal transducer and activator of transcription 5-dependent expression of Bcl-xL. J. Exp. Med. 191977-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu, Y., L. Ding, D. M. Spencer, and G. Nunez. 1998. WD-40 repeat region regulates Apaf-1 self-association and procaspase-9 activation. J. Biol. Chem. 27333489-33494. [DOI] [PubMed] [Google Scholar]
- 22.Isaacs, J. S., W. Xu, and L. Neckers. 2003. Heat shock protein 90 as a molecular target for cancer therapeutics. Cancer Cell 3213-217. [DOI] [PubMed] [Google Scholar]
- 23.Kuo, C. C., C. M. Liang, C. Y. Lai, and S. M. Liang. 2007. Involvement of heat shock protein (Hsp)90β but not Hsp90α in antiapoptotic effect of CpG-B oligodeoxynucleotide. J. Immunol. 1786100-6108. [DOI] [PubMed] [Google Scholar]
- 24.Lees-Miller, S. P., and C. W. Anderson. 1989. Two human 90-kDa heat shock proteins are phosphorylated in vivo at conserved serines that are phosphorylated in vitro by casein kinase II. J. Biol. Chem. 2642431-2437. [PubMed] [Google Scholar]
- 25.Li, P., D. Nijhawan, I. Budihardjo, S. M. Srinivasula, M. Ahmad, E. S. Alnemri, and X. Wang. 1997. Cytochrome c and dATP-dependent formation of Apaf-1/caspase-9 complex initiates an apoptotic protease cascade. Cell 91479-489. [DOI] [PubMed] [Google Scholar]
- 26.Liu, X., C. N. Kim, J. Yang, R. Jemmerson, and X. Wang. 1996. Induction of apoptotic program in cell-free extracts: requirement for dATP and cytochrome c. Cell 86147-157. [DOI] [PubMed] [Google Scholar]
- 27.Michor, F., T. P. Hughes, Y. Iwasa, S. Branford, N. P. Shah, C. L. Sawyers, and M. A. Nowak. 2005. Dynamics of chronic myeloid leukaemia. Nature 4351267-1270. [DOI] [PubMed] [Google Scholar]
- 28.Mimnaugh, E. G., P. J. Worland, L. Whitesell, and L. M. Neckers. 1995. Possible role for serine/threonine phosphorylation in the regulation of the heteroprotein complex between the hsp90 stress protein and the pp60v-src tyrosine kinase. J. Biol. Chem. 27028654-28659. [DOI] [PubMed] [Google Scholar]
- 29.Mishra, S., A. Reichert, J. Cunnick, D. Senadheera, B. Hemmeryckx, N. Heisterkamp, and J. Groffen. 2003. Protein kinase CKIIα interacts with the Bcr moiety of Bcr/Abl and mediates proliferation of Bcr/Abl-expressing cells. Oncogene 228255-8262. [DOI] [PubMed] [Google Scholar]
- 30.Nakao, M., S. Yokota, T. Iwai, H. Kaneko, S. Horiike, K. Kashima, Y. Sonoda, T. Fujimoto, and S. Misawa. 1996. Internal tandem duplication of the flt3 gene found in acute myeloid leukemia. Leukemia 101911-1918. [PubMed] [Google Scholar]
- 31.Neshat, M. S., A. B. Raitano, H. G. Wang, J. C. Reed, and C. L. Sawyers. 2000. The survival function of the Bcr-Abl oncogene is mediated by Bad-dependent and -independent pathways: roles for phosphatidylinositol 3-kinase and Raf. Mol. Cell. Biol. 201179-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nimmanapalli, R., L. Fuino, P. Bali, M. Gasparetto, M. Glozak, J. Tao, L. Moscinski, C. Smith, J. Wu, R. Jove, P. Atadja, and K. Bhalla. 2003. Histone deacetylase inhibitor LAQ824 both lowers expression and promotes proteasomal degradation of Bcr-Abl and induces apoptosis of imatinib mesylate-sensitive or -refractory chronic myelogenous leukemia-blast crisis cells. Cancer Res. 635126-5135. [PubMed] [Google Scholar]
- 33.Ogiso, H., N. Kagi, E. Matsumoto, M. Nishimoto, R. Arai, M. Shirouzu, J. Mimura, Y. Fujii-Kuriyama, and S. Yokoyama. 2004. Phosphorylation analysis of 90 kDa heat shock protein within the cytosolic arylhydrocarbon receptor complex. Biochemistry 4315510-15519. [DOI] [PubMed] [Google Scholar]
- 34.Qin, H., S. M. Srinivasula, G. Wu, T. Fernandes-Alnemri, E. S. Alnemri, and Y. Shi. 1999. Structural basis of procaspase-9 recruitment by the apoptotic protease-activating factor 1. Nature 399549-557. [DOI] [PubMed] [Google Scholar]
- 35.Pandey, P., A. Saleh, A. Nakazawa, S. Kumar, S. M. Srinivasula, V. Kumar, R. Weichselbaum, C. Nalin, E. S. Alnemri, D. Kufe, and S. Kharbanda. 2000. Negative regulation of cytochrome c-mediated oligomerization of Apaf-1 and activation of procaspase-9 by heat shock protein 90. EMBO J. 194310-4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Penserga, E. T., and T. Skorski. 2007. Fusion tyrosine kinases: a result and cause of genomic instability. Oncogene 2611-20. [DOI] [PubMed] [Google Scholar]
- 37.Plas, D. R., S. Talapatra, A. L. Edinger, J. C. Rathmell, and C. B. Thompson. 2001. Akt and Bcl-xL promote growth factor-independent survival through distinct effects on mitochondrial physiology. J. Biol. Chem. 27612041-12048. [DOI] [PubMed] [Google Scholar]
- 38.Raina, D., P. Pandey, R. Ahmad, A. Bharti, J. Ren, S. Kharbanda, R. Weichselbaum, and D. Kufe. 2005. c-Abl tyrosine kinase regulates caspase-9 autocleavage in the apoptotic response to DNA damage. J. Biol. Chem. 28011147-11151. [DOI] [PubMed] [Google Scholar]
- 39.Sanchez-Garcia, I., and G. Grutz. 1995. Tumorigenic activity of the BCR-ABL oncogenes is mediated by BCL2. Proc. Natl. Acad. Sci. USA 925287-5291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schafer, Z. T., and S. Kornbluth. 2006. The apoptosome: physiological, developmental, and pathological modes of regulation. Dev. Cell 10549-561. [DOI] [PubMed] [Google Scholar]
- 41.Scheibel, T., H. I. Siegmund, R. Jaenicke, P. Ganz, H. Lilie, and J. Buchner. 1999. The charged region of Hsp90 modulates the function of the N-terminal domain. Proc. Natl. Acad. Sci. USA 961297-1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scroggins, B. T., K. Robzyk, D. Wang, M. G. Marcu, S. Tsutsumi, K. Beebe, R. J. Cotter, S. Felts, D. Toft, L. Karnitz, N. Rosen, and L. Neckers. 2007. An acetylation site in the middle domain of Hsp90 regulates chaperone function. Mol. Cell 25151-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shuai, K., J. Halpern, J. ten Hoeve, X. Rao, and C. L. Sawyers. 1996. Constitutive activation of STAT5 by the BCR-ABL oncogene in chronic myelogenous leukemia. Oncogene 13247-254. [PubMed] [Google Scholar]
- 44.Silverstein, A. M., M. D. Galigniana, M. S. Chen, J. K. Owens-Grillo, M. Chinkers, and W. B. Pratt. 1997. Protein phosphatase 5 is a major component of glucocorticoid receptor. hsp90 complexes with properties of an FK506-binding immunophilin. J. Biol. Chem. 27216224-16230. [DOI] [PubMed] [Google Scholar]
- 45.Skorski, T., A. Bellacosa, M. Nieborowska-Skorska, M. Majewski, R. Martinez, J. K. Choi, R. Trotta, P. Wlodarski, D. Perrotti, T. O. Chan, M. A. Wasik, P. N. Tsichlis, and B. Calabretta. 1997. Transformation of hematopoietic cells by BCR/ABL requires activation of a PI-3k/Akt-dependent pathway. EMBO J. 166151-6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Srinivasula, S. M., M. Ahmad, T. Fernandes-Alnemri, and E. S. Alnemri. 1998. Autoactivation of procaspase-9 by Apaf-1-mediated oligomerization. Mol. Cell 1949-957. [DOI] [PubMed] [Google Scholar]
- 47.Stennicke, H. R., Q. L. Deveraux, E. W. Humke, J. C. Reed, V. M. Dixit, and G. S. Salvesen. 1999. Caspase-9 can be activated without proteolytic processing. J. Biol. Chem. 2748359-8362. [DOI] [PubMed] [Google Scholar]
- 48.Unwin, R. D., D. W. Sternberg, Y. Lu, A. Pierce, D. G. Gilliland, and A. D. Whetton. 2005. Global effects of BCR/ABL and TEL/PDGFRβ expression on the proteome and phosphoproteome: identification of the Rho pathway as a target of BCR/ABL. J. Biol. Chem. 2806316-6326. [DOI] [PubMed] [Google Scholar]
- 49.Voss, A. K., T. Thomas, and P. Gruss. 2000. Mice lacking HSP90β fail to develop a placental labyrinth. Development 1271-11. [DOI] [PubMed] [Google Scholar]
- 50.Wandinger, S. K., M. H. Suhre, H. Wegele, and J. Buchner. 2006. The phosphatase Ppt1 is a dedicated regulator of the molecular chaperone Hsp90. EMBO J. 25367-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weisberg, E., P. W. Manley, S. W. Cowan-Jacob, A. Hochhaus, and J. D. Griffin. 2007. Second generation inhibitors of BCR-ABL for the treatment of imatinib-resistant chronic myeloid leukemia. Nat. Rev. Cancer 7345-356. [DOI] [PubMed] [Google Scholar]
- 52.Whitesell, L., and S. L. Lindquist. 2005. HSP90 and the chaperoning of cancer. Nat. Rev. Cancer 5761-772. [DOI] [PubMed] [Google Scholar]
- 53.Xu, W., X. Yuan, Z. Xiang, E. Mimnaugh, M. Marcu, and L. Neckers. 2005. Surface charge and hydrophobicity determine ErbB2 binding to the Hsp90 chaperone complex. Nat. Struct. Mol. Biol. 12120-126. [DOI] [PubMed] [Google Scholar]
- 54.Yamamoto, Y., H. Kiyoi, Y. Nakano, R. Suzuki, Y. Kodera, S. Miyawaki, N. Asou, K. Kuriyama, F. Yagasaki, C. Shimazaki, H. Akiyama, K. Saito, M. Nishimura, T. Motoji, K. Shinagawa, A. Takeshita, H. Saito, R. Ueda, R. Ohno, and T. Naoe. 2001. Activating mutation of D835 within the activation loop of FLT3 in human hematologic malignancies. Blood 972434-2439. [DOI] [PubMed] [Google Scholar]
- 55.Yuan, J., and B. A. Yankner. 2000. Apoptosis in the nervous system. Nature 407802-809. [DOI] [PubMed] [Google Scholar]
- 56.Zhao, C., J. Blum, A. Chen, H. Y. Kwon, S. H. Jung, J. M. Cook, A. Lagoo, and T. Reya. 2007. Loss of beta-catenin impairs the renewal of normal and CML stem cells in vivo. Cancer Cell 12528-541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhao, Y. G., R. Gilmore, G. Leone, M. C. Coffey, B. Weber, and P. W. Lee. 2001. Hsp90 phosphorylation is linked to its chaperoning function. Assembly of the reovirus cell attachment protein. J. Biol. Chem. 27632822-32827. [DOI] [PubMed] [Google Scholar]
- 58.Zou, H., W. J. Henzel, X. Liu, A. Lutschg, and X. Wang. 1997. Apaf-1, a human protein homologous to C. elegans CED-4, participates in cytochrome c-dependent activation of caspase-3. Cell 90405-413. [DOI] [PubMed] [Google Scholar]
- 59.Zou, H., Y. Li, X. Liu, and X. Wang. 1999. An APAF-1-cytochrome c multimeric complex is a functional apoptosome that activates procaspase-9. J. Biol. Chem. 27411549-11556. [DOI] [PubMed] [Google Scholar]