Abstract
The genomic architecture of the budding yeast Saccharomyces cerevisiae is typical of other eukaryotes in that genes are spatially organized into discrete and nonoverlapping units. Inherent in this organizational model is the assumption that protein-coding sequences do not overlap completely. Here, we present evidence to the contrary, defining a previously overlooked yeast gene, NAG1 (for nested antisense gene) nested entirely within the coding sequence of the YGR031W open reading frame in an antisense orientation on the opposite strand. NAG1 encodes a 19-kDa protein, detected by Western blotting of hemagglutinin (HA)-tagged Nag1p with anti-HA antibodies and by β-galactosidase analysis of a NAG1-lacZ fusion. NAG1 is evolutionarily conserved as a unit with YGR031W in bacteria and fungi. Unlike the YGR031WP protein product, however, which localizes to the mitochondria, Nag1p localizes to the cell periphery, exhibiting properties consistent with those of a plasma membrane protein. Phenotypic analysis of a site-directed mutant (nag1-1) disruptive for NAG1 but silent with respect to YGR031W, defines a role for NAG1 in yeast cell wall biogenesis; microarray profiling of nag1-1 indicates decreased expression of genes contributing to cell wall organization, and the nag1-1 mutant is hypersensitive to the cell wall-perturbing agent calcofluor white. Furthermore, production of Nag1p is dependent upon the presence of the cell wall integrity pathway mitogen-activated protein kinase Slt2p and its downstream transcription factor Rlm1p. Thus, NAG1 is important for two reasons. First, it contributes to yeast cell wall biogenesis. Second, its genomic context is novel, raising the possibility that other nested protein-coding genes may exist in eukaryotic genomes.
Eukaryotic gene organization is routinely presumed to follow a colinear design, wherein protein-coding genes are ordered at discrete, nonoverlapping points along a given chromosome (19). This organizational model is manifestly evident in the genome of the budding yeast Saccharomyces cerevisiae. The S. cerevisiae genome was sequenced in 1996, and its 13-Mb sequence was subsequently annotated for genes using a combination of existing genetic information and straightforward computational approaches (30). As part of this process, putative protein-coding open reading frames (ORFs) were predicted by gene-finding algorithms, employing a set of criteria based upon ORF size and spatial organization. Specifically, any ORF greater than 100 codons in length was annotated as a gene, provided it did not significantly overlap a longer ORF. If two ORFs overlapped, the longer of the two sequences was annotated as a gene, and the other was discarded (30, 33). Perhaps in part because of this gene prediction strategy, to date, no verified, completely overlapping protein-coding genes have been identified in the yeast genome.
Recently, several lines of evidence have raised doubts concerning the presumed colinear organization of protein-coding genes in eukaryotic genomes. David et al. (4) have employed a high-density oligonucleotide tiling array to profile RNA expression on both DNA strands over the entire genome in the budding yeast; this study highlights a considerable degree of antisense transcription in the yeast genome— that is, transcripts overlapping known genes in an antisense orientation. In addition, Havilio et al. (10) mined microarray expression data in yeast to identify a significant body of transcripts oriented antisense to known genes. This level of antisense transcription, however, may reflect some degree of “leaky” transcription or a potential regulatory mechanism in S. cerevisiae (12) and does not conclusively indicate that protein-coding sequences can exist antisense to other protein-coding genes. In fact, the only confirmed report of entirely overlapping genes in yeast was presented by Coelho et al. (3), describing the mitochondrial protein Tar1p, which is encoded antisense to the 25S rRNA gene in the nuclear ribosomal DNA repeat region of chromosome XII. Tar1p, however, is not oriented opposite a protein-coding gene but is opposite a structural RNA.
Here, we present the first report of a yeast protein-coding gene nested opposite another protein-coding gene. The yeast ORF YGR031C-A is nested antisense and opposite the known gene YGR031W, the latter encoding a mitochondrial protein of unknown function. Genome-wide transposon-tagging studies had previously suggested that the YGR031C-A ORF may encode a protein, and in this study, we established that this ORF (herein renamed NAG1) does encode a 19-kDa protein that localizes to the yeast cell periphery, contains putative transmembrane domains, and cofractionates with known plasma membrane proteins. Sequence analysis revealed that NAG1 is conserved among fungi as a unit oriented opposite an ortholog of YGR031W. Consistent with its conservation in fungi, NAG1 contributes to cell wall synthesis and maintenance in S. cerevisiae. Disruption of NAG1 results in cell sensitivity to calcofluor white and altered transcriptional levels for many cell wall biosynthesis/maintenance genes. Furthermore, Nag1p levels increase upon calcofluor white treatment, and NAG1 expression is dependent upon the Slt2p mitogen-activated protein kinase (MAPK) pathway and its key downstream transcription factor, Rlm1p. In total, these results identify a new protein contributing to cell wall function in yeast, while highlighting both the existence of nested protein-coding genes and the likelihood that other such genes exist in the eukaryotic kingdom.
MATERIALS AND METHODS
S. cerevisiae strains and growth conditions.
S. cerevisiae strains containing the nag1::mTn allele were generated in the genetic background BY4742 (45). The W303 genetic background was obtained from the Yeast Genetics Stock Center (Berkeley, CA). The slt2Δ and rlm1Δ mutants were from the yeast deletion collection (45) generated in the BY4742 background referenced above. Growth media and basic genetic manipulation were as described previously (8). The slt2Δ mutant was grown at 25°C to accommodate its cell wall defect. The nag1::mTn allele was carried on plasmid pHSS6 (39); this plasmid was digested with NotI, and the transposon-mutagenized genomic DNA fragment was introduced into strain BY4742 by standard methods of DNA transformation (15). The BY4742 strain containing NAG1-3×HA was constructed using the PCR-based epitope-tagging method of Longtine et al. (27) using an integration cassette amplified from pFA6a-3HA-kanMX6 with PCR primers containing 40-bp flanking sequence homology.
Sequence alignments.
Sequence similarity searches were performed using tBLASTn with Nag1p amino acid sequence against a six-frame translation of the NCBI nonredundant nucleotide database (46). All searches were repeated with the BLOSUM62 and BLOSUM45 scoring matrices, coupled with default parameters, and optimal sequence alignments were generated using CLUSTAL W (41).
Western blotting.
The hemagglutinin (HA) tag was integrated at the 3′ end of NAG1 using the KanMX6 selection cassette from plasmid pFA6a-3HA-KanMX6 (27). Transformants were selected on yeast extract-peptone-dextrose (YPD) plates containing 200 μg/ml G418. Correct integration was verified by PCR. For Western blotting, yeast strains were grown at 30°C to mid-log phase in YPD medium unless otherwise noted. The cells were then converted into spheroplasts and subjected to subcellular fractionation based on previously described protocols (21). Unlysed spheroplasts were removed by centrifugation at 1,500 × g for 5 min at 4°C. The total lysate was centrifuged at 13,000 × g for 5 min at 4°C to separate supernatant and pellet fractions (S13 and P13, respectively). Aliquots of each fraction were precipitated with 10% trichloroacetic acid on ice for 30 min, washed with 100% acetone, and air dried. The dried pellets were resuspended in sodium dodecyl sulfate sample buffer. Aliquots were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and probed with anti-3×HA antibody (antibody directed against three-hemagglutinin tag) (Santa Cruz Biotechnology).
Fluorescence microscopy.
The NAG1 and YGR031W protein-coding sequences along with 1-kb upstream sequence were cloned into a derivative of the centromeric plasmid YCp50 such that each formed an in-frame 3′ fusion to sequence encoding the Venus variant of yellow fluorescent protein (vYFP), NAG1-vYFP and YGR031W-vYFP, respectively (31). Plasmids carrying NAG1-vYFP and YGR031W-vYFP, respectively, were transformed into strain BY4742. Yeast cultures were grown in synthetic medium lacking uracil but supplemented with dextrose (SD-Ura medium) until mid-log phase before examination. To label mitochondria, MitoFluor Red 594 (Molecular Probes) was added to a final concentration of 5 μM, and the culture was incubated for an additional 30 min prior to microscopy. Cells were washed once before examination using the DeltaVision Spectris inverted microscope (Applied Precision, Issaquah, WA).
Generation of the nag1-1 site-directed mutant.
The nag1-1 mutant contains a nonsense mutation at codon 41 of NAG1 (TAT to TAA) that is silent with respect to the YGR031W ORF, constructed by site-directed mutagenesis of a low-copy plasmid carrying the YGR031W locus. Specifically, we first constructed a Gateway-compatible yeast vector for recombination-based cloning of yeast genomic DNA. This Gateway vector was constructed from the centromeric yeast shuttle vector YCp50. YCp50 was digested with SphI and made blunt with T4 DNA polymerase (New England Biolabs, MA). Gateway cassette A (Invitrogen Corporation, CA) was ligated with the blunt-ended vector, and EcoRI was used to identify the orientation of the cassette. To maintain an intact promoter region for both YGR031W and NAG1, we amplified the YGR031W ORF along with 1 kb of sequence upstream of its start codon and 1 kb downstream of its stop codon; this genomic DNA was introduced into YCp50 by recombination-based cloning (44). The nag1-1 mutant was generated using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) and the following primers: forward primer, 5′-CGTTCTTCGAATGTATAAGACGACACAGACG-3′; reverse primer, 5′-CGTCTGTGTCGTCTTATACATTCGAAGAACG-3′. This construct was subsequently introduced into the yeast deletion strain YGR031WΔ by standard methods of yeast transformation (15).
DNA microarray analysis.
To reduce background variation in gene expression, transcriptional profiles were generated from the YGR031WΔ strain bearing either wild-type NAG1 (in the YCp50 derivative described above) or nag1-1 (the identical plasmid after site-directed mutagenesis). Thus, the single difference between the strains lies in the point mutation described above. Yeast strains were cultured to mid-log phase in SD-Ura medium prior to RNA extraction. RNA was prepared according to standard protocols using the Poly(A)Purist kit (Ambion, Austin, TX). RNA concentration and purity were determined spectrophotometrically and by gel electrophoresis. Microarray hybridization was performed with the yeast genome S98 array using standard protocols (Affymetrix, Inc., Santa Clara, CA). All microarray experiments were performed in quadruplicate (four biological replicates) for each strain. Differentially expressed genes were identified by significance analysis of microarrays (35, 42) according to protocols described by Ma et al. (29).
Calcofluor white sensitivity.
Cell wall-related mutant phenotypes were scored on solid medium as described below. A freshly prepared stock solution of 1% (wt/vol) calcofluor white was added to sterile selection medium at a final concentration of 10 μg/ml. This medium was adjusted to pH 6.0 with NaOH prior to pouring the plates. Wild-type and nag1-1 mutant strains were grown at 30°C in selection medium to log phase. A fivefold dilution series of each cell suspension was made, and 3 μl of each dilution was spotted onto calcofluor white-containing plates. Growth was monitored after 2 days of incubation at 30°C. This phenotypic assay was repeated for the nag1-1 mutant in strains derived from both S288c and W303 strains.
β-Galactosidase assays.
β-Galactosidase assays were performed using the yeast β-galactosidase assay kit (Pierce, Rockford, IL) according to standard methods. Mean activities were averaged from three parallel assays. Strains treated with calcofluor white were grown in liquid medium to mid-log phase; cell cultures were incubated for an additional 5 hours before β-galactosidase activity was measured.
RESULTS
Identification of the NAG1 gene nested antisense and opposite the YGR031W ORF.
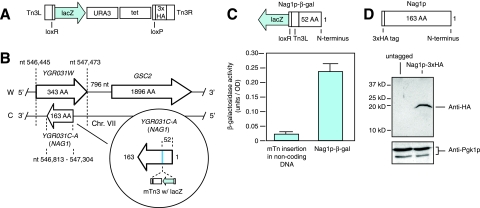
In a previous study, we utilized a transposon-based gene trap to identify putative protein-coding sequences in the S. cerevisiae genome (24). This gene trap is diagrammed in Fig. 1A; it contains a 5′-truncated lacZ reporter lacking its promoter and start codon, such that transposon insertion results in β-galactosidase activity only if the transposon lands in frame with yeast protein-coding sequence (36, 37). By random transposon mutagenesis with this gene trap, we identified a set of previously nonannotated ORFs that putatively encode proteins, including a set of 54 ORFs positioned opposite and antisense of annotated yeast genes (24). Within this gene set, we were particularly interested in the ORF designated YGR031C-A, since it is greater than 100 codons in length, is oriented opposite an ORF that putatively encodes a protein, and exhibited easily detected levels of expression during vegetative growth.
FIG. 1.
The NAG1 gene is nested opposite the YGR031W ORF and encodes a protein product. (A) Schematic of the transposon-based gene trap used to identify YGR031C-A/NAG1. The lacZ reporter lacks its start codon and promoter, so β-galactosidase is produced only if the transposon inserts in genomic DNA such that the lacZ coding sequence is in frame with a host gene. The transposon-encoded lacZ fusion will be separated from host gene coding sequence by the Tn3L terminal sequence and loxR site. (B) Diagrammatic representation of the YGR031W locus, indicating YGR031C-A/NAG1 nested antisense to YGR031W on the opposite strand. The circle inset indicates the exact transposon insertion identifying NAG1. nt, nucleotides; AA, amino acids; Chr., chromosome. (C) β-Galactosidase assay of the Nag1p-β-galactosidase (Nag1p-β-gal) protein product; the Nag1p chimera consists of the N-terminal 52 amino acids (AA) of Nag1p, the Tn3L and loxR sequences, and β-galactosidase from the second amino acid onwards. For comparison, the β-galactosidase level from a transposon insertion in noncoding DNA (chromosome XV, coordinate 216878) is indicated. OD, optical density. (D) Western blot identifying Nag1p tagged at its carboxy terminus with three copies of the HA epitope (3xHA). Protein extract from an untagged strain is also included as a control; both extracts were prepared from cells undergoing vegetative growth. The levels of Pgk1p, 3-phosphoglycerate kinase, were analyzed to ensure comparability between protein samples. The positions of molecular mass markers (in kilodaltons) are shown to the left of the blot.
The YGR031C-A ORF is positioned antisense and opposite the gene YGR031W on yeast chromosome VII as indicated in Fig. 1B. The YGR031W gene is functionally uncharacterized but is known to encode a mitochondrial protein (13, 34). YGR031C-A consists of a single exon 163 codons in length nested on the opposite strand but completely within the YGR031W genomic locus. The YGR031W gene is more than twice the size of YGR031C-A, and both ORFs are relatively well separated from other upstream and downstream genes. Because of this unusual genetic organization, we hereafter refer to YGR031C-A as NAG1 (for nested antisense gene).
To establish that NAG1 encodes a protein, we first sought to confirm expression of NAG1::mTn as a β-galactosidase chimera. By PCR amplification of the transposon insertion junction and DNA sequencing of this PCR product, we verified integration of our mini-transposon gene trap at codon 52 of NAG1 (Fig. 1B and C). Using quantitative liquid assays, we detected an approximately ninefold increase in β-galactosidase activity under conditions of vegetative growth in a diploid yeast strain harboring one transposon-mutagenized copy of NAG1 relative to the same strain containing an integrated transposon insertion in an intergenic region of noncoding DNA (Fig. 1C). To validate further the protein-coding potential of NAG1, we generated a HA-tagged allele of this gene by integration of a 3×HA-G418 drug resistance cassette at the NAG1 3′ terminus. Cell lysates were extracted from this strain under normal growth conditions for Western blotting with anti-HA antibodies, and this analysis revealed a protein product of approximately 19 kDa, corresponding to the predicted molecular mass of Nag1p-3HA. Thus, the NAG1 gene does encode protein, and the size of this protein is consistent with its predicted mass as derived from the NAG1 coding sequence.
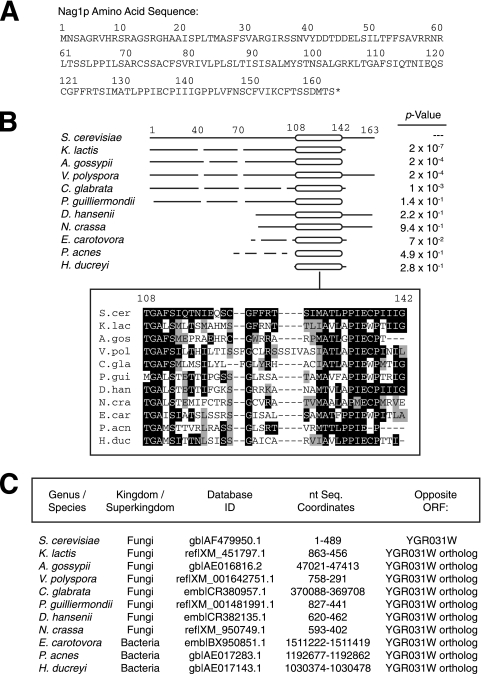
NAG1 is part of an evolutionarily conserved unit in fungi.
The predicted Nag1p sequence (Fig. 2A) is 163 amino acids in length and exhibits no obvious functional motifs. Similarity searches with this sequence indicated putative NAG1 orthologs in several bacterial species and numerous fungi (Fig. 2B). In particular, Nag1p sequence conservation is strongest over a region of 35 residues extending from amino acids 108 to 142. An optimized alignment of this region highlights a strongly conserved Pro-Ile-Glu-Cys-Pro sequence in Nag1p (residues 134 to 138) as well as invariant Gly, Ala, Ser, and Gly residues at positions 109, 110, 112, and 122, respectively. Interestingly, the NAG1 gene is conserved as a unit with the YGR031W ORF: in each organism carrying a putative ortholog of NAG1, the NAG1 sequence is nested antisense an obvious ortholog of YGR031W (Fig. 2C). The YGR031W gene itself is highly conserved in organisms ranging from prokaryotes to humans; however, NAG1 is not present opposite orthologs of YGR031W in higher eukaryotes but instead is specific for prokaryotes and fungi. While these alignments indicate sequence similarity, we cannot conclusively assign functions to any putative NAG1 orthologs without experimental analysis of each sequence in each organism.
FIG. 2.
NAG1 is conserved as a unit with the YGR031W ORF in bacteria and fungi. (A) Amino acid sequence of Nag1p as predicted by conceptual translation. (B) Putative NAG1 orthologs from prokaryotic and fungal species. A schematic diagram illustrating conserved regions within the identified set of putative NAG1 orthologs is shown at the top. Each line represents the full length of the orthologous sequence; gaps in the multiple-sequence alignment are indicated as such in the figure. The prokaryotic and fungal species include Saccharomyces cerevisiae (S.cer), Kluyveromyces lactis (K.lac), Ashbya gossypii (A.gos), Vanderwaltozyma polyspora (V.pol), Candida glabrata (C.gla), Pichia guilliermondii (P.gui), Debaryomyces hansenii (D.han), Neurospora crassa (N.cra), Erwinia carotovora (E.car), Propionibacterium acnes (P.acn), and Haemophilus ducreyi (H.duc). The inset rectangle highlights the most strongly conserved region of the alignment. Identical residues are indicated as white on black, similar residues are shown as black on gray, and gaps introduced to maximize alignment are indicated by dashes. (C) Since Nag1p has not been recognized previously as a protein, neither its sequence nor the sequence of any ortholog is present in a protein database; thus, the Nag1p amino acid sequence was searched against a six-frame translation of genomic DNA sequence. The coordinates of each putative orthologous gene are indicated here, relative to each indicated database accession ID. In each instance, putative orthologs of NAG1 are found opposite orthologs of YGR031W; thus, the nested organization of NAG1 relative to YGR031W is conserved as an evolutionary unit. nt Seq., nucleotide sequence.
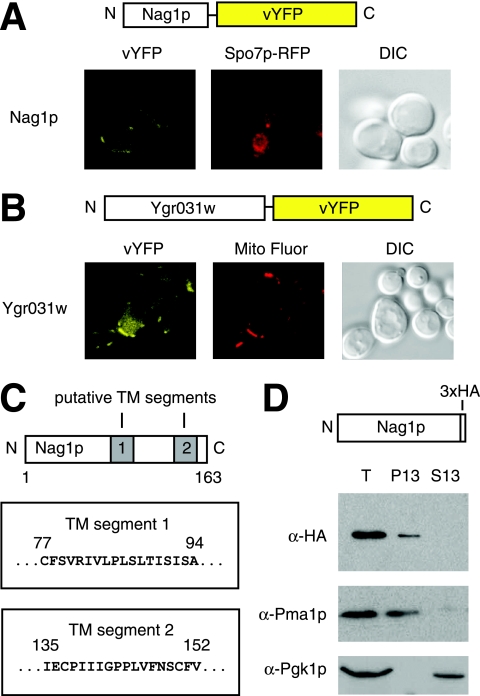
Subcellular localization of Nag1p.
As a means of assessing the subcellular distribution of Nag1p, we cloned the NAG1 gene along with 1 kb of upstream promoter sequence into a low-copy yeast shuttle vector such that the 3′ end of NAG1 forms an in-frame fusion with sequence encoding vYFP. As shown in Fig. 3A, this carboxy-terminal Nag1p-vYFP chimera localized to the yeast cell periphery under conditions of vegetative growth. Nag1p did not localize to the endoplasmic reticulum; the integral membrane protein Spo7p serves as a marker for the nuclear envelope-endoplasmic reticulum network (40), and the carboxy-terminal Spo7p-RFP chimera (Fig. 3A) did not colocalize with Nag1p-vYFP. The localization of Nag1p was also distinct from that of YGR031WP protein. As mentioned above, YGR031W encodes a mitochondrial protein, as determined in a large-scale study of yeast protein localization (13) and in a separate mass spectrometry-based study of yeast mitochondrial proteins (34). To confirm these results, we generated a carboxy-terminal YGR031WP-vYFP chimera and found this protein localized to the mitochondria under conditions of vegetative growth (Fig. 3B).
FIG. 3.
Nag1p localizes to the S. cerevisiae cell periphery, distinct from the YGR031WP protein product, and exhibits properties consistent with those of a plasma membrane protein. (A) Fluorescence microscopy of a Nag1p-vYFP chimera expressed from its native promoter under conditions of vegetative growth. The Spo7p-RFP chimera serves as a marker for the nuclear envelope and endoplasmic reticulum (middle image), and yeast cell morphology was visualized by differential interference contrast (DIC) microscopy (right image). (B) Fluorescence microscopy of YGR031WP-vYFP indicates its localization to the mitochondria. The MitoFluor stain was used to confirm colocalization with mitochondria (middle image), and a DIC image is provided (right). (C) Diagram illustrating predicted Nag1p transmembrane domains. Each putative transmembrane (TM) segment is shaded gray in the Nag1p schematic, and the primary sequence of each segment and its amino acid coordinates are indicated. (D) Western blots indicating the presence of Nag1p-3×HA in the pellet fraction (P13) after centrifugation of total cell lysates (T) at 13,000 rpm (16,000 × g). The known plasma membrane protein Pma1p was also found in the P13 fraction, as indicated by Western blotting with antibody directed against native Pma1p (α-Pma1p). For a further control, we used antibody directed against 3-phosphoglycerate kinase, α-Pgk1p, to confirm the presence of this protein in the supernatant fraction (S13) following centrifugation.
Computational analysis of the Nag1p amino acid sequence revealed two putative transmembrane domains of roughly 20 residues positioned toward the center and carboxy terminus of the protein (Fig. 3C). Predictions of transmembrane segments were obtained from the programs HMMTOP (43), PHDhtm (38), TopPred (2), TMpred (14), and TMHMM (23). Each program highlighted transmembrane segments of slightly different lengths, but residues 77 to 94 and 135 to 152 were identified unanimously (Fig. 3C). These putative transmembrane domains flank the region of strong sequence conservation presented in Fig. 2B, although the carboxy-terminal putative transmembrane segment does overlap this conserved region by 8 bp.
In corollary to the studies above, we also examined the possible membrane association of Nag1p by subcellular fractionation. Lysed spheroplasts were prepared from yeast cells carrying HA-tagged Nag1p expressed from its native promoter under conditions of vegetative growth; cell lysates were subjected to centrifugation at 16,000 × g (13,000 rpm). As visualized by Western blotting, HA-tagged Nag1p was present in the pellet fraction following low-speed centrifugation, cofractionating with other known membrane proteins (Fig. 3D). Thus, Nag1p localizes to the cell periphery, is predicted to contain two transmembrane domains, fractionates with membrane proteins, and does not colocalize with endoplasmic reticulum markers—properties consistent with those of a plasma membrane protein.
Phenotypic characterization of NAG1.
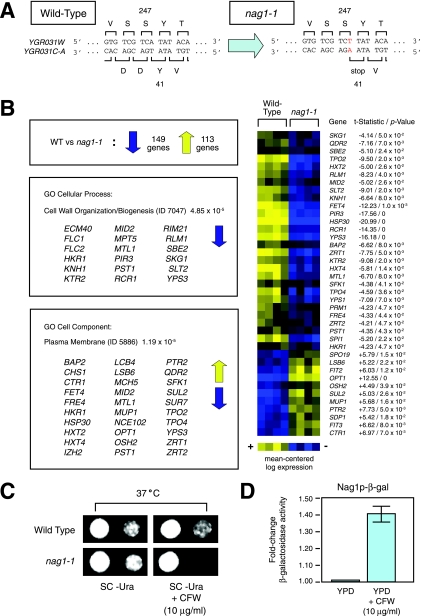
To investigate NAG1 function, we generated a point mutation (nag1-1) disrupting NAG1 but silent with respect to the opposite gene YGR031W (Fig. 4A). Specifically, we mutated NAG1 codon 41 (TAT encoding tyrosine) to a stop codon (TAA); this single base change does not affect the amino acid composition of the YGR031W protein, since the complementary TCA-to-TCT substitution at codon 247 still encodes serine. The nag1-1 mutation truncates NAG1 coding sequence at approximately 25% its full length. Deletion of the entire NAG1 coding sequence in the YGR031WΔ strain generates a phenotype comparable to that of the nag1-1 strain in the assays applied here; thus, nag1-1 mimics a null allele in our studies.
FIG. 4.
NAG1 contributes to yeast cell wall biogenesis. (A) The nag1-1 point mutation introduces a nonsense mutation at codon 41, without altering the predicted amino acid sequence of the YGR031WP protein product. (B) DNA microarray analysis of the nag1-1 mutant under conditions of vegetative growth. Details of this experimental design are presented in Materials and Methods. The total number of differentially expressed genes in the nag1-1 mutant relative to the wild-type (WT) strain is indicated. Genes associated with the GO cellular process term 7047 (cell wall organization/biogenesis) were statistically enriched in the set of genes with decreased transcript levels in nag1-1; these genes are shown under this GO term in the middle box. Genes associated with the GO cell component term 5886 (plasma membrane) were statistically enriched in the total set of genes identified by microarray analysis (both with increased and decreased transcript levels); these genes are also shown below the GO term. The majority of these cell surface-related genes are indicated in the heat map to the left with corresponding t statistics and P values. The full set of differentially expressed genes in the nag1-1 mutant is presented as supplementary data (see file SF1 in the supplemental material). (C) The nag1-1 mutant is sensitive to calcofluor white (CFW). For the assay shown here, we introduced a plasmid carrying the nag1-1 allele in the YGR031W genomic DNA locus into a strain of the W303 genetic background deleted for YGR031W. The resulting nag1-1 mutant was hypersensitive to calcofluor white, with increased severity at elevated temperature (37°C). SC-URA, synthetic complete medium lacking uracil. (D) Nag1p protein levels are increased upon calcofluor white (CFW) treatment, as evidenced by increased β-galactosidase activity from a Nag1p-β-galactosidase (Nag1p-β-gal) chimera.
With no a priori indication of a process to which NAG1 contributes, we decided to implement a global strategy, profiling gene expression in nag1-1 by microarray analysis (Fig. 4B). By this approach, biological processes impaired in the nag1-1 mutant should be evident from altered gene expression profiles. Transcriptional profiling of nag1-1 against a wild-type strain under conditions of vegetative growth revealed differential expression of 262 genes. In particular, 149 genes exhibited decreased transcript levels in nag1-1; this gene set was statistically enriched (P value of 4.85 × 10−5) for genes contributing to cell wall organization and biogenesis (Gene Ontology project [GO] cellular process identification [ID] 7047). Furthermore, the set of 262 genes as a whole was enriched (P value of 1.19 × 10−5) for genes encoding proteins associated with the plasma membrane (GO cell component ID 5886). No other gene subsets were statistically overrepresented in this microarray data set.
The decrease in cell wall-related gene transcription evident in the nag1-1 mutant is unusual for a gene contributing to cell wall biogenesis. More typically, the deletion of a cell wall-related gene leads to an increase in cell wall gene transcription as a compensatory response (16). Of course, most cell wall-associated genes are involved in the biosynthesis or structural organization of the cell wall, and loss of function leads directly to a structural defect that requires a compensatory response to maintain integrity. Since nag1-1 exhibited the opposite effect on cell wall gene transcription, we next asked whether the nag1-1 mutant displayed a phenotype consistent with an altered cell wall structure. Toward this end, we compared the growth of nag1-1 strain with a wild-type strain in the presence of a set of cell wall perturbants, including calcofluor white, Congo red, caffeine, and caspofungin. As indicated in Fig. 4C, the nag1-1 mutant is sensitive to calcofluor white but showed no apparent growth defects with any of the other drugs. Calcofluor white is a negatively charged fluorescent dye that binds nascent chains of chitin and, to a lesser degree, glucan; as a result, calcofluor white prevents microfibril assembly, thereby interfering with cell wall organization (7, 28). Calcofluor white hypersensitivity has been observed as a pleiotropic phenotype associated with many yeast cell wall mutants (9, 28), and the nag1-1 mutant exhibits sensitivity to calcofluor white at 37°C. A similar phenotype is observed for the nag1-1 mutant at 30°C, although the severity is decreased. These phenotypes are consistent across multiple independent transformants in the BY4742 genetic background. To consider further the function of Nag1p in cell wall biogenesis, we introduced the nag1-1 allele into the yeast strain W303. The wild-type W303 strain is defective for the gene SSD1, encoding a protein involved in the maintenance of cellular integrity (18); therefore, cell wall-related phenotypes are often exacerbated in the W303 genetic background. Accordingly, the nag1-1 mutant yields a more pronounced calcofluor white phenotype at both 30°C and 37°C in the W303 background, as assayed in multiple independent transformants. In some calcofluor white-hypersensitive mutants, cell wall chitin is increased, resulting in increased calcofluor white staining. The nag1-1 mutant, however, did not display either an increased amount or abnormal distribution of calcofluor white staining at 25°C or 37°C (data not shown).
Observed nag1-1 phenotypes are specific to cell wall function. The nag1-1 mutant is viable at both 30°C and 37°C, without obvious fitness defects under conditions of vegetative growth at either temperature. In addition, analysis of nag1-1 mutant for growth sensitivity under conditions of nutritional stress, alternative carbon source, nitrogen stress, and high osmolarity did not reveal mutant phenotypes (data not shown).
Nag1p production is regulated by the Slt2p cell wall integrity pathway.
Since many proteins contributing to cell wall function are induced upon treatment with cell wall-perturbing agents, we examined the response of Nag1p upon treatment with calcofluor white in the W303 genetic background (Fig. 4D). Using a Nag1p-β-galactosidase chimera, we investigated Nag1p protein levels under conditions of vegetative growth and in identical growth medium supplemented with calcofluor white. Calcofluor white treatment resulted in a 1.4-fold increase in Nag1p levels relative to those observed during vegetative growth, further supporting its role in cell wall-related processes.
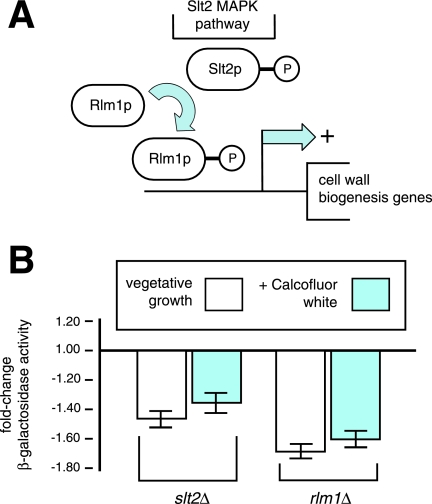
In yeast, cell wall integrity is maintained, in part, through a signaling pathway encompassing the Slt2p/Mpk1p MAPK cascade (Fig. 5A). The Slt2p pathway is activated in response to numerous environmental stimuli, including conditions of hypoosmotic stress, exposure to mating pheromone, and treatment with agents causing cell wall stress (5, 20, 25, 47). These stimuli are transduced into signals activating the GDP/GTP exchange factor Rom2p and the small GTP-binding protein Rho1p (32). Rho1p binds and activates Pkc1p, which in turn elicits serial activation of a MAPK cascade consisting of the MAPK kinase kinase Bck1p, the MAPK kinases Mkk1p and Mkk2p, and the MAPK Slt2p/Mpk1p (11). The transcription factor Rlm1p acts downstream of Slt2p (6), and Rlm1p activates expression of at least 20 genes, the majority of which contribute to yeast cell wall biogenesis (16) (Fig. 5A).
FIG. 5.
Nag1p production is dependent upon the MAPK Slt2p and the transcription factor Rlm1p. (A) Simplified overview of the yeast Slt2p MAPK cell wall integrity pathway. Rlm1p acts as a key transcriptional regulator downstream of Slt2p. P, phosphate. (B) β-Galactosidase assays of the Nag1p-β-galactosidase chimera during vegetative growth and in response to calcofluor white treatment in strains deleted for SLT2 and RLM1, respectively. The change from the wild-type β-galactosidase level is presented for each mutant under the indicated growth conditions. All β-galactosidase assay results were normalized per yeast cell optical density unit.
Interestingly, transcription of both SLT2 and RLM1 was downregulated in the nag1-1 site-directed mutant (Fig. 4B). To investigate a possible role for Nag1p acting downstream of the Slt2p pathway, we assayed protein levels of a Nag1p-β-galactosidase chimera in slt2Δ and rlm1Δ deletion strains under conditions of vegetative growth and calcofluor white treatment (Fig. 5B). Nag1p levels were diminished during vegetative growth in both deletion strains but with a more pronounced decrease evident in the rlm1Δ mutant. Consistent with a role for Rlm1p in the transcriptional activation of Nag1p, a Rlm1p binding site consensus sequence [CTA(T/A)4TA (6, 17)] is present 430 nucleotides upstream of the presumed NAG1 start codon. Interestingly, this putative, palindromic Rlm1p binding site is shared with the promoter of GSC2, the inducible subunit of 1,3-β-glucan synthase and a gene that is upregulated by cell wall stress. Upon exposure to calcofluor white, the overall levels of NAG1 expression were decreased in both slt2Δ and rlm1Δ mutants relative to the wild type. However, exposure to calcofluor white increased NAG1 expression in both mutants relative to the untreated cells, suggesting that Slt2p-independent cell wall response pathways may also contribute to Nag1p expression (26). On the basis of this analysis, we conclude that the cell wall stress-induced production of Nag1p is partially dependent upon Slt2p and Rlm1p.
DISCUSSION
In this paper, we present an unusual orientation of protein-coding genes in the S. cerevisiae genome, identifying a previously overlooked gene, NAG1, nested antisense and opposite another protein-coding gene, YGR031W. This gene superstructure represents an evolutionary unit conserved among many fungal species. The strongly conserved YGR031W gene encodes a mitochondrial protein, while NAG1 encodes a 19-kDa membrane protein localized to the yeast cell periphery. To study NAG1 function, we constructed a point mutation disrupting NAG1 but silent with respect to YGR031W; this mutant exhibited hypersensitivity to calcofluor white and altered transcript levels for a significant subset of genes mediating cell wall biogenesis. Furthermore, Nag1p levels were increased upon calcofluor white treatment and reduced in strains deleted for SLT2 and RLM1, key components of the yeast MAPK cell wall integrity pathway. Collectively, this study highlights a role for Nag1p in maintaining yeast cell wall integrity and function, while validating the protein-coding potential of this nested gene.
In particular, the nested organization of genes at the NAG1 locus holds interesting evolutionary implications. Overlapping genes have been found commonly in viruses and microorganisms, where this type of interleaved and nested gene organization presumably contributes to the maintenance of a compact genome—a beneficial characteristic, since genome size in these organisms is limited by the size of the viral particle or cell (22). Eukaryotic genomes, of course, do not face this constraint, and in this light, two points regarding NAG1 are noteworthy. First, putative NAG1 orthologs are exclusively found opposite an ortholog of YGR031W. Second, YGR031W encodes a mitochondrial protein. Mitochondria are thought to have evolved from purple non-sulfur bacteria (1), wherein this type of nested gene organization might not be uncommon. Extrapolating from this, we can speculate that the NAG1 locus may represent the remnants of an ancient genetic unit, possibly even tracing back to symbiont gene transfer during mitochondrial evolution. YGR031W is strongly conserved in prokaryotes and eukaryotes alike, but over evolutionary time, NAG1 function may have been lost in organisms lacking a cell wall. Consistent with this possibility, putative NAG1 orthologs are present only in prokaryotes and fungi (Fig. 2), although further studies would be necessary to determine whether these orthologs are functional.
The cell wall-related function of Nag1p is supported by three lines of evidence. First, and most striking, is the fact that the nag1-1 mutation leads to a significant decrease in the expression of a large set of cell wall genes during vegetative growth. This is opposite to the more common phenomenon whereby deletion of a cell wall gene causes upregulation of cell wall gene transcription to compensate for the resulting cell wall defects. The negative effect of nag1-1 on cell wall gene expression is more consistent with Nag1p functioning as a type of regulatory protein as opposed to having a direct role in cell wall structure or biosynthesis. This analysis is further supported by the relatively mild cell wall phenotype displayed by the nag1-1 mutant. However, it is important to note that mutation of a number of cell wall-related genes causes calcofluor white hypersensitivity as their only discernible cell wall phenotype; therefore, this second set of observations supporting a cell wall role for Nag1p is consistent with other bona fide cell wall proteins.
Third, the effect of cell wall stress and the cell wall integrity MAPK signaling pathway on NAG1 expression provides compelling support for the cell wall-related function of Nag1p. As is the case for many cell wall-related genes, cell wall stress, such as calcofluor white treatment, induces a modest increase in Nag1p levels. NAG1 may share its promoter region with GSC2 (Fig. 1B), the stress-inducible subunit of 1,3-β-glucan synthase, and therefore, NAG1 expression could be regulated by processes that also regulate GSC2. Indeed, this region contains a consensus binding site for Rlm1p, a transcription factor regulated by the cell wall integrity MAPK signaling pathway. Since the Rlm1p binding site is palindromic, it should control transcription of appropriately oriented ORFs on either the Watson or Crick strand.
Consistent with this analysis, NAG1 expression is dependent on both Slt2p and Rlm1p in a significant but not exclusive fashion. Intriguingly, the effect of Slt2p and Rlm1p on NAG1 expression is quite apparent during vegetative growth. Although the cell wall integrity pathway is more commonly thought of as a stress response cascade, it is activated during specific periods of the cell cycle (26). Therefore, NAG1 expression may be controlled through basal signaling of the cell wall integrity pathway. We speculate that this pattern of expression may relate to the positive effect of Nag1p on the transcription of other cell wall genes during vegetative growth. Obviously, a more extensive characterization of Nag1p will be required to confirm this assertion. However, it is clear from our data that Nag1p is a functional protein involved in yeast cell wall integrity.
Here, we have referred to NAG1 as being unique, but, in fact, NAG1 may actually represent the first identified gene of a larger class: the potential certainly exists for other nested antisense genes in yeast. By transposon mutagenesis using a simple gene trap reporter, we previously identified a set of at least 54 putative nested genes in yeast (24). While we expect that some, and perhaps the majority, of these nested ORFs do not encode protein, additional studies may uncover other nested protein-coding genes previously overlooked in the yeast genome. These overlooked genes potentially represent a wealth of unexplored yeast biology, with implications impacting gene predictions and gene-finding studies in other eukaryotes as well. As a result, the example set by NAG1 may prove useful in refining annotation efforts applied to other genomes, separate from the relevance of this gene as an interesting component of the signaling pathways and networks contributing to yeast cell surface biology.
Supplementary Material
Acknowledgments
We thank Dan Klionsky for providing reagents and for helpful comments regarding the manuscript.
This work was supported by NIH grant K08A1062978 (to D.J.K.) and grants RSG-06-179-01-MBC from the American Cancer Society, DBI 0543017 from the National Science Foundation, and Basil O'Connor Award 5-FY05-1224 from the March of Dimes (to A.K.).
Footnotes
Published ahead of print on 29 February 2008.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Cavalier-Smith, T. 2006. Origin of mitochondria by intracellular enslavement of a photosynthetic purple bacterium. Proc. Biol. Sci. 2731943-1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Claros, M. G., and G. von Heijne. 1994. TopPred II: an improved software for membrane protein structure predictions. Comput. Appl. Biosci. 10685-686. [DOI] [PubMed] [Google Scholar]
- 3.Coelho, P. S., A. C. Bryan, A. Kumar, G. S. Shadel, and M. Snyder. 2002. A novel mitochondrial protein, Tar1p, is encoded on the antisense strand of the nuclear 25S rDNA. Genes Dev. 162755-2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.David, L., W. Huber, M. Granovskaia, J. Toedling, C. J. Palm, L. Bofkin, T. Jones, R. W. Davis, and L. M. Steinmetz. 2006. A high-resolution map of transcription in the yeast genome. Proc. Natl. Acad. Sci. USA 1035320-5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Nobel, H., C. Ruiz, H. Martin, W. Morris, S. Brul, M. Molina, and F. M. Klis. 2000. Cell wall perturbation in yeast results in dual phosphorylation of the Slt2/Mpk1 MAP kinase and in an Slt2-mediated increase in FKS2-lacZ expression, glucanase resistance and thermotolerance. Microbiology 1462121-2132. [DOI] [PubMed] [Google Scholar]
- 6.Dodou, E., and R. Treisman. 1997. The Saccharomyces cerevisiae MADS-box transcription factor Rlm1 is a target for the Mpk1 mitogen-activated protein kinase pathway. Mol. Cell. Biol. 171848-1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elorza, M. V., H. Rico, and R. Sentandreu. 1983. Calcofluor white alters the assembly of chitin fibrils in Saccharomyces cerevisiae and Candida albicans cells. J. Gen. Microbiol. 1291577-1582. [DOI] [PubMed] [Google Scholar]
- 8.Guthrie, C., and G. Fink. 1991. Guide to yeast genetics and molecular biology. Academic Press, San Diego, CA.
- 9.Hampsey, M. 1997. A review of phenotypes in Saccharomyces cerevisiae. Yeast 131099-1133. [DOI] [PubMed] [Google Scholar]
- 10.Havilio, M., E. Y. Levanon, G. Lerman, M. Kupiec, and E. Eisenberg. 2005. Evidence for abundant transcription of non-coding regions in the Saccharomyces cerevisiae genome. BMC Genomics 693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heinisch, J. J., A. Lorberg, H. P. Schmitz, and J. J. Jacoby. 1999. The protein kinase C-mediated MAP kinase pathway involved in the maintenance of cellular integrity in Saccharomyces cerevisiae. Mol. Microbiol. 32671-680. [DOI] [PubMed] [Google Scholar]
- 12.Hongay, C. F., P. L. Grisafi, T. Galitski, and G. R. Fink. 2006. Antisense transcription controls cell fate in Saccharomyces cerevisiae. Cell 127735-745. [DOI] [PubMed] [Google Scholar]
- 13.Huh, W. K., J. V. Falvo, L. C. Gerke, A. S. Carroll, R. W. Howson, J. S. Weissman, and E. K. O'Shea. 2003. Global analysis of protein localization in budding yeast. Nature 425686-691. [DOI] [PubMed] [Google Scholar]
- 14.Ikeda, M., M. Arai, D. M. Lao, and T. Shimizu. 2002. Transmembrane topology prediction methods: a re-assessment and improvement by a consensus method using a dataset of experimentally-characterized transmembrane topologies. In Silico Biol. 219-33. [PubMed] [Google Scholar]
- 15.Ito, H., Y. Fukuda, K. Murata, and A. Kimura. 1983. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153163-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jung, U. S., and D. E. Levin. 1999. Genome-wide analysis of gene expression regulated by the yeast cell wall integrity signalling pathway. Mol. Microbiol. 341049-1057. [DOI] [PubMed] [Google Scholar]
- 17.Jung, U. S., A. K. Sobering, M. J. Romeo, and D. E. Levin. 2002. Regulation of the yeast Rlm1 transcription factor by the Mpk1 cell wall integrity MAP kinase. Mol. Microbiol. 46781-789. [DOI] [PubMed] [Google Scholar]
- 18.Kaeberlein, M., and L. Guarente. 2002. Saccharomyces cerevisiae MPT5 and SSD1 function in parallel pathways to promote cell wall integrity. Genetics 16083-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kapranov, P., A. T. Willingham, and T. R. Gingeras. 2007. Genome-wide transcription and the implications for genomic organization. Nat. Rev. Genet. 8413-423. [DOI] [PubMed] [Google Scholar]
- 20.Ketela, T., R. Green, and H. Bussey. 1999. Saccharomyces cerevisiae Mid2p is a potential cell wall stress sensor and upstream activator of the PKC1-MPK1 cell integrity pathway. J. Bacteriol. 1813330-3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim, J., V. M. Dalton, K. P. Eggerton, S. V. Scott, and D. J. Klionsky. 1999. Apg7p/Cvt2p is required for the cytoplasm-to-vacuole targeting, macroautophagy, and peroxisome degradation pathways. Mol. Biol. Cell 101337-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krakauer, D. C. 2000. Stability and evolution of overlapping genes. Evolution Int. J. Org. Evolution. 54731-739. [DOI] [PubMed] [Google Scholar]
- 23.Krogh, A., B. Larsson, G. von Heijne, and E. L. Sonnhammer. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305567-580. [DOI] [PubMed] [Google Scholar]
- 24.Kumar, A., P. M. Harrison, K. H. Cheung, N. Lan, N. Echols, P. Bertone, P. Miller, M. B. Gerstein, and M. Snyder. 2002. An integrated approach for finding overlooked genes in yeast. Nat. Biotechnol. 2058-63. [DOI] [PubMed] [Google Scholar]
- 25.Lee, K. S., K. Irie, Y. Gotoh, Y. Watanabe, H. Araki, E. Nishida, K. Matsumoto, and D. E. Levin. 1993. A yeast mitogen-activated protein kinase homolog (Mpk1p) mediates signalling by protein kinase C. Mol. Cell. Biol. 133067-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levin, D. E. 2005. Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 69262-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14953-961. [DOI] [PubMed] [Google Scholar]
- 28.Lussier, M., A.-M. White, J. Sheraton, T. di Paulo, J. Treadwell, S. B. Southard, C. I. Horenstein, J. Chen-Weiner, A. F. J. Ram, J. C. Kapteyn, T. W. Roemer, D. H. Vo, D. C. Bondoc, J. Hall, W. W. Zhong, A.-M. Sdicu, J. Davies, F. M. Klis, P. W. Robbins, and H. Bussey. 1997. Large scale identification of genes involved in cell surface biosynthesis and architecture in Saccharomyces cerevisiae. Genetics 147435-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma, J., R. Jin, X. Jia, C. J. Dobry, L. Wang, F. Reggiori, J. Zhu, and A. Kumar. 2007. An interrelationship between autophagy and filamentous growth in budding yeast. Genetics 177205-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mewes, H. W., K. Albermann, M. Bahr, D. Frishman, A. Gleissner, J. Hani, K. Heumann, K. Kleine, A. Maierl, S. G. Oliver, F. Pfeiffer, and A. Zollner. 1997. Overview of the yeast genome. Nature 387(Suppl.):7-65. [DOI] [PubMed] [Google Scholar]
- 31.Nagai, T., K. Ibata, E. S. Park, M. Kubota, K. Mikoshiba, and A. Miyawaki. 2002. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 2087-90. [DOI] [PubMed] [Google Scholar]
- 32.Philip, B., and D. E. Levin. 2001. Wsc1 and Mid2 are cell surface sensors for cell wall integrity signaling that act through Rom2, a guanine nucleotide exchange factor for Rho1. Mol. Cell. Biol. 21271-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Philippsen, P., K. Kleine, R. Pohlmann, A. Dusterhoft, K. Hamberg, J. H. Hegemann, B. Obermaier, L. A. Urrestarazu, R. Aert, K. Albermann, R. Altmann, B. Andre, V. Baladron, J. P. Ballesta, A. M. Becam, J. Beinhauer, J. Boskovic, M. J. Buitrago, F. Bussereau, F. Coster, M. Crouzet, M. D'Angelo, F. Dal Pero, A. De Antoni, and J. Hani. 1997. The nucleotide sequence of Saccharomyces cerevisiae chromosome XIV and its evolutionary implications. Nature 38793-98. [PubMed] [Google Scholar]
- 34.Reinders, J., R. P. Zahedi, N. Pfanner, C. Meisinger, and A. Sickmann. 2006. Toward the complete yeast mitochondrial proteome: multidimensional separation techniques for mitochondrial proteomics. J. Proteome Res. 51543-1554. [DOI] [PubMed] [Google Scholar]
- 35.Rieger, K. E., and G. Chu. 2004. Portrait of transcriptional responses to ultraviolet and ionizing radiation in human cells. Nucleic Acids Res. 324786-4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ross-Macdonald, P., P. S. Coelho, T. Roemer, S. Agarwal, A. Kumar, R. Jansen, K. H. Cheung, A. Sheehan, D. Symoniatis, L. Umansky, M. Heidtman, F. K. Nelson, H. Iwasaki, K. Hager, M. Gerstein, P. Miller, G. S. Roeder, and M. Snyder. 1999. Large-scale analysis of the yeast genome by transposon tagging and gene disruption. Nature 402413-418. [DOI] [PubMed] [Google Scholar]
- 37.Ross-Macdonald, P., A. Sheehan, G. S. Roeder, and M. Snyder. 1997. A multipurpose transposon system for analyzing protein production, localization, and function in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 94190-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rost, B., P. Fariselli, and R. Casadio. 1996. Topology prediction for helical transmembrane proteins at 86% accuracy. Protein Sci. 51704-1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seifert, H. S., E. Y. Chen, M. So, and F. Heffron. 1986. Shuttle mutagenesis: a method of transposon mutagenesis for Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 83735-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Siniossoglou, S., H. Santos-Rosa, J. Rappsilber, M. Mann, and E. Hurt. 1998. A novel complex of membrane proteins required for formation of a spherical nucleus. EMBO J. 176449-6464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 224673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 985116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tusnady, G. E., and I. Simon. 2001. The HMMTOP transmembrane topology prediction server. Bioinformatics 17849-850. [DOI] [PubMed] [Google Scholar]
- 44.Walhout, A. J. M., G. F. Temple, M. A. Brasch, J. L. Hartley, M. A. Lorson, S. van den Heuvel, and M. Vidal. 2000. GATEWAY recombinational cloning: application to the cloning of large numbers of open reading frames or ORFeomes. Methods Enzymol. 328575-592. [DOI] [PubMed] [Google Scholar]
- 45.Winzeler, E. A., D. D. Shoemaker, A. Astromoff, H. Liang, K. Anderson, B. Andre, R. Bangham, R. Benito, J. D. Boeke, H. Bussey, A. M. Chu, C. Connelly, K. Davis, F. Dietrich, S. W. Dow, M. E. Bakkoury, F. Foury, S. H. Friend, E. Gentalen, G. Giaever, J. H. Hegemann, T. J. M. Laub, H. Liao, N. Liebundguth, D. J. Lockhart, A. Lucau-Danila, M. Lussier, N. M'Rabet, P. Menard, M. Mittmann, C. Pai, C. Rebischung, J. L. Revuelta, L. Riles, C. J. Roberts, P. Ross-MacDonald, B. Scherens, M. Snyder, S. Sookhai-Mahadeo, R. K. Storms, S. Véronneau, M. Voet, G. Volckaert, T. R. Ward, R. Wysocki, G. S. Yen, K. Yu, K. Zimmermann, P. Philippsen, M. Johnston, and R. W. Davis. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285901-906. [DOI] [PubMed] [Google Scholar]
- 46.Ye, J., S. McGinnis, and T. L. Madden. 2006. BLAST: improvements for better sequence analysis. Nucleic Acids Res. 34W6-W9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zarzov, P., C. Mazzoni, and C. Mann. 1996. The SLT2(MPK1) MAP kinase is activated during periods of polarized cell growth in yeast. EMBO J. 1583-91. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.