Abstract
The class I myosins play important roles in controlling many different types of actin-based cell movements. Dictyostelium cells either lacking or overexpressing amoeboid myosin Is have significant defects in cortical activities such as pseudopod extension, cell migration, and macropinocytosis. The existence of Dictyostelium null mutants with strong phenotypic defects permits complementation analysis as a means of exploring important functional features of the myosin I heavy chain. Mutant Dictyostelium cells lacking two myosin Is exhibit profound defects in growth, endocytosis, and rearrangement of F-actin. Expression of the full-length myoB heavy chain in these cells fully rescues the double mutant defects. However, mutant forms of the myoB heavy chain in which a serine at the consensus phosphorylation site has been altered to an alanine or in which the C-terminal SH3 domain has been removed fail to complement the null phenotype. The wild-type and mutant forms of the myoB heavy chain appeared to be properly localized when they were expressed in the myosin I null mutants. These results suggest that the amoeboid myosin I consensus phosphorylation site and SH3 domains do not play a role in the localization of myosin I, but are absolutely required for in vivo function.
INTRODUCTION
The myosin Is are a family of ubiquitous actin-based motors that have been implicated in the manipulation of the actin-rich cortex of the cell (Pollard et al., 1991; Mooseker and Cheney, 1995). First discovered in Acanthamoeba castellanii, their biochemical properties and cellular distribution have been well characterized (Pollard et al., 1991). Myosin I localization patterns and their widespread distribution across a range of species and cell types suggest that they play conserved and essential roles in generating movements of membranes along actin filaments. Thus, the myosin Is have been proposed to play important roles in cell migration, endocytosis, and vesicle transport (Mooseker and Cheney, 1995; Ostap and Pollard, 1996b).
The amoeboid myosin Is are comprised of a single heavy chain of approximately 125 kDa and a light chain that is presumably bound to the single IQ motif in the neck region (Pollard et al., 1991). They possess the biochemical properties characteristic of a myosin, including actin-activated Mg-ATPase activity (Pollard et al., 1991) and the ability to generate ATP-dependent motility in vitro (Albanesi et al., 1985; Zot et al., 1992). The in vitro activity of Acanthamoeba and Dictyostelium amoeboid myosin I is tightly regulated by serine/threonine phosphorylation at a consensus phosphorylation site in the N-terminal myosin motor domain (Brzeska and Korn, 1996), referred to as the TEDS rule site (Bement and Mooseker, 1995). The TEDS rule site is in a region of the myosin head that is believed to make a significant contact with actin (Cope et al., 1996) and this interaction is likely to be significantly altered in the absence of a negative charge at this site. Myosin I heavy chain kinases (MIHCKs) have been identified both in Acanthamoeba and Dictyostelium and are members of the Ste20p/PAK family of kinases whose activity is regulated by small G proteins (Brzeska et al., 1996; Lee et al., 1996).
The C-terminal tail region of the amoeboid myosin Is also possesses conserved domains believed to play an important role in determining myosin I function (Pollard et al., 1991). The first is a polybasic domain, adjacent to the IQ motif, that mediates the high-affinity association between membranes and myosin I in vitro (Adams and Pollard, 1989; Miyata et al., 1989; Doberstein and Pollard, 1992). The second is a domain rich in the amino acids glycine, proline, and alanine (or glutamate) referred to as the GPA or GPQ domain. This region has been found to constitute a second, ATP-insensitive actin-binding site in vitro (Lynch et al., 1986; Doberstein and Pollard, 1992; Jung and Hammer, 1994; Rosenfeld and Rener, 1994). Finally, there is a src-homology 3 domain at the C terminus (or close to the C terminus) of the amoeboid myosin Is (Pollard et al., 1991) whose role in myosin I function is unclear. The existence of a membrane-binding site in the tail region as well as an ATP-independent actin-binding site (referred to as the GPA domain) supports the theory that the role of amoeboid myosin Is is to move membranes or actin along actin filaments (Pollard et al., 1991).
Dictyostelium expresses at least three classic amoeboid myosin Is, myoB, C, and D, all of which have been localized to the leading edge of translocating cells (Fukui et al., 1989; Jung et al., 1993, 1996). Additionally, it expresses three “short” myosin Is that are distinguished from the classic forms by the lack of the C-terminal GPA and SH3 domains (Uyeda and Titus, 1997). Loss of myoB from Dictyostelium cells results in defects in pseudopod formation and translocation (Jung and Hammer, 1990; Wessels et al., 1991). Dictyostelium mutants that lack two amoeboid myosin Is, myoA and myoB, were shown to exhibit additional defects in fluid-phase pinocytosis, growth in suspension cultures, and rearrangement of F-actin (Novak et al., 1995). Overexpression of myoB in wild-type Dictyostelium cells also resulted in similar phenotypes, significant decreases in the rate of cellular translocation and fluid-phase pinocytosis, and abnormalities in the normal rearrangement of the actin cytoskeleton (Novak and Titus, 1997). Overexpression of mutant forms of myoB lacking the C-terminal SH3 domain or TEDS rule site (serine 332), however, did not result in cellular cortical defects, suggesting that these elements are required for myoB in vivo function.
Overexpression studies were initially used to determine the importance of myosin I regulatory sequences such as the TEDS rule site or SH3 domain in Dictyostelium myoB function (Novak and Titus, 1997). However, this analysis did not provide unequivocal evidence for the role of these elements in the localization of myoB as the overexpression experiments were performed in wild-type cells, which also possessed endogenous full-length myoB. Furthermore, it did not permit us to detect whether a mutant form of myosin I was only partially inactivated. Therefore, we have undertaken a complementation approach to studying myoB localization and function. The Dictyostelium myoA−/B− double mutant was selected as the host for these studies because its phenotype [defective in cellular translocation, pinocytosis, growth rate, and rearrangement of cortical actin (Novak et al., 1995)] is more severe than that of either the myoA− or myoB− single mutants [only defective in cellular translocation (Jung and Hammer, 1990)]. Complementation analysis in the double mutant allowed us to examine a wider range of myosin I functions to more conclusively test the function of mutant forms. Full-length and mutant forms of myoB (lacking either the SH3 domain or the TEDS rule heavy chain phosphorylation site) were expressed in the myoA−/B− mutant and tested for their ability to undergo proper localization and to restore normal pinocytic activity, growth, and actin distribution to the null mutant cell line.
MATERIALS AND METHODS
Maintenance of Stock Cultures
The parental Dictyostelium discoideum Ax3 axenic strain and myosin I mutant strains were all maintained in HL5, a nutrient medium for axenic stains (Sussman, 1987). All Dictyostelium strains were either carried on bacteriological plastic plates in HL5 or were inoculated into 100 ml of HL5 in 250-ml Erlenmeyer flasks and carried in shaking culture at 240 rpm. “Suspension-grown” cells were inoculated into 100 ml of HL5 in 250-ml Erlenmeyer flasks and carried in shaking culture at 240 rpm for 72 h prior to an experiment. The myoA−/B− myosin I double mutant HTD5–2 (Novak et al., 1995) was maintained in HL5 containing 10 μg/ml G418 (Geneticin, Life Technologies, Gaithersburg, MD). The myoB expressors (myoA−/B− cells expressing full-length myoB), myoB/SH3− expressors (truncated myoB expressing cells), and myoB-S332A expressors (cells expressing a mutant myoB in which serine 332 has been changed to alanine) were all maintained in HL5 in the presence of 20 μg/ml blasticidin (Calbiochem, La Jolla, CA).
Transformation of Dictyostelium
The electrotransformation of Dictyostelium was performed following a slightly modified version (Kuspa and Loomis, 1992) of the original protocol (Howard et al., 1988). The myoB− strain (clone HTD4–4, (Novak et al., 1995) was cotransformed with 10 μg of the plasmid pDTb2 (Novak et al., 1995; Figure 1) that contained the 3.7-kb full-length myoB gene driven by its own 5′ promoter sequence, and the plasmid pLittle (a derivative of pBIG (Patterson and Spudich, 1995) that contained the neomycin resistance gene (Neo). The myoA−/B− double mutant HTD5–2 (Novak et al., 1995) was cotransformed with 10 μg each of the full-length or mutant myoB-bearing plasmids and pUCBsr, a plasmid that carries a gene conferring resistance to blasticidin (Sutoh, 1993). The plasmids used for myoB expression in double mutant cell lines, pDTb18, pDTb39, and pDTb42, have been described previously (Novak and Titus, 1997). The plasmid pDTb18 contained the full-length myoB gene, pDTb42 contained a myoB gene with an altered codon that changed serine 332 to alanine (myoB-S332A), and pDTb39 contained a myoB gene encoding a truncated myoB lacking the C-terminal SH3 domain (myoB/SH3−), each driven by the actin 15 promoter (Cohen et al., 1986). The plasmids pDTb18 and pDTb42 employed the 3′ untranslated region of the myoB gene as a terminator. It should be noted that the actin 6 promoter from the adjacent Neo cassette served as the terminator for the truncated myoB gene in pDTb39.
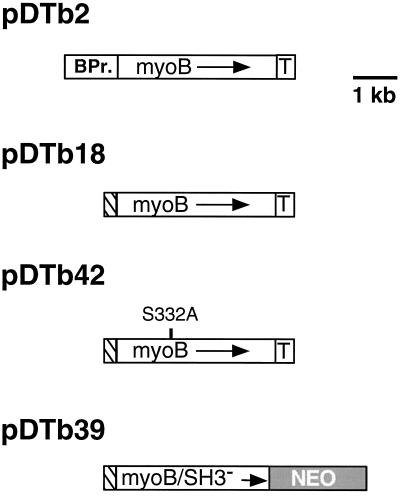
Figure 1.
Schematic diagram of the myoB expression plasmids. The pBluescript-based plasmid pDTb2 contains the 3.6-kb full-length myoB gene along with 1.22 kb of 5′ noncoding region that serves as promoter (BPr.) and 0.45 kb of 3′ noncoding sequences (T) that are likely to encompass the terminator region of the myoB gene. The pGEM7-based plasmid pDTb18 contains the 0.3-kb actin 15 promoter (hatched box) upstream of the full-length 3.6-kb myoB coding sequence and 0.45 kb of 3′ noncoding sequence (T). The plasmid pDTb42 contains the 0.3-kb actin 15 promoter (hatched box) upstream of a mutant 3.7-kb myoB gene containing a serine to alanine change at amino acid 332 (nts. 1361–1363). The vector pDTb39 is a pLittle-based vector that contains the 0.3-kb actin 15 promoter (hatched box) upstream of a truncated form of the myoB gene (myoB/SH3−) that lacks the 3′ 0.16 kb of coding sequence encompassing the SH3 domain and 0.45-kb myoB terminator. The myoB/SH3− gene is immediately adjacent to the 2.2-kb Neo resistance cassette (gray box) whose transcription is driven by the actin 6 promoter. The arrows represent direction of myoB transcription. The vector backbone is not illustrated in these diagrams.
The cells were allowed to recover overnight in HL5 following electroporation, diluted 1:10 into HL5 containing either 10 μg/ml G418 for complementation of the myoB− cell line, or 20 μg/ml blasticidin for complementation of the myoA−/B− cell line. The media were exchanged every third d. Colonies appeared after 9 d under drug selection and were picked in 24-well plates. The transformants were allowed to grow for 4–5 d in the presence of 10 μg/ml G418 or 20 μg/ml blasticidin. Colonies that had grown to confluence were collected, counted, and reinoculated into 10-ml Petri plates containing HL5 and the selection drug. An equal number of cells from each clone were lysed with urea-containing sample buffer and run on a 6% SDS-polyacrylamide gel for quantitative immunoblot analysis (see below).
Quantitative Western blot analysis was performed on whole cell lysates from a total of 20 independent transformants. Lysates were prepared by resuspension of 1 × 106 pelleted cells in urea-containing sample buffer (125 mM Tris, pH 6.8, 6 M urea, 20% glycerol, 4% SDS, 1 mM dithiothreitol). The protein concentration was determined by the Bio-Rad DC assay (Bio-Rad Laboratories, Hercules, CA). Two identical 6% SDS-polyacrylamide gels were loaded with equal amounts of protein for each sample. One gel was stained with Coomassie blue R-250 to confirm that the samples were equally loaded, and the other was transferred to nitrocellulose. The nitrocellulose blot was incubated with a polyclonal antibody generated against myoB (Novak et al., 1995; Novak and Titus, 1997) followed by incubation with a horseradish peroxidase- conjugated goat anti-rabbit secondary antibody (Bio-Rad Laboratories). Antibody reactivity was visualized by either the ECL-enhanced chemiluminescence (Amersham, Arlington Heights, IL) or Super Signal (Pierce Chemical Co., Rockford, IL) detection systems. Films from the Western blots were scanned using an Epson ES1200C (Epson, Pittsburgh, PA) color scanner. The scanned images were then imported into NIH Image 1.54 (Bethesda, MD), and the mean pixel value of the myoB reactive bands were measured. These values were used to determine relative expression levels for each sample. Serial dilutions of Ax3 lysates were included on each Western blot to confirm that autoradiograms used for quantification were within the linear range.
Assays
Pinocytosis by cells in suspension culture was carried out using fluorescein isothiocyanate (FITC)-dextran (Sigma Chemical Co., St. Louis, MO) as described (Klein and Satre, 1986). Fluorescence was measured on a Perkin Elmer-Cetus 650–40 fluorescence spectrophotometer with an excitation wavelength of 470 nm and emission wavelength of 520 nm. A standard curve was used to calculate microliters of FITC-dextran per 106 cells.
The streaming assay was performed as described by Jung and Hammer (1990) with minor modifications (Peterson et al., 1995). Streaming cells were observed on a Zeiss Axiovert using a 10× objective (0.3 aperture) with a 1.25× optivar lens or a 20× objective (0.5 aperture) and a 2× optivar lens. Images were captured into IP Lab software (Signal Analytics Corp., Vienna, VA) using a Star 1 camera (Photometrics Ltd., Tuscon, AZ).
F-actin localization was performed using rhodamine phalloidin (Molecular Probes, Eugene, OR) as described previously (Peterson et al., 1995). Suspension-grown wild-type, double mutant, and complemented strains were allowed to attach to coverslips before processing. Confocal microscopy was performed using a Zeiss Axiovert Laser Scanning Confocal Microscope with a 63× objective (1.25 aperture) with a 2× zoom.
Immunofluorescent localization of myoB was performed using suspension-grown Ax3 and myoB-expressing double mutant cells that were allowed to adhere to coverslips for 15 min as previously described (Novak and Titus, 1997). Immunofluorescent localization of chemotaxing cells was performed using the agar overlay technique as described previously (Fukui et al., 1987, 1989). Briefly, cells were resuspended to 1 × 106 cells/ml in MES starvation buffer (20 mM 2-[N-morpholino] ethane sulfonic acid, pH 6.8, 2 mM MgSO4, 0.2 mM CaCl2) and flattened with a 2% agar sheet in MES buffer after 4-h starvation. They were allowed to continue the starvation response until they reached the preaggregation stage. Ax3 cells required 6 h of total starvation to reach this stage, and reexpressing cells required 8 h. The myoB antibody was preabsorbed using fixed cells (Fukui et al., 1987) and reacts with a single band of approximately 125 kDa in blots of Ax3 cells, but does not cross-react with any other of the Dictyostelium myosin Is, based on the lack of the 125-kDa band in extracts of myoB− cells (Figures 2A and 3). The samples were analyzed by laser scanning confocal microscopy in the same manner as the rhodamine-phalloidin analysis.
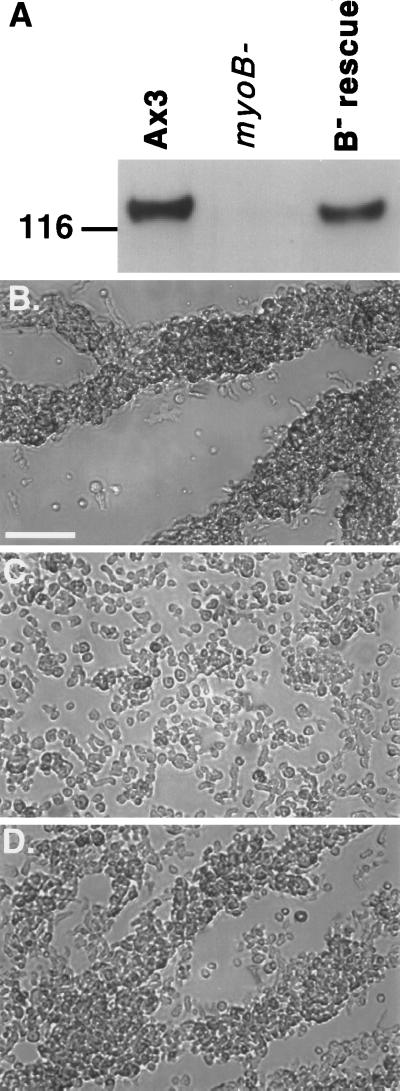
Figure 2.
Expression of myoB in myoB− cells rescues the streaming defect. (A) Western blot analysis shows that myoB is expressed in the B− rescue cell line. The position of the Mr 116,000 marker is indicated on the left of the Western blot. The levels of expression of the 125-kDa myoB heavy chain in equal numbers of Ax3 control (Ax3), myoB− mutant, and the myoB-expressing cell line (B- rescue) are shown. The same cell lines were analyzed in the streaming assay and photographed at 8 h after starvation (B–D). Bar, 50 μm.
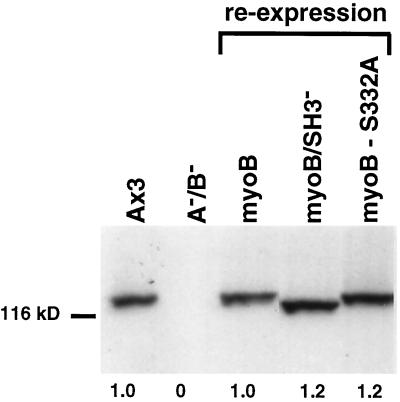
Figure 3.
The complemented myoA−/B− cells express normal levels of the wild-type and mutant myoB heavy chain. Western blot analysis of total cell lysates was used to determine the expression level of the myoB heavy chain in wild-type, null, and reexpression strains. The position of the Mr 116,000 marker is indicated on the left of the Western blot. The levels of expression of the 125-kDa myoB heavy chain in equal numbers of Ax3 control cells (Ax3), myoA−/B− double mutants (A−/B−), and double mutants expressing full-length myoB (myoB), myoB/SH3− (myoB/SH3−), and the myoB serine to alanine mutant (myoB-S332A) are shown. The levels of myoB expression are indicated below each lane, and the values are expressed relative to the Ax3 strain (which is set to 1). Note that the myoB Ab recognizes the endogenous myoB, as well as the smaller truncated myoB in the myoB/SH3− cells, and the mutant form of myoB in the myoB-S332A cells.
Triton-insoluble rigor cytoskeletons were generated as described (Manstein and Hunt, 1995). The cytoskeletons are referred to as “rigor” cytoskeletons because the lysis buffer does not include any Mg-ATP. Ten percent of the Triton lysate was removed prior to the initial centrifugation and spun separately. Gel samples were made from both the supernatant and pellet of this aliquot and these were analyzed by quantitative immunoblotting (described above) to determine the total amount of Triton-soluble and Triton-insoluble myoB. The remainder of the Triton lysate was processed according to the original method.
RESULTS
Rescue of the myoB− Phenotype
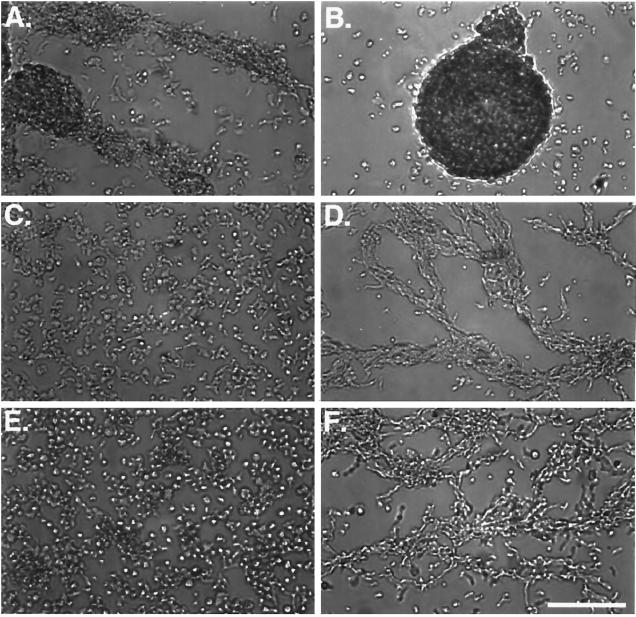
The validity of our complementation strategy was established by first rescuing the myoB− single mutant with wild-type myoB. The plasmid pDTb2 that carries the full-length myoB gene driven by its endogenous promoter (Figure 1) was cotransfected with the plasmid pLittle, which carries neomycin resistance, into a myoB− cell line. A number of clones expressing the myoB heavy chain were identified using Western blot analysis and were named B− rescue cells. Figure 2A shows an example of one clone that expressed wild-type levels of myoB. The B− rescue cell line was assayed for its ability to reverse the myoB− phenotype, a delay in the onset of aggregation after starvation, also known as streaming (Jung and Hammer, 1990). When placed in MES buffer, the wild-type Ax3 cells began the streaming process by 8 h after starvation (Figure 2B) whereas the myoB− cell line had not yet begun to stream (Figure 2C). The myoB− cells did not begin streaming until 10 h after starvation (our unpublished observations). Expression of wild-type levels of myoB in the myoB− null cells rescued the ability of these cells to stream normally at 8 h after starvation (Figure 2D). A similar result was observed when the pDTb18 plasmid was used (our unpublished observations).
Expression of Wild-Type and Mutant Forms of the myoB Heavy Chain in the myoA−/B− Double Mutant
The myoA−/B− mutants are defective in fluid-phase pinocytosis and F-actin rearrangement, in addition to having a streaming defect (Novak et al., 1995). The full-length myoB heavy chain was expressed in these cells. Mutant forms of myoB in which the TEDS rule site at serine 332 (myoB-S332A) had been changed to an alanine and a truncated myoB lacking the 3′ SH3 domain (myoB/SH3−) were also expressed in the myoA−/B− cell line to test their ability to rescue the double mutant phenotype. The myoA−/B− cell line was cotransformed with either the myoB expression plasmids pDTb18, pDTb42, and pDTb39 along with a plasmid carrying the gene for blasticidin resistance (Figure 1). Clones were initially observed to overexpress a three to fivefold excess of either wild-type or mutant forms of myoB (our unpublished observations). The clones were analyzed weekly for the level of expressed wild-type myoB and mutant heavy chains. After 2–3 wk of passage, clones expressing wild-type levels of either the full-length myoB heavy chain, myoB-S332A, or myoB/SH3− were obtained, as determined by quantitative immunoblotting (Figure 3). These clones were named myoB, myoB-S332A, and myoB/SH3− expressors. A total of three independent clones for each cell type was analyzed in detail. Growth rate and pinocytosis results are data averaged from three independent clones, whereas all other results are representative examples of repeated observations.
Full-Length, But Not Mutant Forms of myoB Reverse the Myosin I Double Mutant Pinocytic and Growth Defects
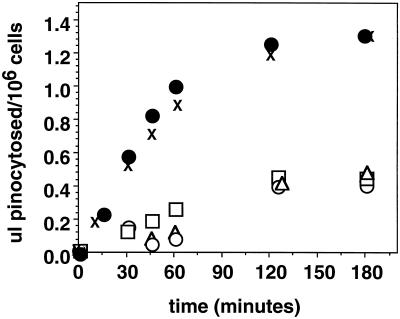
The most striking defect observed in the myoA−/B− double mutant was a decreased rate of pinocytosis in suspension-grown cells (Novak et al., 1995). The fluid-phase marker FITC-dextran was used to measure pinocytosis (Klein and Satre, 1986) in double mutants expressing full-length and mutant forms of myoB (Figure 4). Wild-type cells grown and assayed for pinocytosis in suspension steadily accumulated approximately 0.8–0.9 μl of FITC-dextran/106 cells over the course of 1 h, reaching a fluid internalization plateau of 1.3 μl/106 cells after 2 h (Figure 4). The myoA−/B− mutant, however, only accumulated 0.3 μl of FITC-dextran/106 cells by 60 min, a decrease of approximately 60%, and reached a fluid internalization plateau of 0.45 μl/106 cells after 2.0 h (Figure 4), as previously shown (Novak et al., 1995). Expression of the full-length myoB heavy chain in the myosin I double mutant cells fully rescued the pinocytic defect observed in the suspension-grown double mutant. The myoB-expressing cells internalized 0.9 μl of FITC-dextran/106 cells over the course of 1 h, and 1.3 μl/106 cells after 3 h (Figure 4), values comparable to those observed for wild-type cells. However, the myoB-S332A or myoB/SH3−-expressing cells did not exhibit wild-type levels of fluid-phase pinocytosis, instead they internalized fluid identically to myoA−/B− cells (Figure 4).
Figure 4.
Expression of myoB rescues the double mutant pinocytic defect. The uptake of FITC-dextran by Ax3 (×), myoA−/B− (□), myoB− (•); myoB-S332A (○), and myoB/SH3− expressors (▵) over a 3-h time course.
Expression of full-length myoB also restored the ability of myoA−/B− cells to grow in suspension. The wild-type Ax3 cells grew with a doubling time of 9–11 h at log phase in suspension culture and reached a saturation density of 1 × 107 cells/ml. In contrast, the myoA−/B− double mutant grown in suspension exhibited a doubling time of 22–24 h at log phase and saturated growth at a lower cell density, 2.5 × 106 cells/ml. Expression of the myoB heavy chain in the myoA−/B− double mutant rescued the growth defect, the cell density doubled every 8–11 h, depending on the clone, and saturated growth occurred at 1 × 107 cells/ml. Expression of the myoB/SH3− or myoB-S332A heavy chains in the double mutant had no effect on the double mutant growth rate. The myoB-S332A-expressing cells exhibited a doubling time of 24 h at log phase and their growth reached saturation at a density of 4.25 × 106 cells/ml. The myoB/SH3−-expressing cells exhibited a doubling time of 28 h at log phase and saturated growth at a lower cell density, 3.0 × 106 cells/ml.
Full-Length But Not Mutant Forms of myoB Rescued Defects in F-Actin Rearrangements
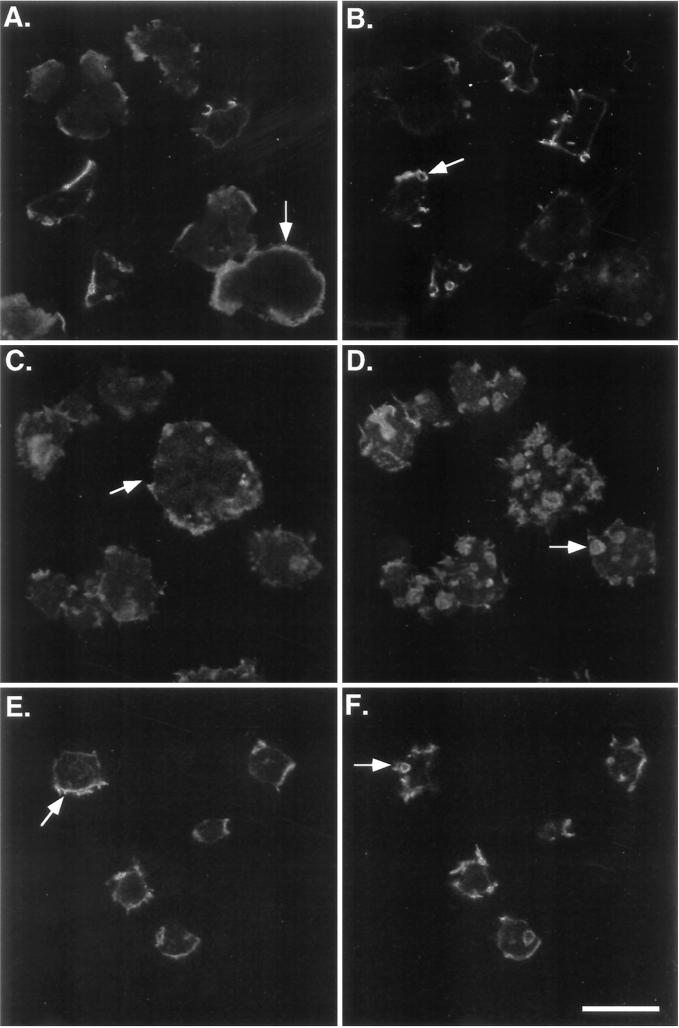
The myoA−/B− cells exhibited delays in the rearrangement of cortical actin that were not observed in either the myoA− or myoB− single mutants (Novak et al., 1995). Expression of the myoB heavy chain in these cells resulted in the recovery of the normal reorganization of F-actin in cells as they attach to a substrate, as demonstrated by rhodamine-phalloidin staining. Ax3 cells taken from suspension cultures and allowed to attach to coverslips for 15 min exhibited a typical pattern of staining, with a thin peripheral band of F-actin present at the base of the cells (Figure 5A, arrow) and a few, small brightly staining projections and one to two crowns per cell at the top (Figure 5B, arrow). In contrast, the myoA−/B− double mutants had few of the basal bands or rings of stain, instead possessing diffuse cytoplasmic staining and bright spots of F-actin at the periphery (Figure 5C, arrow). The myoA−/B− double mutant cells also had numerous brightly staining crowns on their apical surfaces (Figure 5D, arrow) as previously observed (Novak et al., 1995). Expression of full-length myoB heavy chain reversed the defect in F-actin distribution (Figure 5, E and F). Attaching cells localized F-actin at the bases of cells identical to Ax3 cells (compare Figure 5E with 5A) and only formed a few F-actin crowns at the apical surface (Figure 5F). Cells with numerous crowns or other actin-filled projections observed in myoA−/B− double mutant samples were only rarely observed.
Figure 5.
Expression of full-length myoB rescues the myoA−/B− F-actin defect. Shown are confocal images of rhodamine-phalloidin-stained cells that have been allowed to attach to a substrate for 15 min. Images of the bottom (A, C, and E) and top (B, D, and F) of the cells are presented. The pattern of F-actin distribution in the Ax3 (A and B), myoA−/B− (C and D), and double mutant myoB-expressing strains (E and F) are shown. Examples of the peripheral band staining at the base of the cells (A and E) or brightly staining peripheral spots of F-actin on mutant cells (C) and apical crowns (B, D, and F) are indicated with arrows. Bar, 10 μm.
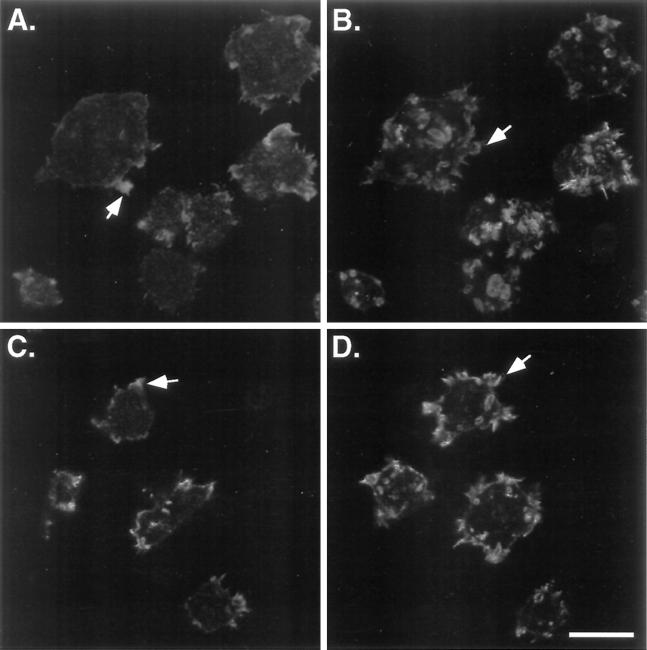
The myoB-S332A and myoB/SH3− expressors did not exhibit the wild-type F-actin distribution. Instead, their F-actin staining pattern closely resembled that of the myoA−/B− cells. The myoB-S332A and myoB/SH3− expressors possessed diffuse F-actin staining at the base of the cells, localizing F-actin only in bright spots at the periphery (Figure 6, A and C, arrows), and not the thin band of peripheral stain observed for Ax3 or myoB-expressing cells (compare Figure 6A and C with Figure 5A and E). The apical surfaces of myoB-S332A (Figure 6B) and myoB/SH3− (Figure 6D)-expressing cells were covered with crowns and other F-actin-filled projections, much like the tops of myoA−/B− cells (compare Figure 6B and D with Figure 5D).
Figure 6.
The F-actin distribution remains abnormal in the myoA−/B− cells expressing mutant forms of myoB. The distribution of F-actin, as determined by rhodamine-phalloidin staining of cells attached to a substrate for 15 min, in the myoA−/B− cells expressing either myoB/SH3− (A and B) or myoB-S332A (C and D) is shown. Confocal images were taken at the bottom (A and C) and top (B and D) of the cells. Examples of the peripheral band staining at the base of the cells (A) or brightly staining peripheral spots of F-actin in mutant cells (A and C) and apical crowns (B and D) are indicated with arrows. Bar, 10 μm.
Wild-Type and Mutant Forms of myoB Are Localized Properly in Axenic and Chemotaxing Cells
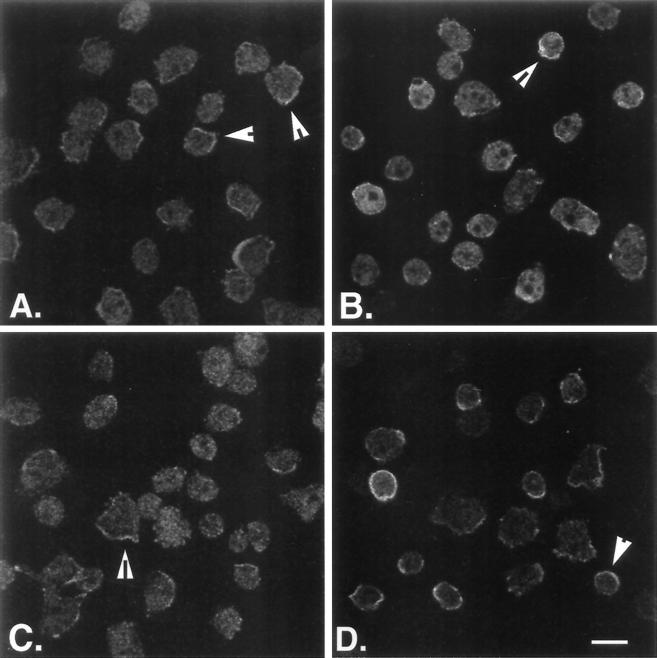
The localization patterns of the wild-type and mutant forms of the myoB heavy chain were also analyzed in the complemented cell lines under both axenic and aggregating conditions. Axenically growing control Ax3 cells were found to have diffuse fluorescence in the cytoplasm, with small amounts of myoB visible at the periphery and in small cellular projections (Figure 7A, arrows). This is in agreement with previously published images of the myoB distribution in axenic cells (Novak et al., 1995). The myoB expressors localized myoB identically to wild-type Ax3 cells (compare Figure 7A and B). The myoB staining in these cells was mostly cytoplasmic, with small areas of peripheral stain (Figure 7B, arrow). The myoB distribution in the myoB-S332A (Figure 7C) or myoB/SH3− (Figure 7D) expressors also closely resembled that of wild-type cells. The mutant forms of the myoB heavy chain were localized primarily to the cytosol, with bright peripheral regions of stain (Figure 7, C and D). Occasionally, myoB/SH3−-expressing cells were observed that exhibited the myoB/SH3-overexpressing staining pattern (compare Figure 7D with 7C in Novak and Titus, 1997).
Figure 7.
myoB, myoB/SH3−, and myoB-S332A are all localized to the periphery of axenic cells. The localization of myoB in Ax3 cells (A) myoA−/B− cells expressing full-length myoB (B), myoB-S332A (C), and myoB/SH3− (D) cells taken from suspension culture and allowed to adhere to a coverslip for 15 min is shown. Confocal images are shown, obtained near the base of cells. The arrowheads in A–D show cells with myoB localized to peripheral regions of the cell. Bar, 10 μm.
Dictyostelium cells undergoing chemotaxis become highly elongated and exhibit concentrated staining of myoB at the leading edge (Fukui et al., 1989; Morita et al., 1996). Although all of the wild-type and mutant forms of the myoB heavy chain appear to be properly localized in axenically growing cells (Figure 7), it was important to observe myoB localization in chemotaxing cells. The signals that cause myosin I to localize to this region of chemotaxing cells are unknown; therefore, it was of interest to determine whether the TEDS rule site or SH3 domain are required for the localization of myoB during this event. The myoB heavy chain was immunolocalized in cells at the preaggregation stage, immediately before the onset of streaming (6 h after starvation for Ax3, 8 h after starvation for cells expressing myoB, myoB/SH3− and myoB-S332A), using the agar overlay technique (Fukui et al., 1987).
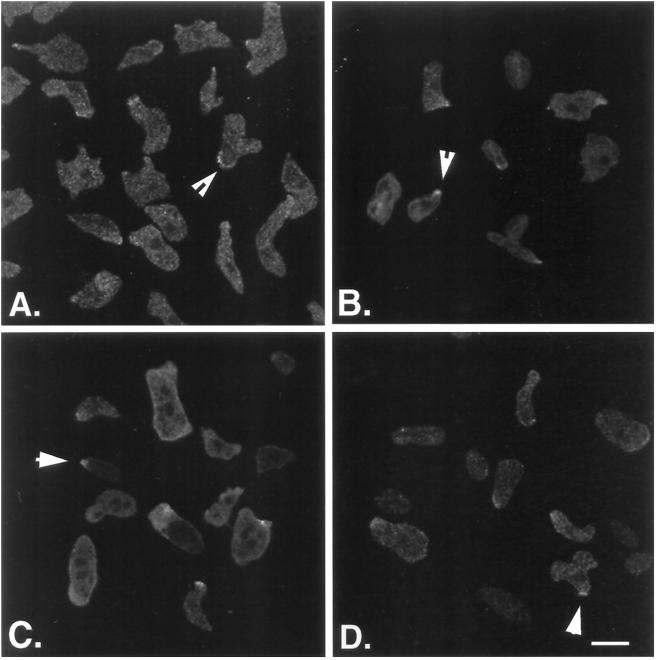
The preaggregation stage Ax3 cells were observed to be highly elongated, with myoB staining in the cytoplasm and also at leading edges (Figure 8A, arrow). Counting at least eight fields of cells for each cell type revealed that 52% of Ax3 cells possessed leading edge myoB staining (n = 125). The myoB-expressing cells were also observed becoming highly elongated, with myoB localized to the cytoplasm and areas of pseudopodial extension (Figure 8B, arrow). Counting the number of myoB-expressing cells with leading edge stain in eight fields of cells revealed that 50% of myoB-expressing cells possessed the leading edge staining pattern (n = 50). The myoB-S332A (Figure 8C) and myoB/SH3− (Figure 8D) expressors exhibited a pattern of myoB heavy chain distribution similar to that of wild-type and myoB-expressing cells (compare Figure 8C and D with 8A and B). Both cytoplasmic and leading edge staining were observed in the cells expressing these mutant forms of myoB (Figure 8, C and D, arrows). Fifty-two percent of the myoB-S332A and 49% of the myoB/SH3− cells localized these mutant forms of myoB to their leading edge (n = 22 and 40, respectively). At this gross level of observation, it appears that neither the phosphorylation site at serine 332 nor the SH3 domain are required for proper localization of myoB in axenic or chemotaxing cells.
Figure 8.
myoB, myoB/SH3−, and myoB-S332A are localized to the leading edge of chemotaxing cells. The localization of myoB in Ax3 cells (A) and myoA−/B− cells expressing full-length myoB (B), myoB/SH3− (C), and myoB-S332A (D) is shown. The cells were taken from suspension culture and allowed to adhere to a coverslip for 15 min prior to processing for immunomicroscopy. Confocal images are shown, obtained near the base of cells. The arrowheads in A–D show cells with myoB localized to the leading edges of migrating cells. Bar, 10 μm.
Expression of myoB Does Not Rescue the Streaming Defect of the myoA−/B− Cells
The reintroduction of the myoB heavy chain into the myoA−/B− cells should result in a cell line that is phenotypically indistinguishable from the myoA− single mutant. The most readily detected defect in the myoA− cells is a delay in streaming in submerged culture (Peterson et al., 1995). Ax3 cells began streaming at 8 h and were aggregated into mounds by 10 h (Figure 9, A and B). The myoA−/B− cells, like the myoA− cells, did not begin streaming until 10 h after starvation (Figure 9, C and D) and required 12 h to fully aggregate into mounds (our unpublished observations; Novak et al., 1995; Peterson et al., 1995). The myoB-expressing cells behaved as predicted. Their streaming was delayed by 2 h, and they did not begin aggregating until 10 h (Figure 9, E and F). Thus, the reintroduction of the myoB heavy chain into the myoA−/B− cells resulted in cells with a phenotype identical to that of the myoA− single mutant cells, a 2-h delay in streaming, but normal growth, pinocytosis, and F-actin distribution (Figures 4 and 5).
Figure 9.
Expression of myoB in myoA−/B− cells has no effect on streaming. The motility of the myoB-expressing cells was assessed using a submerged streaming assay. Shown are Ax3 cells (A and B), myoA−/B− mutant cells (C and D), and myoB expressors (E and F) at 8 (A, C, and E) and 11 h (B, D, and F) after starvation. Bar, 100 μm.
Analysis of S332A-myoB and myoB/SH3− Triton-insoluble Cytoskeletons
Our observations suggested that the mutation of serine 332 to alanine or the removal of the SH3 domain did not grossly alter the localization of these forms of myoB. It was of interest to determine whether or not these mutations affected the ability of myoB to interact normally with F-actin. This was tested by analyzing the fractionation of the myoB heavy chain in Triton-insoluble cytoskeletons and then determining whether or not the mutated forms of the myoB heavy chains could be released from the actin cytoskeleton by the addition of Mg-ATP (Manstein and Hunt, 1995).
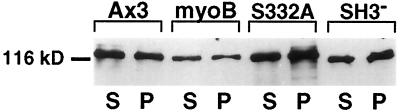
Cells were lysed by the addition of 0.5% Triton X-100, incubated at room temperature for 10 min, and aliquots were collected by centrifugation. Quantitative immunoblotting was used to determine what proportion of the total myoB was present in the Triton-soluble and Triton-insoluble fractions. Nearly equal amounts of the expressed myoB heavy chain were found in the Triton-soluble and insoluble fractions (44.2 ± 14.0% versus 55.8 ± 14.0%; n = 7). A slightly higher amount of the S332A-myoB (54 ± 13.9% in the supernatant and 46 ± 13.9% in the pellet; n = 3) and the myoB/SH3− heavy chains (59 ± 12.1% in the supernatant and 41 ± 12.2% in the pellet; n = 6) were found in the Triton-soluble fractions. Although there appears to be more of the S332A-myoB and myoB/SH3− heavy chains in the supernatant than observed for the myoB heavy chain, these differences were not statistically significant (p > 0.05). Therefore, both the S332A-myoB and myoB/SH3− are present in the actin present in the Triton-insoluble cytoskeleton to the same extent as the wild-type myoB heavy chain.
All myosins share the common property of exhibiting ATP-sensitive binding to actin. This can be quickly assayed in crude lysates and provides a measure of the functional properties of a given myosin. The Triton-insoluble cytoskeletons from each cell line were collected, homogenized in the presence of 10 mM Mg-ATP, and subjected to centrifugation to determine whether a given mutation of the myoB heavy chain altered its interaction with actin. Equal amounts of the Mg-ATP-treated supernatant and pellet were analyzed for the presence of the myoB heavy chain using immunoblotting. The myoB, S332A-myoB, and myoB/SH3− heavy chains were all equally capable of being released from the cytoskeleton by the addition of Mg-ATP (Figure 10).
Figure 10.
Expressed myoB, myoB/SH3−, and myoB-S332A all exhibit ATP-sensitive binding to actin. The Mg-ATP release of the myoB heavy chain from the Triton-insoluble fraction obtained from a rigor cytoskeleton was analyzed using immunoblotting. Shown are equal amounts of the supernatant (S) and pellet (P) fractions derived from the Ax3 myoB, S332A-myoB (S332A), and myoB/SH3− (SH3−)-expressing cells.
DISCUSSION
Complementation of both the myoB− single mutants and myoA−/B− double mutants with the myoB gene has shown that the phenotypes of these mutants are fully reversible. Expression of wild-type levels of the full-length Dictyostelium myoB heavy chain in the myoB− null cell line was able to rescue its delayed streaming phenotype (Figure 2). These results demonstrated that the defects observed in the myoB− single mutant are solely due to the absence of myoB. Consistent with this finding, the expression of myoB in myoA−/B− cells (Figure 3) also rescued the phenotypes unique to these mutant cells, allowing them to grow normally in suspension, undergo normal rates of fluid-phase pinocytosis, and rearrange F-actin properly (Figures 4 and 5). Therefore, the phenotypes of the myoA−/B− cells, resulting from the deletion of myoB in the myoA− single mutant, are solely due to the lack of the myoB heavy chain.
Alteration of potential regulatory elements of the myoB heavy chain, such as the TEDS rule phosphorylation site or the SH3 domain, renders myoB nonfunctional in vivo (Figures 4 and 6). This occurs in spite of the apparently normal localization of both the myoB-S332A and myoB/SH3− heavy chains (Figures 7 and 8), suggesting that neither the SH3 domain nor phosphorylation at the TEDS rule site are required for proper localization of this amoeboid myosin I. These results confirm and extend the previous observations made during overexpression analysis of myoB in Dictyostelium cells (Novak and Titus, 1997). Localization studies in Acanthamoeba revealed that both phosphorylated and nonphosphorylated forms of myosin Is are present in the cytosol and at the plasma membrane (Baines et al., 1995), also demonstrating that phosphorylation is not required for myosin I localization to the plasma membrane.
In contrast to the profound defects observed when the native myoB heavy chain was overexpressed by three to fivefold, overexpression of the myoB-S332A and myoB/SH3− heavy chains did not affect the behavior of the wild-type cells (Novak and Titus, 1997). The results presented in this report, however, more clearly define the extent of the defective function of the two mutant forms of myoB and suggest that the SH3 domain may not be required for the localization of myoB.
The S332A myoB heavy chain was predicted to be nonfunctional in vivo, since both Acanthamoeba and Dictyostelium amoeboid myosin Is absolutely require heavy chain phosphorylation for full activity in vitro (Brzeska and Korn, 1996). Kinetic analysis of dephosphorylated Acanthamoeba myosin I has shown that phosphorylation does not alter the strength of the interaction between myosin I and actin (Ostap and Pollard, 1996a). Rather, it is likely to affect the rate-limiting phosphate release step. Mutation of the TEDS rule phosphorylation site would thus be predicted to alter the ability of myoB to function as a motor, but not affect its localization (Figures 7 and 8) or ability to bind to actin (see RESULTS), as we have observed. Identification and characterization of the kinase responsible for myoB heavy chain phosphorylation would provide more information about how the activity of this motor protein is regulated. A MIHCK specific for the Dictyostelium myoD heavy chain has been identified and characterized, but it does not appear to function as the myoB heavy chain kinase (Lee and Côté, 1995). This is unexpected in light of the finding that the Acanthamoeba MIHCK can phosphorylate both the Dictyostelium myoB and myoD heavy chains (Côtéet al., 1985; Lee and Côté, 1995) and the recent report that Ste20p and ClaIp can phosphorylate the Dictyostelium myoD heavy chain (Wu et al., 1996). The reason for this discrepancy remains to be determined, but it raises the possibility that there is a separate myoB heavy chain kinase that is distinct from the other members of the Ste20/MIHCK family.
The role of the SH3 domain in amoeboid myosin I function is not known. However, removal of the C-terminal myoB SH3 domain renders this myosin I nonfunctional in vivo (Figures 4 and 6, A and B) without affecting its normal localization (Figure 8C). These results demonstrate that the SH3 domain is essential for myosin I function and raise intriguing questions about how it contributes to amoeboid myosin I function. SH3 domains have been found to play key roles in mediating protein–protein interactions important for intracellular signaling events (Pawson and Gish, 1992). Proteins such as Grb2 and Crk are composed almost entirely of SH2 and SH3 domains and function as molecular adapters that nucleate formation of protein complexes important for signaling pathways (Pawson, 1995). The SH3 domains are also found in proteins associated with the actin cytoskeleton, such as α-fodrin (Merilainen et al., 1993) and α-spectrin (Wasenius et al., 1987; Sahr et al., 1990). Therefore, SH3 domains are believed to be involved in bringing together signal transduction proteins and their targets in the membrane cytoskeleton. The SH3 domain could play a role in mediating the interaction between the MIHCK and myoB. The MIHCKs contain a sequence known as the PXXP motif that binds to SH3 domains (Brzeska et al., 1996; Lee et al., 1996). Loss of the SH3 domain from the myoB heavy chain may result in reduced MIHCK binding and less efficient phosphorylation of the TEDS rule site. This could explain our observation that the SH3− myoB behaves in the same manner as myoB-S332A in vivo. However, it should be noted that the Acanthamoeba MIHCK efficiently phosphorylates a nine-amino acid peptide derived from the TEDS rule region in vitro, and that the Km of the kinase for this substrate is not significantly changed when compared with intact myosin I (Brzeska et al., 1990). Future experiments analyzing the in vivo phosphorylation levels of the SH3− myoB heavy chain will allow us to directly determine whether the SH3 domain plays a role in heavy chain phosphorylation.
Alternatively, the SH3 domain may be required for myoB to interact with other proteins necessary for its contractile activity at the cell cortex. The discovery of a complex of proteins that binds specifically to the SH3 domain of Acanthamoeba myosin IC (Xu et al., 1995) suggests that interactions with proteins other than a MIHCK might influence the ability of myoB to function in vivo. The protein that binds to the Acanthamoeba myosin IC SH3 domain, Acan125, contains two tandem consensus PXXP motifs, a leucine-rich repeat (sites for ligand binding), and is related to a protein of unknown function found within the Caenorhabditis elegans cosmid K07G5.1 (Xu et al., 1997). The SH3 domain has also been shown to play a role in the assembly of protein complexes in other systems. For example, the yeast cytoskeletal protein Abp1p has been shown to bind to the adenylyl cyclase-associated protein Srv2p and the actin-associated protein Rvs167p via SH3 domains in vitro (Lila and Drubin, 1997). Through SH3 domains, Abp1p is believed to localize Srv2p and Rvs167p to a complicated protein complex at the cell cortex necessary for proper cortical patch organization (Lila and Drubin, 1997). Such a complex of proteins may be required for the function of myosin I. A kinetic analysis of Acanthamoeba myosin I (Ostap and Pollard, 1996a) strongly suggested that it is necessary for myosin I to be locally concentrated to effectively generate movement along actin. A Dictyostelium homologue of Acan125 may play a role in generating myosin I mulitmers by binding the SH3 domain of several myosin I molecules. Loss of the SH3 domain may not affect the localization of myosin I but could prevent it from forming a complex with other myosin I molecules.
Cytoskeletal proteins such as myosin Is and other actin-binding proteins are likely to work together to organize the location and timing of cellular projections. The composition of these protein complexes, along with their methods of localization and activation, are beginning to be discovered. Demonstration of the in vivo requirement for the myoB TEDS rule phosphorylation site and SH3 domain has offered some clues as to the method by which myosin I activity is controlled. Discovery and characterization of the factors that interact with the myosin I SH3 domains, such as Acan125, will enable us to determine how the activity of these motors is regulated. The complementation system we have established using the Dictyostelium myoA−/B− cell line will also allow us to identify new, functionally important regions of myoB, along with those of other amoeboid myosin Is. Future experiments using this system will provide clues to the exact molecular mechanism by which myosin Is control F-actin reorganization.
ACKNOWLEDGMENTS
The authors thank Drs. Dan Kiehart, Arturo DeLozanne, and Terry O’Halloran for many helpful discussions. The continuing support and encouragement of Dr. Mike Sheetz is also gratefully acknowledged. Thanks also to Dr. Kazuo Sutoh (University of Tokyo, Tokyo, Japan) for supplying us with pUCBsr and its derivatives and to Dr. Yoshio Fukui (Northwestern University, Chicago, IL) for graciously showing us how to perform the agar overlay and myoB immunolocalization techniques. The work described in this article was supported by the March of Dimes, the National Institutes of Health, and the National Science Foundation. M.A.T. is a member of the Duke Comprehensive Cancer Center.
REFERENCES
- Adams RJ, Pollard TD. Binding of myosin I to membrane lipids. Nature. 1989;340:565–568. doi: 10.1038/340565a0. [DOI] [PubMed] [Google Scholar]
- Albanesi JP, Fujisaki H, Hammer JA, III, Korn ED, Jones R, Sheetz MP. Monomeric Acanthamoeba myosins I support movement in vitro. J Biol Chem. 1985;260:8649–8652. [PubMed] [Google Scholar]
- Baines IC, Corigliano-Murphy A, Korn ED. Quantification and localization of phosphorylated myosin I isoforms in Acanthamoeba castellanii. J Cell Biol. 1995;130:591–603. doi: 10.1083/jcb.130.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bement WM, Mooseker MS. TEDS rule: a molecular rationale for differential regulation of myosins by phosphorylation of the heavy chain head. Cell Motil Cytoskeleton. 1995;31:87–92. doi: 10.1002/cm.970310202. [DOI] [PubMed] [Google Scholar]
- Brzeska H, Korn ED. Regulation of class I and class II myosins by heavy chain phosphorylation. J Biol Chem. 1996;271:16983–16986. doi: 10.1074/jbc.271.29.16983. [DOI] [PubMed] [Google Scholar]
- Brzeska H, Lynch TJ, Martin B, Corigliano-Murphy A, Korn ED. Substrate specificity of Acanthamoeba myosin I heavy chain kinase as determined with synthetic peptides. J Biol Chem. 1990;265:16138–16144. [PubMed] [Google Scholar]
- Brzeska H, Szczepanowska J, Hoey J, Korn ED. The catalytic domain of Acanthamoeba myosin I heavy chain kinase. II. Expression of active catalytic domain and sequence homology to p21-activated kinase (PAK) J Biol Chem. 1996;271:27056–27062. doi: 10.1074/jbc.271.43.27056. [DOI] [PubMed] [Google Scholar]
- Cohen SM, Knecht D, Lodish HF, Loomis WF. DNA sequences required for expression of a Dictyostelium actin gene. EMBO J. 1986;5:3361–3366. doi: 10.1002/j.1460-2075.1986.tb04651.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cope MJTV, Whisstock J, Rayment I, Kendrick-Jones J. Conservation within the myosin motor domain: implications for structure and function. Structure. 1996;4:969–987. doi: 10.1016/s0969-2126(96)00103-7. [DOI] [PubMed] [Google Scholar]
- Côté GP, Albanesi JP, Ueno T, Hammer JA, III, Korn ED. Purification from Dictyostelium discoideum of a low-molecular weight myosin that resembles myosin I from Acanthamoeba castellanii. J Biol Chem. 1985;260:4543–4546. [PubMed] [Google Scholar]
- Doberstein SK, Pollard TD. Localization and specificity of the phospholipid and actin binding sites on the tail of Acanthamoeba myosin IC. J Cell Biol. 1992;117:1241–1249. doi: 10.1083/jcb.117.6.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui Y, Lynch TJ, Brzeska H, Korn ED. Myosin I is located at the leading edge of locomoting Dictyostelium amoebae. Nature. 1989;341:328–331. doi: 10.1038/341328a0. [DOI] [PubMed] [Google Scholar]
- Fukui Y, Yumura S, Yumura TK. Agar-overlay immunofluorescence: high-resolution studies of cytoskeletal components and their changes during chemotaxis. Methods Cell Biol. 1987;28:347–356. doi: 10.1016/s0091-679x(08)61655-6. [DOI] [PubMed] [Google Scholar]
- Howard PK, Ahern KG, Firtel RA. Establishment of a transient expression system for Dictyostelium discoideum. Nucleic Acids Res. 1988;16:2613–2623. doi: 10.1093/nar/16.6.2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung G, Fukui Y, Martin B, Hammer JA., III Sequence, expression pattern, intracellular localization and targeted disruption of the Dictyostelium myosin ID heavy chain isoform. J Biol Chem. 1993;268:14981–14990. [PubMed] [Google Scholar]
- Jung G, Hammer JA., III Generation and characterization of Dictyostelium cells deficient in a myosin I heavy chain isoform. J Cell Biol. 1990;110:1955–1964. doi: 10.1083/jcb.110.6.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung G, Hammer JA., III The actin binding site in the tail domain of Dictyostelium myosin IC (myoC) resides within the glycine- and proline-rich sequence (tail homology 2) FEBS Lett. 1994;342:197–202. doi: 10.1016/0014-5793(94)80500-8. [DOI] [PubMed] [Google Scholar]
- Jung G, Wu X, Hammer JA., III Dictyostelium mutants lacking multiple classic myosin I isoforms reveal combinations of shared and distinct functions. J Cell Biol. 1996;133:305–323. doi: 10.1083/jcb.133.2.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein G, Satre M. Kinetics of fluid-phase pinocytosis in Dictyostelium discoideum amoebae. Biochem Biophys Res Commun. 1986;138:1146–1152. doi: 10.1016/s0006-291x(86)80402-8. [DOI] [PubMed] [Google Scholar]
- Kuspa A, Loomis WF. Tagging developmental genes in Dictyostelium by restriction enzyme-mediated integration of plasmid DNA. Proc Natl Acad Sci USA. 1992;89:8803–8807. doi: 10.1073/pnas.89.18.8803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Egelhoff TT, Mahasneh A, Côté GP. Cloning and characterization of a Dictyostelium myosin I heavy chain kinase activated by Cdc42 and Rac. J Biol Chem. 1996;271:27044–27048. doi: 10.1074/jbc.271.43.27044. [DOI] [PubMed] [Google Scholar]
- Lee SF, Côté GP. Purification and characterization of a Dictyostelium protein kinase required for the actin activation of the Mg2+ATPase activity of Dictyostelium myosin ID. J Biol Chem. 1995;270:11776–11782. doi: 10.1074/jbc.270.20.11776. [DOI] [PubMed] [Google Scholar]
- Lila T, Drubin DG. Evidence for physical and functional interactions among two Saccharomyces cerevisiae SH3 domain proteins, an adenylyl cyclase-associated protein and the actin cytoskeleton. Mol Biol Cell. 1997;8:367–385. doi: 10.1091/mbc.8.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch TJ, Albanesi JP, Korn ED, Robinson EA, Bowers BA, Fujisaki H. ATPase activities and actin binding properties of subfragments of Acanthamoeba myosin IA. J Biol Chem. 1986;261:17156–17262. [PubMed] [Google Scholar]
- Manstein DJ, Hunt DM. Overexpression of myosin motor domains in Dictyostelium: screening of transformants and purification of affinity tagged proteins. J Muscle Res Cell Motil. 1995;16:325–332. doi: 10.1007/BF00121141. [DOI] [PubMed] [Google Scholar]
- Merilainen J, Palovuori R, Sormunen R, Wasenius VM, Lehto VP. Binding of the alpha-fodrin SH3 domain to the leading lamellae of locomoting chicken fibroblasts. J Cell Sci. 1993;105:647–654. doi: 10.1242/jcs.105.3.647. [DOI] [PubMed] [Google Scholar]
- Miyata H, Bowers B, Korn ED. Plasma membrane association of Acanthamoeba myosin I. J Cell Biol. 1989;109:1519–1528. doi: 10.1083/jcb.109.4.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooseker MS, Cheney RE. Unconventional myosins. Annu Rev Cell Dev Biol. 1995;11:633–675. doi: 10.1146/annurev.cb.11.110195.003221. [DOI] [PubMed] [Google Scholar]
- Morita YS, Jung G, Hammer JA, III, Fukui Y. Localization of Dictyostelium myoB and myoD to filopodia and cell-cell contact sites using isoform-specific antibodies. Eur J Cell Biol. 1996;71:371–379. [PubMed] [Google Scholar]
- Novak KD, Peterson MD, Reedy MC, Titus MA. Dictyostelium myosin I double mutants exhibit conditional defects in pinocytosis. J Cell Biol. 1995;131:1205–1221. doi: 10.1083/jcb.131.5.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak KD, Titus MA. Myosin I overexpression impairs cell migration. J Cell Biol. 1997;136:633–647. doi: 10.1083/jcb.136.3.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostap EM, Pollard TD. Biochemical kinetic characterization of the Acanthamoeba myosin I ATPase. J Cell Biol. 1996a;132:1053–1060. doi: 10.1083/jcb.132.6.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostap EM, Pollard TD. Overlapping functions of myosin-I isoforms? J Cell Biol. 1996b;133:221–224. doi: 10.1083/jcb.133.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson B, Spudich JA. A novel positive selection for identifying cold-sensitive myosin II mutants in Dictyostelium. Genetics. 1995;140:505–515. doi: 10.1093/genetics/140.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawson T. Protein modules and signalling networks. Nature. 1995;373:573–579. doi: 10.1038/373573a0. [DOI] [PubMed] [Google Scholar]
- Pawson T, Gish G. SH2 and SH3 domains: from structure to function. Cell. 1992;71:359–362. doi: 10.1016/0092-8674(92)90504-6. [DOI] [PubMed] [Google Scholar]
- Peterson MD, Novak KD, Reedy MC, Ruman JI, Titus MA. Molecular genetic analysis of myoC, a Dictyostelium myosin I. J Cell Sci. 1995;108:1093–1103. doi: 10.1242/jcs.108.3.1093. [DOI] [PubMed] [Google Scholar]
- Pollard TD, Doberstein SK, Zot HG. Myosin I. Annu Rev Physiol. 1991;53:653–681. doi: 10.1146/annurev.ph.53.030191.003253. [DOI] [PubMed] [Google Scholar]
- Rosenfeld SS, Rener B. The GPQ-rich segment of Dictyostelium myosin IB contains an actin binding site. Biochemistry. 1994;33:2322–2328. doi: 10.1021/bi00174a045. [DOI] [PubMed] [Google Scholar]
- Sahr KE, Laurila P, Kotula L, et al. The complete cDNA and polypeptide sequences of human erythroid alpha-spectrin. J Biol Chem. 1990;265:4434–4443. [PubMed] [Google Scholar]
- Sussman M. Cultivation and synchronous morphogenesis of Dictyostelium under controlled experimental conditions. Methods Cell Biol. 1987;28:9–29. doi: 10.1016/s0091-679x(08)61635-0. [DOI] [PubMed] [Google Scholar]
- Sutoh K. A transformation vector for Dictyostelium discoideum with a new selectable marker bsr. Plasmid. 1993;30:150–154. doi: 10.1006/plas.1993.1042. [DOI] [PubMed] [Google Scholar]
- Uyeda TQP, Titus MA. The myosins of Dictyostelium. In: Maeda Y, Inouye K, Takeuchi I, editors. Dictyostelium: A Model System for Cell and Developmental Biology. Tokyo, Japan: University Academy Press; 1997. pp. 43–64. [Google Scholar]
- Wasenius VM, Narvanen O, Lehto VP, Saraste M. Alpha-actinin and spectrin have common structural domains. FEBS Lett. 1987;221:73–76. doi: 10.1016/0014-5793(87)80354-x. [DOI] [PubMed] [Google Scholar]
- Wessels D, Murray J, Jung G, Hammer JA, III, Soll DR. Myosin IB null mutants of Dictyostelium exhibit abnormalities in motility. Cell Motil Cytoskeleton. 1991;20:301–315. doi: 10.1002/cm.970200406. [DOI] [PubMed] [Google Scholar]
- Wu C, Lee SF, Furmaniak-Kazmierczak E, Côté GP, Thomas DY, Leberer E. Activation of myosin-I by members of the Ste20p protein kinase family. J Biol Chem. 1996;271:31787–31790. doi: 10.1074/jbc.271.50.31787. [DOI] [PubMed] [Google Scholar]
- Xu P, Mitchelhill KI, Kobe B, Kemp BE, Zot HG. The myosin I binding protein Acan125 binds the SH3 domain and belongs to the superfamily of leucine-rich repeat proteins. Proc Natl Acad Sci USA. 1997;94:3685–3690. doi: 10.1073/pnas.94.8.3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Zot AS, Zot HG. Identification of Acan125 as a myosin-I binding protein present with myosin-I on cellular organelles of Acanthamoeba. J Biol Chem. 1995;270:25316. doi: 10.1074/jbc.270.43.25316. [DOI] [PubMed] [Google Scholar]
- Zot HG, Doberstein SK, Pollard TD. Myosin-I moves actin filaments on a phospholipid substrate: implications for membrane targeting. J Cell Biol. 1992;116:367–376. doi: 10.1083/jcb.116.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]