Abstract
C2-α-Mannosyltryptophan was discovered in human RNase 2, an enzyme that occurs in eosinophils and is involved in host defense. It represents a novel way of attaching carbohydrate to a protein in addition to the well-known N- and O-glycosylations. The reaction is specific, as in RNase 2 Trp-7, but never Trp-10, which is modified. In this article, we address which structural features provide the specificity of the reaction. Expression of chimeras of RNase 2 and nonglycosylated RNase 4 and deletion mutants in HEK293 cells identified residues 1–13 to be sufficient for C-mannosylation. Site-directed mutagenesis revealed the sequence Trp-x-x-Trp, in which the first Trp becomes mannosylated, as the specificity determinant. The Trp residue at position +3 can be replaced by Phe, which reduces the efficiency of the reaction threefold. Interpretation of the data in the context of the three-dimensional structure of RNase 2 strongly suggests that the primary, rather than the tertiary, structure forms the determinant. The sequence motif occurs in 336 mammalian proteins currently present in protein databases. Two of these proteins were analyzed protein chemically, which showed partial C-glycosylation of recombinant human interleukin 12. The frequent occurrence of the protein recognition motif suggests that C-glycosides could be part of the structure of more proteins than assumed so far.
INTRODUCTION
Recently, a new type of protein glycosylation was discovered: the attachment of an α-mannopyranosyl residue to the indole C2 of tryptophan via a C-C link (Hofsteenge et al., 1994; de Beer et al., 1995). This type of linkage is fundamentally different from the ones occurring in protein N- and O-glycosylation. C-mannosylation was originally observed in human ribonuclease (RNase 2) from urine (Hofsteenge et al., 1994). Subsequent studies have shown that Trp-7 in RNase 2 from cellular sources is also C-mannosylated (Löffler et al., 1996; Krieg et al., 1997). The modification reaction in RNase 2 is specific, since Trp-7, but never Trp-10, is modified. The polypeptide chain of RNase 2 comprises 134 residues containing 5 N-linked glycans (Beintema et al., 1988). Its amino acid sequence is identical to that of eosinophil-derived neurotoxin (Rosenberg et al., 1989), which causes the Gordon phenomenon when injected into the cerebellum. This toxicity manifests itself in muscle stiffness, ataxia, and loss of Purkinje cells (Gordon, 1933; Durack et al., 1979; Durack et al., 1981). The three-dimensional structure of the recombinant protein from Escherichia coli has been determined (Mosimann et al., 1996). The overall fold is related to that of bovine pancreatic RNase A, but two regions close to the site of C-mannosylation are different: 1) the N terminus (K-P-P-Q-F-), which forms a type I β-turn that contacts Trp-7; and 2) the large insertion loop (residues 115–123), which packs closely against the N terminus and affects its conformation (Mosimann et al., 1996).
Little is known about the biosynthetic aspects of C-mannosylation. It takes place intracellularly and can be performed by a variety of mammalian cells in culture. In contrast, cells from insects, plant protoplasts, and E. coli do not C-mannosylate recombinant RNase 2 (Krieg et al., 1997). Two major questions are apparent: What are the structural features of RNase 2 that determine the specificity of the reaction? What is the pathway of biosynthesis of (C2-Man-)Trp? Moreover, no experimental evidence exists that shows this reaction to be enzyme catalyzed. RNase 2 is the only example of a C-mannosylated protein so far. Nevertheless, indirect evidence strongly suggests that other proteins carrying this modification exist (Krieg et al., 1997). The search for such proteins would greatly profit from the elucidation of the structural determinant for C-mannosylation.
In this article, we report on the structural features required for the C-mannosylation of Trp-7 in human RNase 2. Toward that aim, recombinant chimeric RNases, consisting of the 17 N-terminal amino acids of human RNase 2 and porcine RNase 4 as well as deletion mutants were analyzed for (C2-Man-)Trp. The exact recognition sequence was determined by site-directed mutagenesis of individual amino acids. The accompanying article (Doucey et al., 1998) addresses the question of the sugar precursors involved in C-mannosylation and the detection of a C-mannosyltransferase. In addition, it confirms the structural determinant found in our research using an in vitro C-mannosylation system and synthetic peptides.
MATERIALS AND METHODS
Taq DNA polymerase, EcoRI, BglII, BamHI, and thermolysin were obtained from Boehringer Mannheim (Mannheim, Germany). Elastase was obtained from Worthington (Freehold, NJ), and endoproteinase Glu-C was purchased from Promega (Madison, WI). The QIAquick Gel Extraction kit was obtained from Qiagen (Hilden, Germany). The C4 and C8 reversed-phase columns were obtained from Vydac (Hesperia, CA). The Sepharose Q and CH-activated Sepharose were purchased from Pharmacia (Uppsala, Sweden). Cell culture media, fetal calf serum, and Lipofectamine reagent were obtained from Life Technologies (Gaithersburg, MD). ECL Western blot analysis reagents were obtained from Amersham Corp. (Arlington Heights, IL).
Antibodies αRNase 2, αRNase 4, and α-(5–10) were made in rabbits and affinity purified as described (Löffler et al., 1996; Krieg et al., 1997).
Construction of Hybrid RNases, SLHV-deleted RNase 2, and Introduction of Point Mutations
To construct the two hybrid RNases, RNase 2.4 and RNase 2 + 4, to introduce the deletion of the four-amino acid sequence SLHV in RNase 2 and the deletion of the nine-amino acid sequence SLHVKPPQF in RNase 2.4, site-directed mutagenesis by overlap extension polymerase chain reaction (PCR) was used as described in Ho et al. (1989). The purified PCR-amplified products were trimmed at the 5′ and 3′ end with EcoRI and BamHI and either ligated into pBluescript vector SK (−) or directly into the expression vector pSMC (Bergwerff et al., 1993). By the same PCR method, residues 1–13 were individually mutated into Ala using the cDNA of RNase 2.4 or RNase 2 as template. Ala-8 was converted into Thr. Because the Ala mutant of Phe-11 was not secreted into the medium, a Leu residue was introduced at this position. The final coding sequence of all constructs was verified by dideoxy sequencing (Sanger et al., 1977).
Cell Culture, Transfection, and Purification of Recombinant RNases
HEK 293 (ATCC CRL 1573) and NIH3T3 (ATCC CCL 92) were cultured and transfected as described (Krieg et al., 1997). Conditioned medium was collected 3 d after transfection and cleared from remaining cells by centrifugation. The medium was passed over a Sepharose Q column equilibrated in 20 mM Tris-HCl (pH 7.5). The recombinant RNase appeared in the flow through and was purified by immunoaffinity chromatography as described previously (Krieg et al., 1997). For the hybrid RNases 2.4 and 2 + 4, an αRNase 4 antibody column was used, whereas ΔSLHV-RNase 2 and point-mutated RNase 2 were isolated by using an αRNase 2 antibody column. Fractions containing recombinant RNase were made 0.1% in trifluoroacetic acid (TFA) and fractionated by reversed-phase high performance liquid chromatography (HPLC) using a 1-mm diameter C4 column equilibrated in 0.1% TFA in water (solvent A). A linear gradient of 0–80% solvent B (70% CH3CN, 0.085% TFA) over 75 min was used to separate the proteins at a flow rate of 0.05 ml/min. Fractions containing RNase were identified by Western blot analysis and pooled.
Protein Chemical and Enzymatic Analysis
For digestion with endoproteinase Glu-C, 5 μg of a particular RNase were lyophilized and dissolved in 5 ml of 0.5 M Tris-HCl (pH 8.6) containing 6 M guanidinium-HCl and 0.2% EDTA. To reduce the protein, 1 ml of 0.5 M dithiothreitol in the same buffer was added, and the solution was incubated for 3 h at 37°C in an argon atmosphere. After cooling to room temperature, 2 ml of 0.55 M iodoacetamide were added to carboxymethylate the protein for 0.5 h in the dark. The mixture was diluted with 120 ml of HEPES-NaOH (pH 7.5) and digested with 0.5 mg of endoproteinase Glu-C overnight at 37°C. Peptides were fractionated and analyzed by liquid chromatography interfaced with electrospray mass spectrometry (LC-ESI-MS) using a Rheos 4000 chromatograph equipped with a 1-mm diameter C8 column and interfaced with a Sciex API 3000 mass spectrometer operating in the multi-ion monitoring mode. The column was equilibrated in 95% solvent A (2% CH3CN, 0.05% TFA), 5% solvent B (80% CH3CN, 0.045% TFA), and a linear gradient was developed from 5 to 40% solvent B in 60 min at a flow rate of 0.05 ml/min. The percentage of modification of a particular RNase was calculated from the ratio of the peak areas of the modified and unmodified peptide −4 to 12. A calibration curve was obtained by digesting and fractionating mixtures containing different ratios of fully C-mannosylated RNase 2/urine and fully unmodified recombinant RNase 2/E. coli. Digestion of the fragment −4 to 12 with elastase (50 ng) was performed in 10 ml of 50 mM NH4HCO3 (pH 8.0). The peptides were fractionated and analyzed as described above but using a gradient of 5–50% buffer B in 20 min. In the case of mutant RNase 2.4 E12A, quantitation of the degree of modification was performed after thermolytic digestion as described (Krieg et al., 1997).
Thermolysin digestion, SDS-PAGE, and Western blotting were performed as described (Löffler et al., 1996). Solid-phase Edman degradation (Pisano et al., 1993) and nanospray ESIMSMS1 (Wilm and Mann, 1996) were performed according to published methods. RNase activity and concentration were determined as described (Vicentini et al., 1994).
Database Searches
The Swiss-Prot (release 34.0) and SP-TrEMBL (release 4.0) databases were searched for mammalian proteins containing the recognition motif W-x-x-W using the program FindPatterns (Genetics Computer Group, Madison, WI). To search for proteins that cross over the endoplasmic reticulum membrane (Doucey et al., 1998), the annotation section of the results was searched with the queries “signal” or “(trans)membrane,” or “precursor,” using the program StringSearch (Genetics Computer Group). The results were inspected manually, one organism was selected in the case of orthologous gene products and false-positives were removed.
RESULTS
C-Mannosylation Does Not Require Entire RNase 2 Molecule
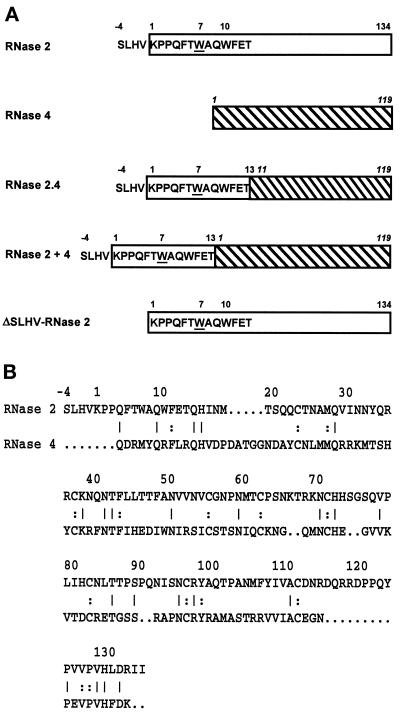
To examine the structural requirements for the specific C-mannosylation of Trp-7 in human RNase 2, a hybrid RNase was constructed in which the signal sequence and the first 17 residues of human RNase 2 were fused to residues 11–119 of porcine RNase 4 (Figure 1A). This RNase is 31% identical to RNase 2 but is not C-mannosylated (Figure 1B). In the RNase 2.4 hybrid, the amino acid sequence following the C-mannosylation site has been changed. Importantly, the insertion loop comprising amino acids 115–125, which in RNase 2 packs closely against the N terminus, is missing. Furthermore, as RNase 4 does not contain N-glycosylation sites in contrast to RNase 2, examination of the hybrid RNase also addresses the question whether N-glycosylation is required for C-mannosylation.
Figure 1.
Structure of hybrid RNases. (A) The RNase 2 portion of a hybrid is depicted as an open rectangle, whereas that of RNase 4 (numbering in italic) is hatched. Trp-7 has been underlined. (B) Comparison of the primary structures of human RNase 2 and porcine RNase 4. Amino acids in common are indicated by a line when they occur at the surface of the protein and by a colon when they are buried.
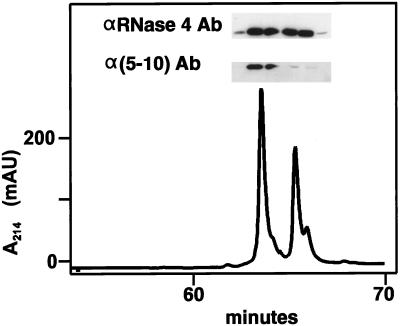
The RNase 2.4 molecule was expressed in HEK 293 cells, which previously have been shown to specifically C-mannosylate Trp-7 in human recombinant RNase 2 (Krieg et al., 1997). The secreted protein was purified from the cell culture medium by a three-step purification procedure using reversed-phase HPLC as the last step. RNase 2.4 eluted from the C4 column in two distinct peaks (Figure 2). The later eluting protein had a mass of 14,515 Da, in agreement with the calculated value for unmodified RNase 2.4, whereas the earlier eluting protein was 162 Da heavier, corresponding to the addition of a hexosyl residue. In Western blot analysis, an αRNase 4-specific antibody bound to the protein in both peaks, whereas the α(5–10) antibody, which specifically recognizes the C-mannosylated form of RNase 2 (Krieg et al., 1997), bound strongly only to the earlier eluting protein. Edman degradation revealed that both proteins start with the sequence SLHV- (position −4 to −1; Figure 1), but only the early eluting one contained (C2-Man-)Trp at position 7. These results demonstrated that RNase 2.4 was partially C-mannosylated at Trp-7 and that the modified and unmodified proteins could be separated by reversed-phase HPLC. In this context, it should be noted that the minor shoulders of the peaks contained RNase 2.4 from which the C-terminal Lys had been cleaved. The C-terminal peptide produced by cleavage with endoproteinase Glu-C at Glu-116 had a mass of 1110 instead of 1238 Da, as revealed by LC-ESIMS analysis. The chemical characterization of the total pool of RNase 2.4 is described below along with that of its mutants.
Figure 2.
Reversed-phase HPLC purification of RNase 2.4. Immunopurified RNase 2.4 was chromatographed on a C4 column (1 mm in diameter) equilibrated in 0.1% TFA. A linear gradient of 0–80% solvent B (70% CH3CN in 0.085% TFA) over 75 min was used at a flow rate of 50 μl/min. The inset shows a Western blot analysis of the fractions using antibodies against RNase 4 or modification-specific antibodies, α(5–10).
To determine whether RNase 2.4 had properly folded, its catalytic properties were examined. The hybrid cleaved yeast RNA at the same rate (ΔA260/ng/ml/min = 1.7 × 10−3) as recombinant porcine native RNase 4 from E. coli (ΔA260/ng/ml/min = 1.6 × 10−3). Also, the second-order rate constants for the cleavage of uridylyl(3′, 5′)adenosine were virtually indistinguishable, 2.6 × 105 M−1s−1 and 2.9 × 105 M−1s−1, respectively.
Residues 1–13 Are Sufficient for C-Mannosylation
Alignment of the sequences of RNase 2.4 and RNase 2 revealed that the two enzymes have, in addition to the 17 N-terminal residues, the same amino acid at several positions scattered throughout the entire length of the polypeptide chain (Figure 1B). To examine whether these amino acids are required for the C-mannosylation of Trp-7, the hybrid RNase 2 + 4 was constructed by fusing the signal peptide and the first 17 amino acids of RNase 2 to the N terminus of RNase 4 (Figure 1). RNase 2, RNase 2.4, and RNase 2 + 4 are only colinear for the first 17 residues. RNase 2 + 4 was transiently expressed in HEK 293 cells and the protein was isolated as outlined above. Western blot analysis using the α(5–10) antibody showed that the RNase 2 + 4 protein is C-mannosylated. In agreement with this, the peptide −4 to 12 generated by digestion with endoproteinase Glu-C had a mass of 2162 Da, as determined by LC-ESIMS. Quantitation of the degree of modification was hampered by the fact that almost half of the protein was proteolytically processed and started at position 8, as determined by Edman degradation. The remaining material started with the sequence SLHV and was C-mannosylated at Trp-7 for 89%. The results obtained with RNases 2.4 and 2 + 4 allow the conclusion that the information for C-mannosylation of Trp-7 lies within residues −4 to 13 of RNase 2.
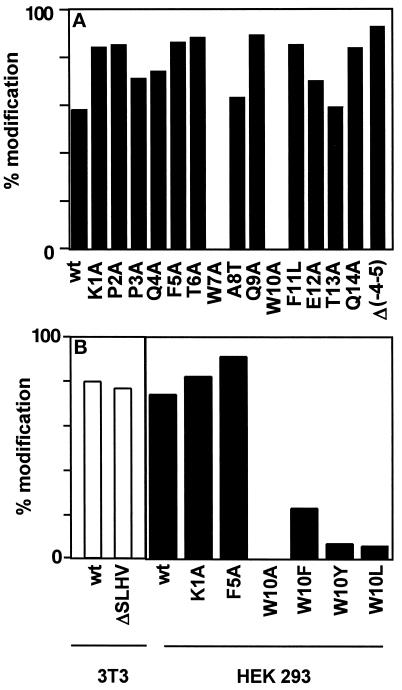
RNase 2 contains a signal sequence for secretion with an unusual C-terminal sequence: SLHV. In nature, both the form with and without this sequence occur and are C-mannosylated (Löffler et al., 1996). Since it is not known when and where the processing of this four-amino acid sequence occurs, it was possible that it is required for the C-mannosylation of Trp-7. To address this possibility, the four amino acids were deleted in RNase 2 and the protein was expressed in NIH 3T3 cells. In Western blot analysis, purified ΔSLHV-RNase 2 protein bound to the α(5–10) antibody, indicating that Trp-7 is modified. Thermolytic digestion and separation of the modified and unmodified peptides by C18 reversed-phase HPLC showed that 79% of the ΔSLHV-RNase 2 is C-mannosylated at Trp-7 compared with 81% in native RNase 2 (Figure 5B). Thus, the information for C-mannosylation of Trp-7 must be contained within residues 1–13 of RNase 2.
Figure 5.
Mutational analysis of RNase 2.4 and RNase 2. (A) The degree of C-mannosylation of Trp-7 in the indicated mutants of hybrid RNase 2.4 was determined from the ratio of modified and unmodified fragments −4 to 12 and is depicted by filled bars. (B) The degree of modification of Trp-7 in the indicated mutants of RNase 2 was determined as in A and are plotted as open bars for the experiment performed in 3T3 cells and as filled bars for the experiment performed in HEK293 cells. The data represent the average of at least two independent experiments. The SD was 1–16% of the mean.
C-Mannosylation of Trp-7 in RNase 2.4 Is Abolished by Mutation of Trp-10 to Ala
Residues 1–13 of RNase 2.4 were mutated individually into Ala (except for Ala-8 and Phe-11, which were mutated into Thr and Leu, respectively) to find out which ones are required for the C-mannosylation of Trp-7. The mutated proteins were expressed in HEK 293 cells and purified as described above. Hybrid RNase 2.4 rather than RNase 2 was used for this analysis to simplify the purification by HPLC. RNase 2 yielded multiple peaks due to heterogeneity in N-glycosylation, whereas RNase 2.4 gave a much simpler pattern. The purity of all mutant proteins was found to be at least 90% by analyzing them on SDS-polyacrylamide gels and scanning of the silver-stained gels. Furthermore, all mutant RNases were catalytically active against yeast RNA, indicating that the enzymes were properly folded.
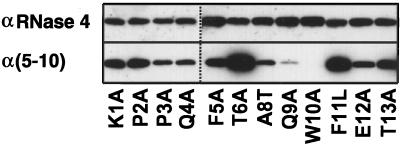
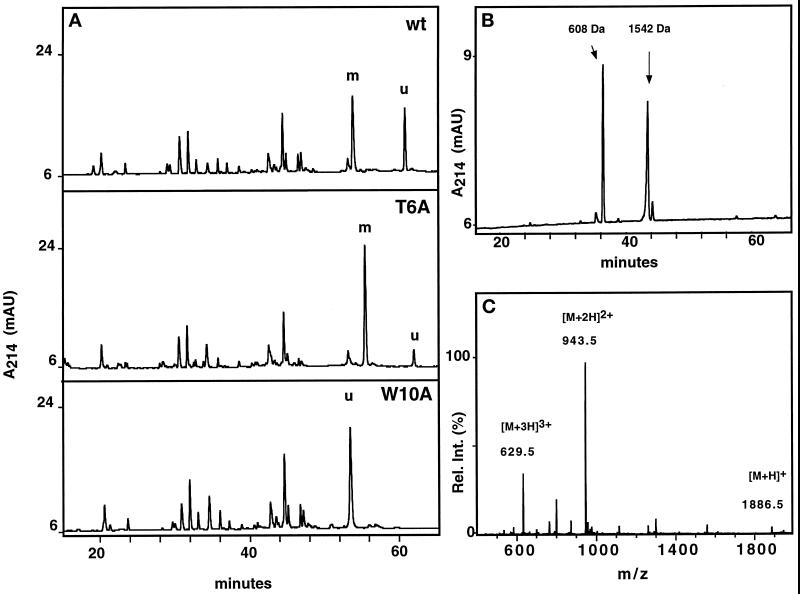
Western blot analysis with α(5–10) antibody revealed that, with the exception of the W10A mutant, all mutants are C-mannosylated (Figure 3), although the mutant Q9A gave a weak signal. To ensure that comparable amounts of protein were loaded, the blots were stripped and reprobed with an αRNase 4 antibody. The degree of C-mannosylation of wild-type RNase 2.4 and each of the mutants, as well as the specificity of the reaction for Trp-7, were determined by peptide mapping using LC-ESIMS. Cleavage with endoproteinase Glu-C yielded two peptides from the region −4 to 12. The results for wild-type RNase 2.4 and a representative example of a mutant (T6A) are shown in Figure 4A (upper and middle panels). The molecular masses of the two fragments readily distinguished them as mannosylated (Figure 4A, m) and unmodified (Figure 4A, “u”). This demonstrated that RNase 2.4 was partially modified, which agrees with the results in Figure 2. The mutant W10A was the only one that yielded a single peptide −4 to 12. Its molecular mass (1885 Da; Figure 4C) showed it not to be mannosylated, which is in agreement with the lack of binding of α(5–10) antibodies to the entire protein (Figure 3). No evidence for the presence of modified peptide could be obtained by extraction of the MS data. The stoichiometry of C-mannosylation of the various RNase 2.4s was calculated from the ratio of the modified and unmodified fragments using a (linear) calibration curve obtained by digestion of mixtures of fully modified RNase 2/urine and fully unmodified RNase 2/E. coli. Compared with wild-type RNase 2.4, which is 58% modified, nearly all mutants showed an increase in modification of Trp-7, which varied from 62% for the mutant T13A up to 89% for the mutant Q9A (Figure 5A). Only the W10A mutation abolished the C-mannosylation of Trp-7 (Figure 5A).
Figure 3.
Western blot analysis of single-site mutants of RNase 2.4. Approximately 0.5 μg of each protein was electrophoresed on a 15% SDS-PAA gel and blotted onto nitrocellulose. The blots were probed with modification-specific antibodies, α(5–10), and, after stripping, with αRNase 4 antibodies.
Figure 4.
Characterization of RNase 2.4 and its mutants. (A) Five micrograms of reduced and carboxymethylated RNase were digested at glutamic acid residues with endoproteinase Glu-C and fractionated by C8 reversed-phase LC-ESIMS. A 1-mm diameter column equilibrated in 95% solvent A (2% CH3CN, 0.05% TFA) and 5% solvent B (80% CH3CN, 0.045% TFA) was used. Peptides were eluted with a linear gradient of 5–40% solvent B at a flow rate of 50 μl/min. C-mannosylated (“m”) and unmodified (“u”) fragment −4 to 12 were assigned based on their molecular masses. The results obtained with wild-type RNase 2.4 (upper panel), mutant T6A (middle panel), and W10A (lower panel) are shown as representative examples. (B) Peptide −4 to 12 was digested with elastase and fractionated by reversed-phase LC-ESIMS. The peptide map obtained from the T6A mutant is shown as a representative example. Peptide 9–12 with unmodified Trp-10, 608 Da; peptide −4 to 8 with C-mannosylated Trp-7, 1542 Da. (C) ESIMS of the fragment −4 to 12 from RNase 2.4, W10A. The molecular mass of 1885 Da corresponds to that of the peptide with unmodified Trp.
To ensure that the specificity for Trp-7 had not changed due to a particular mutation, the modified peptides −4 to 12 were digested with elastase and fractionated by LC-ESIMS. As a representative example, the results for the mutant T6A are presented in Figure 4B. The early eluting peptide (608 Da) comprised residues 9–12, with unmodified Trp-10, whereas the second one (1542 Da) contained residues −4 to 8 with mannosylated Trp-7. These assignments were confirmed by nanospray ESIMSMS analysis. The analysis was performed for all mutants, and in each case it was found that Trp-7, but never Trp-10, was modified. These results were confirmed by solid-phase Edman degradation of the intact proteins. This approach could not be applied to RNase 2.4 containing the E12A mutation, since it eliminated the cleavage site. In this case, the quantitation was obtained from a thermolytic peptide map (Krieg et al., 1997), and the specificity was established by Edman degradation only. Likewise, because elastase cleaves after Ala-8, the specificity of the reaction with the A8T mutant was verified by ESIMSMS analysis of the peptide −4 to 12.
In addition to the single mutations, the mutant Δ(−4 to 5)RNase 2.4, in which the residues forming the type I β-turn of RNase 2 were removed, was analyzed. In this protein Trp was modified for 93% (Figure 5A).
C-Mannosylation of Trp-7 in RNase 2 Is Abolished by Mutation of Trp-10 to Ala But Can Be Partially Rescued by Phe
Because RNase 2.4 is an artificial, albeit enzymatically fully active, enzyme, it was important to verify the effect of some of the mutations in the context of RNase 2. The W10A mutation and two mutations which led to increased C-mannosylation of Trp-7 in RNase 2.4 were also introduced into RNase 2. In addition, the ability of other aromatic or hydrophobic amino acids to function as a signal was examined by the mutations W10F, W10Y, and W10L. The mutated RNase 2s were purified from the conditioned medium of transfected HEK 293 cells and analyzed as described above. In this cell line, wild-type RNase 2 is modified to 73%. An increase in C-mannosylation caused by the K1A and F5A mutations was also found in the RNase 2 context (Figure 5B) with a degree of modification of 84 and 88%, respectively. The substitution of Trp-10 by Ala again abolished the C-mannosylation of Trp-7, whereas replacement of Trp-10 by a Tyr or Leu residue reduced the efficiency of mannosylation to 6%, compared with 23% when Phe was introduced (Figure 5B).
DISCUSSION
The results presented here demonstrate that the structural information for the specific C-mannosylation of Trp-7 in human RNase 2 is located within the region 6–13 and consists of the sequence Trp-x-x-Trp (mannosylated residue in bold). The evidence relies on the observation that RNase 2.4, RNase 2 + 4, ΔSLHV-RNase 2 and Δ(−4 to 5) RNase 2.4 (Figure 1) all were C-mannosylated and that their sequences are only colinear for residues 6–13. This excludes that structural features such as the large insertion loop 115–123, which packs against the N terminus, is crucial for C-mannosylation. Furthermore, since RNase 2.4 does not contain N-glycosylation sites, it can be concluded that C-mannosylation does not require prior N-glycosylation. The identification of exact residues required for recognition is based on mutation of each of the residues in the region 1–13 in RNase 2.4 (Figure 5A). Although RNase 2.4 is an artificial protein, it was secreted by the cells in a fully active form, with kinetic parameters indistinguishable from those of RNase 4. This shows that the catalytic center, and most likely the entire molecule was properly folded, making RNase 2.4 a suitable model system. The purified proteins were examined for C-mannosylation using Western blot analysis (Figure 3), Edman degradation, and LC-ESIMS of peptides obtained with thermolysin, endoproteinase Glu-C and elastase (Figure 4). This allowed the quantitation of the degree of C-mannosylation and the unambiguous assignment of the position of modification. Because the W10A mutation was the only one that abolished C-mannosylation, we conclude that a Trp residue at position +3 forms the signal for C-mannosylation of Trp-7. This result was confirmed in the context of RNase 2 (Figure 5B).
The analysis of some mutants by Western blot analysis deserves comment. RNase 2.4 with Q9A displayed strongly decreased binding to the modification-specific α(5–10) antibodies compared with the wild-type enzyme (Figure 3). Nevertheless, protein chemical analysis demonstrated 84% modification of Trp-7. This would be consistent with Gln-9 forming part of the epitope that is recognized by the antibodies, which were raised against the peptide F-T-(C2-Man-)W-A-Q-W (Krieg et al., 1997). The increased binding of the mutants T6A and F11L, modified 88 and 85% at Trp-7, respectively, is more difficult to explain. LC-ESIMS analysis of the peptide maps excluded the possibility that one of the other two Trp residues in RNase 2.4 had been C-mannosylated. It seems that the mutations affect the presentation of the epitope on the membrane to the polyclonal antibody.
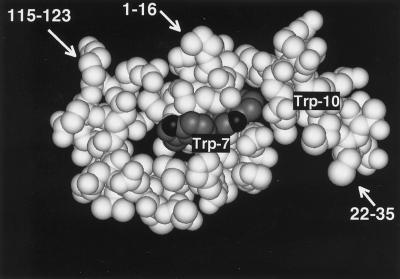
Trp-7 and -10 are located in the N-terminal α-helix of RNase 2 (Mosimann et al., 1996), with the indole moiety of Trp-7 at the surface of the molecule. In contrast, the indole of Trp-10 is largely tucked away between side chains protruding from helices 1 and 2 (Figure 6). This raises the question as to whether Trp-10 interacts directly with the C-mannosyltransferase or whether it is part of a three-dimensional feature in the N terminus of RNase 2 that is being recognized. Trp-10 interacts on the N-terminal side with the type I β-turn formed by residues Pro-2 to Phe-5, and on the C-terminal side with the side chains of Thr-13 and Gln-14 (Mosimann et al., 1996). If the three-dimensional structure was important, presumably the mutation of the residues that contact Trp-10, i.e., Pro-2, Phe-5, Thr-13, and Gln-14 would also decrease the efficiency of C-mannosylation. This was not the case: in fact, removal of all β-turn forming residues, as in Δ(−4 to 5)RNase 2.4, and the mutations P2A, F5A, and Q14A increased the degree of C-mannosylation, whereas the mutation T13A did not have any effect (Figure 5). This is corroborated by the C-mannosylation of the hybrid RNase 2 + 4, where the appended residues are unlikely to have the same ordered structure as present in native RNase 2 or RNase 2.4. Taken together, these results provide strong evidence for a model in which the C-mannosyltransferase interacts directly with the sequence Trp-x-x-Trp in analogy to the Asn-X-Thr/Ser sequon for N-glycosylation.
Figure 6.
Three-dimensional structure around the C-mannosylation site of recombinant RNase 2. The indole moieties of Trp-7 and -10 are shown in dark gray. Since the protein was produced in E. coli, Trp-7 is not C-mannosylated. The coordinates used to produce this figure were obtained from Mosimann et al., 1996.
An alternative model for the specificity of the C-mannosylation reaction would be the existence of a “signal patch” in addition to the sequence Trp-x-x-Trp. A signal patch has been found, among others, for UDP-GlcNAc:lysosomal enzyme N-acetylglucosamine-1-phosphotransferase, where amino acid residues that are separated in the primary structure form a conformation-dependent protein determinant that interacts with the transferase (Reitman and Kornfeld, 1985; Baranski et al., 1990; Baranski et al., 1991). RNase 2 and 4 are 31% identical (Figure 1B) with 18 amino acids in common at the surface that could potentially form a signal patch. Although such a model cannot be excluded based on the in vivo data alone, the data presented in the accompanying article (Doucey et al., 1998) argue strongly against it. The synthetic peptide comprising residues 1–12 of RNase 2 was efficiently C-mannosylated at Trp-7 in vitro, but the mutant peptide with Trp-10 substituted by Ala did not function as a substrate. Importantly, Trp-7 in native recombinant RNase 2 from E. coli was not C-mannosylated in vitro under the same conditions (Doucey et al., 1998). This shows that C-mannosylation can only take place before complete folding of the protein. It excludes the involvement of a conformation-dependent signal patch but confirms that the transferase recognizes the linear determinant Trp-x-x-Trp. In such a model, the improving effect of several of the mutations (Figure 5) could be explained if they decreased the rate of folding. This remains to be tested.
The sequence Trp-x-x-Trp is found in RNase 2 from primates, but does not occur in the enzyme from pig (Iwama et al., 1993) and cow (Irie et al., 1988). Curiously, the residue that forms the signal, i.e., Trp-10, is present in both porcine and bovine RNase 2, but position 7 is occupied by a basic residue. This suggests that C-mannosylation of RNase 2 has evolved relatively recently with the appearance of Trp-7 in the primates (Rosenberg et al., 1995). In contrast, the C-mannosyltransferase activity is not restricted to primates. Recombinant human RNase 2 isolated from porcine, mouse, and other cultured mammalian cells is C-mannosylated (Krieg et al., 1997), and rat liver microsomes can C-mannosylate synthetic peptides in vitro (Doucey et al., 1998). This reinforces the previously formulated hypothesis that other C-mannosylated proteins exist (Krieg et al., 1997).
A search of the Swiss-Prot and TrEMBL databases revealed 12,247 proteins that contain the recognition motif. However, C-mannosyltransferase activity, using RNase 2 as the reporter protein, has only been found in mammalian cells so far (Krieg et al., 1997). This, and the restriction that only proteins that cross the endoplasmic reticulum membrane are likely to be C-mannosylated [see the accompanying article (Doucey et al., 1998], yielded the 336 candidates summarized in Table 1. Two of these have been examined in more detail by peptide mapping using LC-ESIMS. The peptide comprising residues 459–471 from the human fibrinogen Bβ chain, which contains the sequence W-M-N-W, was present in the unmodified form only. Tryptic cleavage of recombinant human interleukin 12β from Chines hamster ovary cells (a generous gift from Dr. A. Stern, Roche Inc., Nutley, NJ) yielded the peptide 314–328 of the 40-kDa subunit, with Trp-319 partially C-glycosylated. The sequence W319-S-E-W present in this peptide is in agreement with that of the recognition motif found in human RNase 2 (Hofsteenge and Hess, unpublished results). This opens the possibility that other proteins containing the motif (Table 1) may be C-mannosylated as well. Their identification would not only establish the generality of this new type of protein glycosylation but also provide ways to determine its possible function.
Table 1.
Summary of mammalian proteins in the Swiss-Prot and SP-TrEMBL protein databases containing the sequence W-x-x-Wa
| Total no. of sequences searched | 163,886 |
| Total no. of proteins containing W-x-x-W | 12,247 |
| Uniqueb mammalian gene products whose cellular localization would allow C-mannosylation: | 336 |
| 1. Receptors | 80 |
| 2. Channels, transporters | 36 |
| 3. Hormones, growth factors | 39 |
| 4. Enzymes: (proteases, glycanohydrolases, lipases, oxidases, glycosyltransferases, kinases) | 70 |
| 5. Cell surface and extracellular matrix proteins | 42 |
| 6. Immunoglobulins, histocompatibility antigens | 22 |
| 7. Others | 47 |
A detailed listing of these proteins is available upon request.
Unique refers to the fact that one organism is counted only in the case of orthologous sequences.
ACKNOWLEDGMENTS
We thank Renate Matthies for sequencing the proteins and Drs. Yoshikuni Nagamine and Jack Rohrer for reading the manuscript.
REFERENCES
- Baranski TJ, Faust PL, Kornfeld S. Generation of a lysosomal targeting signal in the secretory protein pepsinogen. Cell. 1990;63:281–291. doi: 10.1016/0092-8674(90)90161-7. [DOI] [PubMed] [Google Scholar]
- Baranski TJ, Koelsch G, Hartsuck JA, Kornfeld S. Mapping and molecular modeling of a recognition domain for lysosomal enzyme targeting. J Biol Chem. 1991;266:23365–23372. [PubMed] [Google Scholar]
- Beintema JJ, Hofsteenge J, Iwama M, Morita T, Ohgi K, Irie M, Sugiyama RH, Schieven GL, Dekker CA, Glitz DG. Amino acid sequence of the nonsecretory ribonuclease of human urine. Biochemistry. 1988;27:4530–4538. doi: 10.1021/bi00412a046. [DOI] [PubMed] [Google Scholar]
- Bergwerff AA, Oostrum J, Asselbergs AM, Buergi R, Hokke H, Kamerling JP, Vliegenthart FG. Primary structure of N-linked carbohydrate chains of a human chimeric plasminogen activator K2tu-PA expressed in Chinese hamster ovary cells. Eur J Biochem. 1993;212:639–656. doi: 10.1111/j.1432-1033.1993.tb17702.x. [DOI] [PubMed] [Google Scholar]
- de Beer T, Vliegenthart JFG, Löffler A, Hofsteenge J. The hexopyranosyl residue that is C-glycosidically linked to the side chain of tryptophan-7 in human RNase Us is α-mannopyranose. Biochemistry. 1995;34:11785–11789. doi: 10.1021/bi00037a016. [DOI] [PubMed] [Google Scholar]
- Doucey M-A, Hess D, Cacan R, Hofsteenge J. Protein C-mannosylation is enzyme-catalyzed and uses dolichyl-phosphate-mannose as a precursor. Mol Biol Cell. 1998;9:291–300. doi: 10.1091/mbc.9.2.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durack DT, Ackerman SJ, Loegering DA, Gleich GJ. Purification of human eosinophil-derived neurotoxin. Proc Natl Acad Sci USA. 1981;78:5165–5169. doi: 10.1073/pnas.78.8.5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durack DT, Sumi SM, Klebanoff SJ. Neurotoxicity of human eosinophils. Proc Natl Acad Sci USA. 1979;76:1443–1447. doi: 10.1073/pnas.76.3.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon MH. Remarks on Hodgkin’s disease. Br Med J. 1933;1:641–644. doi: 10.1136/bmj.1.3771.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- Hofsteenge J, Müller DR, de Beer T, Löffler A, Richter WJ, Vliegenthart JFG. New type of linkage between a carbohydrate and a protein: C-glycosylation of a specific tryptophan residue in human RNase Us. Biochemistry. 1994;33:13524–13530. doi: 10.1021/bi00250a003. [DOI] [PubMed] [Google Scholar]
- Irie M, Nitta R, Ohgi K, Niwata Y, Watanabe H, Iwama M, Beintema JJ, Sanda A, Takizawa Y. Primary structure of a non-secretory ribonuclease from bovine kidney. J Biochem. 1988;104:289–296. doi: 10.1093/oxfordjournals.jbchem.a122460. [DOI] [PubMed] [Google Scholar]
- Iwama M, Sanda A, Ohgi K, Hofsteenge J, Irie M. Purification and primary structure of a porcine kidney non-secretory ribonuclease. Biosci Biotech Biochem. 1993;57:2133–2138. doi: 10.1271/bbb.57.2133. [DOI] [PubMed] [Google Scholar]
- Krieg J, Gläsner W, Vicentini A, Doucey M-A, Löffler A, Hess D, Hofsteenge J. Protein C-mannosylation is an intracellular process performed by a variety of cultured cells. J Biol Chem. 1997;272:26687–26692. doi: 10.1074/jbc.272.42.26687. [DOI] [PubMed] [Google Scholar]
- Löffler A, Doucey M-A, Jansson AM, Müller DR, de Beer T, Hess D, Meldal M, Richter WJ, Vliegenthart JFG, Hofsteenge J. Spectroscopic and protein chemical analyses demonstrate the presence of C-mannosylated tryptophan in intact human RNase 2 and its isoforms. Biochemistry. 1996;35:12005–12014. doi: 10.1021/bi9610515. [DOI] [PubMed] [Google Scholar]
- Mosimann SC, Newton DL, Youle RJ, James MNG. X-ray crystallographic structure of recombinant eosinophil-derived neurotoxin at 1.83 Å resolution. J Mol Biol. 1996;260:540–552. doi: 10.1006/jmbi.1996.0420. [DOI] [PubMed] [Google Scholar]
- Pisano A, Redmond JW, Williams KL, Gooley AA. Glycosylation sites identified by solid-phase edman degradation: O-linked glycosylation motifs on human glycophorin A. Glycobiology. 1993;3:429–435. doi: 10.1093/glycob/3.5.429. [DOI] [PubMed] [Google Scholar]
- Reitman ML, Kornfeld S. Lysosomal enzyme targeting. J Biol Chem. 1985;256:11977–11980. [PubMed] [Google Scholar]
- Rosenberg HF, Tenen DG, Ackerman SJ. Molecular cloning of the human eosinophil-derived neurotoxin:A member of the ribonuclease gene family. Proc Natl Acad Sci USA. 1989;86:4460–4464. doi: 10.1073/pnas.86.12.4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg HF, Dyer KD, Tiffany HL, Gonzalez M. Rapid evolution of a unique family of primate ribonuclease genes. Nat Genet. 1995;10:219–223. doi: 10.1038/ng0695-219. [DOI] [PubMed] [Google Scholar]
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicentini A, Hemmings BA, Hofsteenge J. Residues 36–42 of liver RNase PL3 contribute to its uridine-preferring substrate specificity. Protein Sci. 1994;3:459–466. doi: 10.1002/pro.5560030311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilm M, Mann M. Analytical properties of the nanoelectrospray ion source. Anal Chem. 1996;68:1–8. doi: 10.1021/ac9509519. [DOI] [PubMed] [Google Scholar]