Abstract
After acute HIV infection, CD8+ T cells are able to control viral replication to a set point. This control is often lost after superinfection, although the mechanism behind this remains unclear. In this study, we illustrate in an HLA-B27+ subject that loss of viral control after HIV superinfection coincides with rapid recombination events within two narrow regions of Gag and Env. Screening for CD8+ T cell responses revealed that each of these recombination sites (∼50 aa) encompassed distinct regions containing two immunodominant CD8 epitopes (B27-KK10 in Gag and Cw1-CL9 in Env). Viral escape and the subsequent development of variant-specific de novo CD8+ T cell responses against both epitopes were illustrative of the significant immune selection pressures exerted by both responses. Comprehensive analysis of the kinetics of CD8 responses and viral evolution indicated that the recombination events quickly facilitated viral escape from both dominant WT- and variant-specific responses. These data suggest that the ability of a superinfecting strain of HIV to overcome preexisting immune control may be related to its ability to rapidly recombine in critical regions under immune selection pressure. These data also support a role for cellular immune pressures in driving the selection of new recombinant forms of HIV.
After acute HIV infection, CD8+ T cell responses contribute to control of viral replication and suppress viral loads to an individual set point (1, 2). Their ability to delay progression to AIDS is strongly associated with specific HLA class I alleles such as HLA-B57 or -B27 (3), which are known to restrict vigorous immunodominant CD8 responses during acute infection against specific regions of HIV (4). Escape from these CD8 responses is associated with a loss of control of viremia, indicating that immune control in such cases can be primarily mediated by a single dominant response (5–7). In the case of HLA-B27, viral control has been strongly associated with responses against the immunodominant KK10 epitope (KRWIILGLNK) in p24 Gag, which may exhibit a unique ability to suppress viral replication (6, 7).
The strong antiviral activity of KK10-specific CD8 responses might be caused by their ability to effectively recognize early viral escape variants. Viral escape typically develops rapidly in the KK10 epitope through a position six L268M escape mutation (7). However, de novo variant-specific CD8 responses against the L268M escape variant are commonly mounted (8, 9), which eventually leads to the subsequent selection of the more potent position 2 escape mutation (R264K) that is associated with the eventual loss of viral control late in chronic infection (7, 10, 11). Therefore, variant-specific responses may play an important role in the control of HIV, enabling prolonged recognition of escaped viruses (12, 13). Unfortunately, the vast diversity of HIV-specific CD8 responses in infected subjects, and the progressive viral escape from these responses, has made it difficult to determine the relative importance of particular responses and escape mutations, either singly or collectively, on viral containment and disease progression.
The ability of the immune system to contain viral replication is also substantially impacted after HIV superinfection. Numerous cases of superinfection have been identified, usually on the basis of a sudden increase in viral loads (14–17). A dramatic shift in the immunodominance patterns of CD8 responses before and after superinfection has also been observed (14); this shift may be related to the transmission of mutations within targeted CD8 epitopes. Although new CD8 responses arise after superinfection, control over viral replication is often lost (14–16), and the factors contributing to the inability of preexisting immune responses to contain the superinfecting strain have as yet not been identified.
HIV superinfection may also enable recombination between two different strains (17), which could facilitate evasion of host immune responses. Recent data suggest that circulating recombinant forms (CRFs) of HIV may be far more common than previously observed (18). New CRFs may have a critical impact on vaccine design as they continue to expand the extensive global diversity of HIV (19). Equally problematic is that there appears to be little or no pattern to the selection of recombination sites within CRFs (15, 20) and the forces governing recombination (15, 21).
In this study, we provide a comprehensive analysis of the forces dictating HIV recombination after superinfection, which rapidly lead to the dramatic loss of viral containment in a subject expressing the otherwise protective MHC class I allele HLA-B27.
RESULTS AND DISCUSSION
Loss of control of viral replication in the setting of HLA-B27
Control of HIV in the presence of HLA-B27 (6, 7, 11, 22) has been attributed to the early and immunodominant targeting of a highly conserved CD8 epitope (KK10; KRWIILGLNK) in Gag. The HLA-B27+ subject AC160, identified during primary HIV infection, rapidly controlled viral replication after a peak viremia of 468,000 copies/ml (Fig. 1). As early as day 22 after presentation, the dominant CD8 response was directed against the KK10 epitope (85 Spot Forming Cells/Mio. [SFC/Mio.]) as measured by IFN-γ ELISpot assay (Table I). Only six other CD8 responses were detected by day 419, with the KK10 response remaining the most immunodominant (Table I). Viral sequencing revealed escape in this epitope through the stereotypic CTL escape mutation L268M, which first developed at day 419 (T3) coincident with a decline in the KK10 WT-specific response (Fig. 2 A and Fig. S1, available at http://www.jem.org/cgi/content/full/jem.20080281/DC1). Viral escape coincided with an initial rise of viral loads to 14,000 copies/ml (Fig. 1), suggesting that loss of this key CD8 response may have partially impaired early viral containment. However, longitudinal testing revealed the subsequent development of an L268M variant-specific response, which expanded substantially by day 419 (Fig. 2 A). These data support not only an important contribution of the B27-KK10 response to early control of HIV, but also a role for variant-specific CD8 responses in limiting the impact of this early escape mutation (13).
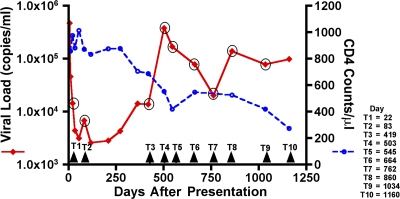
Figure 1.
Loss of viral control in subject AC160. Longitudinal assessment of plasma viral loads (red line) and CD4+ T cell counts (dashed blue line) after acute HIV-1 infection. At day 503 (T4), viral loads dramatically increased to 380,000 copies/ml, followed by a steady decline in CD4+ T cell counts to <300 copies/μl.
Table I.
CD8+ T cell responses after superinfection and recombination
| Epitope | Protein | Sequence | 22 d (SFC/ Mio.) |
419 d (SFC/ Mio.) |
1,160 da (SFC/ Mio.) |
|---|---|---|---|---|---|
| B27-KK10 | p24 | KRWIILGLNK | 85 | 830 | 250 |
| B27-VL9 | Vpr | VRHFPRIWL | - | 550 | 920 |
| B7-IL9 | gp41 | IPRRIRQGL | - | 480 | - |
| A2-AL9 | Vpr | AIIRILQQL | - | 310 | - |
| Cw1-CL9 | gp120 | CAPAGFAIL | - | 240 | 180 |
| B27-IK9 | p17 | IRLRPGGKK | - | 230 | 960 |
| Cw7-RY11 | Nef | RRQDILDLWIY | - | 160 | 70 |
| A1-YT9 | Nef | YFPDWQNYT | - | - | 1,440 |
| B7-RL9 | Nef | RPMTYKAAL | - | - | 1,330 |
| Cw1-VL8 | p24 | VIPMFSAL | - | - | 1,200 |
| B7-TL10 | Nef | TPGPGVRYPL | - | - | 1,070 |
| B7-RV9 | Nef | RPMTYKAAV | - | - | 1,040 |
| A2-SAV10 | gp41 | SLLNATAIAV | - | - | 980 |
6 of the strongest responses, out of a total of 16 new responses developing after superinfection, are shown.
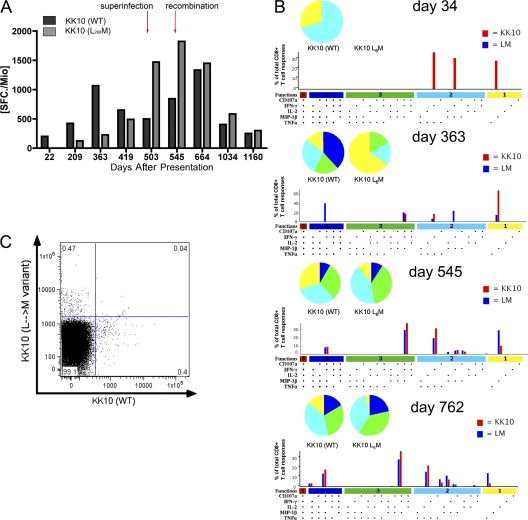
Figure 2.
Loss of the immunodominant HLA-B27–restricted KK10 CD8 response after viral escape, and the development of a variant-specific response. (A) Longitudinal assessment of the WT (black) and L268M variant-specific (gray) KK10 CD8 responses by IFN-γ ELISpot. The WT response declined at day 419, coincident with development of the L268M mutation, after which the L268M-specific response developed. Decline of both responses occurred at day 1,034 after development of the R264K anchor mutation. (B) Assessment of CD8+ T cell polyfunctionality at four different time points. All 32 possible combinations of the 5 antigen-specific functions studied for each epitope are shown on the x axis, and the contributions of each epitope-specific CD8+ T cell population are indicated as bars (WT, red; L268M, blue). Responses are grouped according to the number of functions (1, yellow; 2, cyan; 3, green; 4, blue; 5, red) and summarized by pie charts. (C) Dual tetramer staining for the KK10 WT and L268M-specific response on day 664, illustrating two distinct non–cross-reacting populations of antigen-specific cells.
Functionality of WT and variant-specific CD8 responses
Various studies suggest that the polyfunctionality of a CD8 response may be a critical indicator of vital effector functions and the ability of CD8 responses to actively impair HIV replication (23). Moreover, Almeida et al. have correlated the superior control of HIV replication in subjects expressing HLA-B27 with the polyfunctional capacity of the KK10-specific response (24). To further determine the role of the WT- and L268M-specific B27-KK10 CD8 responses, we examined the polyfunctionality of these responses by assessing five effector functions, including IFN-γ, IL-2, TNF-α, MIP-1β, and CD107a expression by multiparameter flow cytometry. After the L268M-specific CD8 response was first detectable (day 363), it substantially increased in polyfunctional capacity, reaching a similar polyfunctional profile compared with the WT KK10 response (day 545). Both the WT and variant-specific responses exhibited comparable polyfunctional capacities over time (Fig. 2 B), which were not significantly statistically different from one another. To exclude possible cross recognition between the WT and L268M-specific responses, we assessed their antigen-specificity in a dual-tetramer staining experiment (Fig. 2 C), confirming two distinct responses with no detectable cross-reactivity.
The cell surface marker for exhaustion programmed death-1 (PD-1) is up-regulated in CD8+ T cells under high antigen load, but is significantly down-regulated in subjects exhibiting low viral loads or those under antiretroviral therapy (25, 26). Therefore, because it may also be interpreted as an indirect marker of antigen recognition, we assessed the expression of PD-1 on tetramer-specific CD8 cells for the WT KK10 and L268M-specific KK10 responses. Interestingly, both responses revealed similar levels of T cell exhaustion (median fluorescent intensity, WT KK10 530 vs. L268M 518; unpublished data). Collectively, these data support that both the WT and L268M-specific CD8 responses are actively recognizing infected cells, and, in concert, exhibiting immune selection pressures against both forms of the B27-KK10 epitope. More importantly, they suggest that functional variant-specific responses may be contributing to the control of viral replication by tempering the impact of early CTL escape mutations (13).
Superinfection is associated with loss of viral control
Development of the R264K escape mutation in B27-KK10 nearly completely abrogates binding of the epitope to HLA-B27 (11) and has been correlated to a dramatic loss of control over viral replication (6, 7, 11). However, when viral loads rapidly increased to 380,000 copies/ml at day 503 (T4), this specific mutation was notably absent. Rather, a phylogenetic analysis revealed that the virus present at day 503, in fact, represented a distinct strain of HIV (Fig. 3 A and Fig. S1), which is indicative of superinfection with a second strain of HIV. The second strain then dominated over the nearly 3 yr of follow up, with no subsequent detection of the original infecting strain (unpublished data). The shift within the immunodominance pattern of CD8 responses over this same time frame supported this change in the circulating virus (Fig. S2, available at http://www.jem.org/cgi/content/full/jem.20080281/DC1). Remarkably, five of the seven CD8 responses present before superinfection subsequently declined, whereas other novel CD8 responses became immunodominant. A comparison of viral sequences revealed that the superinfecting strain transmitted mutations in five of the seven initially targeted epitopes (Table S1). Similar changes in the patterns of CD8 responses after superinfection have been previously described (14), suggestive of a potentially critical role for transmitted mutations in the evasion of CD8 responses and enabling outgrowth of the new incoming strain.
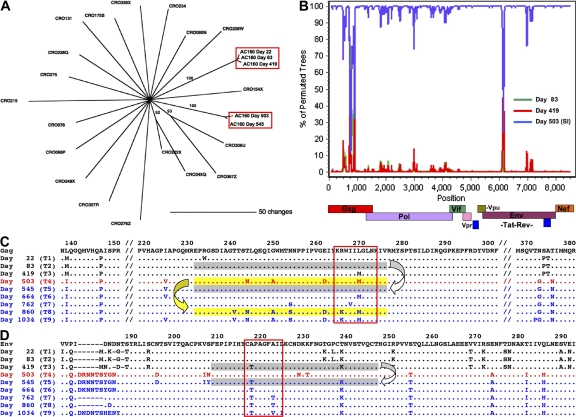
Figure 3.
Superinfection and recombination. (A) HIV gag sequences from subject AC160 and 19 other HIV chronic-infected subjects were compared using a neighbor-joining phylogenetic tree. Sequences from subject AC160 derived from the first year of infection (days 22, 83, and 419) cluster independently from sequences derived later in infection (days 503 and 545). Scale bar indicates the genetic distance along the branches, and bootstrap values >60 are shown. (B) SimPlot recombination analysis of a full-length HIV sequence derived from day 545 compared with viruses derived from pre-superinfection (day 83, green; day 419, red) and the superinfecting strain (day 503, blue) using a window of 100 bp and a step size of 10 bp. Two regions with double recombination breakpoints were observed in Gag and Env and Yates-corrected χ2 values, and P values were calculated for each putative breakpoint. Breakpoints in gag were detected around positions 690 (χ = 5.8; P = 0.0165) and 822 (χ = 50.1; P < 0.0001), and breakpoints in env were detected around positions 6,047 (χ = 50.1; P < 0.0001) and 6,164 (χ = 30.1; P < 0.0001). By day 860 (T8), the breakpoints in gag were no longer detectable, supportive of a possible second recombination event in the recombinant strain that restored this region of the original superinfecting strain. (C) Longitudinal amino acid alignment of Gag sequences in AC160. Sequences derived from the primary infecting strain (black), the superinfecting strain (red), and the new recombinant form (blue) are aligned to clade B consensus. Early viral escape was observed in the B27-KK10 epitope boxed in red. Flanking sequences are deleted (//) to illustrate sequence diversity between the two strains. Sequences suggested to be involved in the first recombination event are shaded gray, whereas those involved in the second recombination event are shaded yellow. (D) Longitudinal amino acid alignment of Env sequences containing the Cw1-CL9 epitope are boxed in red. Sequences suggested to be involved in the third recombination event in Env are shaded gray.
Viral recombination in regions under strong immune selection pressure
Despite the documented importance of the KK10-specific CD8 response, unexpectedly, there was no sequence evolution observed in the KK10 epitope at the time of superinfection. Rather the same preexisting L268M escape mutation was present in the superinfecting strain. Surprisingly, however, only 2 mo after superinfection (day 545), we observed an uncharacteristic reversion of the L268M mutation to the WT sequence in addition to other sequence changes surrounding KK10 (Fig. S1). Strikingly, the comparison of longitudinal sequences revealed a short recombination event in Gag between the original and superinfecting strain at day 545 (T5; Fig. 3 B), which resulted in substitution of a narrow region of Gag from the original strain between amino acid residues 220 and 274 within the superinfecting strain (Fig. 3 C). This short region overlapped with the B27-KK10 epitope and resulted in an unusual replacement of the L268M mutation with the WT form present during acute infection. To evaluate whether immune selection pressures might have influenced this recombination event, we examined the WT and variant-specific KK10 CD8 responses at the time of superinfection. Longitudinal analysis of IFN-γ ELISpot responses revealed that at the time of superinfection the L268M-specific response was actually substantially stronger (1528 SFC/Mio.), and also of higher avidity (IC50 ∼0.26 μg/ml; not depicted), than the WT-specific response (531 SFC/Mio.; IC50 ∼0.88 μg/ml; Fig. 2 A). Therefore, in the setting of a substantially stronger L268M-specific response at day 503, substitution of this region containing the L268M mutation with the WT form of the epitope would have facilitated evasion from a stronger L268M-specific response. Supportive of this hypothesis, levels of the L268M-specific response subsequently declined by as early as day 664 (T6; Fig. 2 A). These data suggest that a recombination event in a narrow region of Gag within 2 mo of superinfection facilitated rapid escape from the dominant variant-specific KK10 response.
Transient regain of viral control is lost through development of a second recombination event in Gag
After recombination to the WT sequence, viral loads declined under the presence of a WT KK10-specific CD8 response to 20,000 copies/ml by day 762, but then rebounded again to 139,000 copies/ml at day 860 (T8; Fig. 1). Viral sequences in the KK10 epitope at this later time point illustrated development of the R264K escape mutation (Fig. 3 C). Moreover, these sequences revealed the likely occurrence of a second recombination event having again substituted a short sequence surrounding KK10 and facilitating development of the R264K mutation (Fig. 3 C), although the narrow length of this second recombination event precluded a critical analysis of breakpoints. The R264K mutation has been shown to substantially impair the recognition of both the WT- and L268M-specific responses (6, 7) and normally requires the specific compensatory mutation S173A (11). In subject AC160, however, the S173A mutation did not arise, but rather two other distinct mutations (T239V and N252S) accompanied R264K. As we have previously observed the development of R264K in conjunction with other rare mutations in its proximity (22), it is likely that one or both of these mutations serve to compensate for the fitness defect of R264K. Therefore, development of R264K through recombination was likely facilitated by suitable compensatory mutations developing within the backbone sequence of the superinfecting strain rather than within the original infecting strain.
This development of R264K resulted in the impaired recognition of both WT and variant-specific responses as indicated by their decline by day 1,034 (T9; Fig. 2 A). Interestingly, although the L268M-specific response was dominant and of higher functional avidity than the WT KK10 response before the first recombination event, both the WT and L268M responses were co-dominant and of similar functional avidity at day 664 (T6) before this second recombination event (1,334 SFC/Mio. and logIC50 ∼0.04 vs. 1,450 SFC/Mio. and logIC50 ∼0.04, respectively; Fig. 2 A). Therefore, at the time the R264K mutation developed in conjunction with a likely second recombination event, both the WT and the variant-specific responses were eliciting similar immune selection pressure.
Recombination in envelope
To determine whether recombination at sites of strong immune selection pressure might represent a more common phenomenon to escape potent CD8 responses after superinfection, full-length viral genome sequences from nine time points over the course of infection were compared. Only one other recombination event of ∼40 aa in length was detected across the whole viral genome. This small recombination event between aa 220 and 260 of envelope arose at day 545 (T5), at the same time as the first recombination event in KK10 (Fig. 3 B). This region resides outside of the V3 loop of the virus, and no switch in the co-receptor tropism was detected using the geno2pheno method (27). Notably, this short region of recombination contained one of the other six CD8 epitopes targeted before superinfection, namely the Cw1-CL9 epitope CAPAGFAIL (Table I and Fig. 4). Similar to the KK10 response, this Cw1-restricted response exerted immune selection pressure against the virus as indicated by early viral escape through an A217T mutation developing at day 419 (T3) that was shown to impair T cell recognition (Fig. 3 D and not depicted). As superinfection had resulted in transmission of the WT form of the Cw1-CL9 epitope, the subsequent recombination event at day 545 (T5) rapidly replaced this region with a sequence containing the A217T escape mutation derived from the virus circulating at day 419 (T3; Fig. 3 D). Thus, viral escape in this epitope after superinfection was also accomplished through recombination rather than through a single amino acid substitution. Similar to the KK10 epitope, an A217T variant-specific response developed, peaking at day 545 (Fig. 4; T5, 730 SFC/Mio.), with the development of additional mutations in this epitope by day 1,034 (Fig. 3 D) observed to completely abrogate both the WT and variant-specific responses (Fig. 4). Collectively, these data revealed the presence of a third recombination event associated with evasion of a CD8 response, which is strongly indicative of successive attempts by the virus to rapidly evade dominant immune selection pressures directly through recombination.
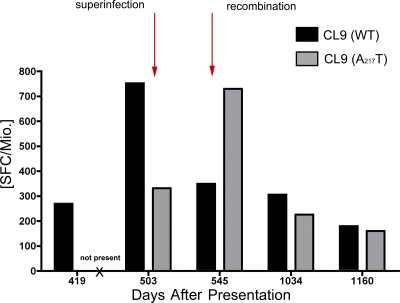
Figure 4.
Abrogation of the Cw1-CL9 response after recombination in Env. Longitudinal development of the WT (black) and A217T variant-specific (gray) CL9 responses as measured by IFN-γ ELISpot. The A217T variant-specific response was not detected before recombination at day 419 (“not present”). At the time of superinfection at day 503, the WT response exhibited the highest magnitude, suggestive of having driven the recombination event to the A217T form of CL9. Decline of both the WT and the A217T -specific CL9 response was observed at day 1,034.
It has been recently suggested that recombination after HIV superinfection occurs more frequently than anticipated (28). Understanding the underlying mechanisms of recombination pertains not only to the impact of greater global HIV diversity (15), but also to the critical events undermining successful immune control of HIV infection. The generation of HIV recombinants is thought to occur by a copy choice mechanism during reverse transcription (20), with breakpoint selection generally being a random event, although it is influenced by sequence identity (20, 21). These data indicate that recombination enabled rapid evasion from the most immunodominant CD8 response in subject AC160, strongly suggesting that the recombination event was driven by immune selective pressures.
The current study indicates that specific selection forces, in this case cellular immune pressures, can strongly influence recombination. The diversity of HLA class I alleles in the population, and the variety of CD8 responses, would explain the complex array of recombination breakpoints between different HIV strains observed to date (19) and the inability to ascribe a strong predictor of recombination hotspots. In conclusion, these data suggest that the ability of a superinfecting strain to overcome immune control may be related to its ability to rapidly recombine in critical regions under immune selection pressure. These data also support a critical role for cellular immune pressures in driving the selection of recombination break points, thus contributing to the selection of new circulating recombinant forms of HIV.
MATERIALS AND METHODS
Subjects.
The HLA-B27+ subject AC160, identified during primary HIV infection, and 19 other chronically infected control subjects, were enrolled at Massachusetts General Hospital in Boston. The study was approved by the institutional review board of the Massachussetts General Hospital.
Sequencing of autologous virus.
Population sequences of autologous full-length viral genomes were derived from proviral DNA, as previously described (13). In brief, genomic DNA extracted from 5 million PBMCs using the QIAamp DNA Blood Mini kit (QIAGEN). Nested PCR protocols with limiting dilution used to amplify full-length HIV genomes using EXL DNA Polymerase (Stratagene). The sequences of primary forward and reverse PCR primers, respectively, are 5′-AAATCTCTAGCAGTGGCGCCCGAACAG-3′ and 5′-TGAGGGATCTCTAGTTACCAGAGTC-3′, whereas the nested forward and reverse primers are 5′-GCGGAGGCTAGAAGGAGAGAGATGG-3′ and 5′-GCACTCAAGGCAAGCTTTA-TTGAGGCTTA-3′. PCR cycling conditions were as follows: 92°C for 2 min; 10 cycles of 10 s at 92°C, 30 s at 60°C, or 10 min at 68°C; and 20 cycles of 10 s at 92°C, 30 s at 55°C, 10 min at 68°C, and a final extension of 10 min at 68°C. Five independent PCR products of each sample were pooled and purified using the QIAquick PCR Purification kit (QIAGEN) and population sequenced bi-directionally on an ABI 3130 automated sequencer (Applied Biosystems) using 70 clade B consensus sequencing primers, as previously described (14).
Autologous clonal gag and env sequences were derived from plasma RNA, as previously described (13). Viral RNA was isolated from plasma, and nested PCR was conducted using a set of described primers specific for HIV (14). First round PCR cycling conditions were as follows: 94°C for 2 min, 35–50 cycles of 30 s at 94°C, 30 s at 56°C, 2 min at 72°C, and a final extension of 68°C for 20 min, and nested PCR reactions were shortened to a 1-min extension time. PCR fragments were then gel purified and sequenced directly or cloned (TOPO TA Cloning kit; Invitrogen). Plasmid DNA was isolated by miniprep (QIAPrep Turbo Miniprep) and sequenced bi-directionally.
Sequence data were manually edited using Sequencher 4.6 (Gene Codes Corporation). In regions where secondary peaks were observed, the dominant base was called. Nucleotide sequences were conceptually translated and aligned using MacVector 7.2.3 (Accelrys). The clade B HIV consensus sequence (2002) from Los Alamos National Laboratory HIV Sequence Database was used as the reference sequence to compare with our sequencing data. All sequence data were deposited in GenBank under accession nos. EU616639–EU616649.
Phylogenetic analysis.
Gag sequences from subject AC160 were aligned and compared with sequences derived from chronically infected subjects within a chronic infection cohort in Boston using Phylip3.6 for constructing neighbor-joining phylogenetic tree.
Recombination.
Recombination break points were determined using SimPlot (see Supplemental materials and methods and Fig. S3, available at http://www.jem.org/cgi/content/full/jem.20080281/DC1). Full-length HIV sequences were analyzed using a 100-bp window and a 10-bp step size, and Yates-corrected χ2 values and P values were calculated for each putative breakpoint.
ELISpot.
Baseline HIV-specific CD8+ T cell responses were quantified on freshly isolated PBMCs by IFN-γ ELISpot assay (4) using 410 overlapping peptides, varying from 15–20 aa in length and overlapping by 10 aa, which spanned the entire clade B consensus sequence 2001, as previously described (4). In addition, described optimal peptides corresponding to HLA-matched epitopes were used to detect responses from frozen cells. Peptides were synthesized commercially (Research Genetics) or at the Massachusetts General Hospital Peptide Synthesis Core Facility. PBMCs were plated at 100,000 cells/well with peptides at a final concentration of 14 μg/ml in 96-well plates and processed as previously described (4). PBMCs were incubated with media alone (negative control) and PHA (positive control). The number of specific IFN-γ–secreting T cells were counted using an automated ELISpot reader (AID), calculated by subtracting the average negative control value and expressed as SFCs per 106 input cells. Negative controls were always ≤ 30 SFCs per 106 input cells. A response was considered positive if ≥ 55 SFCs per 106 and at least 3 times greater than mean background activity. Functional avidity curves for the comparison of recognition of epitope variants were performed by ELISpot assay using serial dilutions of truncated peptides, as previously described (4, 14).
Assessment of CD8+ T cell polyfunctionality.
Cryopreserved PBMCs were thawed, resuspended to 1–2 × 106 cells/ml in R10 media (RPMI 1640 supplemented with 10% heat-inactivated FCS, 100 U/ml penicillin, 100 μg/ml streptomycin sulfate, 5.5 ml Hepes buffer), and rested for 1–2 h at 37°C, 5% CO2. PBMCs were examined for viability by trypan blue exclusion (typically 80–90% viable) and adjusted to 106 cells/ml. Co-stimulatory antibodies (CD28 and 49d; 1 μg/ml; BD Biosciences) and CD107a-PE-Cy5 (BD Biosciences) were added, and the cells were aliquoted at 1 ml to each tube containing 2 μg/ml of each peptide. An unstimulated (R10 only) and a positive control (2 μl of PMA 1 mg/ml and 1 μl of ionomycin 1 mg/ml; AG scientific)) were included in each assay. Cells were incubated for 30 min at 37°C, 5% CO2, and monensin (0.7 μl/ml GolgiStop; BD Biosciences) and brefeldin A (10 μg/ml; Sigma-Aldrich) were added. After a total incubation of 6 h, the cells were washed with PBS and stained for intracellular amine groups to discriminate between live and dead cells (blue viability dye; Invitrogen). Cells were washed again and stained with a panel of anti-CD3–Pacific Blue (BD Biosciences) and anti-CD8-APC-Cy7 (BD Biosciences). Cells were then fixed in 1% paraformaldehyde, washed with PBS, and permeabilized using Fix Perm A and Fix Perm B solution (Caltag Laboratories). Cells were intracellularly stained using a panel of IL-2-FITC (BD Biosciences), IFN-γ-PE-Cy7 (BD Biosciences), TNF-α-Alexa700 (BD Biosciences), and MIP-1β-PE (BD Biosciences). Between 150,000 and 500,000 events were collected per sample. All data were collected on a BD LSRII (BD Biosciences) flow cytometer and analyzed using FlowJo 8.3.3 software (Tree Star, Inc.). Initial gating was on the lymphocyte population, and then used a forward scatter width (FSC-W) versus height (FSC-H) plot to remove doublets. Subsequently, the events were gated through a side scatter (SSC) versus blue viability dye (UV) and sequentially gated on CD3+ and CD8+ events. After identification of CD8+ T cells, a gate was made for each respective function using combinations that provided optimal separation. After the gates for each function were created, we used the Boolean gate platform to create the full array of possible combinations, equating to 32 response patterns when testing 5 functions. Data are reported after background correction, and the percent of epitope-specific CD8+ T cell responses had to be more than twofold higher than background for individual cytokines or CD107a to be considered as a positive response. Data were then analyzed using SPICE software (provided by M. Roederer, National Institute of Allergy and Infectious Disease, Bethesda, MD) and by Wilcoxon rank sum comparison.
Tetramer staining.
Cryopreserved PBMCs were stained with violet viability dye against amine groups, which penetrates and stains dead and dying cells with compromised membrane integrity. After a short wash with 2% FCS/PBS, cells were stained with an APC-labeled tetramer (National Institutes of Health) for WT variant of KK10 and a PE-labeled pentamer (ProImmune) for L→M variant of KK10 for 20 min at room temperature. After an additional wash, cells were stained for surface antibodies against CD3, CD4, CD8 (BD Biosciences), and PD-1 (provided by G. Freeman, Harvard Medical School, Boston, MA) and incubated for 20 min at room temperature. Cells were fixed with 1% paraformaldehyde, and events were acquired on a LSRII (BD Biosciences). Data were analyzed using FlowJo software 8.3.3.
Online supplemental material.
Fig. S1 shows longitudinal amino acid alignment of Gag sequences. Fig. S2 shows that superinfection and recombination is paralleled by a disintegration of the CD8+ T cell immunodominance patterns. Fig. S3 shows the detection of superinfection using the reversible (GTR) model of nucleotide substitution. Table S1 lists the viral sequences of the targeted CD8+ T cell responses before and after superinfection and recombination, with supplemental materials and methods providing a description of the analysis of recombination using clonal gag sequences. The online version of this article is available at http://www.jem.org/cgi/content/full/jem.20080281/DC1.
Supplementary Material
Acknowledgments
We would like to thank Sergei Kosakovsky, Yaoyu E. Wang, and Nicole Frahm for their assistance.
This study was supported by National Institutes of Health grants R01-AI054178 (T.M. Allen), U01-AI052403 (T.M. Allen and M. Altfeld), R01-205672 (M. Altfeld), AI47745 and AI57167 (S.D.W. Frost), and N01-AI-30024 (C. Brander and R. Zuniga).
The authors have no conflicting financial interests.
H. Streeck and B. Li contributed equally to this paper.
References
- 1.Schmitz, J.E., M.J. Kuroda, S. Santra, V.G. Sasseville, M.A. Simon, M.A. Lifton, P. Racz, K. Tenner-Racz, M. Dalesandro, B.J. Scallon, et al. 1999. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science. 283:857–860. [DOI] [PubMed] [Google Scholar]
- 2.Koup, R.A., J.T. Safrit, Y. Cao, C.A. Andrews, G. McLeod, W. Borkowsky, C. Farthing, and D.D. Ho. 1994. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J. Virol. 68:4650–4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carrington, M., and S.J. O'Brien. 2003. The influence of HLA genotype on AIDS. Annu. Rev. Med. 54:535–551. [DOI] [PubMed] [Google Scholar]
- 4.Streeck, H., M. Lichterfeld, G. Alter, A. Meier, N. Teigen, B. Yassine-Diab, H.K. Sidhu, S. Little, A. Kelleher, J.P. Routy, et al. 2007. Recognition of a defined region within p24 gag by CD8+ T cells during primary human immunodeficiency virus type 1 infection in individuals expressing protective HLA class I alleles. J. Virol. 81:7725–7731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leslie, A.J., K.J. Pfafferott, P. Chetty, R. Draenert, M.M. Addo, M. Feeney, Y. Tang, E.C. Holmes, T. Allen, J.G. Prado, et al. 2004. HIV evolution: CTL escape mutation and reversion after transmission. Nat. Med. 10:282–289. [DOI] [PubMed] [Google Scholar]
- 6.Feeney, M.E., Y. Tang, K.A. Roosevelt, A.J. Leslie, K. McIntosh, N. Karthas, B.D. Walker, and P.J. Goulder. 2004. Immune escape precedes breakthrough human immunodeficiency virus type 1 viremia and broadening of the cytotoxic T-lymphocyte response in an HLA-B27-positive long-term-nonprogressing child. J. Virol. 78:8927–8930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goulder, P.J., R.E. Phillips, R.A. Colbert, S. McAdam, G. Ogg, M.A. Nowak, P. Giangrande, G. Luzzi, B. Morgan, A. Edwards, et al. 1997. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat. Med. 3:212–217. [DOI] [PubMed] [Google Scholar]
- 8.Feeney, M.E., Y. Tang, K. Pfafferott, K.A. Roosevelt, R. Draenert, A. Trocha, X.G. Yu, C. Verrill, T. Allen, C. Moore, et al. 2005. HIV-1 viral escape in infancy followed by emergence of a variant-specific CTL response. J. Immunol. 174:7524–7530. [DOI] [PubMed] [Google Scholar]
- 9.Lichterfeld, M., D. Kavanagh, K. Williams, B. Moza, S. Mui, T. Miura, R. Sivamurthy, R. Allgaier, F. Pereyra, A. Trocha, et al. 2007. A viral CTL escape mutation leading to immunoglobulin-like transcript 4–mediated functional inhibition of myelomonocytic cells. J. Exp. Med. 204:2813–2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goulder, P.J., C. Brander, Y. Tang, C. Tremblay, R.A. Colbert, M.M. Addo, E.S. Rosenberg, T. Nguyen, R. Allen, A. Trocha, et al. 2001. Evolution and transmission of stable CTL escape mutations in HIV infection. Nature. 412:334–338. [DOI] [PubMed] [Google Scholar]
- 11.Kelleher, A.D., C. Long, E.C. Holmes, R.L. Allen, J. Wilson, C. Conlon, C. Workman, S. Shaunak, K. Olson, P. Goulder, et al. 2001. Clustered mutations in HIV-1 gag are consistently required for escape from HLA-B27-restricted cytotoxic T lymphocyte responses. J. Exp. Med. 193:375–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Altfeld, M., and T.M. Allen. 2006. Hitting HIV where it hurts: an alternative approach to HIV vaccine design. Trends Immunol. 27:504–510. [DOI] [PubMed] [Google Scholar]
- 13.Allen, T.M., X.G. Yu, E.T. Kalife, L.L. Reyor, M. Lichterfeld, M. John, M. Cheng, R.L. Allgaier, S. Mui, N. Frahm, et al. 2005. De novo generation of escape variant-specific CD8+ T-cell responses following cytotoxic T-lymphocyte escape in chronic human immunodeficiency virus type 1 infection. J. Virol. 79:12952–12960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Altfeld, M., T.M. Allen, X.G. Yu, M.N. Johnston, D. Agrawal, B.T. Korber, D.C. Montefiori, D.H. O'Connor, B.T. Davis, P.K. Lee, et al. 2002. HIV-1 superinfection despite broad CD8+ T-cell responses containing replication of the primary virus. Nature. 420:434–439. [DOI] [PubMed] [Google Scholar]
- 15.Smith, D.M., D.D. Richman, and S.J. Little. 2005. HIV superinfection. J. Infect. Dis. 192:438–444. [DOI] [PubMed] [Google Scholar]
- 16.Yang, O.O., E.S. Daar, B.D. Jamieson, A. Balamurugan, D.M. Smith, J.A. Pitt, C.J. Petropoulos, D.D. Richman, S.J. Little, and A.J. Brown. 2005. Human immunodeficiency virus type 1 clade B superinfection: evidence for differential immune containment of distinct clade B strains. J. Virol. 79:860–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fang, G., B. Weiser, C. Kuiken, S.M. Philpott, S. Rowland-Jones, F. Plummer, J. Kimani, B. Shi, R. Kaul, J. Bwayo, et al. 2004. Recombination following superinfection by HIV-1. AIDS. 18:153–159. [DOI] [PubMed] [Google Scholar]
- 18.Dowling, W.E., B. Kim, C.J. Mason, K.M. Wasunna, U. Alam, L. Elson, D.L. Birx, M.L. Robb, F.E. McCutchan, and J.K. Carr. 2002. Forty-one near full-length HIV-1 sequences from Kenya reveal an epidemic of subtype A and A-containing recombinants. AIDS. 16:1809–1820. [DOI] [PubMed] [Google Scholar]
- 19.Kijak, G.H., and F.E. McCutchan. 2005. HIV diversity, molecular epidemiology, and the role of recombination. Curr. Infect. Dis. Rep. 7:480–488. [DOI] [PubMed] [Google Scholar]
- 20.Negroni, M., and H. Buc. 2001. Retroviral recombination: what drives the switch? Nat. Rev. Mol. Cell Biol. 2:151–155. [DOI] [PubMed] [Google Scholar]
- 21.Baird, H.A., Y. Gao, R. Galetto, M. Lalonde, R.M. Anthony, V. Giacomoni, M. Abreha, J.J. Destefano, M. Negroni, and E.J. Arts. 2006. Influence of sequence identity and unique breakpoints on the frequency of intersubtype HIV-1 recombination. Retrovirology. 3:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schneidewind, A., M.A. Brockman, R. Yang, R.I. Adam, B. Li, S. Le Gall, C.R. Rinaldo, S.L. Craggs, R.L. Allgaier, K.A. Power, et al. 2007. Escape from the dominant HLA-B27-restricted cytotoxic T-lymphocyte response in Gag is associated with a dramatic reduction in human immunodeficiency virus type 1 replication. J. Virol. 81:12382–12393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Betts, M.R., M.C. Nason, S.M. West, S.C. De Rosa, S.A. Migueles, J. Abraham, M.M. Lederman, J.M. Benito, P.A. Goepfert, M. Connors, et al. 2006. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood. 107:4781–4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Almeida, J.R., D.A. Price, L. Papagno, Z.A. Arkoub, D. Sauce, E. Bornstein, T.E. Asher, A. Samri, A. Schnuriger, I. Theodorou, et al. 2007. Superior control of HIV-1 replication by CD8+ T cells is reflected by their avidity, polyfunctionality, and clonal turnover. J. Exp. Med. 204:2473–2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Day, C.L., D.E. Kaufmann, P. Kiepiela, J.A. Brown, E.S. Moodley, S. Reddy, E.W. Mackey, J.D. Miller, A.J. Leslie, C. DePierres, et al. 2006. PD-1 expression on HIV-specific T cells is associated with T-cell exhaustion and disease progression. Nature. 443:350–354. [DOI] [PubMed] [Google Scholar]
- 26.Trautmann, L., L. Janbazian, N. Chomont, E.A. Said, S. Gimmig, B. Bessette, M.R. Boulassel, E. Delwart, H. Sepulveda, R.S. Balderas, et al. 2006. Upregulation of PD-1 expression on HIV-specific CD8+ T cells leads to reversible immune dysfunction. Nat. Med. 12:1198–1202. [DOI] [PubMed] [Google Scholar]
- 27.Low, A.J., W. Dong, D. Chan, T. Sing, R. Swanstrom, M. Jensen, S. Pillai, B. Good, and P.R. Harrigan. 2007. Current V3 genotyping algorithms are inadequate for predicting X4 co-receptor usage in clinical isolates. AIDS. 21:F17–F24. [DOI] [PubMed] [Google Scholar]
- 28.Piantadosi, A., B. Chohan, V. Chohan, R.S. McClelland, and J. Overbaugh. 2007. Chronic HIV-1 Infection Frequently Fails to Protect against Superinfection. PLoS Pathog. 3:e177. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.