Abstract
In this paper we describe denitrification at extremely high salt and pH in sediments from hypersaline alkaline soda lakes and soda soils. Experiments with sediment slurries demonstrated the presence of acetate-utilizing denitrifying populations active at in situ conditions. Anaerobic enrichment cultures at pH 10 and 4 M total Na+ with acetate as electron donor and nitrate, nitrite and N2O as electron acceptors resulted in the dominance of Gammaproteobacteria belonging to the genus Halomonas. Both mixed and pure culture studies identified nitrite and N2O reduction as rate-limiting steps in the denitrification process at extremely haloalkaline conditions.
Electronic supplementary material
The online version of this article (doi:10.1007/s00792-008-0166-6) contains supplementary material, which is available to authorized users.
Keywords: Denitrification, Soda lakes, Haloalkaliphilic, Halomonas, Alkalispirillum
Introduction
Denitrification is an important process of oxidation of organic and inorganic compounds in natural and engineered anoxic environments. Regarding its energy efficiency, it is following aerobic respiration and, therefore should be well represented at extreme conditions demanding the efficient energy conservation, such as haloalkaline lakes, saline soils and highly saline industrial wastewater (Oren 1999). While it is indeed well documented for moderate haloalkaline conditions (i.e., a pH around 9 and a salt concentration up to 2 M Na+), little is known on the possibility of denitrification at extremely haloalkaline conditions, such as those present in hypersaline alkaline soda lakes (i.e., a pH up to 11 and a salt concentration up to 4 M Na+). Denitrification at moderate haloalkaline conditions has been shown previously for several members of the Gammaproteobacteria, both with chemolithoautotrophic metabolism, such as sulfur-oxidizing Thioalkalivibrio spp. (Sorokin et al. 2001; Sorokin and Kuenen 2005) and obligate heterotrophs, such as Halomonas spp. (Berendes et al. 1996; Mormille et al. 1999; Peyton et al. 2001; Romano et al. 2005; Boltyanskaya et al. 2004, 2007). The only example of active denitrification at extremely haloalkaline conditions has recently been demonstrated in experiments with sediment slurries from Mono Lake (CA, USA). The maximum activity of nitrate reduction with glucose was found at a salt concentration between 1.6 and 2.3 M NaCl, pH 9.8 (Kulp et al. 2007), which was above the natural salinity of the lake (i.e., 1.5 M).
Here we report the results of denitrification at extremely haloalkaline conditions in sediments of hypersaline alkaline lakes and soda soils and the characterization of the bacteria responsible for this process.
Methods
Soda lake characteristics and sampling
Surface sediment samples (0–10 cm) were obtained from hypersaline soda lakes in northeastern Mongolia, Kulunda Steppe (Altai, Russia), Kenya (Lakes Magadi and Bogoria) and Wadi al Natrun in Egypt. The pH of the brines varied from 9.2 to 10.6, the total salt concentration, from 20 to 400 g l−1, and the total (soluble) alkalinity, from 0.05 to 3.0 M. More information about the sites can be found in Sorokin and Kuenen (2005), Taher (1999) and Duckworth et al. (1996). In addition, several samples of saline soda soils were collected in Kulunda Steppe and NE Mongolia. Measurements of the potential denitrifacation rates (DR) in the sediment slurry experiments were conducted in July 2007 for three lakes in Kulunda Steppe. Lake Picturesque is a low productivity lake with gray clay sediments without free sulfide (total salt = 102 g l−1; pH = 10.20; soluble carbonate alkalinity = 1.34 M). Lakes Tanatar-5 and Bitter-1 are high productivity lakes with black sulfidic sediments (total salt content of the brines = 65 and 180 g l−1, pH = 10.35 and 10.53, and total alkalinity = 0.77 and 2.95 M, respectively). For the enrichments, we mixed 5–10 samples from each lake or soil to form a single inoculum for each geographic region.
The potential DR was measured in the laboratory with sediment slurries. For this, the sediments were either diluted with two parts of the lake water (i.e., “natural slurry”) or first were separated from the pore brines by centrifugation and suspended in various buffers to study the effect of pH and salt (see below) on the potential DR of the natural denitrifying populations. In some cases (i.e., at the lowest and highest pH range), it was necessary to adjust the pH to the desired values before the experiment. The experiments were conducted in 12-ml serum bottles closed with butyl rubber stoppers with 3 ml slurry and 9 ml gas phase consisted of 90% (v/v) argon/10% (v/v) acetylene. In experiments with N2O as the electron acceptor, acetylene was omitted. After the end of each experiment, the final pH values in the slurries were measured.
Cultivation of bacteria
Anaerobic enrichments and subsequent cultivation of isolated strains were performed at 30°C using a highly buffered alkaline saline mineral base medium containing 0.6–4.0 M Na+, of which 50% Na+ was present as NaCl and 50% as sodium carbonate/bicarbonate buffer pH 10.1 (2 M total Na+). A measure of 0.5 g l−1 of K2HPO4 served as P and K source. After sterilization, the medium was supplemented with 1 mM MgSO4·7H2O, 1 ml l−1 trace metal solution (Pfenning and Lippert 1966), 20 mM sodium acetate, 0.1 g l−1 yeast extract and 5–20 mM nitrate or nitrite or 40 mM N2O. Routine anaerobic cultivation was performed in 100-ml serum bottles with 80 ml medium/20 ml argon for nitrate/nitrite cultures, and 50 ml medium/50 ml N2O for N2O cultures. Solid alkaline medium with a final salt concentration of 2 M Na+ was prepared by 1:1 mixing of 4% (w/v) agar (Noble, Difco) and the 4 M Na+ liquid medium (see above) at 50°C. After inoculation, the plates were incubated in closed jars under 100% (v/v) argon or 90% (v/v) argon/10% N2O atmosphere in presence of the oxygen-scavenging catalyzer (Oxoid).
The pH dependence of denitrifying activity was examined at 0.6–2.0 M Na+/0.05 M K+, using the following buffers: for pH 7–8, 0.1 M HEPES and NaCl; for pH 8.5–11, a mixture of sodium bicarbonate/sodium carbonate containing 0.1 M NaCl. Incubation resulted in a shift of initial pH values, especially in highly alkaline region. Therefore, final pH values were taken to indicate suitable range for growth and activity. To study the influence of salt concentration on growth, mineral bases containing 0.1 and 4.0 M of total Na+ at pH 10 were mixed in different proportions.
Analytical procedures
Nitrite was analyzed by diazotation method according to Gries-Romijn-van Eck (1966). Nitrate analysis was complicated by extremely high alkalinity of the samples. Therefore, a most sensitive colorimetric method was chosen for the analysis on the basis of Szechrome NAS reagent (Polysciences, Inc., Warrington, PA) with the detection limit of 10 μM. This allowed the dilution of the brines by at least 100 times. N2O in the gas phase was analyzed by the gas chromatography using Fison Instruments chromatograph, Poropaq Q column and 63Ni-electron capture detector with a 6-port valve assembly activated pneumatically to vent column effluent after elution of nitrous oxide to avoid contamination of the detector by acetylene. The temperature of injector, column and detector was 100, 40 and 350°C, respectively, the carrier gas was N2 at 10 ml min−1. Cell protein was measured after alkaline hydrolysis by the Lowry method (Lowry 1951). Phase contrast microphotographs were obtained with a Zeiss Axioplan Imaging 2 microscope (Göttingen, Germany). Comparison of the total cell proteins by SDS-PAGE was done according to Laemmli (1970).
Genetic and phylogenetic analysis
The isolation of the DNA and determination of the G + C content of the DNA was performed according to Marmur (1961) and Marmur and Doty (1962). The DNA–DNA hybridization was performed by optical reassociation according to De Ley (1970). Genomic DNA for phylogenetic analysis was extracted from the cells using the UltraClean Soil DNA Extraction Kit (MoBio Laboratories, USA), following the manufacturer’s instructions. The nearly complete 16S rRNA gene was obtained from pure cultures using general bacterial primers 11F-1492R (Lane 1991). The 16S rRNA gene PCR products were purified from low-melting agarose using the Wizard PCR-Prep kit (Promega, USA). Sequencing was performed using Big Dye Terminator v.3.1 sequencing reaction kit on an ABI 3730 DNA automatic sequencer (Applied Biosystems, Inc.,USA). The sequences were aligned with those from GenBank using CLUSTALW. The 16S rRNA gene sequences were first compared to sequences stored in the GenBank database using BLAST search tool. Phylogenetic trees were reconstructed with four different algorithms using the TREECONW software package (van de Peer and de Wachter 1994). Pair wise evolutionary distances (expressed as estimated changes per 100 nucleotides) were computed using Jukes and Cantor (1969) method. A resulting phylogenetic tree was constructed by the neighbor-joining method.
Results and discussion
Potential DR in sediments of hypersaline soda lakes from Kulunda Steppe
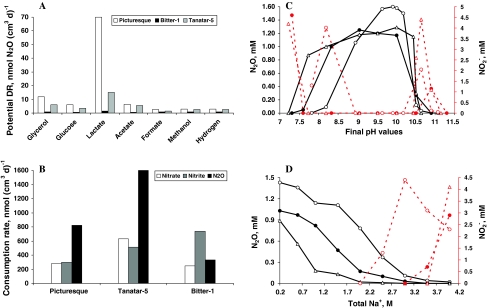
The potential DR measurements in the sediment slurries amended with nitrogen oxides and electron donors revealed highest N2O formation in a less productive lake with lactate as the electron donor (Fig. 1a), although nitrate consumption was faster in the sediments from the other two, highly productive, lakes. It is possible, that the acetylene block was not efficient in the latter, because of high sulfide concentrations in the sediments (Knowles 1990). At least the potential for nitrite and N2O reduction was either equal or even higher in the eutrophic lake sediments (Fig. 1b). The influence of pH and salt on the potential DR was examined with acetate as electron donor and nitrate as electron acceptor. The experiments clearly demonstrated an obligate alkaliphilic nature (Fig. 1c), but only a moderate salt tolerance (Fig. 1d) of the denitrifying populations in all three lakes. A common trend can also be seen for both pH and salt extremes where most of the nitrate added was reduced only to the level of nitrite. This happened at a pH below 8 and above 10.5 and at salinity above 2.5–3 M Na+.
Fig. 1.
Potential denitrification activity in sediments of hypersaline soda lakes in Kulunda Steppe (Altai, Russia). a Potential nitrate (2 mM) reduction to N2O in natural sediment slurries with different electron donors (4 mM). b Reduction of different nitrogen oxides in the presence of 4 mM acetate in natural sediments. c, d Influence of pH (at 0.6 M total Na+) and soda (at pH 10), respectively, on nitrate reduction with 4 mM acetate in sediments freed from the pore waters and suspended in salt buffers. The solid lines depict N2O formation in the presence of acetylene and the dashed lines depict nitrite formation; open circles lake Picturesque; closed circles lake Tanatar-5; open triangles lake Bitter-1
Enrichment and isolation of bacterial strains
Anaerobic enrichments with acetate as electron donor from six combined sediment and soil samples at pH 10 and 4 M Na+ were most active with nitrate as electron acceptor, although significant accumulation of nitrite occurred at nitrate concentrations above 5 mM, similar to that found in the sediment slurry experiments (see above). In several cases a lower salinity was used, resulting in a less prominent (at 2.3 M Na+) or complete absence (at 0.6 M Na+) of nitrite accumulation. Incubations with nitrite also gave positive results when the nitrite concentration was below 10 mM. With N2O as an acceptor, activity was observed in all cultures, but only in two cases it was stable during further transfers. Overall, 15 pure cultures of acetate-utilizing haloalkaliphilic denitrifiers were obtained from six mixed samples (Table 1). Fourteen strains were motile rods of variable cell size and a single strain AGDZ, enriched and isolated with N2O, was a motile spirillum (Supplementary Figure 1). A commonly observed feature of the cell morphology in most of the rod-shaped isolates was the formation of large empty patches (see for example Supplementary Fig. 1a).
Table 1.
Isolates from hypersaline alkaline habitats capable of denitrification at 2–4 M total Na+ and pH 10
| Strains | Source | Salinity (M Na+) | Acceptor | Phylogenetic affiliation |
|---|---|---|---|---|
| AGD 1 | Wadi Natrun lakes | 2.3 | NO3 − | Halomonas |
| AGD 15 | 4.0 | NO3 − | Halomonas | |
| AGD 12 | 4.0 | NO2 − | Halomonas | |
| AGDZ | 4.0 | N2O | Alkalispirillum | |
| AGJ 1–3 | Kenyan lakes | 0.6 | NO3 − | Halomonas |
| AGD 5 | 4.0 | NO2 − | Halomonas | |
| AGD 7 | 4.0 | NO3 − | Halomonas | |
| AGD 2 | Mongolian lakes | 4.0 | NO3 − | Halomonas |
| AGD 3 | 4.0 | NO2 − | Halomonas | |
| AGD 13 | 4.0 | N2O | Halomonas | |
| AGD 8–1 | Mongolian soils | 2.3 | NO3 − | Halomonas |
| AGD 8–2 | 2.3 | NO2 − | Halomonas | |
| AGD 8–3 | 4.0 | NO3 − | Halomonas | |
| AGDK 5 | Kulunda Steppe lakes | 4.0 | NO3 − | Halomonas |
| AGDKS | Kulunda Steppe soils | 4.0 | NO3 − | Halomonas |
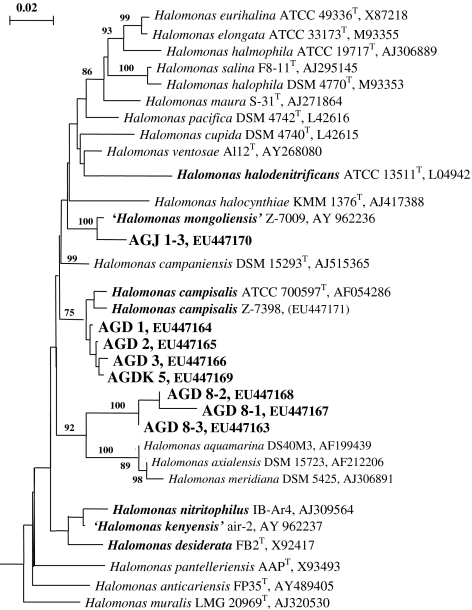
The DNA–DNA hybridization between and the total protein profiling of the rod-shaped isolates (Supplementary Figure 2) demonstrated the presence of three different clusters, two of them including the lake isolates and the third one the Mongolian soil isolates. The DNA relatedness within the clusters was 50–90% and between the clusters was below 25%. The phylogenetic analysis based on 16S rRNA gene sequences placed the rod-shaped isolates into the genus Halomonas and the spiral-shaped strain AGDZ into the genus Alkalispirillum, both in the Gammaproteobacteria (Fig. 2). The properties of Alkalispirillum sp. strain AGDZ have been described in detail elsewhere (Sorokin et al. 2006). The rod-shaped lake isolates clustered with two haloalkaliphilic Halomonas species (H. mongoliensis and H. campisalis) capable of denitrification at moderate salinity, while the soil isolates formed another, well separated group, which probably represents a novel species.
Fig. 2.
Phylogenetic affiliation based on 16S rDNA gene sequence analysis of the haloalkaliphilic denitrifying isolates from hypersaline soda lakes within the genus Halomonas. The tree was reconstructed using maximum likelihood method and filter. The percentage of bootstraps was derived from 1,000 resampling using neighbor-joining algorithm, only values greater than 70 are given. Denitrifying species are printed in bold type
Influence of salts and pH on the growth and activity of Halomonas sp. strain AGD 3
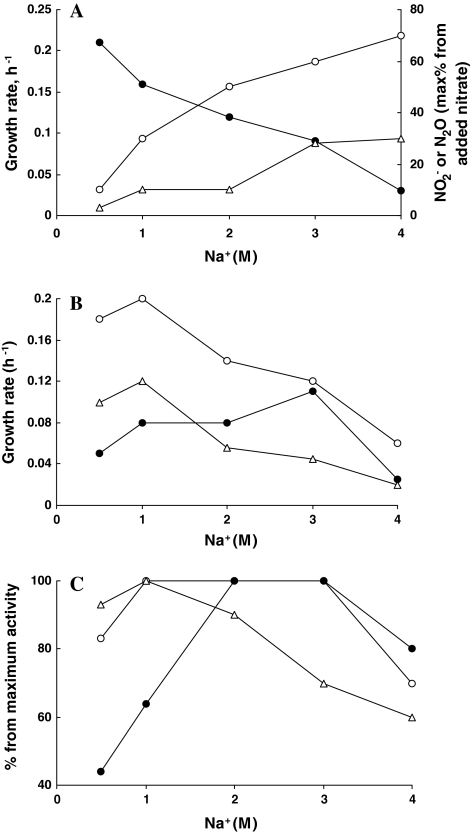
All isolated Halomonas strains, in contrast to any previously described haloalkaliphilic denitrifiers, were capable of anaerobic growth with nitrate and nitrite, and some also with N2O in saturated sodium carbonate brines (4 M total Na+) at pH 10. However, at such high salinity, the anaerobic growth with nitrate was very slow and high intermediate product accumulation was evident, which was similar to that observed in the sediment slurry incubations. This is illustrated with an example of strain AGD 3 (Fig. 3a). Nitrite and N2O did not accumulate at significant level only at salinity below 1 M Na+. Similar to anaerobic growth with nitrate, a sharp decrease in the growth rate was also observed with N2O and oxygen as the electron acceptors but, strangely, not with nitrite (Fig. 3b). High level of nitrite accumulation during anaerobic growth with nitrate at high salt suggested that the nitrite reduction step was a bottleneck in the denitrification process. However, when nitrate was not present, strain AGD 3 was growing optimally with nitrite at 3 M Na+ (Fig. 3b). To clarify the situation, activity experiments with washed cells of strain AGD 3 grown at 3 M Na+ with nitrate were conducted. The results confirmed a high salt dependence and a high salt optimum of the nitrite reduction step. In contrast, nitrate reductase was active within a very broad salinity range, while N2O reductase was the least salt tolerant (Fig. 3c). From these it can be concluded that difficulties in anaerobic growth with N2O and accumulation of the latter at extreme salinity in haloalkaliphilic Halomonas isolates were clearly connected to a low salt tolerance of the N2O reductase. In contrast, high nitrite accumulation during the anaerobic growth with nitrate might rather indicate a disconnected action of nitrate and nitrite reductases at extremely haloalkaline conditions (for example, by a competition for electrons).
Fig. 3.
Influence of soda concentration on anaerobic growth (a, b) and on activity of washed cells (c) in strain AGD 3 at pH 10 with acetate as the electron donor. a Anaerobic growth with nitrate: closed circles growth rate, open circles and triangles formation of nitrite and N2O, respectively. b Influence of soda on aerobic (open circles), and anaerobic growth with nitrite (closed circles) and N2O (triangles). c Influence of soda on activity of reduction of nitrate (open circles), nitrite (closed circles) and N2O (triangles) by washed cells, grown anaerobically with nitrate at 3 M Na+
Dithionite-reduced minus air-oxidized difference spectroscopy of the cell-free extract obtained from the cells of strain AGD 3 grown anaerobically with nitrate or nitrite indicated a possible presence of a cytochrome cd 1-type nitrite-reductase in its soluble fraction (alpha peaks at 551, 615 and 670 nm; data not shown). In the cells grown either aerobically or anaerobically with N2O, this component was not detectable.
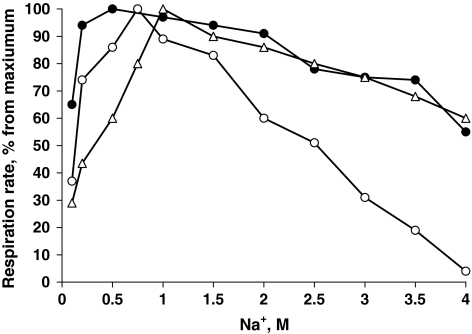
Another interesting question was whether strain AGD 3, isolated from a typical soda lake habitat with domination of sodium carbonates in its brines would react differently on different sodium salts. This was tested with washed cells grown aerobically with acetate at 3 M total Na+. Respiration tests clearly demonstrated a preference for sodium carbonate and sodium sulfate over sodium chloride (Fig. 4). Sodium carbonate and sodium sulfate are very similar in their physico-chemical properties (weakly dissociated electrolytes) in contrast to highly dissociated strong electrolyte sodium chloride. Apparently, this difference of ca. 2 times is important for bacteria thriving in soda brines. One of the reasons might be a lower osmotic demand for compatible solutes as has been demonstrated recently for chemolithoautotrophic “soda-philic” sulfur-oxidizing bacteria from soda lakes (Banciu et al. 2004, 2008). However, this cannot explain such a preference on the level of activity of non-growing cells. Apparently, catabolic respiratory enzymes in the soda lake bacteria are better “tuned” to sodium carbonate.
Fig. 4.
Influence of sodium salts at pH 10 on acetate-dependent respiration activity of washed cells of strain AGD 3. Open circles respiration in NaCl, closed circles respiration in Na2SO4, triangles respiration in soda. One hundred percent activity was 250–350 nmol O2/(mg protein min)
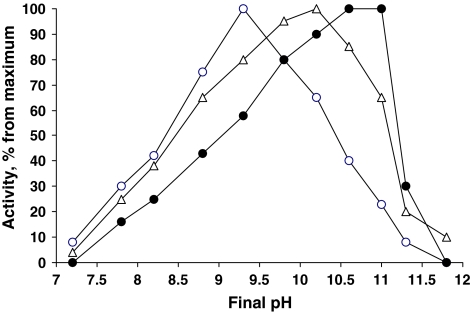
The pH effect on different denitrification steps was investigated in washed cells of strain AGD 3 grown anaerobically with three different nitrogen oxides. The general response was very characteristic of obligate alkaliphiles, with virtually no activity at neutral pH. On the other hand, the denitrifying activity was still possible within the extremely alkaline region (pH 11–11.5), where growth was not possible, similar to that observed in various groups of chemolithoautotrophic bacteria from soda lakes (Sorokin and Kuenen 2005). From the three different reductases, the nitrite-reductase was the most alkali-dependent with a pH optimum of 10.6–11.0. The N2O reductase was also extremely alkali-dependent with an optimum of pH 10, and the nitrate reductase was only moderately alkaliphilic with an optimum of pH 9 (Fig. 5).
Fig. 5.
Influence of pH on activity of denitrifying enzymes in washed cells of Halomonas strain AGD 3 grown at 2 M Na+, pH 10. The cells were tested at 2 M Na+ with 10 mM acetate as the electron donor and 5 mM of nitrate (nitrate-grown cells open circles), nitrite (nitrite-grown cells closed circles) or N2O (N2O-grown cells open triangles). The following buffer systems were used: for pH 7–8, HEPES/NaCl; pH 8.5–11.5, NaHCO3/Na2CO3; pH 12, Na2CO3/NaOH. Incubation time, 4 h; cell concentration, 0.6 mg protein/ml. One hundred percent activity: nitrate reductase = 240, nitrite reductase = 190 and N2O reductase = 150 nmol/(mg protein min)
Concluding, we were able to demonstrate denitrification by the native microbial populations in the sediments of hypersaline soda lakes, as well as by the pure cultures of haloalkaliphilic Gammaproteobacteria isolated from the hypersaline soda habitats. Among the cultivated strains, the facultatively anaerobic bacteria of the genus Halomonas were obviously dominating. They are alkaliphilic and extremely salt tolerant with a preference of sodium carbonate over sodium chloride. The anaerobic growth at extremely haloalkaline conditions was constrained mainly at the level of N2O reduction and also resulted in high intermediate nitrite accumulation despite the fact of high salt tolerance of nitrite reduction step. Such bacteria might have a role as a sink for acetate accumulating in anaerobic sediments at hypersaline conditions (Oren 1995, 2001), assuming that nitrogen oxides are available. The source of nitrate in hypersaline habitats is still a question, since nitrification is apparently arrested at salinity above 1 M Na+ (Oren 1999; Sorokin and Kuenen 2005). Most probably, nitrate is entering from the surrounding soils and with the ground waters. The extremely salt tolerant alkaliphilic denitrifiers are also interesting as a possible tool in biological decontamination of acetate/nitrate-containing alkaline wastewater produced during the regeneration of ion-exchange columns (Clifford and Liu 1993).
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
This work was supported by NWO-RFBR (47.011.2004.010), RFBR (07-04-00153) and by the Program of the Russian Academy of Sciences “Molecular and Cell Biology”. We are grateful to A. Lysenko for conducting the DNA–DNA hybridization experiments and to B. Jones for supplying samples from Kenyan soda lakes.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Footnotes
The GenBank accession numbers of the 16S rRNA gene sequences obtained in this study are EU447163-EU447170.
References
- Banciu H, Sorokin DY, Muyzer G, Kleerebezem R, Galinski EA, Kuenen JG. Thioalkalivibrio halophilus sp. nov, a novel obligately chemolithoautotrophic facultatively alkaliphilic and extremely salt-tolerant sulfur-oxidizing bacterium from a hypersaline alkaline lake. Extremophiles. 2004;8:325–334. doi: 10.1007/s00792-004-0391-6. [DOI] [PubMed] [Google Scholar]
- Banciu H, Sorokin DY, Tourova TP, Galinski EA, Muntyan MS, Kuenen JG, Muyzer G (2008) Influence of salts and pH on growth and activity of a novel facultatively alkaliphilic, extremely salt-tolerant, obligately chemolithoautotrophic sufur-oxidizing Gammaproteobacterium Thioalkalibacter halophilus gen. nov., sp. nov. from south-eastern Siberian soda lakes. Extremophilies 12 (in press) [DOI] [PubMed]
- Berendes F, Gottschalk G, Heine-Dobbernack E, Moore ERB, Tindall BJ. Halomonas desiderata sp. nov., a new alkaliphilic, halotolerant and denitrifying bacterium isolated from a municipal sewage works. Syst Appl Microbiol. 1996;19:158–167. [Google Scholar]
- Boltyanskaya YuV, Antipov AN, Kolganova TV, Lysenko AM, Kostrikina NA, Zhilina TN. Halomonas campisalis, an obligatorily alkaliphilic, nitrous oxide-reducing denitrifier with a molybdenum cofactor-lacking nitrate reductase. Mikrobiologia (Moscow Engl Transl.) 2004;73:271–278. [PubMed] [Google Scholar]
- Boltyanskaya YuV, Kevbrin VV, Lysenko AM, Kolganova TV, Tourova TP, Osipov GA, Zhilina TN. Halomonas mongoliensis sp. nov. and Halomonas kenyensis sp. nov., new haloalkaliphilic denitrifiers capable of N2O reduction, isolated from soda lakes. Microbiology (Moscow, Engl Transl) 2007;76:739–747. [PubMed] [Google Scholar]
- Clifford D, Liu X. Biological denitrification of spent regenerant brine using a sequencing batch reactor. Water Res. 1993;27:1477–1484. doi: 10.1016/0043-1354(93)90028-G. [DOI] [Google Scholar]
- De Ley P, Cattoir H, Reynaerts A. The quantitative measurement of DNA hybridization from renaturation rates. Eur J Biochem. 1970;12:133–142. doi: 10.1111/j.1432-1033.1970.tb00830.x. [DOI] [PubMed] [Google Scholar]
- Duckworth AW, Grant WD, Jones BE, Van Steenbergen R (1996) Phylogenetic diversity of soda lake alkaliphiles. FEMS Microbiol Ecol 19:181–191
- Gries-Romijn-van Eck (1966) Physiological and chemical test for drinking water. NEN 1056, IY-2 Nederlandse Normalisatie Instituut Rijswijk
- Jukes TH, Cantor CR. Evolution of protein molecules. In: Munro HN, editor. Mammalian protein metabolism. New York: Academic Press; 1969. pp. 21–123. [Google Scholar]
- Knowles R. Acetylene inhibition technique: development, advantages, and potential problems. FEMS Symp. 1990;56:151–166. [Google Scholar]
- Kulp TR, Han S, Saltikov CW, Lanoil BD, Zargar K, Oremland RS. Effects of imposed salinity gradients on dissimilatory arsenate reduction, sulfate reduction, and other microbial processes in sediments from two California soda lakes. Appl Environ Microbiol. 2007;73:5130–5137. doi: 10.1128/AEM.00771-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lane DJ. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. Chichester: John Wiley & Sons; 1991. pp. 115–177. [Google Scholar]
- Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- Marmur J. A procedure for isolation of DNA from microorganisms. J Mol Biol. 1961;3:208–214. [Google Scholar]
- Marmur J, Doty P. Determination of the base composition of deoxyribonucleic acid from microorganisms. J Mol Biol. 1962;5:109–118. doi: 10.1016/s0022-2836(62)80066-7. [DOI] [PubMed] [Google Scholar]
- Mormile MR, Romine MF, Garcia MT, Ventosa A, Bailey TJ, Peyton BM. Halomonas campisalis sp. nov., a denitrifying, moderately haloalkaliphilic bacterium. Syst Appl Microbiol. 1999;22:551–558. doi: 10.1016/S0723-2020(99)80008-3. [DOI] [PubMed] [Google Scholar]
- Oren A. Uptake and turnover of acetate in hypersaline environments. FEMS Microbiol Ecol. 1995;18:75–84. doi: 10.1111/j.1574-6941.1995.tb00165.x. [DOI] [Google Scholar]
- Oren A. Bioenergetic aspects of halophilism. Microbiol Mol Biol Rev. 1999;63:334–348. doi: 10.1128/mmbr.63.2.334-348.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oren A. The bioenergetic basis for the decrease in metabolic diversity at increasing salt concentrations: Implications for the functioning of salt lake ecosystems. Hydrobiologia. 2001;466:61–72. doi: 10.1023/A:1014557116838. [DOI] [Google Scholar]
- Peyton BM, Mormile MR, Petersen JN. Nitrate reduction with Halomonas campisalis: kinetics of denitrification at pH 9 and 12.5% NaCl. Water Res. 2001;35:4237–4242. doi: 10.1016/S0043-1354(01)00149-X. [DOI] [PubMed] [Google Scholar]
- Pfennig N, Lippert KD. Über das Vitamin B12-Bedürfnis phototropher Schwefelbakterien. Arch Microbiol. 1966;55:245–256. [Google Scholar]
- Romano I, Giordano A, Lama L, Nicolaus B, Gambacorta A. Halomonas campaniensis sp. nov., a haloalkaliphilic bacterium isolated from a mineral pool of Campania region, Italy. Syst Appl Microbiol. 2005;28:610–618. doi: 10.1016/j.syapm.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Sorokin DY, Kuenen JG. Alkaliphilic chemolithotrophs from sodas lakes. FEMS Microbiol Ecol. 2005;52:287–295. doi: 10.1016/j.femsec.2005.02.012. [DOI] [PubMed] [Google Scholar]
- Sorokin DY, Kuenen JG, Jetten M. Denitrification at extremely alkaline conditions in obligately autotrophic alkaliphilic sulfur-oxidizing bacterium Thioalkalivibrio denitrificans. Arch Microbiol. 2001;175:94–101. doi: 10.1007/s002030000210. [DOI] [PubMed] [Google Scholar]
- Sorokin DY, Zhilina TN, Spiridonova EM, Tourova TP, Lysenko AM. Increased metabolic versatility of haloalkaliphilic bacteria belonging to the Alkalispirillum-Alkalilimnicola group from soda lakes. Extremophiles. 2006;10:213–220. doi: 10.1007/s00792-005-0487-7. [DOI] [PubMed] [Google Scholar]
- Taher AG. Inland saline lakes of Wadi El Natrun depression, Egypt. Int J Salt Lake Res. 1999;8:149–170. [Google Scholar]
- Van de Peer Y, De Wachter R. TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput Appl Biosci. 1994;10:569–570. doi: 10.1093/bioinformatics/10.5.569. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.