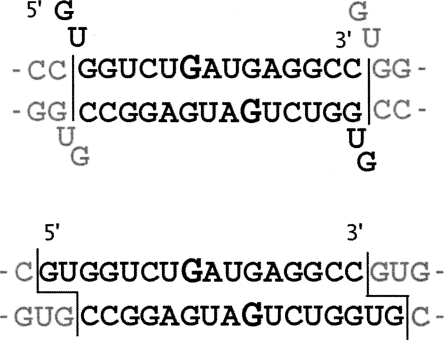Abstract
The structures of two crystal forms of the RNA 16-mer with the sequence GUGGUCUGAUGAGGCC, grown in the presence of a high concentration of sulphate ions, have been determined using synchrotron radiation at 1.4- and 2.0-Å resolution. RNA with this sequence is known as one of the two strands of the noncleavable form of the hammerhead ribozyme. In both crystal structures, two G(syn)–G(anti) noncanonical base pairs are observed in the middle of a 14 base-pair (bp) duplex having 5′-dangling GU residues. Both structures contain sulphate anions interacting with the G–G bp stabilizing G in its syn conformation and bridging the two RNA strands. In both cases the interactions take place in the major groove, although the anions are accommodated within different helix geometries, most pronounced in the changing width of the major groove. In one structure, where a single sulphate spans both G–G pairs, the major groove is closed around the anion, while in the other structure, where each of the two G–G pairs is associated with a separate sulphate, the groove is open. This work provides the first examples of a G–G pair in syn-anti conformation, which minimizes the purine–purine clash in the center of the duplex, while utilizing its residual hydrogen bonding potential in specific interactions with sulphate anions.
Keywords: RNA, duplex, crystal structure, base pairing, G(syn)–G(anti), sulphate anion
INTRODUCTION
RNA forms a wide range of functionally important structural domains containing both single- and double-stranded regions such as hairpins, bulge loop duplexes, or pseudoknots. The tendency to form double-helical regions plays a crucial role in the RNA folding. Double-stranded RNA helices with Watson–Crick base pairs exist principally in the right-handed A-form, the averaged helical parameters of which have been deduced from fiber diffraction data (Arnott et al. 1972; Saenger 1984). The polyanionic structure of RNA and its folding is highly dependent on the environmental conditions—the salt type, its concentration, its pH, and the presence of small molecules or protein ligands. In the native RNA molecules, canonical UA and CG base pairs are often accompanied by different noncanonical base–base interactions, the most frequently found being GU, UU, AA, or GG (Leontis and Westhof 2001). These pairs, located in a certain structural context, often play an important role as recognition sites for cations, especially magnesium, small ligands, or differently charged protein side chains. As recently indicated, they also attract anions, like sulphates (Masquida et al. 1999; Auffinger et al. 2004).
X-ray crystallography and NMR spectroscopy are the principal methods to study RNA fragments at high resolution. Thanks to recent advances in chemical and enzymatic oligoribonucleotide synthesis, the number of X-ray and NMR RNA structures is rapidly increasing; around 1100 of them have been deposited in the PDB and NBD. It appeared that some of the RNA structures depend strongly on the environmental conditions chosen, either to measure the sample (NMR) or used to promote crystal growth. This is especially pronounced for certain oligoribonucleotide chains that are capable of salt dependent hairpin/duplex equilibria (Antao and Tinoco 1992). Moreover, the presence of small ligands used in X-ray crystallography as precipitation and cryo-protective agents (Adamiak et al. 2001; Rypniewski et al. 2006) can influence the RNA structure. RNA crystallization frequently requires high salt conditions far exceeding those typical for a living cell. This often makes comparison of crystallographic structures with those delivered by high resolution NMR, taken under low salt conditions, complicated and puzzling.
Our study concerns the 16-mer RNA with the sequence r(GUGGUCUGAUGAGGCC). This sequence is known as one of the two strands of the noncleavable hammerhead ribozyme (16-mer + 25-mer), the X-ray crystal structure of which was solved in the Klug laboratory (Scott et al. 1995). The hammerhead crystal was grown in 1.8 M Li2SO4. Former NMR solution studies applying salt concentrations up to 0.5 M Li2SO4 and 0.3 M NaCl show that both the 16-mer and the 25-mer strands cannot associate to form the hammerhead structure at lower salt concentrations (Zamaratski et al. 2001) and that the 16-mer RNA alone forms a stable hairpin structure (Z. Gdaniec, J. Milecki, and R.W. Adamiak, unpubl.).
Here we show that the 16-mer r(GUGGUCUGAUGAGGCC) crystallized in high salt (1.3 M Li2SO4 or 1.8 M (NH4)2SO4) forms self-complementary duplexes with two noncanonical G(syn)–G(anti) base pairs stabilized by sulphate di-anions. The two structures analyzed at 1.4-Å and 2.0-Å resolution provide the first view of the interactions of sulphate di-anions with G–G noncanonical base pairs.
RESULTS
The refined models and crystal packing
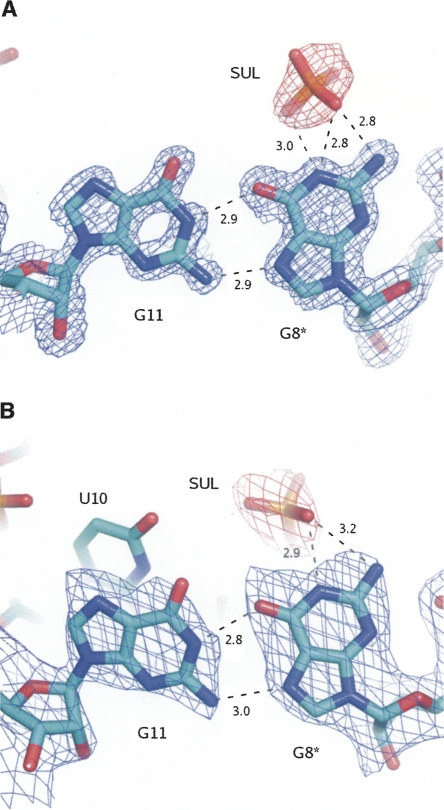
Two crystal forms, monoclinic and rhombohedral, of the RNA 16-mer with the sequence GUGGUCUGAUGAGGCC, grown in the presence of a high concentration of sulphate ions, have been analyzed using synchrotron radiation at 1.4- and 2.0-Å resolution (Table 1). The final model of the monoclinic structure consists of 14-nt residues in one strand, forming a self-complementary right-handed double helix with a symmetry-related strand. The helix starts and ends with a base pair G3–C16 (Fig. 1). The dangling two residues on the 5′ side are disordered and are not visible in the electron density map. The position of the phosphate group of G3 indicates that the disordered G1 and U2 extend away from the helix axis and into the solvent space. The other residues form helices that stack end to end in the crystal lattice. The monoclinic model also contains 69 ordered water molecules and one sulphate ion with the occupancy factor 0.5 located near a crystallographic twofold axis.
TABLE 1.
Summary of X-ray data and the final models
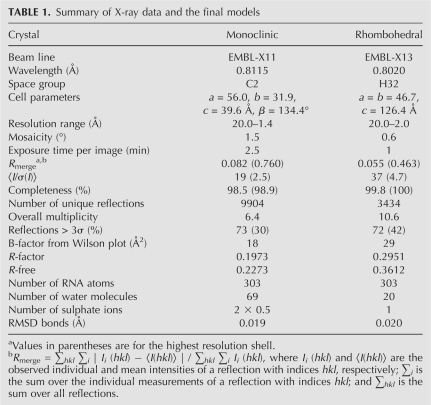
FIGURE 1.
Pairing-up scheme of the 16-mer RNA strands in the monoclinic (top) and the rhombohedral crystal forms (bottom), including the G–G cis Watson–Crick/Hoogsteen pairs (according to Leontis and Westhof 2001). Residues in the syn conformation are emphasized. Dangling ends are indicated in the monoclinic form. Residues belonging to neighboring duplexes in the crystal lattice are indicated in gray.
The rhombohedral model consists of 16 ordered nucleotide residues in one strand forming a double helix with two symmetry-related strands. The first two residues, G1 and U2, base-pair with the first two residues of a symmetry mate: G1 with U2* and U2 with G1* (asterisks denote residues from symmetry-related strands), and residues G3 to C16 pair up with residues C16** to G3** from another symmetry mate (Fig. 1). The model of the rhombohedral form also contains 20 ordered water molecules and one sulphate ion located on the crystallographic twofold axis. The electron density for the middle region of the rhombohedral structure shows considerable disorder, particularly for the sugar–phosphate backbone. This is reflected in considerably poorer R-factor statistics (Table 1).
Both crystal lattices consist of pseudoinfinite parallel columns of coaxially stacked duplexes, but in the monoclinic structure the dangling ends are frayed out, while in the rhombohedral lattice they pair up as sticky ends.
The RNA duplex conformation and the noncanonical base pairs
In both duplexes, most of the conformational parameters describing the phospho-sugar backbone are typical of the RNA A-form (Saenger 1984). However, the middle base-pair region (sequence 5′-GAUG-3′), containing two noncanonical G–G pairs, departs from the A-helix for residues U7, G8, A9, U10, and G11 (Supplemental Table 1). All but one of the sugar residues have a C3′-endo pucker with average PN value of 15° and a high pucker. In both crystal structures the G8 residues are characterized by the syn conformation, all others have anti glycosidic bond angles. The γ torsion angles in the monoclinic structure are in the (+) gauche conformation, typical of the A-form, except for G8, which is trans, whereas in the rhombohedral structure the γ angle is elevated for residues G8 to G11.
With the exception of the disordered G1 and U2 in the monoclinic structure (see below), the base-pairing scheme is similar in both the crystal forms, but the pairing details differ. In the monoclinic crystal standard Watson–Crick interactions are observed for G3–C16*, G4–C15*, and C6–G14* (and, naturally, for the symmetric interactions: G3*–C16, G4*–C15, and C6*–G14).
U5 and G14* form wobble pairs in both structures. The 5′ ends of coaxially stacked duplexes in the rhombohedral structure also form wobble pairs: G1–U2** and U2–G1**. The corresponding residues in the monoclinic structure are disordered. In this form well-defined electron density ends at the G3 5′ phosphate group, whose position, away from the helical axis, indicates that the GU 5′ end of the RNA strand points into the solvent region. Thus, the end-to-end duplex stacking involves pairs G3–C16* and their symmetry-equivalent residues.
In both crystal structures the center of the duplex contains the characteristic two G–G base pairs: G8(syn)–G11*(anti) and G8*(syn)–G11(anti). Each pair has two hydrogen bonds: G8(carbonyl)–G11(N1H) and G(8)N7–G11(exo-amino). All the H-bonds are in the range 2.8–3.0 Å. The conformation of the G8(syn) residue is additionally stabilized by a hydrogen bond between the exo-amino function and its 5′ phosphate oxygen (2.8 Å).
The sequence-independent helical parameters, based on interstrand vectors between the C1′ atoms (Lu and Olson 2003), were found to be a convenient, although simplified, measure of the helix properties (Supplemental Table 2). The helical twist of the rhombohedral form is systematically lower than for the monoclinic structure, giving on average 12.8 and 12.1 base pairs (bp) per turn, respectively. Both helices are less tightly wound than a canonical A-RNA (11 bp/turn). The interstrand stacking of purine residues is observed. Such a pattern of purine interstrand stacking in pyrimidine–purine steps is a typical feature of A-RNA helices.
One outstanding difference between the monoclinic and the rhombohedral structures is in the width of the major groove. Whereas in the monoclinic structure the grooves are rather regular throughout the length of the duplex, in the rhombohedral structure the major groove is narrower in the middle of the duplex (Fig. 2). The widths of the grooves, as defined by El Hassan and Calladine (1998) are given in Supplemental Table 3.
FIGURE 2.
The sulphate ions in the major groove. Omit electron density (red contours) is shown at the level of 2.8 σ. In the monoclinic structure (A) the major groove accommodates two partially occupied sulphate sites, whereas in the rhombohedral structure (B) the major groove is closed around the single sulphate.
Sulphate ions and hydration in the major groove
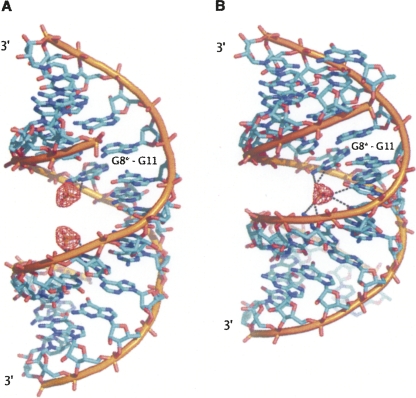
In the monoclinic form two partially occupied (0.5) sulphate ion sites are located in the major groove (Fig. 3A). The sulphates interact with guanosine residues that are in the syn conformation. One of the sulphate oxygen atoms is positioned such that it can accept H-bonds from the exo-NH2 (acceptor–donor distance 2.8 Å) or from the N1H (2.8 Å). Another sulphate oxygen atom is also within H-bonding distance of the latter imino group (3.0 Å). Three distinct water sites are located near each sulphate ion, at 2.7 (W37), 2.6 (W38), and 2.2 Å (W67) from sulphate oxygen atoms. The latter close distance implies disorder where two partial sites are occupied alternatively rather than concurrently. In addition, W38 stabilizes the G8(syn)–G11(anti) pair, interacting with both O6 sites (2.8 and 3.0 Å). Two further water molecules interact with the G(anti) residue: at the N7 and the phosphate sites, respectively. Two U–A pairs flank the G–G pair and are hydrated involving U residues at the O4 and A residues at the exo-amino sites. Thus, their hydration is similar to the general scheme observed previously in RNA crystal structures (Auffinger and Hashem 2007). The same is true of the C–G pairs: The ordered water molecules interact with the exo-amino functions of C residues and O6, N7 sites of G. The wobble base pair G–U is stabilized by a water molecule interacting with the O6 and O4 acceptors.
FIGURE 3.
The interactions within the G11(anti)–G8*(syn) pair and the sulphate ion, for the monoclinic (A) and the rhombohedral structures (B). The blue contours are for the 2Fo-Fc electron density maps at 1σ level and the red contours are for the omit difference maps at 2.8σ level. The star denotes the symmetry related RNA strand.
In the rhombohedral form, a single, fully occupied sulphate anion site is observed on the crystallographic twofold axis where it bridges the two symmetry related RNA strands (Fig. 3B). The sulphate is accepting a H-bond from N1H (2.9 Å) and exo-NH2 (3.2 Å) of the G8 residue, which is in the syn conformation. These interactions are accompanied by a weaker hydrogen bond between another O sulphate atom and the exo-NH2 function of A9 residue (3.6 Å). Interactions symmetric to these also occur between the remaining O sulphate atoms and the symmetry-related G8* and A9* residues. No water molecules were found associated with the sulphate ion. The major groove is narrowed around the sulphate site, as shown in Supplemental Table 2.
Minor groove hydration
In the monoclinic structure, the majority of the 69 ordered water molecules are located in the minor groove. The overall scheme of hydration is such that interacting solvent molecules span the sugar 2′-hydroxyl groups between the duplex strands. The interactions within the groove involve carbonyl O2 atoms of pyrimidines, exo-amino function of G and N3 of both purines. Typically, three water molecules are involved in spanning the width of the groove between the sugar residues unless the network is interrupted by an intrusion of a neighboring duplex. There are four such intrusions involving phosphate–sugar interactions and two involving sugar–sugar contacts along the 14 bp of the duplex. An ordered water molecule stabilizes the G14–U5 wobble by interacting with the carbonyl and the 2′ oxygen atoms of U and with the exo-amino function of G. This hydration pattern is similar to that observed in the case of G8–U12 wobble by Masquida et al. (1999) and other RNA structures.
In the rhombohedral structure, most of the 20 ordered water molecules are also associated with the minor groove and with the phosphate oxygen atoms of the backbone.
DISCUSSION
The 16-mer oligoribonucleotide, having the sequence r(GUGGUCUGAUGAGGCC), comprises the shorter strand of the hammerhead ribozyme (the other strand being a 25-mer). The structure of the ribozyme, solved by Scott et al. (1995), was obtained using crystals grown under high salt, 1.8 M Li2SO4. However, it was found by NMR that under physiological salt concentrations the association of the two strands is incomplete (Zamaratski et al. 2001). The isolated 16-mer shows a preference for self-folding as a hairpin even at salt concentrations up to 0.5 M Li2SO4 and 0.3 M NaCl (Zamaratski et al. 2001). This has been confirmed by our UV measurements, which also showed that in higher salt the RNA hairpin/duplex equilibrium is shifted toward the duplex (data not shown).
Here we show that the r(GUGGUCUGAUGAGGCC) forms right-handed duplexes when crystallized in high salt: 1.3 M lithium sulphate (monoclinic form) or 1.7 M ammonium sulphate (rhombohedral). In both cases the helices have 5′-dangling ends consisting of GU residues. The structures contain two G(syn)–G(anti) noncanonical base pairs within the central region 5′-GAUG-3′. Although the formation of the G(syn)–G(anti) pairs considerably diminishes purine–purine clashes in the middle of the duplex, the conformational features of that region differ considerably from the canonical A-RNA (Supplemental Table 1). Most of the sugar residues have typical C3′-endo puckers. All residues except G8 are characterized by anti glycosidic bond. Apart from the middle of the duplex, the Watson–Crick base pairing and G–U wobble, standard geometry is observed.
The G(syn)–G(anti) base pair has been observed in RNA in only a few cases: in the 16S rRNA of the Escherichia coli ribosome (Schuwirth et al. 2005), in the HIV Rev response element hairpin (Ippolito and Steitz 2000) and its duplex model (Hung et al. 2000), and in synthetic RNA duplexes studied by X-ray (Timsit and Bombard 2007) and by NMR methods (Burkard and Turner 2000). Despite being embedded in different sequence and structural contexts, the above structures share similar G–G base-pair geometry, which includes G(carbonyl)–G(N1) and G(N7)–G(exo-amino) hydrogen bonds. In addition, the G(syn) conformation is stabilized by a hydrogen bond between the exo-amino function of that residue and its 5′ phosphate oxygen. The 16-mer high-salt duplex structures presented here share those features. However, the well-ordered G(syn)–G(anti) pairs within the segment 5′-GAUG-3′ in the crystal can be contrasted with the NMR structure of r(GCAGGCGUGC)2, analyzed by Burkard and Turner (2000). In the latter case, two distinct populations of duplexes have been observed in low-salt solution, showing a preference for G4(anti)–G7(syn) over G4(syn)–G7(anti).
The polyanionic structure of RNA is usually discussed in terms of its dependence on the presence of mono- and divalent cations, especially magnesium (Misra and Draper 1998). More recently, several crystallographic reports have been published pointing to specific interactions between RNA and anions, like sulphate or chloride (Auffinger et al. 2004 and references therein; Auffinger and Hashem 2007). The reported interactions with sulphate encompass nucleobases and ribose residues. Sulphate anions have been observed to interact with guanine residues in different structural contexts, e.g., binding to two neighboring guanosines in the minor groove (Jovine et al. 2000) or binding to the guanosine exo-amino function in the G–U wobble pairs while spanning three interacting helices forming the so-called sulphate pocket (Masquida et al. 1999).
Our work provides the first examples of sulphate anions interacting with G–G base pairs in an RNA duplex. The syn-anti geometry of this base pair (G/G cis Watson–Crick/Hoogsteen, according to the nomenclature proposed by Leontis and Westhof 2001) seems to be well suited for accommodating the di-anions. In both structures the G8 in syn conformation engages its Hoogsteen edge in pairing with G11 while it exposes its Watson–Crick face (–N1H, –NH2) to the solvent. The predominantly proton-donating character of the Watson–Crick edge shows its affinity for electro-negative ligands, such as the observed sulphate di-anions. This may be an indication of the potential role of this base-pairing arrangement in specific recognition of ligands, such as acidic protein residues. Our results extend a proposition of Masquida et al. (1999) based on the sulphate affinity for the exo-amino group of the wobble G–U base pair.
The presence of two G–G base pairs together with the associated sulphate anions have considerable impact on the duplex geometry, especially in the vicinity of the mismatch. Comparison of our two structures reveals how the sulphate anions stabilize the duplex by saturating the hydrogen bonding potential of the G(syn)–G(anti) pair. The comparison also shows how the anions are accommodated within different helix geometries. The effect is most pronounced in the changing width of the major groove. In the rhombohedral structure, where a single sulphate spans both G–G pairs, the major groove is closed around the anion, while in the monoclinic structure, where each of the two G–G pairs is associated with a separate sulphate, the groove is open. The very marked differences in the widths of the major grooves are apparent to the eye (Fig. 2), but less so if one only examines the commonly used conformational parameters. The distances between phosphorus atoms are not necessarily a good indication of groove width because they depend on the juxtaposition of the atoms. The best measure of the groove width in the examined structures was the “refined P–P distance” (Supplemental Table 3) calculated according to El Hassan and Calladine (1998).
The reported crystal structures confirm the current opinion that under high salt the RNA hairpin/duplex equilibrium is shifted toward the duplex. The two RNA molecules, being chemically identical, crystallize in different crystal forms showing both similarities and differences. In both cases the duplexes form bundles of semi-infinite helices that stack end to end. For unclear reasons the 5′ dangling ends in the monoclinic structure are directed away from the helix axis and disordered, whereas in the rhombohedral structure they participate in the helix-to-helix interactions. The major common features are the G(syn)–G(anti) pairs and their affinity for sulphate anions. In both cases the interactions take place in the major groove, although the geometry of the sulphate binding differs. In the monoclinic structure the conformation of the duplex is such that the two G–G pairs are so close that a single sulphate interacts with both, whereas in the rhombohedral structure the G–G pairs are further apart and accordingly the sulphate site is split in two.
Noncanonical base pairs are common features of RNA molecules, being part of their structural repertoire. To understand their role in RNA function one needs to describe both the structure of the molecules themselves and their potential to interact with the various elements of the cellular environment. Crystallographic structures of RNA contain very detailed information about the molecules themselves and their interactions with the solvent and, although the high salt concentrations often used in crystallography are far from physiological, they point to a propensity of the RNA to engage specific ligands. The two structures presented here show how, under high salt conditions, G–G pairs embedded in a helical context adopt a stable syn-anti conformation well accommodated within right-handed RNA while utilizing their residual hydrogen bonding potential in ordered interactions with sulphate anions.
MATERIALS AND METHODS
Crystallization and data processing
RNA 16-mer of the r(GUGGUCUGAUGAGGCC) sequence was prepared by solid-support aided phosphoramidite chemistry applying the 2′-O-TOM protection (5 × 1 μmol scale; 5′-O-DMTr off option), all according to the manufacturer's protocol (Glen Research). Oligoribonucleotide was purified using HPLC (reverse-phase X-Terra Waters column, acetonitrile gradient in 10 mM ammonium carbonate at pH 6.5), desalted on Sephadex-G10, and lyophilized. Crystals were grown at 20°C by the hanging drop/vapor diffusion method. Crystals of the monoclinic form were prepared under the following conditions: The reservoir contained 0.5 mL of solution containing 1.3 M lithium sulphate, 100 mM cacodylate buffer (pH 6.0). Crystals of the rhombohedral form were obtained under the following conditions: The reservoir contained 0.5 mL of solution containing 1.7 M ammonium sulphate, 50 mM cacodylate buffer (pH 6.0) and 15 mM magnesium acetate. The crystallization drops initially consisted of 3 μL of the 5 mg/mL RNA solution and 3 μL of the reservoir solution. X-ray diffraction data were obtained on the EMBL X11 and X13 beam lines at the DORIS storage ring, DESY, Hamburg. Prior to mounting, the crystals were transferred to cryoprotectant solutions similar to the reservoir solution but containing in addition 20% (v/v) glycerol. The crystals remained in the cryoprotectant solution for ∼1 min., i.e., only the time that was needed to pick them up again in the cryo-loop. The crystals were transferred in cryo-loops directly to the goniostat and vitrified in the stream of cold nitrogen gas. The diffraction images were recorded on a MAR CCD 165-mm detector. The diffraction intensities were integrated and scaled using the program suite DENZO/SCALEPACK (Otwinowski and Minor 1997). The crystals and X-ray data statistics are summarized in Table 1.
Structure solution and crystallographic refinement
Both crystal structures were solved by molecular replacement (MR). The rhombohedral structure was solved with the program AMoRe (Navaza 1994) using as the search model the atomic coordinates of the RNA duplex formed by the strands 5′-GUCU-3′ and 5′-AGGC-3′, excised from the X-ray model of the 50S ribosomal subunit (NDB id: rr0033) (Klein et al. 2001). Electron density maps, 2Fo-Fc and Fo-Fc, calculated using phases derived from the initial MR model were initially poor but sufficient for refining and extending the model until it was complete. The monoclinic structure was solved with the program Phaser (McCoy et al. 2005), using as the search model the atomic coordinates of RNA from the rhombohedral structure. Although the asymmetric unit contained one 16-mer strand of the RNA duplex, no MR solution was obtained with the single strand as the search model. The model that was used successfully comprised the first seven and the last five nucleotide residues in a duplex (including the overhang). Afterward, the model could be easily corrected and extended based on electron density maps. The models were refined using the program Refmac5 (Murshudov et al. 1997) from the CCP4 program suite (Collaborative Computational Project 4 1994). The atomic coordinates and the diffraction data have been deposited under PDB codes 3CZW (monoclinic) and 3D0M (rhombohedral).
Analysis of the helical parameters
Helical parameters based on C1′–C1′ have been calculated for both the crystal structures using the 3DNA program package (Lu and Olson 2003). Unlike the conventional local base-pair parameters, they are sequence independent and therefore do not suffer from computational artifacts arising from the reverse direction of the local z-axis due to the “reverse face” of the G(syn). The standard output of the 3DNA program showed apparently negative rise and “strange” values of tilt and several other local helical parameters.
SUPPLEMENTAL DATA
Supplemental material contains tables detailing (a) the phosphosugar backbone conformation (Supplemental Table 1); (b) the helical parameters (Supplemental Table 2); and (c) the duplex groove analysis (Supplemental Table 3). Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
This work was supported by a grant from the Polish Ministry of Science and Education to R.W.A. (7T09A09720) and by the European Community Research Infrastructure Action under the F6P “Structuring the European Research Area Programme” (contract number RII3/CT/2004/5060008) to W.R. The authors are grateful to M. Kluge for assistance with crystallization.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1164308.
REFERENCES
- Adamiak D.A., Rypniewski W.R., Milecki J., Adamiak R.W. The 1.19 Å X-ray structure of 2′-O-Me(CGCGCG)2 duplex shows dehydrated RNA with 2-methyl-2,4-pentanediol in the minor groove. Nucleic Acids Res. 2001;29:4144–4153. doi: 10.1093/nar/29.20.4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antao V.P., Tinoco I., Jr Thermodynamic parameters for loop formation in RNA and DNA hairpin tetraloops. Nucleic Acids Res. 1992;20:819–824. doi: 10.1093/nar/20.4.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnott S., Hukins D.W.L., Dover S.D. Optimized parameters for RNA double-helices. Biochem. Biophys. Res. Commun. 1972;48:1392–1399. doi: 10.1016/0006-291x(72)90867-4. [DOI] [PubMed] [Google Scholar]
- Auffinger P., Hashem Y. SwS: A solvation web service for nucleic acids. Bioinformatics. 2007;23:1035–1037. doi: 10.1093/bioinformatics/btm067. [DOI] [PubMed] [Google Scholar]
- Auffinger P., Bielecki L., Westhof E. Anion binding to nucleic acids. Structure. 2004;12:379–388. doi: 10.1016/j.str.2004.02.015. [DOI] [PubMed] [Google Scholar]
- Burkard M.E., Turner D.H. NMR structures of r(GCAGGCGUGC)2 and determinants of stability for single guanosine–guanosine base pairs. Biochemistry. 2000;39:11748–11762. doi: 10.1021/bi000720i. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project 4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- El Hassan M.A., Calladine C.R. Two distinct modes of protein-induced bending in DNA. J. Mol. Biol. 1998;282:331–343. doi: 10.1006/jmbi.1998.1994. [DOI] [PubMed] [Google Scholar]
- Hung L.-W., Holbrook E.L., Holbrook S.R. The crystal structure of the Rev binding element of HIV-1 reveals novel base pairing and conformational variability. Proc. Natl. Acad. Sci. 2000;97:5107–5112. doi: 10.1073/pnas.090588197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ippolito J.A., Steitz T. The structure of the HIV-1 RRE high affinity Rev binding site at 1.6 Å resolution. J. Mol. Biol. 2000;295:711–717. doi: 10.1006/jmbi.1999.3405. [DOI] [PubMed] [Google Scholar]
- Jovine L., Hainzl T., Oubridge C., Scott W.G., Li J., Sixma T.K., Wonacott A., Skarzynski T., Nagai K. Crystal structure of the ffh and EF-G binding sites in the conserved domain IV of Escherichia coli 4.5S RNA. Structure. 2000;8:527–540. doi: 10.1016/s0969-2126(00)00137-4. [DOI] [PubMed] [Google Scholar]
- Klein D.J., Schmeing T.M., Moore P.B., Steitz T.A. The kink-turn: A new RNA secondary structure motif. EMBO J. 2001;20:4214–4221. doi: 10.1093/emboj/20.15.4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leontis N.B., Westhof E. Geometric nomenclature and classification of RNA base pairs. RNA. 2001;7:499–512. doi: 10.1017/s1355838201002515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X.-J., Olson W.K. 3DNA: A software for the analysis, the rebuilding and the vizualisation of three-dimensional nucleic acid structures. Nucleic Acids Res. 2003;31:5108–5121. doi: 10.1093/nar/gkg680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masquida B., Sauter C., Westhof E. A sulfate pocket formed by three GoU pairs in the 0.97 Å resolution X-ray structure of a nonameric RNA. RNA. 1999;5:1384–1395. doi: 10.1017/s1355838299991173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy A.J., Grosse-Kunstleve R.W., Storoni L.C., Read R.J. Likelihood-enhanced fast translation functions. Acta Crystallogr. D Biol. Crystallogr. 2005;61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- Misra V.K., Draper D.E. On the role of magnesium ions in RNA stability. Biopolymers. 1998;48:113–135. doi: 10.1002/(SICI)1097-0282(1998)48:2<113::AID-BIP3>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Murshudov G.N., Vagin A.A., Dodson E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Navaza J. AMoRe: An automated package for molecular replacement. Acta Crystallogr. A. 1994;50:157–163. [Google Scholar]
- Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. In: Carter C.W., Sweet R.M., editors. Methods in enzymology. Vol. 276. Academic Press; New York: 1997. pp. 307–325. [DOI] [PubMed] [Google Scholar]
- Rypniewski W., Vallazza M., Perbandt M., Klussmann S., Delucas L.J., Betzel C., Erdmann V.A. The first crystal structure of an RNA racemate. Acta Crystallogr. D Biol. Crystallogr. 2006;62:659–664. doi: 10.1107/S090744490601359X. [DOI] [PubMed] [Google Scholar]
- Saenger W. Principles of nucleic acids structure. Springer; Berlin, Heidelberg: 1984. [Google Scholar]
- Schuwirth B.S., Borovinskaya M.A., Hau C.W., Zhang W., Vila-Sanjurjo A., Holton J.M., Cate J.H. Structures of the bacterial ribosome at 3.5 Å resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- Scott W.G., Finch J.T., Klug A. The crystal structure of an all-RNA hammerhead ribozyme: A proposed mechanism for RNA catalytic cleavage. Cell. 1995;81:991–1002. doi: 10.1016/s0092-8674(05)80004-2. [DOI] [PubMed] [Google Scholar]
- Timsit Y., Bombard S. The 1.3 Å resolution structure of the RNA tridecamer r(GCGUUUGAAACGC): Metal ion binding correlates with base unstacking and groove contraction. RNA. 2007;13:2098–2107. doi: 10.1261/rna.730207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamaratski E., Trifonova A., Acharya P., Isaksson J., Maltseva T., Chattopadhayaya J. Do the 16 mer, 5′-GUGGUCUGAUGAGGCC-3′ and the 25 mer, 5′-GGCCGAAACUCGUAAGAGUCACCAC-3′, form a hammerhead ribozyme structure in physiological conditions? An NMR and UV thermodynamic study. Nucleosides Nucleotides Nucleic Acids. 2001;20:1219–1223. doi: 10.1081/NCN-100002522. [DOI] [PubMed] [Google Scholar]