Abstract
Rho G proteins and their regulators are critical for cytoskeleton organization and cell morphology in all eukaryotes. In the opportunistic pathogen Candida albicans, the Rho G proteins Cdc42 and Rac1 are required for the switch from budding to filamentous growth in response to different stimuli. We show that Dck1, a protein with homology to the Ced-5, Dock180, myoblast city family of guanine nucleotide exchange factors, is necessary for filamentous growth in solid media, similar to Rac1. Our results indicate that Dck1 and Rac1 do not function in the same pathway as the transcription factor Czf1, which is also required for embedded filamentous growth. The conserved catalytic region of Dck1 is required for such filamentous growth, and in vitro this region directly binds a Rac1 mutant, which mimics the nucleotide-free state. In vivo overexpression of a constitutively active Rac1 mutant, but not wild-type Rac1, in a dck1 deletion mutant restores filamentous growth. These results indicate that the Dock180 guanine nucleotide exchange factor homologue, Dck1 activates Rac1 during invasive filamentous growth. We conclude that specific exchange factors, together with the G proteins they activate, are required for morphological changes in response to different stimuli.
INTRODUCTION
Rho G proteins, which are part of the Ras G protein superfamily, function as molecular switches required for transducing a large range of signals. They are critical regulators of several key biological processes, including cytoskeleton organization and cell polarity (Jaffe and Hall, 2005). Rho G proteins are found in eukaryotic cells from yeast to humans, and they are divided into three subfamilies: Cdc42, Rac, and Rho. In humans, >20 Rho G proteins have been identified (Hall, 2005), whereas in yeast such as Saccharomyces cerevisiae, only six Rho G proteins have been described (Park and Bi, 2007). Rho G proteins are highly conserved (Jaffe and Hall, 2005), for example S. cerevisiae Cdc42 has >80% overall sequence identity with its human counterpart. All Rho G proteins cycle between an inactive guanosine diphosphate (GDP)-bound form and an active guanosine triphosphate (GTP)-bound form. These GTPases are temporally and spatially regulated by activators or guanine nucleotide exchange factors (GEFs) and inactivators such as GTPase-activating proteins (GAPs) or guanine nucleotide dissociation inhibitors (DerMardirossian and Bokoch, 2005; Buchsbaum, 2007; Tcherkezian and Lamarche-Vane, 2007).
In humans, there are two major classes of Rho G protein GEFs, which together have >85 members (Hall, 2005). The principal class of GEFs, the Dbl family (Rossman et al., 2005) is made up of several transforming factors or proto-oncogenes, including Dbl, Vav, Tiam1, and LARG, which are involved in a variety of cancers (Karnoub et al., 2004). Cdc24, the sole Cdc42 activator in S. cerevisiae (Gulli and Peter, 2001), is one of the founding members of this class of GEFs, which are characterized by a conserved catalytic domain called a Dbl Homology (DH) domain (Erickson and Cerione, 2004; Rossman and Sondek, 2005). Recently, a new class of GEFs has been identified that has a common catalytic domain, referred to as a DOCKER domain, with no apparent sequence homology to the DH domain (Brugnera et al., 2002; Cote and Vuori, 2002). This class of GEFs, referred to as the CDM zizimin Homologs (CZH) family (Meller et al., 2005) is made up of the CDM (Caenorhabditis elegans Ced-5, Homo sapiens Dock180, and Drosophila melanogaster myoblast city) family (Hasegawa et al., 1996; Erickson et al., 1997; Wu and Horvitz, 1998) and the zizimin family (Meller et al., 2002). These GEFs either activate Rac (typically CDM family) or Cdc42 (typically zizimin family). For example, human Dock180 has been shown to bind nucleotide-free Rac1 (Brugnera et al., 2002; Cote and Vuori, 2002). Homologues of this CZH class of GEFs have also been identified in Dictyostelium discoideum, fungi, and, interestingly, plants such as Arabidopsis thaliana, in which no Dbl family GEF has been identified (Meller et al., 2005). Given their recent identification, much less is known about the CZH GEF family compared with the Dbl family. Nonetheless, the CZH GEFs such as myoblast city have been shown to be involved in myoblast fusion, dorsal closure, cytoskeletal organization, and border cells migration (Erickson et al., 1997; Bianco et al., 2007), whereas Ced-5 plays a critical role in phagocytosis and cell migration (Wu and Horvitz, 1998) and Dock180 is required for cell morphogenesis (Hasegawa et al., 1996) and phagocytosis (Albert et al., 2000; Park et al., 2007). Very recently, Dock180 has been shown to have a critical role in netrin-dependent axon outgrowth and attraction (Li et al., 2008). In S. cerevisiae, where there is no Rac1 homologue, the protein Ylr422wp, which has a region similar to the catalytic DOCKER domain, has been reported to bind human Rac1 (Brugnera et al., 2002), yet its function is unknown. A gene encoding Rac1 is present in virtually all other fungal genomes yet little is known about its activation, in particular the role of CDM family GEFs.
Candida albicans is an opportunistic human fungal pathogen that can cause superficial mucosal infections and systemic infections, in particular when the host immune system is compromised (Odds et al., 1988; Calderone, 2002). The capacity of C. albicans to reversibly switch from “yeast” to filamentous growth is associated with pathogenicity. Several different signals can induce this morphogenesis, including serum, nutrients, or entrapment in an agar matrix (Kumamoto and Vinces, 2005). Both mitogen-activated protein kinase and cAMP-dependent protein kinase A pathways are required for the induction of a range of genes that are critical for this morphological transition (Biswas et al., 2007). This has also been observed for invasive pseudohyphal growth of S. cerevisiae (Park and Bi, 2007). Nonetheless, the cytoskeleton reorganization that is necessary for this morphogenesis in C. albicans can be uncoupled from hyphal specific gene induction (Sinha et al., 2007). Two critical cytoskeleton regulators, Cdc42 and its activator Cdc24 are required for serum-induced invasive filamentous growth and pathogenicity (Michel et al., 2002; Ushinsky et al., 2002; Bassilana et al., 2003; VandenBerg et al., 2004). Recently, we identified an additional Rho G protein in C. albicans, Rac1, which is required for filamentous growth in an agar matrix (Bassilana and Arkowitz, 2006). Despite ∼60% sequence identity between C. albicans Rac1 and Cdc42, these two G proteins are required for filamentous growth in response to distinct signals. Furthermore, overexpression of either G protein is unable to complement the function of the other G protein. These results suggest that Cdc42 and Rac1 function in distinct signaling pathways. In contrast to Cdc42, nothing is known about how Rac1 is regulated in C. albicans. Here, we identify Dck1, which contains a region with similarity to the DOCKER catalytic domain, as the Rac1 activator during C. albicans filamentous growth. Our results indicate that CDM family GEF function has been conserved in fungi.
MATERIALS AND METHODS
Growth Conditions
Yeast extract-peptone dextrose (YEPD) or synthetic complete (SC) medium was used, and strains were grown at 30°C, unless indicated otherwise. Filamentous growth induction was carried out in liquid or solid media as described previously either in 50% serum (Bassilana et al., 2003), Spider medium (Calera et al., 2000) or N-acetyl glucosamine (GlcNAc) (Castilla et al., 1998). For filamentous growth induction on solid media, 2% agar was included. Filamentous growth induction in embedded media was carried out in YEP containing 2% sucrose as described previously (Brown et al., 1999) and for quantification, colonies with five or more filaments were scored as filamentous.
Strains
The strains and primers used in this study are listed in Tables 1 and 2, respectively. The DCK1 and DCK2 Tn7-UAU1 strains (PY284 and PY286) were obtained by transforming an auxotrophic his−ura−arg− strain (BWP17) with the respective Tn7-UAU1 plasmids (CAGE530 and CAGDV28, a gift from A. Mitchell), digested with NotI. ARG+URA+ prototrophs were isolated and verified as described previously (Davis et al., 2002). Dck1Δ/dck1Δ mutants were obtained by initially transforming BWP17 with a dck1Δ::HIS1 polymerase chain reaction (PCR) product generated from pGemHis1, by using DCK1.P1 and DCK1.P2 primers, yielding the heterozygote strain PY646. This deletion removed essentially all of the DCK1 open reading frames (ORF), with only 74 base pairs 3′ of the ATG and 68 base pairs 5′ of the stop codon remaining. Subsequently, a dck1Δ::URA3 PCR product was generated using the same primers and pGemUra3, transformed into PY646 and URA+HIS+ prototrophs were selected, yielding PY704. Correct insertion into the DCK1 locus was confirmed by PCR. Czf1Δ/czf1Δ mutants were obtained by initially transforming BWP17 with a czf1Δ::HIS1 PCR product generated from pGemHis1, by using CZF1.P1 and CZF1.P2 primers, yielding the heterozygote strain PY523. This deletion removed all of the CZF1 ORF. Subsequently, a czf1Δ::URA3 PCR product was generated using CZF1.P3 and CZF1.P4 primers and pGemUra3, transformed into PY523, yielding the homozygote strain PY897. This deletion removed essentially all of the second CZF1 ORF with 78 base pairs 3′ of the ATG and 74 base pairs 5′ of the stop codon remaining. Correct insertion into the CZF1 locus was confirmed by PCR. The pExpArg constructs were digested with StuI and targeted to the RP10 locus in the appropriate strains. Similar results were obtained with two independent isolates of each strain.
Table 1.
Yeast strains used in this study
| Yeast strain | Relevant genotype | Reference |
|---|---|---|
| BWP17 | ura3Δ:: imm434/ura3Δ:: imm434 his1Δ::hisG/his1Δ::hisG arg4Δ::hisG/arg4Δ::hisG | Wilson et al. (1999) |
| PY82 | ura3Δ:: imm434/ura3Δ:: imm434 his1::hisG/HIS1::his1::hisG arg4::hisG/URA3::ARG4::arg4::hisG | Bassilana et al. (2003) |
| PY92 | cdc24Δ::HIS1/cdc24::URA3PMETCDC24 arg4::hisG/arg4::hisG RP10::ARG4 | Bassilana et al. (2003) |
| PY101 | cdc24Δ::HIS1/cdc24::URA3PMETCDC24 arg4Δ::hisG/arg4Δ::hisG | Bassilana et al. (2003) |
| PY123 | cdc42Δ::HIS1/cdc42::URA3PMETCDC42 arg4::hisG/arg4::hisG RP10::ARG4 | Bassilana et al. (2005) |
| PY189 | rac1Δ::URA3/rac1Δ::HIS1 arg4Δ::hisG/arg4Δ::hisG | Bassilana and Arkowitz (2006) |
| PY191 | Same as PY189 with RP10::ARG4 | Bassilana and Arkowitz (2006) |
| PY284 | Same as BWP17 with dck2::Tn7-UAU1/dck2::Tn7-URA3 | This study |
| PY286 | Same as BWP17 with dck1::Tn7-UAU1/dck1::Tn7URA3 | This study |
| PY309 | Same as PY189 with RP10::ARG4-PADHRAC1 | This study |
| PY352 | Same as BWP17 with RP10::ARG4 | This study |
| PY376 | Same as BWP17 with RP10::ARG4-PADHRAC1 | This study |
| PY378 | Same as BWP17 with RP10::ARG4-PADHrac1[T17N] | This study |
| PY379 | Same as BWP17 with RP10::ARG4-PADHrac[Q61L] | This study |
| PY387 | Same as PY189 with RP10::ARG4-PADHrac1[Q61L] | This study |
| PY389 | Same as PY189 with RP10::ARG4-PADHrac1[T17N] | This study |
| PY456 | Same as PY189 with RP10::ARG4-PADHCZF1 | This study |
| PY523 | czf11::HIS1/CZF1 ura3Δ:: imm434/ura3Δ:: imm434 arg4::hisG/arg4::hisG | This study |
| PY569 | Same as PY189 with RP10::ARG4-PADHDCK1 | This study |
| PY580 | Same as BWP17 with RP10::ARG4-PADHDCK1 | This study |
| PY646 | dck1Δ::HIS1/DCK1 ura3Δ:: imm434/ura3Δ:: imm434 arg4::hisG/arg4::hisG | This study |
| PY704 | dck1Δ::HIS1/dck1Δ::URA3 arg4::hisG/arg4::hisG | This study |
| PY706 | Same as PY704 with RP10::ARG4 | This study |
| PY710 | Same as PY704 with RP10::ARG4-PDCK1DCK1 | This study |
| PY714 | Same as PY704 with RP10::ARG4-PADHDCK1 | This study |
| PY718 | Same as PY704 with RP10::ARG4-PADHdck1 ΔDHR | This study |
| PY723 | Same as PY704 with RP10::ARG4-PADHGFPDCK1 | This study |
| PY727 | Same as PY704 with RP10::ARG-PADHGFPdck1 ΔDHR | This study |
| PY757 | Same as PY704 with RP10::ARG4-PADHRAC1 | This study |
| PY761 | Same as PY704 with RP10::ARG4-PADHrac1[Q61L] | This study |
| PY763 | Same as PY704 with RP10::ARG4-PADHrac1[T17N] | This study |
| PY785 | Same as PY704 with RP10::ARG4-PADHCDC24GFP | This study |
| PY787 | Same as PY101 with RP10::ARG4-PADHDCK1 | This study |
| PY838 | Same as PY704 with RP10::ARG4-PDCK1DCK1GFP | This study |
| PY897 | czf1::HIS1/czf1::URA3 arg4::hisG/arg4::hisG | This study |
| PY901 | Same as PY897 with RP10::ARG4 | This study |
| PY904 | Same as PY897 with RP10::ARG4-PADHCZF1 | This study |
| PY910 | Same as PY897 with RP10::ARG4-PADHrac1[Q61L] | This study |
Table 2.
Primers used in this study
| Primer | Sequence |
|---|---|
| DCK1.P1 | ACTTGGATCAGAACTGATTTTTTGATAAAGGGGAAAATTATCAAACCTTTTTTACCCAATGATAAACATCC-TGTGGAATTGTGAGCGGATA |
| DCK1.P2 | CCATTTTGTATTTATTGCATTTGTTTTACCGGATCCGTTTATTGATTTAGAAGTGACAGATCCATTACC-TTTCCCAGTCACGACGTT |
| DCK1.P3 | TATACGCGTGTAACCAAGCATTTGGATGCG |
| DCK1.P4 | CATCTACTACTTTCCTCTTTG |
| DCK1.P5 | CGGTTTTCGGTAAACAAGATC |
| DCK1.P6 | TATCTCGAGGTCACAGTAGTTTGGGTCCC |
| DCK1.P7 | ACGTCATTGGATCAAGAAAATTTCATTGAAAGAATGAAAAAGAATTTCCAAGAAGAAATTGTTGCTTTAC-GTTTAGGTGA |
| DCK1.P8 | TCACCTAAACGTAAAGCAACAATTTCTTCTTGGAAATTCTTTTTCATTCTTTCAATGAAATTTTCTTGAT-CCAATGACGT |
| DCK1.P9 | TTCGGATCCATGATTTCCGAATATATTTTAAG |
| DCK1.P10 | TCAGGTCGACCTAAAAATGGAAACACGACTTGAG |
| VECTOR.P1 | CAACAAATACAAAAACAAAGATCCGAATTCGGACCGAATGTCTAAGGAGAAGAATTATT-CACTGG |
| VECTOR.P2 | CCAGTGAATAATTCTTCTCCTTAGACATTCGGTCCGAATTCGGATCTTTGTTTTTGTAT-TTGTTG |
| VECTOR.P3 | GATCCTACCCATACGATGTCCCAGACTACGCACGGACCGTGATGAGAAGCATTAAATCA-GTCGTAGTTGGAGATGG |
| VECTOR.P4 | CCATCTCCAACTACGACTGATTTAATGCTTCTCATCACGGTCCGTGCGTAGTCTGGGAC-ATCGTATGGGTAGGATC |
| RAC1.P1 | CGAGGATCCATGAGAAGCATTAAATCAGTCG |
| RAC1.P2 | TCTGCGGCCGCCCCATGATTATAATATAGTACATTTTTTAGCTCTC |
| RAC1.P3 | TCAGGTCGACCCATGATTATAATATAGTACATTTTTTAGCTCTC |
| CZF1.P1 | CAAACATTCAAACGAAAACTATCTGGTATTTAATTTTCCATTTTCAACATCAAGTGTTTAAATATCAACAT-GTGGAATTGTGAGCGGATA |
| CZF1.P2 | TAATACAAGAGTGGTGATTTACCTTTTTATTGAGTTTTTTTTAAACGATATCCCTCCAACACAGAGAAGCT-TTCCCAGTCACGACGTT |
| CZF1.P3 | AGTTCAATACCCAATATCAATTGGAATGACCCTAACAATGGTAAATCCAATACCTCTCGTCAATCACAA-CCACTGTGGAATTGTGAGCGGATA |
| CZF1.P4 | TTATTTACTTCTGTATTCAACAATACCTCTCAAAACTTTGATTCTACCAAACTCGGCATGTTCAATTGTATT-CCAGTCACGACGTT |
| CaActETMpa | ATGTTCCCAGGTATTGCTGA |
| CaActETMma | ACATTTGTGGTGAACAATGG |
| CaDck1TMpa | TTGAAACCATCGGCTAACAA |
| CaDck1TMma | GCCACCATACCAGCAAATC |
| CaDck2TMpa | AGCTCTGGAAGGCTTACCTGTT |
| CaDck2TMma | CTCCTAAACGTCCCCAGACTTT |
| Czf1pTMa | TAGGCGGTCCACCATTTCC |
| Czf1mTMa | GGATAACCAGTGTTTGGCATATACC |
a Primers used for qRT-PCR.
Plasmids
Initially, the DCK1 ORF, with 987 base pairs 5′ of the ATG and 914 base pairs 3′ of the stop codon (∼7.7 kb) was amplified by PCR from genomic DNA in two fragments and each was cloned into pCR2.1 TA (Invitrogen, Cergy Pontoise, France). The primers DCK1.P3 (with a unique MluI site at the 5′ end) and DCK1.P4 were used to amplify a fragment from 987 base pairs 5′ of the ATG to 2855 base pairs 3′ of the ATG, which was cloned into pCR2.1 TA, yielding pCR-PDCK1DCK1-5′. The primers DCK1.P5 and DCK1.P6 (with a unique XhoI site at the 3′ end) were used to amplify a fragment from 2300 base pairs 3′ of the ATG to 914 base pairs 3′ of the stop codon, which was also cloned into pCR2.1 TA, yielding pCR-DCK1-3′. Subsequently, pCR-PDCK1DCK1-5′ was digested with MluI, blunted, and digested with AatII. The released PDCK1DCK1-5′ fragment was cloned into pCR-DCK1-3′, which was digested with NotI, blunted and digested with AatII, yielding pCR-PDCK1DCK1. A unique NotI site was subsequently introduced by site-directed mutagenesis with the Pfu polymerase (Promega, Charbonnieres Les Bains, France), by using the DpnI method (Weiner et al., 1994), in pCR-PDCK1DCK1, 5′ of the DCK1 promoter. The NotI/XhoI PDCK1DCK1 fragment was then cloned into the respective sites in pExpArg (derived from pExp-PADHGFPRAC1) (Bassilana and Arkowitz, 2006). The sequence of all PCR products and mutations were confirmed (ABI [Courtaboeuf, France] PRISM big-dye terminator cycle sequencing kit). Two clones of pCR-DCK1-3′ along with DCK1-3′ amplified from genomic DNA were sequenced to confirm the difference with the published genome sequence (http://www.candidagenome.org/).
For Rac1 and Cdc24 overexpression, pExp-PADHRAC1 and pExp-PADHCDC24GFP plasmids were used (Bassilana et al., 2005; Bassilana and Arkowitz, 2006). Constitutively active [G12V] and [Q61L] and inactive [T17N] Rac1 mutants were generated by site-directed mutagenesis by using pExp-PADHRAC1 as a template. For DCK1 overexpression a pExp-PADH plasmid, which was derived from pExp-PADHGFPRAC1, was used. To generate pExp-PADHDCK1 and pExp-PADHGFPDCK1 plasmids, the primer pairs VECTOR.P1, VECTOR.P2, and VECTOR.P3, VECTOR.P4 were used to introduce an RsrII site into pExp-PADHGFPRAC1, either 3′of the ADH1 promoter or 3′ of green fluorescent protein (GFP), respectively, yielding pExp-PADH-R-GFPRAC1 and pExp-PADHGFP-R-RAC1. In parallel, an RsrII site was inserted, two base pairs 5′ of the ATG, and an MluI site, 15 base pairs 3′ of the stop codon in pCR-PDCK1DCK1, yielding pCR-PDCK1-R-DCK1-M. This plasmid was then digested with RsrII and MluI and the DCK1 ORF was cloned into the respective sites of pExp-PADH-R-GFPRAC1 and pExp-PADHGFP-R-RAC1, yielding pExp-PADHDCK1 and pExp-PADHGFPDCK1. To generate pExp-PDCK1DCK1GFP, site-directed mutagenesis was used to introduce a unique SacI site 5′ and a unique MluI site 3′ of the stop codon in pExp-PDCK1DCK1, resulting in pExp-PDCK1DCK1-S-M. Subsequently, GFP from pAW6-GFP (Nishikawa et al., 2002) was cloned into pExp-PDCK1DCK1-S-M, resulting in pExp-PDCK1DCK1GFP. To generate DCK1 constructs lacking the DOCKER Homology Region (DHR) domain, the region between base pairs 3998 and 5349 3′ of the ATG (encoding amino acids [aa] 1332-1783) was removed by site-directed mutagenesis, using pCR-DCK1-3′ as a template and the primers DCK1.P7 and DCK1.P8, yielding pCR-dck1ΔDHR. The AatII/SacI fragment from this plasmid was then cloned into the respective sites in pCR-PDCK1DCK1-5′, resulting in pCR-PDCK1dck1ΔDHR. Subsequently, an RsrII site was inserted 5′ of the ATG and an MluI site, 3′ of the stop codon as described above, resulting in pCR-PDCK1-R-dck1ΔDHR-M. The dck1ΔDHR ORF was then cloned behind either the ADH1 promoter or ADHGFP, as described above.
For overexpression of CZF1, both the MAL2 and ADH1 promoters were used. PMALCZF1 originated from pDB212 (Brown et al., 1999). Initially, a unique XhoI site was introduced 3′ of the stop codon by site-directed mutagenesis and subsequently a PMALCZF1 fragment was obtained by PacI digestion, followed by blunting and XhoI digestion. This fragment was then cloned into pExp-PADHGFPRAC1 that was digested with NotI, blunted and digested with XhoI to yield pExp-PMALCZF1. For CZF1 expression behind the ADH1 promoter, a unique BamHI site 5′ of the ATG and an MluI site 3′ of the stop codon were introduced by site directed mutagenesis, using pDB212. The BamHI/MluI CZF1 ORF was then cloned into compatible sites in plasmid pAW6 resulting in pAW6-PADHCZF1, which was subsequently digested with NotI and XhoI. The NotI-PADHCZF1-XhoI fragment released was finally cloned into respective sites in pExp-PADHGFPRAC1, resulting in pExp-PADHCZF1. The sequence of all site directed mutagenesis products was confirmed (ABI PRISM Big Dye terminator cycle sequencing kit; Applied Biosystems, Foster City, CA).
To generate proteins for in vitro binding experiments, antibody production, and antibody purification, Rac1 from pExp-PADHRAC1 was amplified using the primer pairs RAC1.P1 and RAC1.P2, which introduced a BamHI site 5′ of the ATG and a NotI site 3′ of the Stop codon. The BamHI/NotI RAC1 ORF was then cloned in pGEX 6P-2 (Amersham, Orsay, France), resulting in pGEX-RAC1. Constitutively active [G12V] and inactive [T17N] GST-Rac1 were generated using the same primers (RAC1.P1 and RAC1.P2), with pExp-PADHrac1[G12V] and pExp-PADHrac1[T17N] as templates. To generate maltose-binding protein (MBP)-Rac1, RAC1 from pGEX-RAC1 was amplified using the primer pairs RAC1.P1 and RAC1.P3, which introduced a BamHI site 5′ of the ATG and a SalI site 3′ of the Stop codon. The BamHI/SalI RAC1 ORF was then cloned in pMal-c2 (New England Biolabs, Saint Quentin en Yvelines, France), resulting in pMal-RAC1. To generate MBP-DHR, the region encoding aa 1288-1812 of Dck1 was amplified, by using pExp-PADHDCK1 and the primer pairs DCK1.P9 and DCK1.P10. These primers introduced a BamHI site 5′ of codon for aa 1288 and a Stop codon, followed by a SalI site, 3′ of the codon for aa 1812. The BamHI/SalI DHR fragment was then cloned in pMal-c2 resulting in pMal-DHR. Subsequently, one CTG codon in the DHR domain (residue at position 1773), which codes for a serine in Candida albicans was changed to AGC by site-directed mutagenesis. The sequences of all cloned PCR products and site-directed mutagenesis products were confirmed (ABI PRISM Big Dye terminator cycle sequencing kit).
General Techniques
Quantitative reverse transcription (qRT)-PCR analyses were carried out as described previously (Bassilana et al., 2005), and the primers used are listed in Table 2.
Microscopy
For colony morphology, after growth on or in agar containing media for 3–6 d, colonies were imaged with an MZ6 dissection scope (Leica, Rueil-Malmaison, France) at a magnification of 10× or 20×, as described previously (Bassilana et al., 2005). For cell morphology, cells were fixed with formaldehyde after incubation in the different inducing media and imaged with a Leica DMR epifluorescence microscope with a numerical aperture 1.35 63× objective by differential interference contrast optics, as described previously (Bassilana et al., 2005).
Immunoblot Analyses
Yeast protein extracts were prepared from logarithmically growing cells. Western blot analyses were carried out as described previously (Bassilana et al., 2003). Equal amounts of cells or fusion proteins (for in vitro binding studies) were used in each experiment, and proteins in each lane were visualized by Ponceau S staining of nitrocellulose membranes. Blots were probed with either a rabbit polyclonal serum against GFP (1:5000; Nern and Arkowitz, 2000), MBP (1:1000; a gift from W. Wickner), S. cerevisiae Pma1p (1:10,000; a gift from R. Kelly; Kelly et al., 2000), S. cerevisiae Cdc11p (sc-7170, 1:200; Santa Cruz Biotechnology, Heidelberg, Germany), or C. albicans Rac1 (1:1000). Immunoblots were then visualized by enhanced chemiluminescence (luminol-coumaric acid) on a Fuji-Las3000 (Fuji, Clichy, France).
To generate polyclonal sera against C. albicans Rac1, rabbits were immunized with glutathione transferase (GST)-Rac1 purified from Escherichia coli BL21 cells, by using pGEX-RAC1. MBP-Rac1 was purified from E. coli BL21 cells, by using pMal-RAC1, and it was bound to amylose resin. This fusion protein was cross-linked to the resin with dimethyl pimelimidate and used to purify sera as described previously (Harlow and Lane, 1988). The resin was extensively washed with buffer A (10 mM Tris-Cl, pH 7.5) followed by buffer A containing 0.5, 1, and 2 M NaCl. Subsequently, the resin was washed with 100 mM glycine, pH 2.5, followed by 10 mM Tris-Cl, pH 8.8, and then antibodies were eluted with 100 mM triethylamine, pH 11.5, and neutralized as described (Harlow and Lane, 1988). These purified antibodies were stored at 4°C in the presence of 0.5 mg/ml gelatin.
Protein Binding Studies
GST-Rac1 (wild-type and mutants), MBP-Rac1, and MBP-DHR fusion proteins were expressed in BL21 E. coli cells grown at 30°C and induced with 0.1 mM isopropyl β-d-thiogalactoside for 3–4 h. After centrifugation, cells were resuspended in 10% sucrose, 50 mM Tris-Cl, pH 8, and frozen in liquid nitrogen. All subsequent steps were carried out at 4°C. Fusion proteins were purified using glutathione (GSH)-Sepharose 4B (Amersham) or amylose resin (New England Biolabs, Ipswich, MA), as described previously (Barale et al., 2006) in buffer B (50 mM Tris-Cl, pH 7.5, 100 mM NaCl, 2.5 mM MgCl2, 1 mM dithiothreitol [DTT], 1 mM phenylmethylsulfonyl fluoride [PMSF], 10% glycerol, and protease inhibitor cocktail [Roche Diagnostics, Mannheim, Germany]) for G protein fusions and buffer C (20 mM Tris-Cl, pH 7.5, 100 mM NaCl, 2.5 mM MgCl2, 1 mM DTT, and 1 mM PMSF) for the DHR fusion. Protein bound to the respective resin was washed extensively in the same buffer (buffer B or C) containing 250 mM potassium acetate, 500 mM potassium acetate and for MBP-DHR fusions, additionally 1 M potassium acetate. MBP and MBP-DHR were then eluted with buffer C containing 10 mM maltose, and GST-Rac1 was eluted in buffer D (50 mM Tris-Cl, pH 8, 100 mM NaCl, 2.5 mM MgCl2, 1 mM DTT, 1 mM PMSF, 10% glycerol, and 10 mM GSH). For binding assays, ∼5 μg of GST-Rac1 fusion proteins or GST, bound to GSH-Sepharose, was incubated with ∼2 μg of MBP-DHR (or MBP) in 50 μl of buffer E (50 mM Tris-Cl pH 7.5, 100 mM NaCl, 5 mM MgCl2, 1 mM DTT, 1 mM PMSF, and 10% glycerol) for 1 h at 25°C. GSH-Sepharose samples were then washed eight times with 200 μl of buffer E, and proteins were eluted in SDS-polyacrylamide gel electrophoresis (PAGE) sample buffer, analyzed by SDS-PAGE, followed by immunoblotting.
Cell Fractionation
Logarithmically growing PY838 cells expressing Dck1-GFP were collected by centrifugation and resuspended in GPL buffer lacking NP-40. All subsequent steps were carried out at 4°C. Cells were broken with glass beads in a RiboLyser (MP Biomedicals, Illkirch, France) for 45 s at maximum power. Cell extracts were clarified by centrifugation at 200 × g for 5 min. This supernatant was then centrifuged for 10 min at 10,000 × g, resulting in P10 pellet and S10 supernatant fractions. The P10 fraction was suspended in a volume equivalent to the S10 fraction. The S10 fraction was subsequently centrifuged for 1 h at 100,000 g in a TL-100 (Beckman, Villepinte, France), resulting in P100 pellet and S100 supernatant fractions. The P100 fraction was resuspended in the same volume as the S100 supernatant fraction. Fractions were subsequently analyzed by 7% (for Dck1-GFP and Pma1) and 12% SDS-PAGE (for Rac1) followed by immunoblotting and probing with anti-GFP, anti-Pma1, and anti-Rac1 sera.
RESULTS
Rac1-GTP Is Required for C. albicans-embedded Filamentous Growth
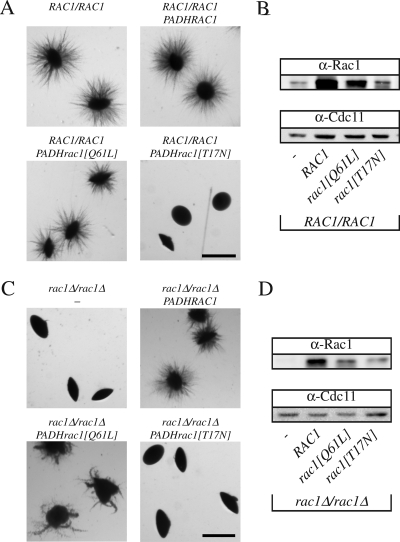
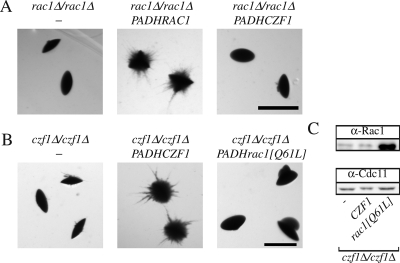
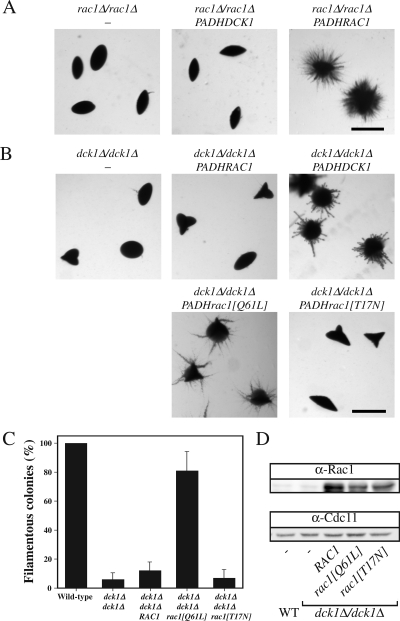
We have shown previously that the Dbl GEF Cdc24 (Bassilana et al., 2003) and the Rho G protein it activates, Cdc42 (Michel et al., 2002; Ushinsky et al., 2002; Bassilana et al., 2003; VandenBerg et al., 2004), are required for viability and serum-induced hyphal growth. Conversely, cdc42 mutants that were defective in serum-induced hyphal growth were able to form filaments under embedded conditions (VandenBerg et al., 2004). The Rho G protein Rac1, which is 57% identical to Cdc42, is required for filamentous growth when cells are embedded in an agar matrix (Bassilana and Arkowitz, 2006). These responses to different filamentous growth stimuli, together with the inability of overexpressed Rac1 to complement cdc42 mutants, suggest that these two similar G proteins function in distinct signaling pathways. Typically, the GTP bound form of a G protein binds effectors; hence, we examined whether overexpression of a Rac1 mutant, which mimics the GTP-bound state (rac1[Q61L]) or the nucleotide-free state (rac1[T17N]), alters embedded filamentous growth in both wild-type or rac1 deletion strains. Overexpression of the nucleotide-free form of a G protein results in a dominant-negative effect due to sequestration of its GEF (Feig, 1999). Overexpression of rac1[T17N] did not substantially affect growth or serum-dependent hyphal growth of either strain (data not shown), indicating that Rac1 does not sequester Cdc24, suggesting the existence of a distinct Rac1 GEF. In contrast, overexpression of the nucleotide-free Rac1 form in a wild-type strain completely blocked filamentous growth under embedded conditions (Figure 1A). Overexpression of the GTP bound Rac1 form resulted in embedded filamentous growth that was similar, albeit somewhat reduced, compared with strains that overexpressed wild-type Rac1. In a rac1Δ/rac1Δ strain, overexpression of the GTP bound Rac1 form partially complemented the filamentous growth defect compared with overexpression of wild-type Rac1, whereas that of a nucleotide-free form did not (Figure 1C). Similar results were observed in wild-type and rac1 mutant strains by using different GFPRac1 fusions (RAC1, rac1[Q61L], and rac1[T17N]). Quantitative real-time RT-PCR measurements indicated that the mRNA transcript levels of the different Rac1 forms were expressed at ∼50-fold higher levels than that of endogenous Rac1 in these strains. To confirm that the Rac1 proteins were overexpressed, we generated specific Rac1 polyclonal antibodies. These antibodies revealed a band corresponding to the expected size of Rac1 in a wild-type strain but not in a rac1 deletion strain and did not cross-react with Cdc42 (Figure 1, B and D, and Supplemental Figure S1). The Rac1 proteins (wild-type and mutants) were overexpressed both in the wild-type strain (Figure 1B) and the rac1 deletion strain (Figure 1D), respectively. In both strain backgrounds, wild-type Rac1 protein overexpression was higher than that of Rac1[Q61L] and Rac1[T17N], the latter was overexpressed the least. Expression of rac1[T17N] was nonetheless sufficient to block embedded filamentous growth in a strain containing both copies of endogenous RAC1 (Figure 1A). Together, these results indicate that the GTP-bound form of Rac1 is required for embedded filamentous growth; yet, given that such a Rac1 mutant, which is blocked for GTP hydrolysis, only partially complements a rac1 deletion strain, it is likely that GDP–GTP cycling is important for efficient filamentous growth in embedded media.
Figure 1.
Activated Rac1 is required for filamentous growth in embedded media. (A) Overexpression of a nucleotide-free form of Rac1 blocks embedded filamentous growth in a wild-type strain. BWP17 cells carrying either an empty plasmid (pExpArg; PY352), a plasmid with PADHRAC1 (PY376), with PADHrac1[Q61L] (PY379) or with PADHrac1[T17N] (PY378) were embedded in YEPS uridine. Exponentially growing cells (∼8 × 102) were added to 25 ml of YEPS uridine containing 2% molten agar and poured into Petri dishes. After 5 d at 25°C, images of colonies were taken. Similar results were observed in three independent experiments. Bar, 1 mm. (B) Rac1 protein levels in the different strains. Extracts of exponentially growing strains indicated in Figure 1A were analyzed by SDS-PAGE, followed by immunoblotting and probing with anti-Rac1 and anti-Cdc11 sera. (C) Overexpression of the GTP bound form of Rac1 partially complements the embedded filamentous growth defect of rac1 mutant cells. PY189 (rac1Δ/rac1Δ) cells carrying either an empty plasmid (pExpArg; PY191), a plasmid with PADHRAC1 (PY309), with PADHrac1[Q61L] (PY387) or with PADHrac1[T17N] (PY389) were embedded in YEPS and imaged as described above. Similar results were observed in three independent experiments. (D) Rac1 protein levels in the different strains. Extracts of exponentially growing strains indicated in Figure 1C were analyzed by SDS-PAGE, followed by immunoblotting and probing with anti-Rac1 and anti-Cdc11 sera.
Identification of a C. albicans Rac1 Activator
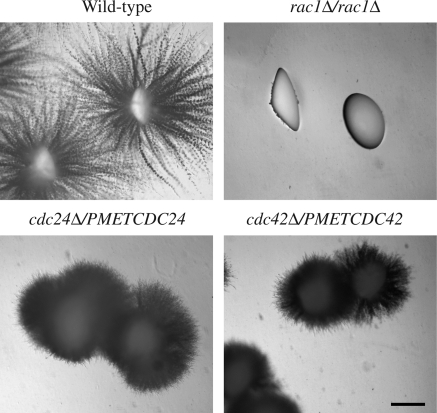
In humans, Rac1 is activated by either Dbl family GEFs, such as Tiam1 (Michiels et al., 1995), or CDM family GEFs, such as Dock180 (Brugnera et al., 2002; Cote and Vuori, 2002). To address whether the Dbl family member Cdc24 could activate Rac1, in addition to Cdc42, we examined whether Cdc24 was required for embedded filamentous growth. Because Cdc42 and Cdc24 are essential for viability, we used strains (cdc42Δ/PMETCDC42 and cdc24Δ/PMETCDC24) with reduced levels of these proteins, which are defective in serum-dependent invasive filamentous growth (Bassilana et al., 2003; Bassilana et al., 2005). Figure 2 shows that although the rac1Δ/rac1Δ strain formed nonfilamentous smooth colonies when embedded in YEPS, the cdc24Δ/PMETCDC24 strain formed filamentous colonies, similar to that of the cdc42Δ/PMETCDC42 strain (Bassilana and Arkowitz, 2006). It should be noted that colonies of both cdc24 and cdc42 mutant strains are morphologically different from the wild-type strain; they seem to have an increased number of shorter filaments that result in a fur ball-like morphology. Although we cannot rule out the possibility that the reduced level of Cdc24 expression in the cdc24Δ/PMETCDC24 strain (Bassilana et al., 2005) is sufficient for embedded filamentous growth, because the cdc24 mutant is filamentous in this media similar to the cdc42 mutant, we consider it unlikely that Cdc24 activates Rac1 in this process.
Figure 2.
Cdc24 is not the Rac1 activator for embedded medium-induced filamentous growth. Wild-type cells (PY82), rac1Δ/rac1Δ cells (PY191), cdc24Δ/PMETCDC24 cells (PY92) and cdc42Δ/PMETCDC42 cells (PY123), were embedded in YEPS and imaged as described in Figure 1. Similar results were observed in 4 independent experiments. Bar, 1 mm.
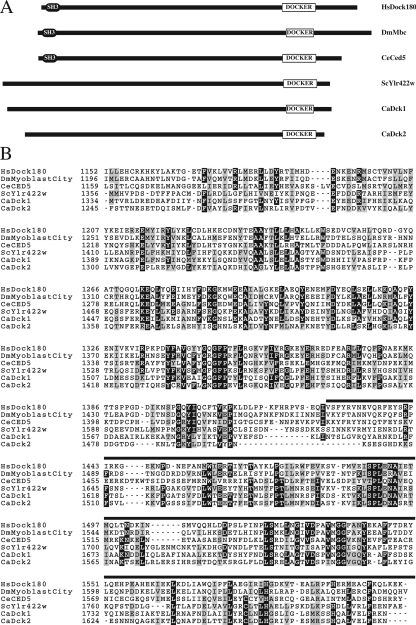
Members of the GEF CDM family have a conserved domain, referred to as a DOCKER domain (Cote and Vuori, 2007). Given that members of the CDM family can activate Rac1 in vitro, we used the BLAST algorithm to search the C. albicans genome for proteins with such a domain and found two ORFs (19.815 and 19.816), which will be hereafter referred to as DCK1 and DCK2, respectively. These two ORFs, located on chromosome 2 <1 kb apart, are 5745 base pairs and 5307 base pairs, respectively. Dck1 and Dck2 are 1915 and 1769 aa, respectively, with a region of 452 and 422 aa with similarity to the DOCKER domains (DHR) of HsDock180, CeCed5, DmMyoblast city, and ScYlr422w (Figure 3, A and B). The DHR of Dck1 and Dck2 are similar (26 and 23% identity, respectively) with that of HsDock180. C. albicans Dck1 and Dck2 share 35% overall sequence identity, with 42% identity between their DHRs. HsDock180, CeCed5, DmMyoblast City, have in addition to the DOCKER domain a second homology region, DHR1 and an amino-terminal Src homology 3 (SH3) domain, which are not apparent in Dck1 or Dck2. Dck1 homologues are found in all other fungal species; however, only in Candida species closest to C. albicans, i.e., C. dubliniensis and C. tropicalis is a second Dck protein found, suggesting that DCK2 was generated by a gene duplication (Fitzpatrick and Butler, personal communication).
Figure 3.
Comparison of different CDM family members. (A) Schematic representation of different CDM family members. Indicated proteins from Hs, Homo sapiens; Dm, Drosophila melanogaster; Ce, Caenorhabditis elegans; Sc, S. cerevisiae and Ca, Candida albicans are shown. The DOCKER domain, together with the SH3 (Src Homology 3) domain are indicated. (B) Alignment of the putative DOCKER Homology Region (DHR) of C. albicans Dck1 and Dck2 with different CDM family members. DHR domains, according to Brugnera et al. (Brugnera et al., 2002), from indicated proteins were aligned with C. albicans Dck1 and Dck2. Residues boxed in black and gray are identical and similar in ≥4 sequences, respectively. The black line above the sequence represents the DOCKER domain (Dedicator of cytokinesis; pfam06920, http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml), which is ∼200 amino acids. The sequence of the DCK1 ORF revealed one base change compared with the sequence in the Candida genome database (http://www.candidagenome.org/), which resulted in a Leu at position 1760.
Dck1 Is Required for Invasive Filamentous Growth
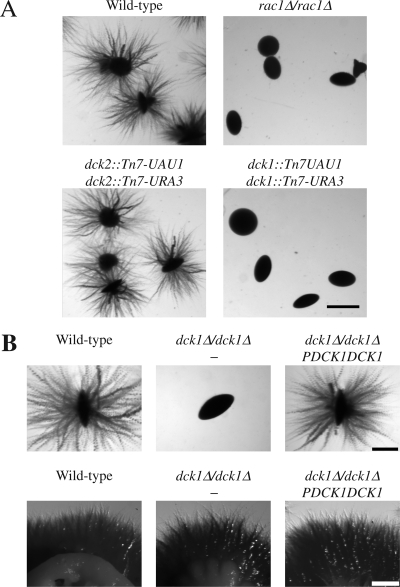
To investigate the function of these two potential C. albicans CDM members, we have used the Tn7-UAU1 transposon insertion approach described by Davis et al. (2002). We generated strains in which the Tn7-UAU1 cassette (dck1::Tn7-UAU1 and dck2::Tn7-UAU1) was inserted into both copies of either DCK1 or DCK2, respectively, suggesting that neither of these genes is required for viability. Quantitative real-time RT-PCR confirmed that neither DCK1 nor DCK2 mRNA transcripts were detectable in such dck1 and dck2 mutants, in contrast to the wild-type strain, in which both DCK1 and DCK2 transcripts were present (0.2 ± 0.07 for DCK1 and 0.05 ± 0.02 for DCK2, relative to actin). Figure 4A shows that colonies of the dck2 mutant are filamentous in agar, similar to wild-type colonies. Strikingly, in the same conditions, the dck1 mutant was unable to filament, instead forming smooth colonies, identical to that of the rac1 deletion mutant. These results suggest that DCK1, but not DCK2, is important for matrix-embedded-dependent filamentous growth.
Figure 4.
Dck1 is required for matrix-induced filamentous growth. (A) Dck1 but not Dck2 is required for filamentous growth in embedded medium. Wild-type cells (PY82), rac1Δ/rac1Δ cells (PY191), together with homozygous insertion mutant strains dck2::Tn7- UAU1/dck2::Tn7-URA3 (PY284), and dck1::Tn7-UAU1/ dck1::Tn7-URA3 (PY286) were embedded in YEPS (as described in Figure 1). Similar results were observed in four independent experiments. Bar, 1 mm. (B) Dck1 is not required for serum-dependent filamentous growth. Top, wild-type cells (PY82), dck1Δ/dck1Δ cells (PY706) and dck1Δ/dck1Δ PDCK1DCK1 cells (PY710) were embedded in YEPS as described in Figure 1. Similar results were observed in five independent experiments. Bar, 0.5 mm. Bottom, exponentially growing cells were spotted in serial dilution on YEPD FCS and after 5 d, images were taken. Similar results were observed in three independent experiments. Bar, 1 mm.
To further substantiate the requirement for DCK1 in filamentous growth in an agar matrix, we generated a strain in which both copies of DCK1 were deleted. Such a dck1Δ/dck1Δ strain was viable and its doubling time in rich media comparable with that of the wild-type strain. DCK1 mRNA transcripts were not detectable in this deletion strain by qRT-PCR. However, neither deletion nor overexpression of DCK1 affected Rac1 protein levels (Supplemental Figure S1). Figure 4B (top) shows that colonies of the dck1Δ/dck1Δ strain are nonfilamentous when grown in YEPS compared with wild-type colonies. Reintroduction of a copy of DCK1 behind its own promoter, at the RP10 locus restored filamentous growth in these conditions, demonstrating that the dck1Δ/dck1Δ filamentous growth defect is due to deletion of the DCK1 gene. In contrast, dck1Δ/dck1Δ colonies are filamentous when grown on serum containing media (Figure 4B, bottom), indicating that Dck1, like Rac1, is specifically required for matrix-induced filamentous growth.
Given that the transcription factor Czf1 is also specifically required for matrix-induced filamentous growth (Brown et al., 1999), we investigated whether overexpression of CZF1 could suppress the defect of the rac1 and dck1 deletion strains. We generated strains in which CZF1 was overexpressed via either the MAL2 or ADH1 promoters, resulting in an increase in CZF1 transcript levels by 9- and 30-fold (compared with the wild-type level), respectively. Such CZF1 overexpression did not suppress the embedded filamentous growth defect of either the rac1 deletion strain (Figure 5A) or the dck1 deletion strain (data not shown), suggesting that this transcription factor does not function downstream of Rac1 and Dck1. To investigate whether CZF1 could function upstream of Rac1, we generated a czf1 deletion mutant, isogenic to the rac1 and dck1 deletion mutants. This czf1 deletion strain was nonfilamentous in embedded media, similar to the czf1 deletion strain generated by Brown et al. (1999), and reintroduction of CZF1 under the control of the ADH1 promoter (Figure 5B) complemented this filamentous growth defect. We examined whether overexpression of a constitutively active Rac1 mutant could restore such filamentous growth in this czf1 mutant (because overexpression of this mutant restores embedded filamentous growth in a dck1 mutant; Figure 10). Overexpression of constitutively active Rac1, rac1[Q61L] (Figure 5B), did not restore filamentous growth in the czf1 deletion mutant. Overexpression of rac1[Q61L] was confirmed both at the mRNA transcript level by qRT-PCR (data not shown) and at the protein level (Figure 5C). Altogether, these results suggest that Rac1 and Dck1 function in a novel signaling pathway, different from that in which Czf1 functions, necessary for matrix-induced filamentous growth.
Figure 5.
CZF1 and RAC1 do not function in the same pathway required for embedded filamentous growth. (A) Overexpression of Czf1 does not restore the embedded filamentous growth defect of rac1 mutant cells. rac1Δ/rac1Δ (PY191) cells, rac1Δ/rac1Δ PADHRAC1 (PY309) cells and rac1Δ/rac1Δ PADHCZF1 (PY456) cells were embedded in YEPS and imaged as described in Figure 1. Similar results were observed in three independent experiments. Bar, 1 mm. (B) Overexpression of constitutively active Rac1 does not restore the embedded filamentous growth defect of czf1 mutant cells. czf1Δ/czf1Δ (PY901) cells, czf1Δ/czf1Δ PADHCZF1 (PY904) cells and czf1Δ/czf1Δ PADHRAC1[Q61L] (PY910) cells were embedded in YEPS and imaged as described in Figure 1. Similar results were observed in three independent experiments. Bar, 1 mm. (C) Rac1[Q61L] protein is overexpressed compared with endogenous Rac1. Extracts of exponentially growing cells indicated in Figure 5B were analyzed by SDS-PAGE, followed by immunoblotting and probing with anti-Rac1 and anti-Cdc11 sera.
Figure 10.
Constitutively active Rac1 restores embedded filamentous growth in dck1 mutant cells. (A) Overexpression of Dck1 does not restore embedded filamentous growth in rac1 mutant cells. Rac1Δ/rac1Δ (PY191) cells, rac1Δ/rac1Δ PADHDCK1 (PY569) cells, and rac1Δ/rac1Δ PADHRAC1 (PY309) cells were embedded in YEPS and imaged as described in Figure 1. Similar results were observed in three independent experiments. Bar, 1 mm. (B) Overexpression of a constitutive active Rac1 mutant partially suppresses the embedded filamentous growth defect of a dck1 deletion mutant. dck1Δ/dck1Δ (PY706) cells, dck1Δ/dck1Δ PADHRAC1 (PY757) cells, dck1Δ/dck1Δ PADHDCK1 (PY714) cells, dck1Δ/dck1Δ PADHrac1[Q61L] (PY761) cells, and dck1Δ/dck1Δ PADHrac1[T17N] (PY763) cells were embedded in YEPS and imaged as described in Figure 1. Similar results were observed in 4 independent experiments. Bar, 1 mm. (C) Percentage of filamentous colonies in the indicated strains. Averages of four experiments (n = 150 colonies each) are shown with SD. (D) Rac1 expression levels in the indicated strains. Extracts of exponentially growing strains indicated in C were analyzed by SDS-PAGE, followed by immunoblotting and probing with anti-Rac1 and anti-Cdc11 sera.
To determine whether Dck1 and Rac1 are only required for embedded filamentous growth, we analyzed the ability of the corresponding deletion mutants to filament in response to low nutrients, both in Spider media and GlcNAc. When grown on Spider media containing agar for 6 d, colonies of both dck1Δ/dck1Δ and rac1Δ/rac1Δ strains were filamentous, although the length of the filaments was shorter than that of the wild-type strain (Supplemental Figure S2A). At earlier times, the dck1Δ/dck1Δ and rac1Δ/rac1Δ strains were less crenelated than the corresponding wild-type strain. When grown in Spider liquid media dck1Δ/dck1Δ and rac1Δ/rac1Δ cells formed hyphae similar to wild-type cells, albeit somewhat reduced in length (Supplemental Figure S2B). On GlcNAc containing solid media both dck1Δ/dck1Δ and rac1Δ/rac1Δ strains had a strong filamentous growth defect compared with the wild-type strain (Supplemental Figure S3A). Strikingly, in liquid media containing GlcNAc both deletion strains were filamentous similar to the wild-type strain (Supplemental Figure S3B). Together, these data indicate that Dck1 and Rac1 are important for invasive filamentous growth in solid media.
The DOCKER Domain Is Required for Dck1 Function
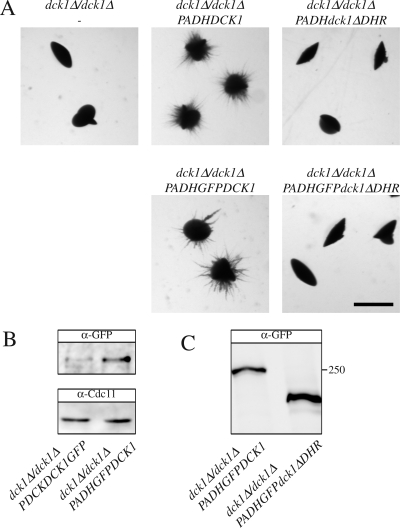
The DOCKER domain is required for HsDock180 function (Brugnera et al., 2002; Cote and Vuori, 2002). To determine the importance of the homologous DOCKER domain in Dck1, we generated a strain that overexpressed, as a sole DCK1 copy, a version in which the region encoding amino acid residues 1332-1783 (homologous to HsDock180 aa 1152-1607, termed DHR) was removed. This strain overexpressed dck1ΔDHR mRNA ∼90-fold, similar to the overexpression of DCK1 mRNA, as determined by qRT-PCR. Figure 6A shows that the dck1ΔDHR overexpression strain is unable to form filamentous colonies when embedded in agar, in contrast to the control strain. To verify that the Dck1ΔDHR protein is expressed, we constructed GFP fusions and generated strains containing GFPDck1ΔDHR or GFPDck1 as the sole Dck1 copy. In addition, we confirmed that the expression of these fusion proteins via the ADH1 promoter resulted in a substantial increase in protein level compared with expression via its endogenous promoter (Figure 6B). The immunoblot illustrated in Figure 6C shows that GFPDck1 and GFPDck1ΔDHR migrated consistent with their expected molecular weights (approximately at 250 and 220 kDa, respectively). We did not observe substantial degradation of either fusion. Furthermore, we confirmed that the GFPDck1 fusion, but not GFPDck1ΔDHR, was functional as overexpression of GFPDck1, but not GFPDck1ΔDHR, complemented the filamentous growth defect of the dck1 deletion strain (Figure 6A). These results suggest that the catalytic DOCKER domain is required for Dck1 function in vivo.
Figure 6.
The Dck1 DHR is required for embedded filamentous growth. (A) Overexpression of Dck1 lacking DHR does not complement the embedded filamentous growth defect of the dck1 mutant cells. dck1Δ/dck1Δ (PY706) cells, dck1Δ/dck1Δ PADHDCK1 (PY714) cells, dck1Δ/dck1Δ PADHdck1ΔDHR (PY718) cells, dck1Δ/dck1Δ PADHGFPDCK1 (PY723) cells, and dck1Δ/dck1Δ PADHGFPdck1ΔDHR (PY727) cells were embedded in YEPS and imaged as described in Figure 1. Similar results were observed in three independent experiments. Bar, 1 mm. (B) Dck1 protein levels in the indicated strains. Extracts from exponentially growing dck1Δ/dck1Δ PDCK1DCK1GFP (PY838) cells and dck1Δ/dck1Δ PADHGFPDCK1 (PY723) were analyzed by SDS-PAGE, followed by immunoblotting and probing with anti-GFP and anti-Cdc11 sera. (C) Dck1 lacking DHR is expressed at similar levels compared with Dck1. Extracts of exponentially growing dck1Δ/dck1Δ PADHGFPDCK1 (PY723) and dck1Δ/dck1Δ PADHGFPdck1ΔDHR (PY727) cells were analyzed by SDS-PAGE, followed by immunoblotting and probing with anti-GFP sera. Similar results were observed in three different experiments.
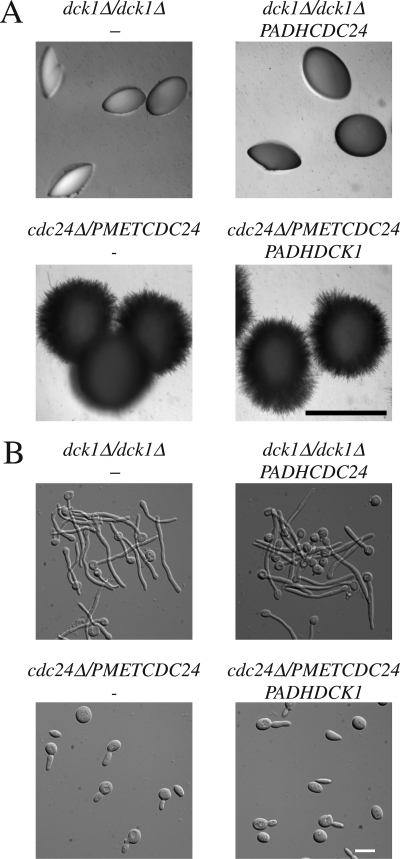
Dck1 and Cdc24 Function in Different Pathways
To investigate the specificity of Dck1 and Cdc24, we examined whether overexpression of Cdc24 can complement the filamentous growth defect of dck1 mutants and vice versa. This overexpression was confirmed by qRT-PCR. Figure 7A shows that overexpression of Cdc24 in the dck1Δ/dck1Δ strain does not restore filamentous growth in embedded media, whereas overexpression of Dck1 complements the filamentous growth defect of this mutant (Figure 6A). Conversely, we examined whether Dck1 overexpression affected the matrix-dependent filamentous growth of the cdc24Δ/PMETCDC24 strain, and no difference in colony morphology was observed (Figure 7A). We also determined whether Dck1 overexpression could restore serum-dependent filamentous growth of cdc24Δ/PMETCDC24 cells and Figure 7B shows that this strain is still defective. Together, our results indicate that Dck1 and Cdc24 function in response to different inducers of filamentous growth and likely activate the distinct Rho G proteins Rac1 and Cdc42, respectively.
Figure 7.
Dck1 and Cdc24 do not have overlapping functions. (A) Overexpression of Cdc24 does not complement the embedded matrix-dependent filamentous growth defect of dck1 mutant cells. dck1Δ/dck1Δ cells (PY706), dck1Δ/dck1Δ PADHCDC24GFP (PY785) cells, cdc24Δ/PMETCDC24 (PY92) cells, and cdc24Δ/PMETCDC24 PADHDCK1 (PY787) cells were embedded in YEPS and imaged as described in Figure 1. Similar results were observed in two independent experiments. Bar, 1 mm. (B) Overexpression of Dck1 does not complement serum dependent filamentous growth defect of cdc24 mutant cells. Exponentially growing cells were added to equal volume of fetal calf serum and incubated at 37°C for 3 h. Images of fixed cells were taken. Bar, 10 μm. Similar results were observed in two independent experiments.
Dck1 Functions As a Rac1 Activator In Vivo
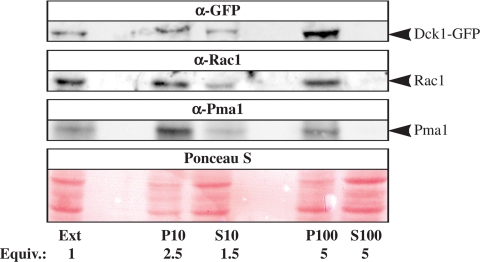
We examined the cellular distribution of Dck1 and Rac1 to determine whether these two proteins were present in the same cellular compartment. Subcellular fractionation of Dck1 and Rac1 was carried out in a strain expressing Dck1GFP from its endogenous promoter and proteins were detected using GFP and Rac1 antibodies, respectively. As illustrated in Figure 8, both Rac1 and Dck1 were found primarily in the membrane fractions (10,000 × g and 100,000 × g pellets) and not in the cytosolic fraction (100,000 × g supernatant), consistent with these two proteins being present in the same cellular compartment.
Figure 8.
Rac1 and Dck1 are present in the same membrane fraction. Extracts (Ext) from exponentially growing dck1Δ/dck1Δ PDCK1DCK1GFP (PY838) (∼1 × 108 cells) were centrifuged at 10,000 × g, resulting in the P10 pellet and S10 supernatant fractions. The S10 fraction was then centrifuged at 100,000 × g resulting in the P100 pellet and the S100 supernatant fraction. Fractions were analyzed by SDS-PAGE followed by immunoblotting and probing with anti-GFP, anti-Rac1, and anti-Pma1 sera. Ponceau S staining revealed proteins in all lanes, including the S100 fraction. Similar results were observed in three different experiments.
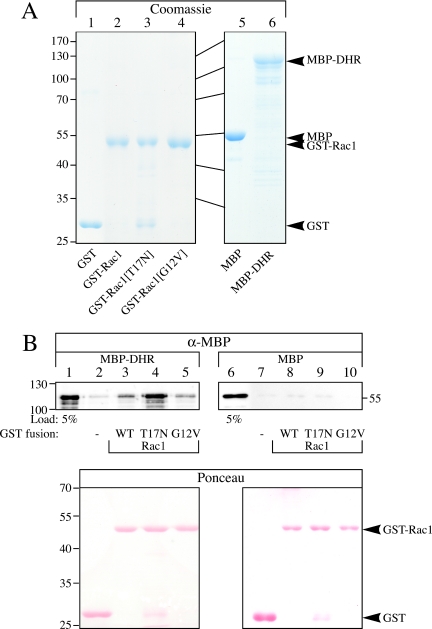
To investigate whether Dck1 and Rac1 can interact directly, we analyzed the in vitro binding of the Dck1 DHR (1288 aa to 1812 aa, fused to MBP), which includes the putative DOCKER catalytic domain, to different forms of Rac1, fused to GST. The fusion proteins, MBP-DHR, GST-Rac1, GST-Rac1[T17N], GST-Rac1[G12V], along with MBP and GST were expressed in bacteria and purified as illustrated in the Coomassie gel shown in Figure 9A. Either MBP or MBP-DHR was added to the different Rac1 forms or GST alone immobilized on GSH-agarose resin and bound MBP or MBP-DHR was analyzed by SDS-PAGE and immunoblotting. Figure 9B (top) shows that DHR can directly bind to Rac1, with DHR binding preferentially to the nucleotide-free form of Rac1 (Rac1[T17N]). Similar amounts of these GST fusions were present in the different binding reactions, as illustrated by Ponceau S staining (Figure 9B, bottom). MBP alone did bind low levels of the GST fusions, with little to no binding preference for the different GST proteins observed. These results are consistent with Dck1 functioning as a guanine nucleotide exchange factor, because GEFs typically bind the nucleotide-free form of the G protein (Feig, 1999).
Figure 9.
The Dck1 DHR preferentially binds Rac1[T17N] in vitro. (A) Rac1 and Dck1 DHR fusion proteins used for in vitro binding assays. GST and MBP fusion proteins purified from E. coli (described in Materials and Methods) were analyzed by SDS-PAGE and stained with Coomassie Blue. Approximately 5–10 μg each of purified GST (lane 1), GST-Rac1 (lane 2), GST-Rac1[T17N] (lane 3), GST-Rac1[G12V] (lane 4), MBP (lane 5), and MBP-DHR (lane 6) fusion proteins were loaded. (B) The Dck1 DHR binds Rac1[T17N] directly. Either MBP-DHR (lanes 1–5) or MBP (lanes 6–10) was incubated with GST (lanes 2 and 7), GST-Rac1 (lanes 3 and 8), GST-Rac1[T17N] (lanes 4 and 9), or GST-Rac1[G12V] (lanes 5 and 10) bound to GSH-agarose, followed by SDS-PAGE analysis. Immunoblots were probed with anti-MBP sera (top) or stained with Ponceau S (bottom). Lanes 1 and 6 are 5% of the total input of MBP-DHR (lane 1) and MBP (lane 6) used in each binding experiment. Similar results were observed in two independent experiments.
If Dck1 were the sole Rac1 activator during matrix-induced filamentous growth, one would predict that overexpression of a constitutively active Rac1 mutant should complement the defect of a dck1 deletion mutant whereas conversely, overexpression of Dck1 should not complement the defect of a rac1 deletion mutant. Figure 10A shows that Dck1 overexpression in a rac1Δ/rac1Δ strain was unable to restore filamentous growth, in contrast to Rac1 overexpression. Quantitative RT-PCR revealed that Dck1 was overexpressed 70-fold. Dck1 overexpression complemented the filamentous growth defect of a dck1Δ/dck1Δ strain, whereas overexpression of Rac1 did not (Figure 10B). Strikingly, overexpression of an activated form of Rac1, Rac1[Q61L], partially restored embedded filamentous growth in the dck1 deletion strain (Figure 10B). These results contrast those observed for rac1 deletion strain (Figure 1C) in which overexpression of activated form of Rac1 results in less extensive filamentous growth compared with Rac1. In contrast, overexpression of the nucleotide-free form of Rac1 ([T17N]) did not restore embedded filamentous growth in all strains examined (Figures 1, A and C, and 10B). Quantification indicated that ∼80% of the colonies of the dck1 deletion strain overexpressing Rac1[Q61L] were filamentous compared with ∼10% filamentous colonies in the same strain overexpressing wild-type Rac1 or Rac1[T17N] (Figure 10C). The different Rac1 forms were overexpressed 30- to 50-fold in this dck1 deletion mutant as determined by qRT-PCR; furthermore, Figure 10D shows overexpression of these different forms of Rac1 protein. These results, together with the similar phenotypes of rac1 and dck1 mutants in response to different inducers, indicate that Dck1 activates Rac1 in vivo during invasive filamentous growth.
DISCUSSION
We have identified a specific activator for Rac1 in C. albicans, which is a member of the CDM GEF family, called Dck1. This protein has a domain with similarity to the DOCKER catalytic domain of human Dock180. Our results indicate that Dck1 is necessary for invasive filamentous growth but not for filamentous growth in response to serum. In contrast, Cdc24, a member of the Dbl GEF family, is required for serum-induced filamentous growth, and we show that Cdc24 and Dck1 do not have overlapping functions. The conserved DHR is required for Dck1 function, and this region binds nucleotide-free Rac1 in vitro. Overexpression of a GTP-bound Rac1 mutant partially complements the embedded filamentous growth defect of a dck1 mutant. Together, our results indicate that the CDM family guanine nucleotide exchange factor Dck1 specifically activates Rac1 for invasive filamentous growth in C. albicans.
The Dock180 related GEF subfamily, which is currently made up of 11 mammalian members, has been shown to specifically activate Rac1 in vitro (Cote and Vuori, 2007). Very recent studies suggest that Dock180 is also important for Cdc42 activation in neurons in vivo (Li et al., 2008). In C. albicans, Dck1 does not complement Cdc24 function, suggesting that Dck1 does not activate Cdc42. Proteins with similarity to CDM GEFs are present in a range of fungal species, including Neurospora crassa, Cryptococcus neoformans, Aspergillus nidulans, Magnaporthe grisea, Ustilago maydis, Candida albicans, Ashbya gossypii, and S. cerevisiae (Meller et al., 2005). Interestingly, the latter two fungal species do not possess a discernible Rac1 homologue. In each of these fungal species, there is one protein with similarity to CDM GEFs, except for C. albicans, where two such proteins, Dck1 and Dck2, exist. In a large-scale haploinsufficiency-based transposon mutagenesis for defects in filamentous growth, Uhl et al. (2003) identified insertion mutations in the ORFs corresponding to Dck1 and Dck2 as having reduced filamentous growth on Spider medium. However, we did not detect any defect in dck2 mutants. In contrast to other CDM GEFs, no apparent SH3 domain or conserved DHR1 region is present in Dck1. Nonetheless, we observed weak homology of the amino-terminal half of C. albicans Dck1 to the DHR1 regions of mammalian Dock proteins. DHR1 has been shown to have some similarity to lipid binding modules and in particular binds phosphatidylinositol 3,4,5-triphosphate (Cote et al., 2005). Whether C. albicans Dck1 binds phospholipids and this region plays a role in lipid binding remains to be elucidated. The SH3 domain is critical for Dock180 function in flies and humans (Brugnera et al., 2002; Balagopalan et al., 2006). For example, it has been reported that this domain autoinhibits GEF activity by binding the DOCKER catalytic domain and that it is involved in interactions with ELMO (Lu et al., 2005). ELMO is critical for efficient Rac1 signaling by Dock180 via alteration of the affinity of Dock180 for Rac1 and targeting Dock180 to the plasma membrane (Lu et al., 2004; Park et al., 2007). A protein with similarity to ELMO is present in the C. albicans genome. It remains to be determined whether this protein functions together with Dck1 and is required for Dck1 membrane association.
Our results indicate that Cdc42 and its activator Cdc24 are required for filamentous growth in response to serum, whereas Rac1 and its activator Dck1 are required for invasive filamentous growth in solid media. The specificity of these two highly similar G proteins to distinct filamentous growth stimuli could be due to regulation of GEFs, regulation of G protein expression, and/or association of each activated G protein with specific effectors. Even when overexpressed, Cdc24 is unable to replace Dck1 in Rac1 activation and conversely Dck1 is unable to replace Cdc24 in Cdc42 activation. The amino acid at position 56 in Rac1 has been shown to be required for interaction with Dbl family exchange factors (Gao et al., 2001; Karnoub et al., 2001; Hlubek et al., 2008), and mutation of this residue does not substantially impair binding to HsDock180 (Brugnera et al., 2002). We mutated C. albicans Rac1 Trp 56 to Phe, which is present in Cdc42, and this mutant when overexpressed was able to partially complement a rac1 deletion strain for embedded filamentous growth (Arkowitz and Bassilana, unpublished data). In addition, overexpression of the Rac1[W56F] mutant, which has been shown to be activated by other DH family GEFs such as Cdc24, as has been recently shown in U. maydis (Hlubek et al., 2008), did not restore embedded filamentous growth in a dck1Δ/dck1Δ strain, further consistent with the specific requirement of Dck1 for Rac1 activation during embedded filamentous growth. These results suggest that specificity in part is conferred by G protein–GEF interactions. Although Cdc42 and Rac1 are very similar at the amino acid level, the latter G protein has a 43-amino acid insertion before the carboxy terminus, which may be critical for specific interactions with the Dck1 GEF. Our Cdc42/Rac interactive binding (CRIB) binding experiments indicate that both activated Cdc42 and Rac1 can bind CRIB domain containing effectors (Bassilana and Arkowitz, 2006), consistent with the notion that signaling specificity is achieved by upstream components. The components upstream of Dck1 and Rac1 are unknown; however, recent studies in mammalian systems have identified two membrane proteins that can recruit Dock180, the adhesion-type G protein-coupled receptor brain-specific angiogenesis inhibitor 1 (BAI1), which recruits Dock180 via its association with ELMO (Park et al., 2007) and the transmembrane netrin receptor, deleted in colorectal carcinoma, which binds Dock180 directly (Li et al., 2008). We speculate that Dck1 in C. albicans is recruited to the plasma membrane by a presently unidentified membrane protein, which would be necessary for localized Rac1 activation.
Our results indicate that a GTP bound form of Rac1 partially recovers invasive filamentous growth in both rac1 and dck1 mutants; however, the extent of filamentation in these strains is reduced compared with the wild-type strain. Therefore, it is likely that Rac1 must cycle between GDP-bound and GTP-bound states to fully restore filamentous growth. Ectopic expression of the GTP bound form of Cdc42 was highly detrimental to C. albicans cells, suggesting that cycling of this G protein between GDP and GTP states is also critical (Ushinsky et al., 2002). In addition to Cdc24, the Cdc42 activator, GTPase-activating proteins are also important in filamentous growth (Court and Sudbery, 2007; Zheng et al., 2007). It is unknown whether such Cdc42 GAPs can also stimulate Rac1 GTP hydrolysis or whether specific Rac1 GAPs are required.
We propose that specific stimuli, such as contact with solid agar or presence of serum, result in recruitment and/or activation of Rac1 or Cdc42 GEFs, respectively. It is likely that locally activated Rac1 or Cdc42 recruits effector proteins that are critical for morphological changes. Understanding what determines the specificity for small G protein and GEF interactions will be critical for revealing how these morphological changes occur in response to distinct stimuli. Given that these two classes of exchange factors and the G proteins they activate are conserved from humans to fungi, the specificity determinants are likely to be similar in these organisms.
Supplementary Material
ACKNOWLEDGMENTS
We thank Q. Zhao and W. Nierman (TIGR), F. Smith and A. Mitchell (Columbia University), and National Institutes of Health grant 1R01AI057804 for the Tn7-UAU1 transposon insertion plasmids; N. Dean (Stony Brook University) and C. Kumamoto (Tufts University) for plasmids and strains; and R. Kelly (Merck) and B. Wickner (Dartmouth University) for antibodies. We also thank D. Fitzpatrick and G. Butler (University College Dublin) for phylogeny analysis. This work was supported by the Centre National de la Recherche Scientifique and Fondation pour la Recherche Médicale-BNP-Paribas. H. H. was supported by EU Marie Curie early stage training fellowship (International Ph.D. Program in Developmental and Cellular Decisions).
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-12-1272) on June 25, 2008.
REFERENCES
- Albert M. L., Kim J. I., Birge R. B. alphavbeta5 integrin recruits the CrkII-Dock180-Rac1 complex for phagocytosis of apoptotic cells. Nat. Cell Biol. 2000;2:899–905. doi: 10.1038/35046549. [DOI] [PubMed] [Google Scholar]
- Balagopalan L., Chen M. H., Geisbrecht E. R., Abmayr S. M. The CDM superfamily protein MBC directs myoblast fusion through a mechanism that requires phosphatidylinositol 3,4,5-triphosphate binding but is independent of direct interaction with DCrk. Mol. Cell. Biol. 2006;26:9442–9455. doi: 10.1128/MCB.00016-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barale S., McCusker D., Arkowitz R. A. Cdc42p GDP/GTP cycling is necessary for efficient cell fusion during yeast mating. Mol. Biol. Cell. 2006;17:2824–2838. doi: 10.1091/mbc.E05-11-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassilana M., Arkowitz R. A. Rac1 and Cdc42 have different roles in Candida albicans development. Eukaryot. Cell. 2006;5:321–329. doi: 10.1128/EC.5.2.321-329.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassilana M., Blyth J., Arkowitz R. A. Cdc24, the GDP-GTP exchange factor for Cdc42, is required for invasive hyphal growth of Candida albicans. Eukaryot. Cell. 2003;2:9–18. doi: 10.1128/EC.2.1.9-18.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassilana M., Hopkins J., Arkowitz R. A. Regulation of the Cdc42/Cdc24 GTPase module during Candida albicans hyphal growth. Eukaryot. Cell. 2005;4:588–603. doi: 10.1128/EC.4.3.588-603.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco A., Poukkula M., Cliffe A., Mathieu J., Luque C. M., Fulga T. A., Rorth P. Two distinct modes of guidance signalling during collective migration of border cells. Nature. 2007;448:362–365. doi: 10.1038/nature05965. [DOI] [PubMed] [Google Scholar]
- Biswas S., Van Dijck P., Datta A. Environmental sensing and signal transduction pathways regulating morphopathogenic determinants of Candida albicans. Microbiol. Mol. Biol. Rev. 2007;71:348–376. doi: 10.1128/MMBR.00009-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown D. H., Jr, Giusani A. D., Chen X., Kumamoto C. A. Filamentous growth of Candida albicans in response to physical environmental cues and its regulation by the unique CZF1 gene. Mol. Microbiol. 1999;34:651–662. doi: 10.1046/j.1365-2958.1999.01619.x. [DOI] [PubMed] [Google Scholar]
- Brugnera E., Haney L., Grimsley C., Lu M., Walk S. F., Tosello-Trampont A. C., Macara I. G., Madhani H., Fink G. R., Ravichandran K. S. Unconventional Rac-GEF activity is mediated through the Dock180-ELMO complex. Nat. Cell Biol. 2002;4:574–582. doi: 10.1038/ncb824. [DOI] [PubMed] [Google Scholar]
- Buchsbaum R. J. Rho activation at a glance. J. Cell Sci. 2007;120:1149–1152. doi: 10.1242/jcs.03428. [DOI] [PubMed] [Google Scholar]
- Calderone R. A. Washington, D.C: ASM Press; 2002. Candida and candidiasis. [Google Scholar]
- Calera J. A., Zhao X. J., Calderone R. Defective hyphal development and avirulence caused by a deletion of the SSK1 response regulator gene in Candida albicans. Infect. Immun. 2000;68:518–525. doi: 10.1128/iai.68.2.518-525.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castilla R., Passeron S., Cantore M. L. N-acetyl-D-glucosamine induces germination in Candida albicans through a mechanism sensitive to inhibitors of cAMP-dependent protein kinase. Cell Signal. 1998;10:713–719. doi: 10.1016/s0898-6568(98)00015-1. [DOI] [PubMed] [Google Scholar]
- Cote J. F., Motoyama A. B., Bush J. A., Vuori K. A novel and evolutionarily conserved PtdIns(3,4,5)P3-binding domain is necessary for DOCK180 signalling. Nat. Cell Biol. 2005;7:797–807. doi: 10.1038/ncb1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cote J. F., Vuori K. Identification of an evolutionarily conserved superfamily of DOCK180-related proteins with guanine nucleotide exchange activity. J. Cell Sci. 2002;115:4901–4913. doi: 10.1242/jcs.00219. [DOI] [PubMed] [Google Scholar]
- Cote J. F., Vuori K. GEF what? Dock180 and related proteins help Rac to polarize cells in new ways. Trends Cell Biol. 2007;17:383–393. doi: 10.1016/j.tcb.2007.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Court H., Sudbery P. Regulation of Cdc42 GTPase activity in the formation of hyphae in Candida albicans. Mol. Biol. Cell. 2007;18:265–281. doi: 10.1091/mbc.E06-05-0411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis D. A., Bruno V. M., Loza L., Filler S. G., Mitchell A. P. Candida albicans Mds3p, a conserved regulator of pH responses and virulence identified through insertional mutagenesis. Genetics. 2002;162:1573–1581. doi: 10.1093/genetics/162.4.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DerMardirossian C., Bokoch G. M. GDIs: central regulatory molecules in Rho GTPase activation. Trends Cell Biol. 2005;15:356–363. doi: 10.1016/j.tcb.2005.05.001. [DOI] [PubMed] [Google Scholar]
- Erickson J. W., Cerione R. A. Structural elements, mechanism, and evolutionary convergence of Rho protein-guanine nucleotide exchange factor complexes. Biochemistry. 2004;43:837–842. doi: 10.1021/bi036026v. [DOI] [PubMed] [Google Scholar]
- Erickson M. R., Galletta B. J., Abmayr S. M. Drosophila myoblast city encodes a conserved protein that is essential for myoblast fusion, dorsal closure, and cytoskeletal organization. J. Cell Biol. 1997;138:589–603. doi: 10.1083/jcb.138.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feig L. A. Tools of the trade: use of dominant-inhibitory mutants of Ras-family GTPases. Nat. Cell Biol. 1999;1:E25–E27. doi: 10.1038/10018. [DOI] [PubMed] [Google Scholar]
- Gao Y., Xing J., Streuli M., Leto T. L., Zheng Y. Trp(56) of Rac1 specifies interaction with a subset of guanine nucleotide exchange factors. J. Biol. Chem. 2001;276:47530–47541. doi: 10.1074/jbc.M108865200. [DOI] [PubMed] [Google Scholar]
- Gulli M. P., Peter M. Temporal and spatial regulation of Rho-type guanine-nucleotide exchange factors: the yeast perspective. Genes Dev. 2001;15:365–379. doi: 10.1101/gad.876901. [DOI] [PubMed] [Google Scholar]
- Hall A. Rho GTPases and the control of cell behaviour. Biochem. Soc. Trans. 2005;33:891–895. doi: 10.1042/BST20050891. [DOI] [PubMed] [Google Scholar]
- Harlow E., Lane D. Antibodies: A Laboratory Manual. Cold Spring Harbor Laboratory, NY: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- Hasegawa H., Kiyokawa E., Tanaka S., Nagashima K., Gotoh N., Shibuya M., Kurata T., Matsuda M. DOCK180, a major CRK-binding protein, alters cell morphology upon translocation to the cell membrane. Mol. Cell. Biol. 1996;16:1770–1776. doi: 10.1128/mcb.16.4.1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hlubek A., Schink K. O., Mahlert M., Sandrock B., Bolker M. Selective activation by the guanine nucleotide exchange factor Don1 is a main determinant of Cdc42 signalling specificity in Ustilago maydis. Mol. Microbiol. 2008;68:615–623. doi: 10.1111/j.1365-2958.2008.06177.x. [DOI] [PubMed] [Google Scholar]
- Jaffe A. B., Hall A. Rho GTPases: biochemistry and biology. Annu. Rev. Cell Dev. Biol. 2005;21:247–269. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- Karnoub A. E., Worthylake D. K., Rossman K. L., Pruitt W. M., Campbell S. L., Sondek J., Der C. J. Molecular basis for Rac1 recognition by guanine nucleotide exchange factors. Nat. Struct. Biol. 2001;8:1037–1041. doi: 10.1038/nsb719. [DOI] [PubMed] [Google Scholar]
- Karnoub A. E., Symons M., Campbell S. L., Der C. J. Molecular basis for Rho GTPase signaling specificity. Breast Cancer Res. Treat. 2004;84:61–71. doi: 10.1023/B:BREA.0000018427.84929.5c. [DOI] [PubMed] [Google Scholar]
- Kelly R., Card D., Register E., Mazur P., Kelly T., Tanaka K. I., Onishi J., Williamson J. M., Fan H., Satoh T., Kurtz M. Geranylgeranyltransferase I of Candida albicans: null mutants or enzyme inhibitors produce unexpected phenotypes. J. Bacteriol. 2000;182:704–713. doi: 10.1128/jb.182.3.704-713.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumamoto C. A., Vinces M. D. Alternative Candida albicans lifestyles: growth on surfaces. Annu. Rev. Microbiol. 2005;59:113–133. doi: 10.1146/annurev.micro.59.030804.121034. [DOI] [PubMed] [Google Scholar]
- Li X., Gao X., Liu G., Xiong W., Wu J., Rao Y. Netrin signal transduction and the guanine nucleotide exchange factor DOCK180 in attractive signaling. Nat. Neurosci. 2008;11:28–35. doi: 10.1038/nn2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M., et al. PH domain of ELMO functions in trans to regulate Rac activation via Dock180. Nat. Struct. Mol. Biol. 2004;11:756–762. doi: 10.1038/nsmb800. [DOI] [PubMed] [Google Scholar]
- Lu M., Kinchen J. M., Rossman K. L., Grimsley C., Hall M., Sondek J., Hengartner M. O., Yajnik V., Ravichandran K. S. A Steric-inhibition model for regulation of nucleotide exchange via the Dock180 family of GEFs. Curr. Biol. 2005;15:371–377. doi: 10.1016/j.cub.2005.01.050. [DOI] [PubMed] [Google Scholar]
- Meller N., Irani-Tehrani M., Kiosses W. B., Del Pozo M. A., Schwartz M. A. Zizimin1, a novel Cdc42 activator, reveals a new GEF domain for Rho proteins. Nat. Cell Biol. 2002;4:639–647. doi: 10.1038/ncb835. [DOI] [PubMed] [Google Scholar]
- Meller N., Merlot S., Guda C. CZH proteins: a new family of Rho-GEFs. J. Cell Sci. 2005;118:4937–4946. doi: 10.1242/jcs.02671. [DOI] [PubMed] [Google Scholar]
- Michel S., Ushinsky S., Klebl B., Leberer E., Thomas D., Whiteway M., Morschhauser J. Generation of conditional lethal Candida albicans mutants by inducible deletion of essential genes. Mol. Microbiol. 2002;46:269–280. doi: 10.1046/j.1365-2958.2002.03167.x. [DOI] [PubMed] [Google Scholar]
- Michiels F., Habets G. G., Stam J. C., van der Kammen R. A., Collard J. G. A role for Rac in Tiam1-induced membrane ruffling and invasion. Nature. 1995;375:338–340. doi: 10.1038/375338a0. [DOI] [PubMed] [Google Scholar]
- Nern A., Arkowitz R. A. G proteins mediate changes in cell shape by stabilizing the axis of polarity. Mol. Cell. 2000;5:853–864. doi: 10.1016/s1097-2765(00)80325-1. [DOI] [PubMed] [Google Scholar]
- Nishikawa A., Poster J. B., Jigami Y., Dean N. Molecular and phenotypic analysis of CaVRG4, encoding an essential Golgi apparatus GDP-mannose transporter. J. Bacteriol. 2002;184:29–42. doi: 10.1128/JB.184.1.29-42.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odds F. C., Webster C. E., Mayuranathan P., Simmons P. D. Candida concentrations in the vagina and their association with signs and symptoms of vaginal candidosis. J. Med. Vet. Mycol. 1988;26:277–283. doi: 10.1080/02681218880000391. [DOI] [PubMed] [Google Scholar]
- Park D., Tosello-Trampont A. C., Elliott M. R., Lu M., Haney L. B., Ma Z., Klibanov A. L., Mandell J. W., Ravichandran K. S. BAI1 is an engulfment receptor for apoptotic cells upstream of the ELMO/Dock180/Rac module. Nature. 2007;450:430–434. doi: 10.1038/nature06329. [DOI] [PubMed] [Google Scholar]
- Park H. O., Bi E. Central roles of small GTPases in the development of cell polarity in yeast and beyond. Microbiol. Mol. Biol. Rev. 2007;71:48–96. doi: 10.1128/MMBR.00028-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossman K. L., Der C. J., Sondek J. GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat. Rev. Mol. Cell Biol. 2005;6:167–180. doi: 10.1038/nrm1587. [DOI] [PubMed] [Google Scholar]
- Rossman K. L., Sondek J. Larger than Dbl: new structural insights into RhoA activation. Trends Biochem. Sci. 2005;30:163–165. doi: 10.1016/j.tibs.2005.02.002. [DOI] [PubMed] [Google Scholar]
- Sinha I., Wang Y. M., Philp R., Li C. R., Yap W. H., Wang Y. Cyclin-dependent kinases control septin phosphorylation in Candida albicans hyphal development. Dev. Cell. 2007;13:421–432. doi: 10.1016/j.devcel.2007.06.011. [DOI] [PubMed] [Google Scholar]
- Tcherkezian J., Lamarche-Vane N. Current knowledge of the large RhoGAP family of proteins. Biol. Cell. 2007;99:67–86. doi: 10.1042/BC20060086. [DOI] [PubMed] [Google Scholar]
- Uhl M. A., Biery M., Craig N., Johnson A. D. Haploinsufficiency-based large-scale forward genetic analysis of filamentous growth in the diploid human fungal pathogen C. albicans. EMBO J. 2003;22:2668–2678. doi: 10.1093/emboj/cdg256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushinsky S. C., Harcus D., Ash J., Dignard D., Marcil A., Morchhauser J., Thomas D. Y., Whiteway M., Leberer E. CDC42 is required for polarized growth in human pathogen Candida albicans. Eukaryot. Cell. 2002;1:95–104. doi: 10.1128/EC.1.1.95-104.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VandenBerg A. L., Ibrahim A. S., Edwards J. E., Jr, Toenjes K. A., Johnson D. I. Cdc42p GTPase regulates the budded-to-hyphal-form transition and expression of hypha-specific transcripts in Candida albicans. Eukaryot. Cell. 2004;3:724–734. doi: 10.1128/EC.3.3.724-734.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner M. P., Costa G. L., Schoettlin W., Cline J., Mathur E., Bauer J. C. Site-directed mutagenesis of double-stranded DNA by the polymerase chain reaction. Gene. 1994;151:119–123. doi: 10.1016/0378-1119(94)90641-6. [DOI] [PubMed] [Google Scholar]
- Wu Y. C., Horvitz H. R. C. elegans phagocytosis and cell-migration protein CED-5 is similar to human DOCK180. Nature. 1998;392:501–504. doi: 10.1038/33163. [DOI] [PubMed] [Google Scholar]
- Zheng X. D., Lee R. T., Wang Y. M., Lin Q. S., Wang Y. Phosphorylation of Rga2, a Cdc42 GAP, by CDK/Hgc1 is crucial for Candida albicans hyphal growth. EMBO J. 2007;26:3760–3769. doi: 10.1038/sj.emboj.7601814. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.