Abstract
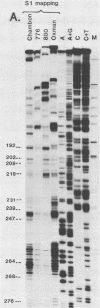
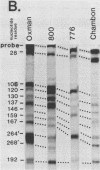
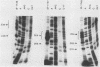
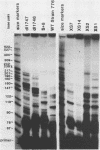
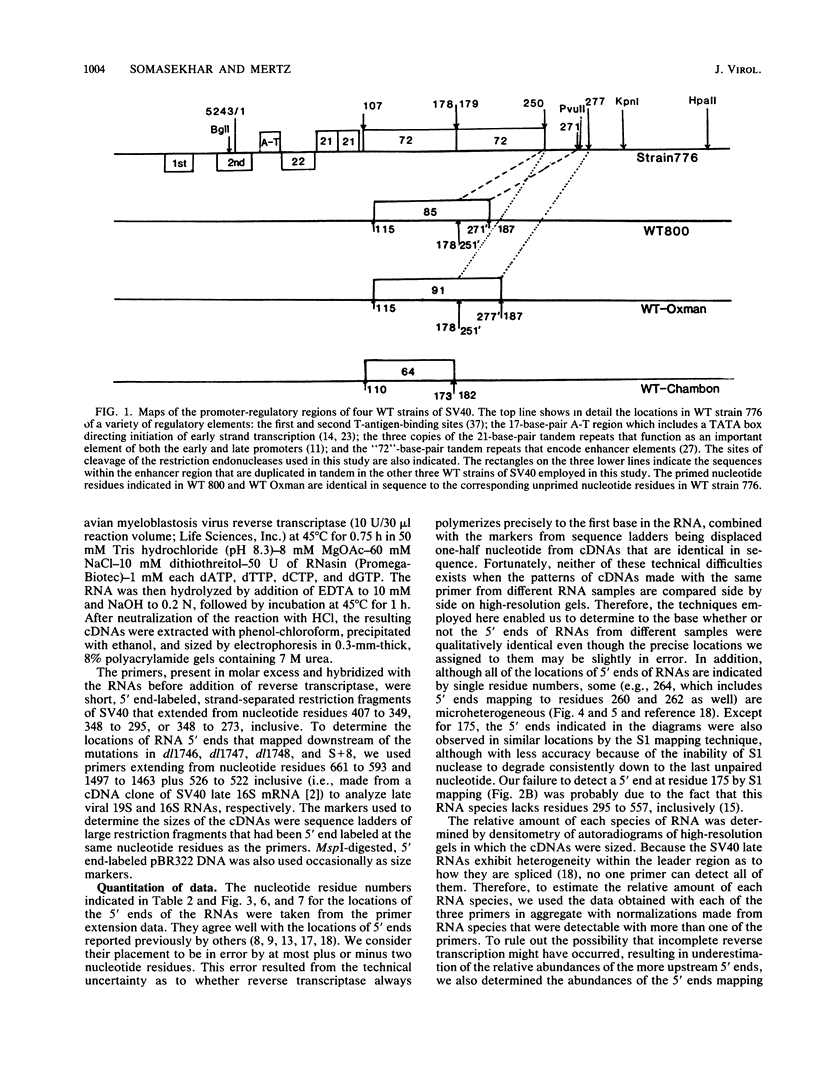
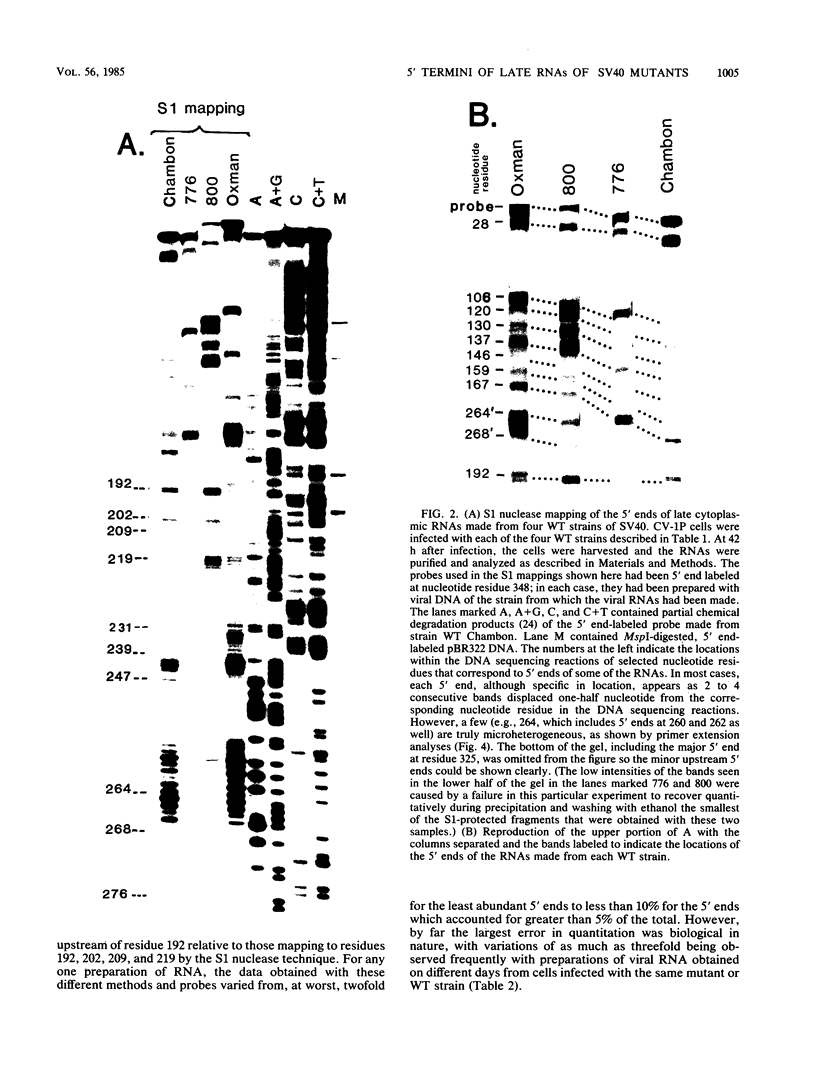
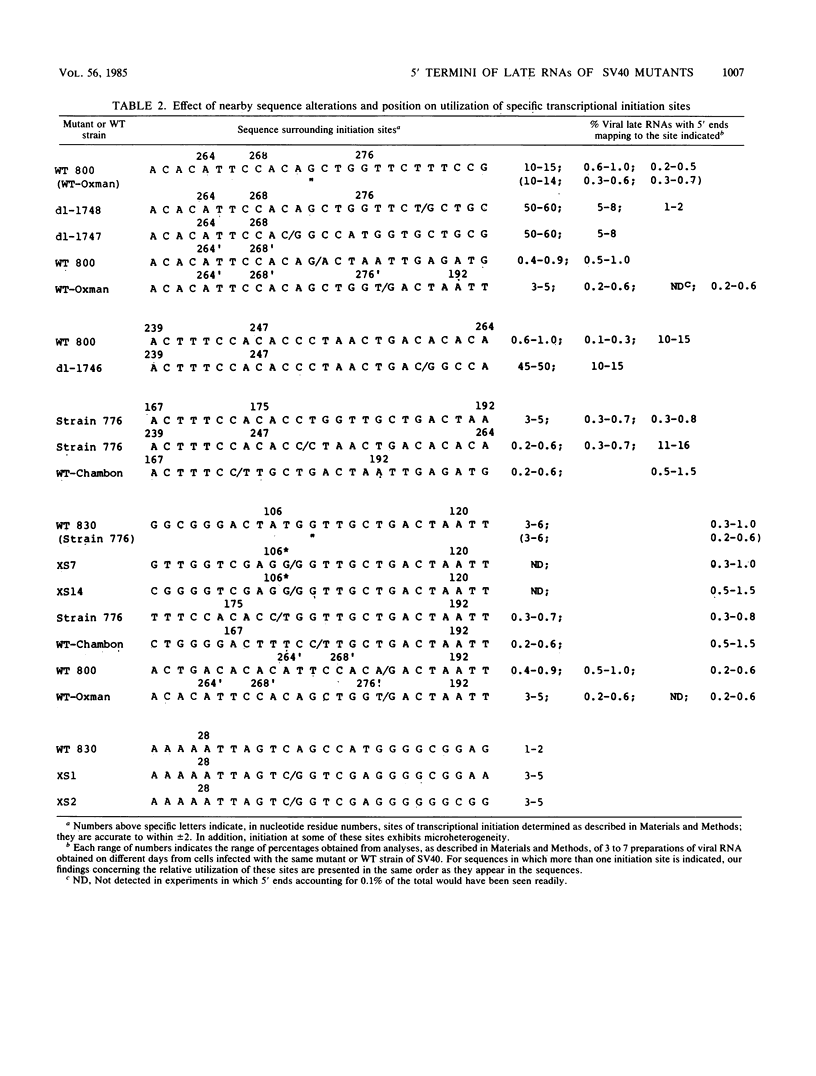
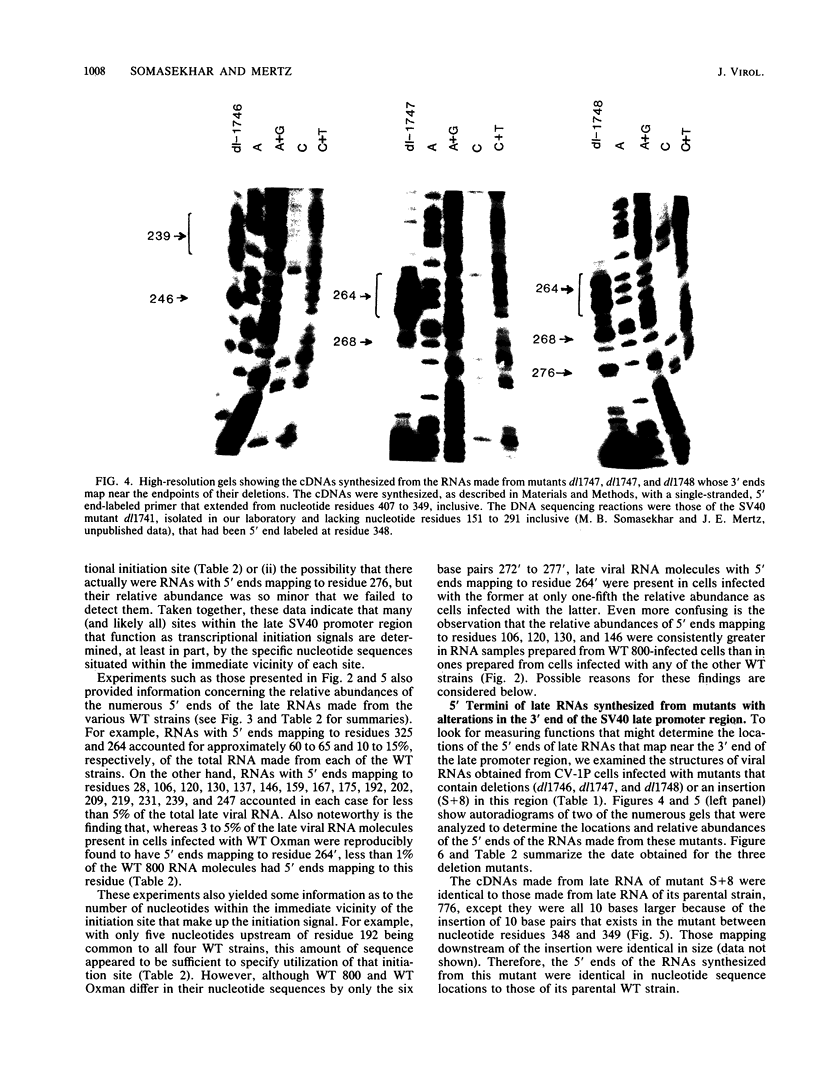
The 5' ends of the simian virus 40 (SV40) late RNAs are heterogeneous in location, spanning a 300-nucleotide region from residues 28 to 325. To examine whether upstream or downstream measuring functions analogous to the TATA box play roles in positioning the 5' ends of these RNAs, we determined by S1 and primer extension mapping the locations of the 5' ends of the late viral RNAs made in monkey cells infected with: (i) three wild-type strains of SV40 that contain tandem duplications of the enhancer region that are 64, 85, and 91, rather than 72, base pairs in length; (ii) four viable mutants that contain alterations in the 21-base-pair tandem repeats; and (iii) four viable mutants that possess small deletions or insertions at or near the major cap site at residue 325. Most of the 5' ends of the RNAs were identical in location to those seen with wild-type strain 776. The only exceptions were the absence of RNAs whose 5' ends mapped to within three bases upstream or downstream of a sequence alteration. In addition, the sequences within residues 251 to 277 that function as transcriptional initiation sites in wild-type strain 776 also did so in their second locations in the wild-type strains in which these sequences are duplicated. Differences were noted in the relative abundances of the numerous 5' ends of the late RNAs, even among the wild-type strains. These findings indicate that many (and likely all) of the approximately two dozen locations of 5' ends of SV40 late RNAs are each determined largely by sequences within their immediate vicinity. However, sequences somewhat removed from these transcriptional initiation sites may modulate the efficiencies with which they are utilized.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alwine J. C. Evidence for simian virus 40 late transcriptional control: mixed infections of wild-type simian virus 40 and a late leader deletion mutant exhibit trans effects on late viral RNA synthesis. J Virol. 1982 Jun;42(3):798–803. doi: 10.1128/jvi.42.3.798-803.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A., Mertz J. E. The number of ribosomes on simian virus 40 late 16S mRNA is determined in part by the nucleotide sequence of its leader. Mol Cell Biol. 1984 Apr;4(4):813–816. doi: 10.1128/mcb.4.4.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoist C., Chambon P. In vivo sequence requirements of the SV40 early promotor region. Nature. 1981 Mar 26;290(5804):304–310. doi: 10.1038/290304a0. [DOI] [PubMed] [Google Scholar]
- Benoist C., O'Hare K., Breathnach R., Chambon P. The ovalbumin gene-sequence of putative control regions. Nucleic Acids Res. 1980 Jan 11;8(1):127–142. doi: 10.1093/nar/8.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady J., Radonovich M., Vodkin M., Natarajan V., Thoren M., Das G., Janik J., Salzman N. P. Site-specific base substitution and deletion mutations that enhance or suppress transcription of the SV40 major late RNA. Cell. 1982 Dec;31(3 Pt 2):625–633. doi: 10.1016/0092-8674(82)90318-x. [DOI] [PubMed] [Google Scholar]
- Breathnach R., Chambon P. Organization and expression of eucaryotic split genes coding for proteins. Annu Rev Biochem. 1981;50:349–383. doi: 10.1146/annurev.bi.50.070181.002025. [DOI] [PubMed] [Google Scholar]
- Contreras R., Fiers W. Initiation of transcription by RNA polymerase II in permeable, SV40-infected or noninfected, CVI cells; evidence for multiple promoters of SV40 late transcription. Nucleic Acids Res. 1981 Jan 24;9(2):215–236. doi: 10.1093/nar/9.2.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras R., Gheysen D., Knowland J., van de Voorde A., Fiers W. Evidence for the direct involvement of DNA replication origin in synthesis of late SV40 RNA. Nature. 1982 Dec 9;300(5892):500–505. doi: 10.1038/300500a0. [DOI] [PubMed] [Google Scholar]
- Fiers W., Contreras R., Haegemann G., Rogiers R., Van de Voorde A., Van Heuverswyn H., Van Herreweghe J., Volckaert G., Ysebaert M. Complete nucleotide sequence of SV40 DNA. Nature. 1978 May 11;273(5658):113–120. doi: 10.1038/273113a0. [DOI] [PubMed] [Google Scholar]
- Fromm M., Berg P. Deletion mapping of DNA regions required for SV40 early region promoter function in vivo. J Mol Appl Genet. 1982;1(5):457–481. [PubMed] [Google Scholar]
- Fromm M., Berg P. Transcription in vivo from SV40 early promoter deletion mutants without repression by large T antigen. J Mol Appl Genet. 1983;2(1):127–135. [PubMed] [Google Scholar]
- Gheysen D., van de Voorde A., Contreras R., Vanderheyden J., Duerinck F., Fiers W. Simian virus 40 mutants carrying extensive deletions in the 72-base-pair repeat region. J Virol. 1983 Jul;47(1):1–14. doi: 10.1128/jvi.47.1.1-14.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh P. K., Lebowitz P., Frisque R. J., Gluzman Y. Identification of a promoter component involved in positioning the 5' termini of simian virus 40 early mRNAs. Proc Natl Acad Sci U S A. 1981 Jan;78(1):100–104. doi: 10.1073/pnas.78.1.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh P. K., Piatak M., Mertz J. E., Weissman S. M., Lebowitz P. Altered utilization of splice sites and 5' termini in late RNAs produced by leader region mutants of simian virus 40. J Virol. 1982 Nov;44(2):610–624. doi: 10.1128/jvi.44.2.610-624.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh P. K., Reddy V. B., Piatak M., Lebowitz P., Weissman S. M. Determination of RNA sequences by primer directed synthesis and sequencing of their cDNA transcripts. Methods Enzymol. 1980;65(1):580–595. doi: 10.1016/s0076-6879(80)65061-7. [DOI] [PubMed] [Google Scholar]
- Ghosh P. K., Reddy V. B., Swinscoe J., Choudary P. V., Lebowitz P., Weissman S. M. The 5'-terminal leader sequence of late 16 S mRNA from cells infected with simian virus 40. J Biol Chem. 1978 May 25;253(10):3643–3647. [PubMed] [Google Scholar]
- Ghosh P. K., Reddy V. B., Swinscoe J., Lebowitz P., Weissman S. M. Heterogeneity and 5'-terminal structures of the late RNAs of simian virus 40. J Mol Biol. 1978 Dec 25;126(4):813–846. doi: 10.1016/0022-2836(78)90022-0. [DOI] [PubMed] [Google Scholar]
- Gidoni D., Kahana C., Canaani D., Groner Y. Specific in vitro initiation of transcription of simian virus 40 early and late genes occurs at the various cap nucleotides including cytidine. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2174–2178. doi: 10.1073/pnas.78.4.2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosveld G. C., de Boer E., Shewmaker C. K., Flavell R. A. DNA sequences necessary for transcription of the rabbit beta-globin gene in vivo. Nature. 1982 Jan 14;295(5845):120–126. doi: 10.1038/295120a0. [DOI] [PubMed] [Google Scholar]
- Haegeman G., van Heuverswyn H., Gheysen D., Fiers W. Heterogeneity of the 5' terminus of late mRNA induced by a viable simian virus 40 deletion mutant. J Virol. 1979 Aug;31(2):484–493. doi: 10.1128/jvi.31.2.484-493.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura G., Dulbecco R. Isolation and characterization of temperature-sensitive mutants of simian virus 40. Virology. 1972 Aug;49(2):394–403. doi: 10.1016/0042-6822(72)90492-8. [DOI] [PubMed] [Google Scholar]
- Mathis D. J., Chambon P. The SV40 early region TATA box is required for accurate in vitro initiation of transcription. Nature. 1981 Mar 26;290(5804):310–315. doi: 10.1038/290310a0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Mertz J. E., Berg P. Defective simian virus 40 genomes: isolation and growth of individual clones. Virology. 1974 Nov;62(1):112–124. doi: 10.1016/0042-6822(74)90307-9. [DOI] [PubMed] [Google Scholar]
- Mertz J. E., Carbon J., Herzberg M., Davis R. W., Berg P. Isolation and characterization of individual clones of simian virus 40 mutants containing deletions duplications and insertions in their DNA. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 1):69–84. doi: 10.1101/sqb.1974.039.01.012. [DOI] [PubMed] [Google Scholar]
- Moreau P., Hen R., Wasylyk B., Everett R., Gaub M. P., Chambon P. The SV40 72 base repair repeat has a striking effect on gene expression both in SV40 and other chimeric recombinants. Nucleic Acids Res. 1981 Nov 25;9(22):6047–6068. doi: 10.1093/nar/9.22.6047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatak M., Ghosh P. K., Norkin L. C., Weissman S. M. Sequences locating the 5' ends of the major simian virus 40 late mRNA forms. J Virol. 1983 Nov;48(2):503–520. doi: 10.1128/jvi.48.2.503-520.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatak M., Subramanian K. N., Roy P., Weissman S. M. Late messenger RNA production by viable simian virus 40 mutants with deletions in the leader region. J Mol Biol. 1981 Dec 15;153(3):589–618. doi: 10.1016/0022-2836(81)90409-5. [DOI] [PubMed] [Google Scholar]
- Reddy V. B., Ghosh P. K., Lebowitz P., Piatak M., Weissman S. M. Simian virus 40 early mRNA's. I. Genomic localization of 3' and 5' termini and two major splices in mRNA from transformed and lytically infected cells. J Virol. 1979 Apr;30(1):279–296. doi: 10.1128/jvi.30.1.279-296.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy V. B., Thimmappaya B., Dhar R., Subramanian K. N., Zain B. S., Pan J., Ghosh P. K., Celma M. L., Weissman S. M. The genome of simian virus 40. Science. 1978 May 5;200(4341):494–502. doi: 10.1126/science.205947. [DOI] [PubMed] [Google Scholar]
- Reynolds G. A., Basu S. K., Osborne T. F., Chin D. J., Gil G., Brown M. S., Goldstein J. L., Luskey K. L. HMG CoA reductase: a negatively regulated gene with unusual promoter and 5' untranslated regions. Cell. 1984 Aug;38(1):275–285. doi: 10.1016/0092-8674(84)90549-x. [DOI] [PubMed] [Google Scholar]
- Ross J. A precursor of globin messenger RNA. J Mol Biol. 1976 Sep 15;106(2):403–420. doi: 10.1016/0022-2836(76)90093-0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R. The use of thin acrylamide gels for DNA sequencing. FEBS Lett. 1978 Mar 1;87(1):107–110. doi: 10.1016/0014-5793(78)80145-8. [DOI] [PubMed] [Google Scholar]
- Shenk T. Transcriptional control regions: nucleotide sequence requirements for initiation by RNA polymerase II and III. Curr Top Microbiol Immunol. 1981;93:25–46. doi: 10.1007/978-3-642-68123-3_3. [DOI] [PubMed] [Google Scholar]
- Tjian R. The binding site on SV40 DNA for a T antigen-related protein. Cell. 1978 Jan;13(1):165–179. doi: 10.1016/0092-8674(78)90147-2. [DOI] [PubMed] [Google Scholar]