Abstract
Aldose-1-epimerase (mutarotase) catalyzes the interconversion of α and β hexoses, which is essential for normal carbohydrate metabolism and the production of complex oligosaccharides. Galactose mutarotase (GALM) has been well characterized at the protein level but information is lacking on the regulation of GALM gene expression. We report herein that all-trans-retinoic acid (RA), an active metabolite of vitamin A that is known to induce myeloid lineage cell differentiation into macrophage-like cells, induces a rapid and robust regulation of GALM mRNA expression in human myeloid cells. All-trans-RA at a physiological concentration (20 nM), or Am580, a ligand selective for the nuclear retinoid receptor RARα, increased GALM mRNA in THP-1 cells, with significantly increased expression in 2 h, increasing further to an ~8-fold elevation after 6–40 h (P< 0.005). In contrast, tumor necrosis factor-α did not increase GALM mRNA expression, although it is capable of inducing cell differentiation. RA also increased GALM mRNA in U937 and HL-60 cells. The increase in GALM mRNA by RA was blocked by pretreating THP-1 cells with actinomycin D, but not by cycloheximide. GALM protein and mutarotase activity were also increased time dependently in RA-treated THP-1 cells. In addition to GALM, several other genes in the biosynthetic pathway of galactosyl-containing complex oligosaccharides were more highly expressed in RA-treated THP-1 cells, including B4GALT5, ST3GAL3, ST6GALNAC5, and GALNAC4S-6ST. Thus, the results of this study identify RA as a significant regulator of GALM and other galactose-related genes in myeloid-monocytic cells, which could affect energy utilization, synthesis of cell-surface glycoproteins or glycolipids involved in cell motility, adhesion, and/or functional properties.
Monosaccharides such as glucose and galactose exist in two anomeric forms, α and β, which differ only in the stereochemical configuration of the pyranose ring at carbon 1. Enzymes that metabolize glucose and galactose often preferentially use either the alpha or beta anomer as substrate (1). For example, galactokinase, which produces galactose-1-phosphate, selectively metabolizes α-galactose, while glucose dehydrogenase is selective for the β anomeric form of these hexoses. Aldose-1-epimerases (mutarotases) increase the efficiency of anomer interconversion in vivo, catalyzing the transformation of β-D-galactose into α-D-galactose used by hexokinase (2). Mutarotases are present throughout prokaryotic and eukaryotic phyla, and the proteins have been purified from various sources, including several bacteria and animal and human tissues [reviewed in (1)]. The 3-dimensional structure of galactose mutarotase from Lactococcus lactis has been determined (3) and mechanisms of catalysis of aldose-1-epimerase have been elucidated (4). Timson and Reese (5) used information from a human gene database to identify and clone the cDNA for human aldose-1-epimerase (EC 5.1.3.3). The expressed enzyme exhibited a 4-fold preference for galactose over glucose and, hence, they suggested that the gene and its product should be named galactomutarotase (GALM). This nomenclature has now been approved by the Human Gene Nomenclature Committee (6). Although the mutorotation reaction catalyzed by GALM is reversible, in normal galactose metabolism the production of α-D-galactose predominates (1, 5). GALM activity produces α-galactose which can be oxidized or modified and used in the biosynthesis of various complex oligosaccharides present in glycoproteins and glycolipids (5, 7). Although GALM protein has been well studied, the regulation of GALM gene expression has not been reported.
THP-1 cells have been studied extensively for their ability to differentiate into macrophage-like cells (8, 9). This immortalized cell line is considered to behave more like native monocyte-derived macrophages, and therefore to provide a useful model for studying the mechanisms involved in macrophage differentiation, and the regulation of genes related to the physiological functions displayed by these cells (10). Several nutrients, cytokines and other biological compounds are effective in inducing THP-1 cell differentiation, including retinoic acid (RA), activated vitamin D, phorbol esters, and tumor necrosis factor (TNF)-α (11–13). However, these agents may affect cell differentiation through different, or only partly overlapping, pathways. All-trans-RA, the most active carboxylic acid metabolite of vitamin A (retinol), is the natural ligand for nuclear hormone receptors of the RAR gene family, RARα, RARβ and RAR. These nuclear proteins bind with RXR proteins to form heterodimers that function as transcription factors for numerous genes, and thus participate in the regulation of nearly all physiological processes (14–16). The addition of physiologically relevant concentrations of RA (20–100 nM) to cultures of THP-1 cells has been demonstrated to alter several cell cycle-related genes while increasing the proportion of cells in the G1 phase of the cell cycle (13). Both RA and TNFα induced the differentiation of THP-1 cells, as observed by an increase in expression of cell-surface CD11b, a molecule associated with macrophage functions, and increased phagocytosis of opsonized bacterial particles (13).
The present investigation began with a gene expression profiling study that we conducted in THP-1 cells treated with RA and TNFα to identify gene expression changes in the early stages of cell differentiation. Based on observations that GALM mRNA is increased rapidly and robustly by RA in THP-1 cells, we then focused on examining GALM expression, protein and mutarotase functional activity in THP-1 cells and other myelocytic cell lines. The results provide evidence that the increase in GALM expression is selective for RA, while it does not appear to be a general result of cell differentiation. Additionally, RA up-regulated the expression of several other genes that may act downstream of GALM in the formation of galactose-containing glycoproteins and glycolipids. These changes suggest that RA could significantly regulate the functionality of myeloid cells through changes in the expression of GALM and other galactose-related genes.
Materials and Methods
Reagents
All-trans-retinoic acid, alpha-D-glucose, NAD, 1,25-dihydroxyvitamin D, actinomycin D, and cycloheximide, and glucose dehydrogenase (from Thermoplasma acidophilum) were purchased from Sigma-Aldrich (St. Louis, MO). TNFα was from R&D Systems (Minneapolis, MN). Am580 (RARα agonist) (17) was kindly provided by S. Kagechika (Tokyo, Japan).
Cell culture and treatment
THP-1 cells purchased from the American Type Culture Collection (ATCC, Rockville, MD) were maintained in RPMI-1640 medium containing 10% heat-inactivated fetal bovine serum (FBS, Invitrogen, Carlsbad, CA), 10−5 M 2-mercaptoethanol, 100 units/ml penicillin and 100 μg/ml streptomycin (Life Technologies, Rockville, MD) at 37°C in an atmosphere of 95% air and 5% CO2. Cells were passed for no more than 3 months before renewal from frozen stocks. For each experiment, the cells were resuspended in the medium supplemented with 3% FBS and 2-ml aliquots of cell suspension (0.7 × 106 cells/ml) were transferred to each well (35-mm diameter) of a 6-well culture plate and incubated for 6 h with or without RA and other compounds. RA was added in a small volume of ethanol (vehicle), which did not exceed 0.1% in the final medium. Other human myeloid cell lines, U937 and HL60, and human hepatoma cells, HepG2, were obtained from ATCC and cultured as previously described (18).
Gene analysis by microarray
THP-1 cell RNA prepared in our laboratory was analyzed for gene expression by microarray at the National Cancer Institute (NCI) Microarray Facility (Frederick, MD) as part of the Bio-Active Nutrient Gene Expression Omnibus (BANGEO) project. Each RNA sample represented an individual well of THP-1 cells. Experiment 1 included 21 arrays from 4 groups of THP-1 cells (n=5–6 arrays each representing an independent well of cultured cells): control; 20 nM RA for 16 h, followed by re-treatment after 24 h for a total time of 40 h; TNFα, 5 ng/ml for 24 h; RA, 20 nM for 16 h followed by treatment with TNFα and RA for 24 h. Experiment 2 was a RA time course study, without TNFα, which included 17 arrays from THP-1 cells treated with RA, 20 nM, for times of 0 (ethanol control, n=6), 2 h (n=2), 6 h (n=2), and 16 h (n=3). After 16 h, RA was added again to the remaining wells, which were harvested at 40 h (n=2). After completion of treatments, the cells were collected by centrifugation and total RNA was prepared using RNeasy kit (Qiagen) and checked for quality by agarose gel electrophoresis. The identities of RNA samples from the various treatment groups were coded so that each batch of n=6–8 arrays processed together contained RNA from different treatments. RNA samples were shipped on dry ice to the NCI facility. cDNA targets for the Affymetrix U133 PLUS 2 arrays were synthesized, labeled, and purified according to vendor’s (Affymetrix Inc., Santa Clara, CA) instructions. Briefly, for each sample, 1 μg of RNA was used to generate double-stranded cDNA using the T7-oligo(dT) primer according to the GeneChip Eukaryotic Small Sample Target Labeling Assay Version II (19). After second-strand synthesis, in vitro transcription after second strand synthesis was performed using the MEGAscript T7 kit (Ambion, Austin, TX) and the cRNA thus synthesized was used as a template in a second cycle of first-strand cDNA synthesis using random primers (Invitrogen Life Technologies, Carlsbad, CA) performed in the presence of the T7-oligo(dT) primer. A second round of in vitro transcription was performed similarly to the Standard Protocol. The arrays were hybridized overnight at 45°C and washed using the fluidics station, then stained with streptavidin-phycoerythrin using the standard antibody amplification protocol (GeneChip Expression Analysis Technical Manual, Affymetrix, Santa Clara, CA), and scanned with an Affymetrix GeneArray scanner at 488 nm at 11-micron resolution. Data were acquired using Genechip Operating Software (GCOS, Affymetrix) and uploaded onto the NCI website mAdb (http://nciarray.nci.nih.gov/).
Data were analyzed in our laboratory after setting the signal floor value to 1 and centering to the signal median for each array. The results were expressed as log(2)-transformed values, with the median intensity represented by a log(2) value of 0. An analysis of global gene expression profiling of the two studies, to be reported separately, was conducted using the Statistical Analysis for Microarrays (SAM) feature of the mAdb website, which computes an F-like statistic (20, 21). There were no missing values and ≥1000 permutations were performed using a fixed random seed. SAM analysis identified GALM (Affymetrix feature IDs 234974_at and 235256_s-at) as a significantly regulated gene, and further studies to examine the regulation of GALM expression were then designed using other methods. After these studies were completed, we queried the full database (54,675 gene targets) for both experiments 1 and 2 using the ad hoc query feature in mAdb to identify other genes related to galactose metabolism that showed significant regulation as detected by 1-way ANOVA. Other web resources used included mAdb Feature Reports (available by entering the Feature ID or Gene Bank accession number at http://nciarray.nci.nih.gov/index.shtml), GeneCards (Version 2.36, Wiezmann Institute of Science, URL: www.genecards.org), MATCH (http://www.gene-regulation.com/pubprograms.html#match), and The Meme/Mast System for motif discovery and search (http://meme.nbcr.net/meme/intro.html).
Real-time RT-PCR
GALM mRNA was quantified by real-time PCR. Total cellular RNA was isolated from THP-1, U937, HL-60, and HepG2 cells after treatment using an RNeasy Kit (Qiagen, Valencia, CA). One μg of total RNA was subjected to reverse transcription as previously described (22) and one-twentieth of the diluted reaction mixture was used for real time PCR analysis using Real Time PCR iQ™ SYBR Green Supermix from BioRad (Emory, CA) in a final volume of 25 μl (DNA Engine Opticon 2 System, MJ Research, Inc., Reno, NV). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was employed as the internal control for RNA analysis. A pair of primers spanning two exons was designed from human GALM mRNA (NM_138801) in the GenBank database. The sequences of forward and reverse primers are 5′-GTGACCAGGGCCGTGTTTG-3′ and 5′-TCTGGACTGATGCGCGAGAA-3′, respectively, giving a 395 bp PCR product. The protocol for the RT-PCR was as follows: 95· for 3 min; 40 cycles of 94· for 30 sec, 60· for 30 sec, and 72· for 30 sec. After completion of 40 PCR cycles, the products from representative wells were analyzed for a single band by agarose gel electrophoresis.
GALM protein expression
Western blot analysis was performed using an affinity-purified rabbit polyclonal IgG raised against porcine GALM (NB600-921, Novus Biologicals, Inc. Littleton, CO). Cell lysate samples from THP-1 cells treated with RA and TNFα were prepared for denaturing polyacrylamide gel electrophoresis (10% gel in the presence of 0.1% SDS), then transferred to nitrocellulose prior to blocking and incubation with a 1:2000 dilution of antibody. After washing, the nitrocellulose blot was incubated with ECL reagent (Pierce Chemicals, Rockland, IL) and exposed to X-ray film.
Mutarotase activity assay
Enzyme activity was indirectly determined by using a coupled enzyme assay involving GALM and β-D-glucose dehydrogenase, and a freshly prepared solution of α-D-glucose, as previously described by Beebe and Frey (7). When α-D-glucose is converted to β-D-glucose by GALM in the presence of β-D-glucose dehydrogenase and NAD+, β-D-glucose is oxidized to D-glucono-β-lactone with equimolar reduction of NAD+ to NADH. Therefore, optical density measurement of NADH at 340 nm was used for indirect determination of mutorotase activity. In brief, 50 μl of cell extract (representing 7.0 × 104 THP-1 cells) was added to 940 μl of a solution of 50 mM Tris-HCl (pH 7.2) and 3 mM β-NAD with 10 units of α-D-glucose dehydrogenase. The reaction was initiated by the addition of 10 μl of freshly prepared 50 mM α-D-glucose to the enzyme-cell extract mixture. NADH formation was measured by monitoring 340 nm absorbance at time 0 and after 0.5, 1, 2, and 5 min of incubation. A blank was determined by using the same enzyme mixture described above without cell extract, and subtracted from the samples. For calculating activity, the protein content was determined using a dye-binding assay (Bio-Rad, Hercules, CA) to be 91 μg per 1 × 106 THP-1 cells. Enzyme specific activity was expressed as μmole substrate converted/min/mg protein and converted to relative specific activity compared to control cells.
Statistical analyses
Data are expressed as mean ± SEM. One-way analysis of variance (ANOVA) followed by Fisher’s Protected Least Significant Difference Test to determine P values between groups, and linear regression analysis were used to statistically analyze the data for GALM and individual genes (Minitab Statistical Software, Minitab Inc., State College, PA, and SuperANOVA, Abacus, Berkeley, CA). P values less than 0.05 were considered statistically significant.
Results
GALM gene expression is robustly regulated by RA in THP-1 cells
We first performed two experiments in which THP-1 cells were treated with a physiological concentration of RA (20 nM) and TNFα (5 ng/ml), added alone and together in combination for 40 h (experiment 1), or with only RA (20 nM) in a time-course study from 0 to 40 h (experiment 2). GALM emerged as a significantly regulated gene in both analyses (Table 1). Using SAM, GALM was highly regulated (q.value <0.001 for each of two GALM target sequences). For expt. 1, when the FDR was set to 0.0001 one of two GALM targets was present in the set of 60 significantly regulated genes, out of a total of 54,675 targets present on the array. When the FDR was relaxed to 0.0012, both GALM target sequences were identified in the set of 147 significantly regulated genes (Table 1). In expt 2, the two target sequences for GALM ranked 3rd and 8th, respectively, among the 10 most significantly regulated genes. Thus, by a stringent statistical analysis the probability of these changes in GALM mRNA occurring by chance alone was very low.
Table 1.
Significance analysis of microarray (SAM) identifying GALM gene as regulated by retinoic acid in THP-1 cellsa
| Array experiment numberb | False discovery rate (FDR) | SAM Delta value | # genes identified | Affymetrix Feature ID | d.valuec | q.valuec |
|---|---|---|---|---|---|---|
| Expt. 1 | 0.0001 | 29.7 | 60 | 234974_at | 43.3197 | 0.0000429 |
| Expt. 1 | 0.0012 | 16.7 | 147 | 235256_s_at | 27.0897 | 0.0005073 |
| Expt. 2 | 0.0024 | 18.1 | 10 | 235256_s_at | 44.8792 | 0.0001182 |
| Expt. 2 | 0.0024 | 18.1 | 10 | 234974_at | 32.7839 | 0.0017131 |
GALM, Entrez Gene number 130589.
Data designated Expt 1 included 21 arrays from a 2 × 2 design which included: control (n = 6), RA alone (n = 5), TNFα alone (n = 5) and RA and TNFα combined (n = 5). Data designated Expt 2 included 17 arrays from a time course: t=0 (n = 6), and treatment with 20 nM RA for 2h (n = 2), 6 h (n = 2), 10 h (n = 3), 16 h (n = 2) and 40 h (n = 2) (see Materials and Methods for culture conditions).
For SAM (20) [also see http://www-stat.stanford.edu/~tibs/SAM/index.html], the d value is an F-like statistic that represents the normalized distance between group means; the q value represents the lowest FDR for this gene.
The signal intensity for GALM mRNA evaluated by microarray was greater than the median intensity for all of the genes present (log2 of signal intensity > 0), even for cells that did not receive treatment. In expt. 1, GALM expression was significantly up-regulated by RA (P < 0.0001), but it was not increased by TNFα (Fig. 1A). Therefore, the up-regulation of GALM expression could not be attributed to the induction of differentiation in general, as both of these compounds have been shown previously to increase expression of the cell differentiation marker CD11b within the time frame of the 24–40 h treatment period used in expt. 1 (13). Moreover, expt. 2 showed that GALM mRNA increased rapidly after the addition of RA, rising to approximately 4 times the basal level after just 2 h and reaching a plateau after 6–40 h at a level more than 8 times that in the control cells (Fig. 1B). These changes preceded those reported for markers of cell differentiation (13).
Figure 1.
GALM mRNA expression is increased by RA, but not by TNFα, in THP-1 monocytic cells. A, THP-1 cells were treated with control medium (n=6), retinoic acid (20 nM at time 0 and again after overnight incubation (24 h, n=5), TNFα (5 ng/ml, for 24 h, n=5), or RA overnight, followed by RA and TNFα for the final 24 h (n=6). The level of GALM mRNA was detected by microarray analysis of total RNA prepared from each well of cells. B, Time course of GALM mRNA expression in THP-1 cells. RA (20 nM) was applied at time 0 (n=6), and cells were analyzed after 2 h (n=2), 6 h (n=2), 16 h (n=3); after overnight incubation at 37°C, RA was added again (indicated by black triangles), up to 40 h (n=2). The mean ± SEM of the log(2) values of the array signal intensities are shown. Bars or points that do not share a superscript letter differed significantly (P values as shown).
Other myeloid cells, but not HepG2 cells, respond to RA with increased GALM
Next, we used real-time PCR to quantify GALM mRNA levels in several independent experiments conducted with THP-1 cells and two additional human myeloid cell lines, U937 and HL-60. In THP-1 cells (Figure 2A), all-trans-RA (20 nM) significantly increased GALM mRNA, which was not further increased at a RA concentration of 100 nM. A synthetic analogue of RA, Am580, shown to bind selectively to the nuclear receptor RAR (23) also significantly increased GALM mRNA. GALM mRNA expression was further studied in THP-1 cells treated with RA or other lipophilic substances which were selected because they are either structurally related to RA or have been shown previously to induce THP-1 cell differentiation. While GALM mRNA was again increased by all-trans-RA and Am580, there was no significant effect of 4HPR, a retinoid not known to activate nuclear retinoid receptors (data not shown), but nonetheless capable of inducing cell differentiation or apoptosis (24–27). GALM mRNA also was not induced in THP-1 cells treated with 1,25-dihyroxy-vitamin D, tested as another nuclear receptor ligand with differentiation-promoting properties (12). These results further support that the effect of RA on GALM is not due to a general effect on cell differentiation. The fatty acid, oleic acid, which was tested as a general acidic lipophilic substance of similar molecular weight and physiochemical properties as RA, also did not alter GALM mRNA in THP-1 cells (data not shown).
Figure 2.

Retinoic acid and Am580 increase GALM mRNA in myeloid cell lines, but not in human hepatoma HepG2 cells. A, GALM mRNA expression (relative to GAPDH) determined by real-time PCR in THP1 cells that were treated either with <0.01% ethanol (control), RA, or Am580 (20 or 100 nM as shown) for 6 h. HepG2 cells (not shown) that were treated similarly did not show significant regulation. Inset shows agarose gel electrophoresis of PCR products after the end of 40 cycles, indicating the appropriate 395 bp size of the amplified product. B, GALM mRNA relative to GAPDH (left axis) and relative to control cells (right axis) in U937 cells and, C, HL-60 cells that were treated RA (20 nM) for 24 h. Data are means ± SEM, n≥3/treatment; * are significantly different from control within each panel, P<0.05.
GALM mRNA was also increased by RA in two other human myeloid cells, but not in a hepatocellular carcinoma cell line, HepG2. In U937 cells (Fig. 2B), GALM expression was increased nearly 20-fold by RA. This large relative change was due in part to a very low level of basal expression of GALM in untreated cells (Fig. 2A). In HL-60 cells (Fig. 2C), GALM expression was higher in the absence of RA, as compared to that in U937 cells, but RA still significantly increased GALM mRNA. In contrast, while GALM expression was readily detectable in HepG2 cells without the addition of RA, it was increased by RA only about 50% over the control level, nor was it increased by Am580 (data not shown), even though RARα is the most abundant form of RAR in the liver (28, 29).
Because the increase in GALM expression in THP-1 cells occurs rapidly, we next tested the ability of actinomycin D, a general inhibitor of new RNA synthesis, or cycloheximide, an inhibitor of protein synthesis, to inhibit the induction of GALM by RA in THP-1 cells. Actinomycin D completely abrogated the induction of GALM expression by RA (Fig. 3A). Cycloheximide had no effect alone and it did not prevent an increase in GALM mRNA by RA (Fig. 3B), although the increase was somewhat less than with RA alone, possibly due to general changes in cellular proteins in the 24-h study.
Figure 3.
GALM mRNA is induced by RA, and completely inhibited in cells pretreated with actinomycin D, but not with cycloheximide. A, THP-1 cells were treated with either RA (20 nM), actinomycin D (20 nM), RA + actinomycin D, or vehicle (control) for 24 h. B. THP-1 cells were treated with either RA (20 nM), cycloheximide (20 nM), RA+ cycloheximide, or vehicle (control) for 24 h. After incubation, cells were harvested and GALM and GAPDH mRNAwas measured by real-time RT-PCR as described in Materials and Methods. Data are means (with control set to 1) ± SEM, n=3/treatment; * denotes different from control, P<0.05.
GALM protein expression is increased in RA-treated THP-1 cells
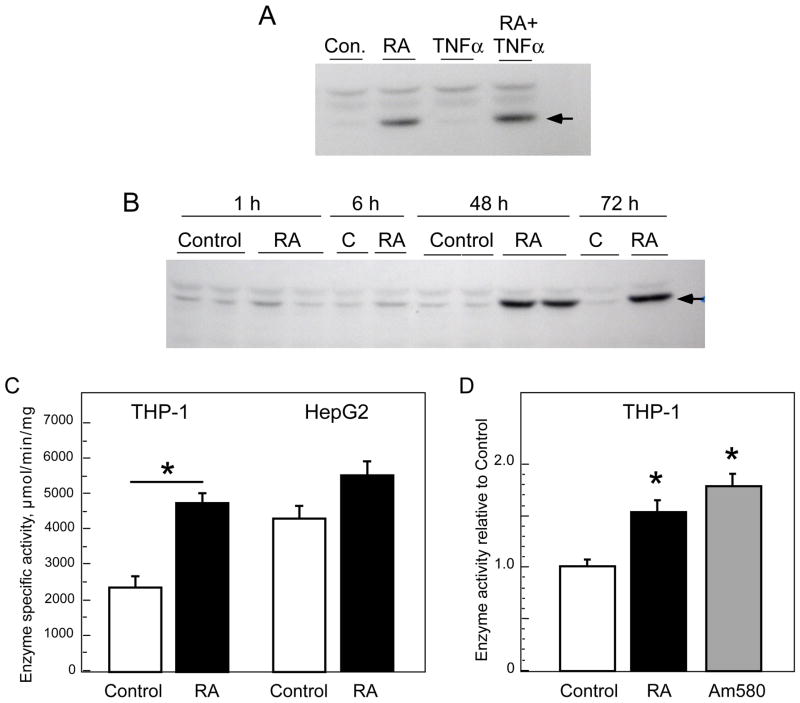
To determine if GALM protein is also increased by RA, THP-1 cell extracts from RA and TNFα-treated THP-1 cells were subjected to immunoblot analysis using an antibody raised against GALM protein purified from porcine kidney. Based on our comparison of the predicted protein sequences for porcine GALM (NP 999571) and human GALM (Q96C23), the two proteins have 89% amino acid identity and both have a predicted size of 37 kDa (342 amino acids). Immunoblot analysis of THP-1 cell lysates showed a band slightly above the 35 kDa molecular weight marker, consistent with the expected size of GALM protein. In each of two separate experiments, this band was significantly more intense in RA-treated THP-1 cells. THP-1 cells that had been treated with TNFα did not differ from the control cells (Fig. 4A). The increase in GALM protein in RA-treated cells was stable up to 48 h (Fig. 4B). Thus, GALM mRNA and protein were both increased by RA in THP-1 cells, while neither GALM mRNA nor protein was altered by TNFα.
Figure 4.
Regulation of GALM protein and mutorotase enzyme activity by RA. A, GALM protein levels were detected by immunoblot analysis in lysates of THP-1 cells that had been treated for 24 h with vehicle (control, C), RA (20 nM), TNFα (5ng/ml), or both RA and TNFα. B. GALM protein in THP-1 cells treated with vehicle or RA (20 nM) for 24 and 48 h, showing persistent increase in RA-treated cells. The uniform non-specific band above GALM (38 kDa) demonstrates equal protein loading in all lanes. C. Mutorotase assay in THP-1 cells and HepG2 cells (1.4 × 106 cells/well) treated without (control, C) or with RA (100 nM) for 6 h, after which cells were harvested and homogenized. Bars show mean ± SEM, n=4/condition. The * denotes a significant difference between control and RA-treated cells (P < 0.002). D. THP-1 cells were treated with either RA or Am580 (20 nM) and incubated for 6 h. Results for each of n=3 independent experiments totaling n=12 for control cells, n=12 for RA-treated cells, and n=8 for Am580-treated cells were pooled and compared by setting the mean value for the control group to 1.0. Values shown are the mean ± SEM; * denotes differences from control, P < 0.001.
Mutarotase enzymatic activity is increased by RA and Am580 in THP-1 cells
Mutarotase activity was assayed using a coupled enzymatic assay in which the β-D-glucose formed by epimerization of β-D-glucose, added as the primary substrate, is rapidly and stoichiometrically used by an NAD-dependent, β-D-hexose-specific glucose dehydrogenase. Both THP-1 cells and HepG2 cells (1.4 × 106 cells/well) were treated with RA (100 nM) or vehicle only for 6 h, after which cell homogenates were prepared and used for the in vitro assay. Retinoic acid significantly increased mutarotase activity in THP-1 cells (Fig. 4C). Basal GALM activity was higher in HepG2 cells than in THP-1 cells, and activity was increased slightly by RA in HepG2 cells. Mutorotase activity was assayed in 3 independent experiments in THP-1 cells treated with RA or Am580 (Fig. 4D). Overall, both retinoids increased mutarotase activity significantly in THP-1 but not HepG2 cells.
Several other galactose-related genes in THP-1 cells exhibit regulation by RA
To date, no chemicals other than galactose have been reported as regulators of GALM, and RA is not especially known as a regulator of galactose-related metabolism or gene expression. To investigate if other genes related to GALM activity may also be regulated by RA, we used the ad hoc query/filtering options to query the mAdb database from the microarray time course (expt. 2) for other genes of galactose metabolism. Four other genes showed a significant time-dependent increase in RA-treated cells (Fig. 5): B4GALT5 (UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, Fig. 5A); ST3GAL3 (ST3 beta-galactoside alpha-2,3-sialyltransferase 3, Fig. 5B); ST6GALNAC5 [ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1, 3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5, Fig. 5C); and GALNAC4S-6ST [B cell RAG associated protein (GALNAC4S-6ST, Fig. 5D). B4GALT5 and ST3GAL3 increased significantly by 2 h, similar to GALM, and the levels of GALM and B4GALT5 mRNA were highly correlated (R2 = 0.86; P < 0.0001). ST6GALNAC5 and GALNAC4S-6ST differed significantly from control values after 6 h of cell exposure to RA, and then continued to increase over the 40-h time course to approximately >8-fold above the control level. To determine if these 5 genes may contain binding sites for common transcription factors, we conducted a database search using MATCH software of the 2000 nucleotides upstream of the transcription start site for each of these 5 genes [GALM: NM_138801; ST3GAL3: NM_174963; B4GALT5: NM_004776; ST6GALNAC5: NM_030965; GALNAC4S-6ST: NM_015892]. This analysis revealed the presence of the eight base-pair nucleotide target of Oct-1, a member of the POU-domain family of conformationally flexible transcription factors, and variants of this octamer site (30) as the only factor common to all 5 of these genes.
Figure 5.
Time dependent up-regulation of galactose-related genes in THP-1 cells treated with RA. THP-1 cells were treated with RA (20 nM) at t=0 and again after overnight incubation (shown by black triangles); microarray analysis was conducted (as described in Fig. 1B and Materials and Methods). Median signal intensity for the arrays is 0 on a log(2) scale and the Y-axis displays the mean ± SEM of the log(2) values of the array signal intensities at each time. Bars or points that do not share a superscript letter differed significantly (P values as shown). Genes illustrated are A, B4GALT5 (Entrez Gene ID 9334; Feature IDs 221484_at, and 221485_at); B, ST3GAL3 (Entrez Gene ID 6487; Feature ID 1555702_a_at); C, ST6GALNAC5 (Entrez Gene ID 81849; Feature ID 220979_s_at); and D, GALNAC4S-6ST (Entrez Gene ID 51363; Feature ID 203066_at).
Discussion
GALM protein has been studied in molecular detail, including by in vitro expression of bacterial and human GALM protein (5), by mechanistic studies of the catalytic activity of GALM from L. lactis (4), and by crystallographic structural studies of GALM protein from both L. lactis and human sources (3, 6). In the present study, we first observed in a microarray study, from which data were subjected to a stringent statistical analysis, that GALM is highly regulated by RA in the human monocytic cell line THP-1. A search of mAdb and GeneCards databases (as referenced in Methods and Materials) revealed no previously reported relationships of GALM with chemicals or compounds other than hexoses. Thus, RA may be the first identified non-sugar molecular regulator that is capable of modulating GALM gene and protein expression.
RA is well known as a pleiotropic hormone that plays a regulatory role in many biological processes (14–16). The concept that vitamin A and RA may alter cell surface or secreted glycoproteins was hypothesized several decades ago, based on observations in vitamin A-deficient animals (31, 32) and retinoid actions on normal cells (33) and tumor cells (34, 35). A nutritional deficiency of vitamin A has long been associated with abnormalities of cellular differentiation in many types of cells, including epithelial cells of the respiratory tract (36) and mucous-secreting goblet cells of the cornea and gastrointestinal tract (37–39). More recently, retinoid insufficiency in mouse embryos resulted in alterations in several genes associated with protein galactosylation (40). RA has been shown to modify the synthesis and distribution of glycolipids in Xenopus embryos (41), and to alter the incorporation of labeled galactose, and other sugars, into glycoconjugates in carcinoma (42) and melanoma cell lines (43). The genes reported in these studies, when genes were identified, were not those we have identified in the present study, but they could be part of larger galactose-related pathways that affect protein or lipid modification, which possibly may be coordinately regulated during RA-induced cell differentiation (44). Recently, clinical data have suggested a link between vitamin A’s transport protein, glucose homeostasis and type 2 diabetes, suggesting another potential link between retinoids and monosaccharides [reviewed in (45)]. The results of our present study in the THP-1 myeloid cell model suggest that RA, at physiological concentrations, can increase the cell’s ability to interconvert hexose anomers through the up-regulation of GALM expression and mutarotase activity. These results suggest the idea that retinoids that are used therapeutically, such as all-trans-RA and potentially Am580, may alter monocytic cell differentiation, migration, and transformation through galactose-related protein and/or lipid glycosylation. The regulation of GALM and/or additional enzymes required for the biosynthesis of complex glycosylated proteins and lipids by RA could be part of this process.
The regulation of GALM expression by RA may be cell-type or cell lineage specific. We observed strong regulation by RA in three human myeloid cell lines, THP-1, U937 and HL-60 cells. GALM expression was higher in untreated HepG2 cells than in the myeloid cells, but HepG2 cells did not respond to RA. Maintaining a high basal level of GALM activity in liver may be important for the metabolism of hexoses absorbed from the diet. The rapid up-regulation of GALM by RA in myeloid cells suggests that RA could have a special role in the glycobiology of these cells, possibly related to their differentiation. In addition to the present results, we recently reported that CD1d, an MHC-class I-like antigen receptor that binds α-galactosylceramide (46), is also rapidly and highly up-regulated by RA (18). Thus, several genes related to galactose metabolism and/or the functions of galactose-containing compounds may be significantly regulated by RA in myeloid lineage cells.
In the present studies, evidence of a specific effect of RA was obtained in studies comparing all-trans-RA, Am580, and 4-HPR, all of which are classified as retinoids but each of which possesses different structural features that affects its functionality, as well as activated vitamin D and a non-specific fatty acid, oleic acid. Of these, only RA and Am580 regulated GALM mRNA and protein levels. Although further data are required to demonstrate transcription regulation, the rapid and high level of induction by all-trans-RA is typical of other genes, such as RAR-β, that have been shown to respond directly through the binding of RA to a RAR-RXR heterodimer associated with a direct repeat (DR)-5 or DR-2 element (16). Additionally, the increase in GALM mRNA by RA was completely blocked by actinomycin D, but not by cycloheximide. The increase in GALM mRNA by Am580 suggests the possible involvement of RARα. Am580 is a retinobenzoic acid analog of RA with high affinity for RARα, and much weaker, if any, affinity for RARβ or RARγ (47). At the low concentrations of Am 580 we used with THP-1 cells, 20 and 100 nM, Am580 would be expected to be selective for RARα. The ability of Am580 to mimic all-trans-RA suggests that RARα, which THP-1 cells express along with RARβ, RARγ, and RXRs (QC, unpublished data), could be involved in GALM gene regulation. The lack of an effect of 4HPR is not surprising given that this retinoid is unlikely to act through binding to nuclear retinoid receptors, but it is still an interesting finding because 4HPR has been shown to induce cell differentiation in a number of cell types and to be anticarcinogenic in vivo [reviewed in (48, 49)]. While our data are still indirect, they provide evidence suggesting that increases in GALM mRNA in RA-treated cells could be due to increased RARα-RXR-mediated gene transcription. Nevertheless, many genes that are physiologically regulated by RA have not been demonstrated to be transcriptionally regulated by an RAR-RXR dependent mechanism (15).
Because mutarotase activity does not modify the structure of the substrate, except geometrically, the mutarotase reaction cannot be detected by sensitive assay techniques, such as by isotope incorporation or chromatographic separation of substrate and product. The mutorotation of sugars in solution can be detected by optical rotation, but this method is not sensitive enough to be suitable for use with the small amounts of cellular extracts material that were available as enzyme source from cultured cells. For our studies, we used an indirect coupled mutarotase assay (7), with α-D-glucose as substrate for GALM, coupled to the oxidation of β-D-glucose by β-D-glucose dehydrogenase, which converts NAD to NADH. This pyridine nucleotide-coupled assay revealed significant regulation of mutarotase activity by RA and Am580 in THP-1 cells. Nevertheless, it is possible that mutarotase activity/ies besides GALM are present in the cells, and may have contributed to the activity we determined, both in the untreated control cells and RA-treated cells. Therefore, we prefer to call this activity mutarotase activity. However, we also tested the level of GALM protein expression by a specific immunoassay and detected a band of expected molecular mass, ~37 kDa, that exhibited very similar quantitative changes in response to RA, but not TNFα, as was observed for GALM mRNA. Overall, these data support a model in which the increase in GALM gene expression by RA is by translated into GALM protein, with a concomitant increase in functional mutorotase activity, which is likely to be due at least in part to GALM.
Finally, we explored our microarray data for other galactose-related genes that might also be regulated by RA. We identified four genes besides GALM that were also increased in RA-treated THP-1 cells, each of which has some role in the formation of complex oligosaccharides and their transfer to proteins or lipids (Fig. 5). B4GALT5 is a member of the beta-4 galactosyl transferase gene family of Golgi-associated proteins (50). B4GALT5 is involved in the pathways of glycosphingolipid and glycan biosynthesis; however, no drugs or compounds are currently listed as being related to B4GALT5. ST3GAL3 mRNA was increased transiently after exposure of THP-1 cells to RA (Fig. 5). This enzyme transfers acetylneuraminate from CMP to galactosylated oligosaccharides in the biosynthesis of glycoproteins. ST6GALNAC5 (alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5, alias SIAT7e) is one of several Golgi-associated sialic acid transferases that function downstream of galactose metabolism and maps onto the pathways of ganglioside and glycan biosynthesis. ST6GALNAC5 exhibits higher activity with glycolipids than with glycoproteins. GALNAC4S-6ST (alias BRAG, B-cell RAG associated protein) is a sulfotransferase that modifies galactosyl-containing peptides to form chondroitin sulfates (KEGG). This enzyme is also principally Golgi associated, although a small portion may be present on the cell surface where, in B cells, it acts as a B-cell receptor or co-receptor. The expression of GALNAC4S-6ST in myeloid cells, which lack a B-cell receptor, has not previously been reported, nor has its regulation by RA. If GALNAC4S-6ST/BRAG has a similar function in myeloid cells, which do not undergo Ig gene rearrangement in response to antigen as do B cells, then it could possibly be involved in other gene rearrangements/transpositions, or have other unanticipated functions. The up-regulation of GALNAC4S-6ST by RA was less dramatic than that of GALM, but expression was still increased 8-fold over the 40-h time course. Previous studies have shown that RA is a potent regulator of B-cell receptor function and B-cell activation (51–53). Oct-1, the only transcription factor we could identify by binding site analysis in the 2000 nucleotides upstream of the transcription start site for all 5 of these genes, is a relatively common transcription factor involved in many biological processes, including the development of specialized tissues, as well as in more general cellular housekeeping functions (54). It does not appear to have a specific relationship to the functions of retinoids or nuclear retinoid receptors. Thus, at present, we cannot speculate as to how these genes are regulated by RA, but it is worth noting that only a small minority of genes that have been shown to respond physiologically to RA have also been shown conclusively to be regulated by a direct RA-activated nuclear retinoid hormone receptor-mediated transcriptional mechanism (15).
In summary, the results of this study in myeloid cells provide new insight into the regulation of GALM and the possible mechanisms by which retinoids can affect glycoprotein and glycolipid synthesis. It is interesting and plausible that the regulation of GALM gene expression by RA, and the regulation of several other genes that function downstream of GALM, could have an important regulatory role in the formation of complex glycoproteins or glycolipids that, in turn, have been shown to function in several biological processes, including cell adhesion, migration, and antigen presentation.
Figure 6.
Acknowledgments
We are grateful to Dr. Chang-Hee Kim, NCI Microarray Facility, Frederick, MD, for microarray analysis as part of the Bioactive Nutrients Gene Omnibus (BANGEO) project.
Abbreviations
- GALM
galactomutarotase
- RA
all-trans-retinoic acid
- TNF
tumor necrosis factor
Footnotes
Funding: Supported by National Cancer Institute grant CA-90214 and BANGEO supplement provided as CA-90214-S. TP was supported in part by the Sabbatical Leave Program of Anyang University and YZ was supported in part by the Department of Nutritional Sciences.
References
- 1.Holden HM, Rayment I, Thoden JB. Structure and function of enzymes of the Leloir pathway for galactose metabolism. J Biol Chem. 2003;278:43885–43888. doi: 10.1074/jbc.R300025200. [DOI] [PubMed] [Google Scholar]
- 2.Frey PA. The Leloir pathway: a mechanistic imperative for three enzymes to change the stereochemical configuration of a single carbon in galactose. FASEB J. 1996;10:461–470. [PubMed] [Google Scholar]
- 3.Thoden JB, Holden HM. High resolution X-ray structure of galactose mutarotase from Lactococcus lactis. J Biol Chem. 2002;277:20854–20861. doi: 10.1074/jbc.M201415200. [DOI] [PubMed] [Google Scholar]
- 4.Thoden JB, Kim J, Raushel FM, Holden HM. Structural and kinetic studies of sugar binding to galactose mutarotase from Lactococcus lactis. J Biol Chem. 2002;277:45458–45465. doi: 10.1074/jbc.M208395200. [DOI] [PubMed] [Google Scholar]
- 5.Timson DJ, Reece RJ. Identification and characterisation of human aldose 1-epimerase. FEBS Lett. 2003;543:21–24. doi: 10.1016/s0014-5793(03)00364-8. [DOI] [PubMed] [Google Scholar]
- 6.Thoden JB, Timson DJ, Reece RJ, Holden HM. Molecular structure of human galactose mutarotase. J Biol Chem. 2004;279:23431–23437. doi: 10.1074/jbc.M402347200. [DOI] [PubMed] [Google Scholar]
- 7.Beebe JA, Frey PA. Galactose mutarotase: purification, characterization, and investigations of two important histidine residues. Biochemistry. 1998;37:14989–14997. doi: 10.1021/bi9816047. [DOI] [PubMed] [Google Scholar]
- 8.Tsuchiya S, Yamabe M, Yamaguchi Y, Kobayashi Y, Konno T, Tada K. Establishment and characterization of a human acute monocytic leukemia cell line (THP-1) Int J Cancer. 1980;26:171–176. doi: 10.1002/ijc.2910260208. [DOI] [PubMed] [Google Scholar]
- 9.Tsuchiya S, Kobayashi Y, Goto Y, Okumura H, Nakae S, Konno T, Tada K. Induction of maturation in cultured human monocytic leukemia cells by a phorbol diester. Cancer Res. 1982;42:1530–1536. [PubMed] [Google Scholar]
- 10.Auwerx J. The human leukemia cell line, THP-1: a multifacetted model for the study of monocyte-macrophage differentiation. Experientia. 1991;47:22–31. doi: 10.1007/BF02041244. [DOI] [PubMed] [Google Scholar]
- 11.Ferret PJ, Soum E, Negre O, Wollman EE, Fradelizi D. Protective effect of thioredoxin upon NO-mediated cell injury in THP1 monocytic human cells. Biochem J 346 Pt. 2000;3:759–765. [PMC free article] [PubMed] [Google Scholar]
- 12.Defacque H, Dornand J, Commes T, Cabane S, Sevilla C, Marti J. Different combinations of retinoids and vitamin D3 analogs efficiently promote growth inhibition and differentiation of myelomonocytic leukemia cell lines. J Pharmacol Exp Ther. 1994;271:193–199. [PubMed] [Google Scholar]
- 13.Chen QY, Ross AC. Retinoic acid regulates cell cycle progression and cell differentiation in human monocytic THP-1 cells. Exp Cell Res. 2004;297:68–81. doi: 10.1016/j.yexcr.2004.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Altucci L, Gronemeyer H. Nuclear receptors in cell life and death. Trends Endocrinol Metab. 2001;12:460–468. doi: 10.1016/s1043-2760(01)00502-1. [DOI] [PubMed] [Google Scholar]
- 15.Balmer JE, Blomhoff R. Gene expression regulation by retinoic acid. J Lipid Res. 2002;43:1773–1808. doi: 10.1194/jlr.r100015-jlr200. [DOI] [PubMed] [Google Scholar]
- 16.Wei LN. Retinoid receptors and their coregulators. Annual Review of Pharmacol Toxicol. 2003;43:47–72. doi: 10.1146/annurev.pharmtox.43.100901.140301. [DOI] [PubMed] [Google Scholar]
- 17.Kagechika H, Kawachi E, Fukasawa H, Saito G, Iwanami N, Umemiya H, Hashimoto Y, Shudo K. Inhibition of IL-1-induced IL-6 production by synthetic retinoids. Biochem Biophys Res Commun. 1997;231:243–248. doi: 10.1006/bbrc.1997.6087. [DOI] [PubMed] [Google Scholar]
- 18.Chen Q, Ross AC. Retinoic acid regulates CD1d gene expression at the transcriptional level in human and rodent monocytic cells. Exp Biol Med (Maywood) 2007;232:488–494. [PMC free article] [PubMed] [Google Scholar]
- 19.Dumur CI, Garrett CT, Archer KJ, Nasim S, Wilkinson DS, Ferreira-Gonzalez A. Evaluation of a linear amplification method for small samples used on high-density oligonucleotide microarray analysis. Anal Biochem. 2004;331:314–321. doi: 10.1016/j.ab.2004.03.040. [DOI] [PubMed] [Google Scholar]
- 20.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Efron B, Tibshirani R. On testing the significance of sets of genes. 2007 http://www-stat.stanford.edu/~tibs/SAM/index.html (to appear in Ann. Appl. Stat. vol. 1)
- 22.Zolfaghari R, Cifelli CJ, Lieu SO, Chen Q, Li NQ, Ross AC. Lipopolysaccharide opposes the induction of CYP26A1 and CYP26B1 gene expression by retinoic acid in the rat liver in vivo. Am J Physiol Gastrointest Liver Physiol. 2007;292:G1029–G1036. doi: 10.1152/ajpgi.00494.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tamura K, Kagechika H, Hashimoto Y, Shudo K, Ohsugi K, Ide H. Synthetic retinoids, retinobenzoic acids, Am80, Am580, and Ch55 regulate morphogenesis in chick limb bud. Cell Diff Devel. 1990;32:17–26. doi: 10.1016/0922-3371(90)90095-e. [DOI] [PubMed] [Google Scholar]
- 24.Sheikh MS, Shao ZM, Li XS, Ordonez JV, Conley BA, Wu SL, Dawson MI, Han QX, Chao WR, Quick T, Niles RM, Fontana JA. N-(4-hydroxyphenyl)retinamide (4-HPR)-mediated biological actions involve retinoid receptor-independent pathways in human breast carcinoma. Carcinogenesis. 1995;16:2477–2486. doi: 10.1093/carcin/16.10.2477. [DOI] [PubMed] [Google Scholar]
- 25.Hsieh TC, Ng CY, Wu JM. The synthetic retinoid N-(4-hydroxyphenyl) retinamide (4-HPR) exerts antiproliferative and apoptosis-inducing effects in the androgen-independent human prostatic JCA-1 cells. Biochem Mol Biol Int. 1995;37:499–506. [PubMed] [Google Scholar]
- 26.Holmes WF, Soprano DR, Soprano KJ. Comparison of the mechanism of induction of apoptosis in ovarian carcinoma cells by the conformationally restricted synthetic retinoids CD437 and 4-HPR. J Cell Biochem. 2003;89:262–278. doi: 10.1002/jcb.10505. [DOI] [PubMed] [Google Scholar]
- 27.Golubkov V, Garcia A, Markland FS. Action of fenretinide (4-HPR) on ovarian cancer and endothelial cells. Anticancer Res. 2005;25:249–253. [PubMed] [Google Scholar]
- 28.Kato S, Mano H, Kumazawa T, Yoshizawa Y, Kojima R, Masushige S. Effect of retinoid status on α, β and g retinoic acid receptor mRNA levels in various rat tissues. Biochem J. 1992;286:755–760. doi: 10.1042/bj2860755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zolfaghari R, Ross AC. Chronic vitamin A intake affects the expression of mRNA for apolipoprotein A-I, but not for nuclear retinoid receptors, in liver of young and aging Lewis rats. Arch Biochem Biophys. 1995;323:258–264. doi: 10.1006/abbi.1995.9966. [DOI] [PubMed] [Google Scholar]
- 30.Remenyi A, Tomilin A, Pohl E, Lins K, Philippsen A, Reinbold R, Scholer HR, Wilmanns M. Differential dimer activities of the transcription factor Oct-1 by DNA-induced interface swapping. Mol Cell. 2001;8:569–580. doi: 10.1016/s1097-2765(01)00336-7. [DOI] [PubMed] [Google Scholar]
- 31.Kirven MJ, Wolf G. Synthesis and glycosylation of fibronectin in hepatocytes from vitamin A-deficient rats. Mol Cell Biochem. 1991;101:101–114. doi: 10.1007/BF00229528. [DOI] [PubMed] [Google Scholar]
- 32.Shankar S, Creek KE, De Luca LM. The effect of the progression of vitamin A deficiency on glucose, galactose and mannose incorporation into sugar phosphates and sugar nucleotides in hamster liver. J Nutr. 1990;120:361–374. doi: 10.1093/jn/120.4.361. [DOI] [PubMed] [Google Scholar]
- 33.King IA, Pope FM. Retinoids increase the incorporation of D-[3H]galactose into epidermal glycoproteins. Biochem Biophys Res Commun. 1984;121:364–371. doi: 10.1016/0006-291x(84)90731-9. [DOI] [PubMed] [Google Scholar]
- 34.Levin LV, Clark JN, Quill HR, Newberne PM, Wolf G. Effect of retinoic acid on the synthesis of glycoproteins of mouse skin tumors during progression from promoted skin through papillomas to carcinomas. Cancer Res. 1983;43:1724–1732. [PubMed] [Google Scholar]
- 35.Lotan R, Lotan D, Meromsky L. Correlation of retinoic acid-enhanced sialyltransferase activity and glycosylation of specific cell surface sialoglycoproteins with growth inhibition in a murine melanoma cell system. Cancer Res. 1984;44:5805–5812. [PubMed] [Google Scholar]
- 36.Gray TE, Guzman K, Davis CW, Abdullah LH, Nettesheim P. Mucociliary differentiation of serially passaged normal human tracheobronchial epithelial cells. Am J Respir Cell Mol Biol. 1996;14:104–112. doi: 10.1165/ajrcmb.14.1.8534481. [DOI] [PubMed] [Google Scholar]
- 37.DeLuca L, Schumacher M, Wolf G. Biosynthesis of a fucose-containing glycopeptide from rat small intestine in normal and vitamin A-deficient conditions. J Biol Chem. 1970;245:4551–4558. [PubMed] [Google Scholar]
- 38.Sullivan WR, McCulley JP, Dohlman CH. Return of goblet cells after vitamin A therapy in xerosis of the conjunctiva. Am J Ophthalmol. 1973;75:720–725. doi: 10.1016/0002-9394(73)90828-3. [DOI] [PubMed] [Google Scholar]
- 39.Rojanapo W, Lamb AJ, Olson JA. The prevalence, metabolism and migration of goblet cells in rat intestine following the induction of rapid, synchronous vitamin A deficiency. J Nutr. 1980;110:178–188. doi: 10.1093/jn/110.1.178. [DOI] [PubMed] [Google Scholar]
- 40.Flentke GR, Baker MW, Docterman KE, Power S, Lough J, Smith SM. Microarray analysis of retinoid-dependent gene activity during rat embryogenesis: increased collagen fibril production in a model of retinoid insufficiency. Dev Dyn. 2004;229:886–898. doi: 10.1002/dvdy.10489. [DOI] [PubMed] [Google Scholar]
- 41.Rossi F, Gornati R, Rizzo AM, Venturini L, Bernardini G, Berra B. Glycolipid glycosyltransferase activities during early development of Xenopus: effect of retinoic acid. Cell Biol Int. 1999;23:91–95. doi: 10.1006/cbir.1999.0351. [DOI] [PubMed] [Google Scholar]
- 42.Sacks PG, Amos B, Lotan R. Enhancement of glycosylation of cellular glycoconjugates in the squamous carcinoma cell line MDA886Ln by beta-all-trans retinoic acid. Glycoconj J. 1996;13:791–796. doi: 10.1007/BF00702343. [DOI] [PubMed] [Google Scholar]
- 43.Amos B, Deutsch V, Lotan R. Modulation by all-trans retinoic acid of glycoprotein glycosylation in murine melanoma cells: enhancement of fucosyl- and galactosyltransferase activities. Cancer Biochem Biophys. 1990;11:31–43. [PubMed] [Google Scholar]
- 44.Chen C, Fenderson BA, Andrews PW, Hakomori S. Glycolipid glycosyltransferases in human embryonal carcinoma cells during retinoic acid induced differentiation. Biochemistry. 1989;28:2229–2238. doi: 10.1021/bi00431a039. [DOI] [PubMed] [Google Scholar]
- 45.Wolf G. Serum retinol-binding protein: a link between obesity, insulin resistance, and type 2 diabetes. Nutr Rev. 2007 May 2007;:251–256. doi: 10.1111/j.1753-4887.2007.tb00302.x. [DOI] [PubMed] [Google Scholar]
- 46.Dougan SK, Kaser A, Blumberg RS. CD1 expression on antigen-presenting cells. Curr Top Microbiol Immunol. 2007;314:113–141. doi: 10.1007/978-3-540-69511-0_5. [DOI] [PubMed] [Google Scholar]
- 47.Hashimoto Y, Kagechika H, Shudo K. Expression of retinoic acid receptor genes and the ligand-binding selectivity of retinoic acid receptors (RAR’S) Biochem Biophys Res Commun. 1990;166:1300–1307. doi: 10.1016/0006-291x(90)91007-f. [DOI] [PubMed] [Google Scholar]
- 48.Holmes WF, Soprano DR, Soprano KJ. Synthetic retinoids as inducers of apoptosis in ovarian carcinoma cell lines. J Cell Physiol. 2004;199:317–329. doi: 10.1002/jcp.10338. [DOI] [PubMed] [Google Scholar]
- 49.Hail N, Jr, Kim HJ, Lotan R. Mechanisms of fenretinide-induced apoptosis. Apoptosis. 2006;11:1677–1694. doi: 10.1007/s10495-006-9289-3. [DOI] [PubMed] [Google Scholar]
- 50.Lo NW, Shaper JH, Pevsner J, Shaper NL. The expanding beta 4-galactosyltransferase gene family: messages from the databanks. Glycobiology. 1998;8:517–526. doi: 10.1093/glycob/8.5.517. [DOI] [PubMed] [Google Scholar]
- 51.Chen Q, Ross AC. Vitamin A and immune function: Retinoic acid modulates population dynamics in antigen receptor and CD38-stimulated splenic B cells. Proc Natl Acad Sci USA. 2005;102:14142–14149. doi: 10.1073/pnas.0505018102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Blomhoff HK, Smeland EB, Erikstein B, Rasmussen AM, Skrede B, Skjonsberg C, Blomhoff R. Vitamin A is a key regulator for cell growth, cytokine production, and differentiation in normal B cells. J Biol Chem. 1992;267:23988–23992. [PubMed] [Google Scholar]
- 53.Blomhoff HK. Vitamin A regulates proliferation and apoptosis of human T- and B-cells. Biochem Soc Trans. 2004;32:982–984. doi: 10.1042/BST0320982. [DOI] [PubMed] [Google Scholar]
- 54.Phillips K, Luisi B. The virtuoso of versatility: POU proteins that flex to fit. J Mol Biol. 2000;302:1023–1039. doi: 10.1006/jmbi.2000.4107. [DOI] [PubMed] [Google Scholar]