Abstract
Studies of the biochemistry of Listeria monocytogenes virulence protein ActA have typically focused on the behavior of bacteria in complex systems or on the characterization of the protein after expression and purification. Although prior in vivo work has proposed that ActA forms dimers on the surface of L. monocytogenes, dimerization has not been demonstrated in vitro, and little consideration has been given to the surface environment where ActA performs its pivotal role in bacterial actin-based motility. We have synthesized and characterized an ActA dimer and provide evidence that the two ActA molecules do not interact with each other even when tethered together. However, we also demonstrate that artificial dimers provide superior activation of actin nucleation by the Arp2/3 complex compared with monomers and that increased activation of the Arp2/3 complex by dimers may be a general property of Arp2/3 activators. It appears that the close packing (∼19 nm) of ActA molecules on the surface of L. monocytogenes is so dense that the kinetics of actin nucleation mimic that of synthetic ActA dimers. We also present observations indicating that ActA is a natively unfolded protein, largely random coil that is responsible for many of the unique physical properties of ActA including its extended structure, aberrant mobility during SDS-PAGE, and ability to resist irreversible denaturation upon heating.
Listeria monocytogenes is a Gram-positive intracellular bacterial pathogen that is the causative agent of listeriosis, a potentially fatal food-borne infection that can result in meningitis or septicemia or late-term spontaneous abortion in pregnant women. Listeriosis primarily affects the elderly, the very young, and people who are immunocompromised, whereas healthy individuals can tolerate food with relatively high amounts of L. monocytogenes, making outbreaks involving this bacterium infrequent. L. monocytogenes is unusual in that it resists heat better than many food-borne bacteria and continues to multiply at ActAs, so even cooked and refrigerated foods pose an infection risk (1).
After ingestion, L. monocytogenes is taken up by the enterocytic epithelial cells lining the intestine (2, 3) where it escapes from the phagocytic vacuole and undergoes several rounds of cell division within the host cell cytoplasm. Once in the cytoplasm, L. monocytogenes begins expression of a virulence factor, the surface-associated protein ActA (4). The ActA protein enables the bacteria to harness the actin-based machinery of the host cell and generate an actin-rich “comet tail,” allowing the bacteria to become motile and spread directly from cell to cell (5), thereby avoiding the host humoral immune system (6). The most serious sequelae of infection including meningitis and abortion depend on the ability of the bacterium to use actin-based motility and direct cell-to-cell spread to cross intact epithelial barriers (the blood-brain barrier and the placenta, respectively).
ActA was first identified as a gene product necessary for actin-based motility by mapping of a transposon-induced mutation that resulted in a mutant strain incapable of direct cell-to-cell spread (7). Subsequent work demonstrated that ActA is asymmetrically distributed on the surface of L. monocytogenes (8) and that ActA is sufficient to impart actin-based motility to nonpathogenic Listeria innocua (9), to Streptococcus pneumoniae (10), and to latex microspheres when nonspecifically absorbed to the surface (11). Although other factors are required for a successful and sustained infection, ActA is the only bacterial component necessary for actin-based motility.
The biochemical functions of ActA have been systematically dissected using three kinds of assays; 1) site-directed mutagenesis followed by expression of altered proteins in L. monocytogenes (12–15) or in eukaryotic cells (16, 17), 2) purification of a soluble truncated form of ActA followed by in vitro characterization of its biochemical properties and ability to interact with other proteins (18) (19–21), and 3) immobilization of purified ActA on the surface of polystyrene beads or other small objects followed by assessment for the ability to recruit large-scale clouds of actin filaments and comet tails (11, 22–24). The N-terminal domain of ActA is sufficient to support actin-based motility; its critical subdomains bind to and activate the Arp2/3 complex, which catalyzes nucleation of actin filament growth (19, 21, 25), and also bind monomeric actin (19, 21). The central domain of ActA includes four proline-rich repeat sequences that are responsible for binding proteins of the Ena/VASP2 family (13, 20, 26, 27) and regulate the speed and directional persistence of motile bacteria (14, 28). The C-terminal domain of native ActA contains a transmembrane anchor and a spacer long enough to traverse the cell wall, a structure that is on the order of tens of nanometers in thickness (29–31). Soluble ActA has also been reported to bind to phosphoinositides (32, 33), although it is unclear what the role of such binding might be.
Relatively little is known about the structural characteristics of ActA in solution or on the surface of L. monocytogenes. Analytical ultracentrifugation has demonstrated that ActA exists as a monomer in solution (20, 32) with ambiguous results regarding higher order multimers (32). ActA has been shown to behave as an anomalously large species by gel filtration, suggesting that it may be an elongated molecule (32, 34). Dimer formation has been reported for ActA (35) based on its ability to interact with itself in a yeast two-hybrid assay and on cross-linking in situ on the bacterial surface to a dimeric form but not to higher-order multimers. Because ActA is found at a high density on the bacterial surface (8), it is possible that a weak tendency to form lateral dimers might affect its biochemical activities in situ. However, all of the data describing the biochemistry of soluble ActA in vitro to date has utilized monomeric protein. To explore possible contributions of ActA dimerization to its function in actin-based motility, we have synthesized an ActA dimer and compared its ability to nucleate actin filament growth with the standard soluble monomeric protein both in solution and on the surface of latex microspheres. Although we find no evidence for specific protein-protein interaction between the two tethered ActA moieties, we nonetheless observe an enhancement of its nucleation function that is probably due to physical proximity alone. This kind of local enhancement of actin assembly due to local cooperation may contribute generally to the spatial regulation of actin network assembly.
EXPERIMENTAL PROCEDURES
Iodoacetamide Cross-linker Synthesis—2 mmol of bis[2-(3-aminopropoxy)ethyl] ether in 10 ml of 1 m sodium carbonate with 5 ml of dioxane was mixed all at once while stirring with 4.4 mmol of iodoacetyl chloride in 5 ml of dioxane. After a 1-h incubation at room temperature, the mixture was extracted with methylene chloride. The organic phase was extracted three times each with cold saturated sodium bicarbonate, saturated sodium chloride brine, and 1 m hydrochloric acid followed by drying over anhydrous sodium sulfate. After drying, the solvent was removed by rotary evaporation, and the resulting oil was dissolved in TLC solvent, 92% chloroform, 8% methanol. The cross-linker was further purified by preparative thin layer chromatography with TLC solvent. The molecular weight of the N,N′-bis(iodoacetyl)-bis[2-(3-aminopropoxy)ethyl] ether and its ability to dimerize glutathione were confirmed by mass spectrometry.
Protein Purifications—All proteins were characterized by SDS-PAGE (36) and assayed for activity according to established protocols. ActA-His6 (ActA) and ActA-Cys-His6 (ActA-Cys) were purified as described (19, 37) with an additional purification step of quaternary ion exchange chromatography as in previous reports (11). Arp2/3 was purified from bovine thymus according to the method of Higgs et al. (38) followed by affinity purification with a glutathione S-transferase (GST)-VCA column (39). Actin was purified from rabbit skeletal muscle (40). All proteins were quantitated using their extinction coefficients (41) with the exception of ActA, which was quantitated by quantitative amino acid analysis.
The VCA domain of neuronal Wiskott-Aldrich syndrome protein was purified as a GST fusion (42) expressed in Escherichia coli by glutathione-Sepharose chromatography followed by quaternary anion exchange chromatography essentially according to published procedures (43). GST-VCA is a dimer due to the GST domain. To investigate the effects of monomeric VCA, we used the Factor Xa cleavage site in the GST-VCA fusion to generate monomeric VCA. GST-VCA at 263 μm was incubated at 37 °C with Factor Xa at 0.2 μm in 10 mm Tris, pH 8.0, 50 mm sodium chloride, and 0.67 mm calcium chloride. After 1 h the Factor Xa protease inhibitor 1,5-dansyl-Glu-Gly-Arg-chloromethyl ketone dihydrochloride was added to 1 μm and incubated for an additional 30 min at 21 °C followed by the addition of EGTA to 3 mm. The resulting mixture of GST, VCA, Factor Xa, and inhibitor was used without further purification as “monomeric” VCA. A second sample of GST-VCA was treated identically except the Factor Xa was added after it had already been inactivated by the inhibitor. This second sample, identical in composition to the first sample, except uncleaved, was used as “dimeric” VCA. SDS-PAGE analysis confirmed that the monomer VCA was separated from the GST and that the dimer VCA remained fused to GST. Concentration of the VCA dimer was expressed in terms of the number of VCA units.
The Listeria-specific endolysin HPL511 was purified essentially according to published procedures (44) except that with the addition of cation exchange chromatography in 25 mm sodium phosphate, pH 7.4, before the metal chelate chromatography. Growing the E. coli expressing HPL511 at 30 °C with 0.5 mm isopropyl 1-thio-β-d-galactopyranoside greatly facilitated the production of soluble HPL511.
ActA Dimer Synthesis—The bisiodoacetamide cross-linker, at 10 mm in DMSO, was added to ActACys in 200 mm sodium phosphate, pH 7, 0.1 mm Tris(2-carboxyethyl)phosphine at a 1:2 molar ratio while stirring at room temperature for 1 h. Typical reaction conditions used 100 μm bisiodoacetamide and 200 μm ActACys. Under these reaction conditions the yield of ActA dimer was around 30%. The reaction was quenched with 1 mm dithiothreitol, and the monomer was separated from the dimer by quaternary anion exchange chromatography on a ResourceQ column (GE Healthcare) using the method for purification of ActA or by gel filtration with a Superose 6 (GE Healthcare) column. Concentration of the ActA dimer was expressed in terms of the number of individual ActA units.
Size Exclusion Chromatography—We used a Superose 6 column calibrated with the appropriate standards to determine the size of ActA and ActA dimer. For native runs the column was developed with 20 mm sodium phosphate, pH 7.4, and 0.15 m sodium chloride. For denatured runs the column was developed with 20 mm sodium phosphate, pH 6.5, 6 m guanidine hydrochloride. For the denatured gel filtration runs the cysteines on the protein standards were blocked by reduction with 10 mm dithiothreitol in 8 m urea, 100 mm Tris base, pH 8.0, for 1 h at 21 °C followed by irreversible blocking with the addition of 100 mm iodoacetamide at 21 C for 16 h. The standards were repurified from the reactants by dialysis and gel filtration.
Mass Spectrometry—Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry was performed as described (45) in a Voyager-Elite Biospectrometry Work station.
Motility Assays—The methods for L. monocytogenes motility assays and ActA-coated bead assays have been previously described (11, 46). For assays with heat-treated ActA, the ActA was incubated at 90 °C for 10 min before bead coating. For motility assays with L. monocytogenes, the bacteria were either used as described (46) or heated to 90 °C for 3 min.
Pyrene Actin Assays—Pyrene actin assays were performed essentially as described (25). The concentrations of Arp2/3 and actin were held constant at 20 nm and 2 μm, respectively. Concentrations of VCA and ActA ranged from 0 to 100 nm. The buffer composition was 50 mm KCl, 1 mm MgCl2, 1 mm EGTA, 10 mm imidazole, pH 7.0, 0.2 mm ATP, 0.5 mm dithiothreitol.
F-actin Cloud Formation Assays—For observing isolated beads over time, 1.0 μm carboxylate-modified latex beads (Polysciences, Warrington, PA) were cleaned by incubating with mixed bead ion exchange resin (AG501-X8, Bio-Rad) and then saturated with either ActA monomer or dimer as described (11). The beads were added to Xenopus egg extract doped with rhodamine actin as for the motility assays and immediately mounted onto microscope slides for imaging.
For the large scale time course, 0.7- and 1.0-μm carboxylate modified latex beads (Polysciences) were saturated with either ActA monomer or dimer as above. The beads bind identical amounts of either ActA monomer or dimer, 2.1 × 10-9 μg/μm2, as determined by SDS-PAGE of the protein remaining in the supernatant. To prevent bias in the imaging or analysis, ActA monomer and dimer were immobilized separately onto 0.7- and 1.0-μm beads and then blindly mixed in all four pairwise combinations along with fixed L. monocytogenes as a positive control. The beads were added to the Xenopus egg extract motility assay and mounted on microscope slides for imaging. Phase/fluorescence image pairs were collected over the next 30 min using only the phase image to select beads for imaging. The imaging field was changed constantly to allow for sampling a wide area of the slide, and whenever possible, a bead of both sizes along with a bacterium was included in each image.
For analysis, the intensity data were unblinded and normalized to the values expected for a 1-μm bead utilizing data from the experiments with the same form of ActA on both bead sizes. The data points were then binned into 30-s bins (average of 8 data points per bin) and plotted as a function of time versus normalized fluorescence intensity with the S.D. of the binned intensity values.
Sedimentation Equilibrium Experiments—A Beckman XL-A analytical ultracentrifuge with a 4-place An-60Ti rotor and 6 sector centerpieces was used for all experiments. Partial specific volume (v) of ActA was calculated to be vActA = 0.727, using the method of Cohn and Edsall (47). Solvent density was calculated using the equation for water density from the CRC Handbook of Chemistry and Physics (48) and the buffer density increments as published (49), such that rhoElution buffer = 1.004 g/ml. The values used in calculations were temperature-corrected (50) such that at 4 °C, vActA = 0.718. Data analysis was only performed on data from experiments where sedimentation equilibrium had been reached throughout the cell. All readings were taken at 20, 30, and 40 h at speeds of 14,000, 12,000, and 9,000 rpm and concentrations of ActA from A280 = 0.015 to 0.500. The data were analyzed using Beckman XL-A data analysis software and fitted using the algorithm for ideal fit allowing molecular weight, Co, and base line to float.
Circular Dichroism—Circular dichroism (CD) spectra were collected on an AVIV model 62DS CD Spectrometer using a 1-mm quartz cell with monomer and dimer ActA below 0.4 mg/ml in CD buffer (1 mm potassium phosphate monobasic, 4 mm potassium phosphate dibasic). Wavelength scans from 190 to 260 nanometers were collected sequentially at 4, 20, 37, 65, 90, 65, 37, 20, and ActAs using a 1-nm bandwidth and 1-nm step size. For melts at 222 nm, the temperature was increased in 5° steps with a 4.5-min equilibration time between steps. CD signal was converted to mean residue ellipticity in the usual manner (51).
ActA Surface Density on Listeria Monocytogenes—The L. monocytogenes strain Mackaness SLCC 5764 (Mack) exhibits robust motility in Xenopus egg cytoplasmic extracts (46). Mack was grown up to an A600 of 0.3–0.5 with shaking at 37 °C in LB medium and used for quantitation of ActA only after confirming they were capable of motility in Xenopus extract. ActA was extracted from the bacteria by boiling them in SDS-PAGE gel loading buffer. The number of bacteria was determined by colony counts on BHI agar plates. Complete lysis of the bacteria was accomplished using the Listeria-specific endolysin HPL511 (44).
RESULTS
Characterization of the Soluble ActA Protein and Synthetic Dimer—Mass spectrometry of purified soluble ActA showed that it has a molecular mass of 65.2 kDa, close to its expected molecular mass of 65.5 kDa. We did not detect any species of higher mass, although it is unlikely we could detect higher order multimers using mass spectrometry if the protein associations are weak. Prior work on soluble ActA using analytical ultracentrifugation has demonstrated that ActA behaves as a monomer in solution with conflicting results regarding higher order multimers (20, 32). We conducted analytical ultracentrifugation experiments on our highly purified ActA preparation using speeds from 9,000 to 14,000 rpm and determined that ActA behaves as a monomer in solution with a median reduced molecular mass of 69.7 kDa (S.D. = 9.8, 37 trials). This molecular weight is in close agreement with the calculated molecular weight and is significantly less than that expected for a dimer. At ActA concentrations of greater than 2 mg/ml, the residuals to ideal fit exhibited a complicated behavior inconsistent with dimer formation and suggestive of aggregate formation along with non-ideality. In addition, portions of cells with very high concentrations of ActA did not reach equilibrium even after 40 h, whereas experiments with lower concentrations reached equilibrium after less than 20 h.
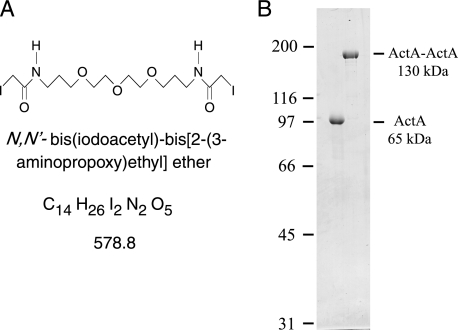
To examine the effects of possible lateral self-association, we constructed and purified a synthetic ActA dimer. To form ActA dimers we chose a strategy that would force the ActA molecules into close proximity while allowing them enough degrees of freedom to self-associate as they might on the surface of L. monocytogenes. To that end we synthesized a cross-linker with two sulfhydryl-reactive groups separated by a backbone that is 2.4-nm long and was chosen to be very soluble and flexible (Fig. 1A). Native ActA contains no cysteines, so we used a soluble ActA with a single cysteine close to the C-terminal end of the truncated protein just before the hexahistidine tag (37). After cross-linking and repurification, we were able to obtain pure covalently cross-linked ActA dimers (Fig. 1B). Forming a covalent link between the C-terminal ends of two monomers, raising the effective local concentration to more than 30 mm, should permit intrinsic lateral self-association even with very low affinity. The site of cross-linking should allow association between the two monomers in a parallel fashion but might not permit antiparallel associations. With this optimal cross-linking and high local concentration we would expect to be able to observe the structural as well as biochemical signatures of self-association for the parallel dimer.
FIGURE 1.
Homobifunctional cross-linker and dimeric ActA. A, structural formula for N,N′-bis(iodoacetyl)-bis[2-(3-aminopropoxy)ethyl] ether, a flexible hydrophilic cysteine-reactive cross-linker. B, purified soluble ActA migrates with an apparent mobility of 97 kDa, whereas synthetic ActA dimer migrates with an apparent mobility of 185 kDa by SDS-PAGE. Molecular weight marker positions are indicated to the left of the Coomassie-stained gel.
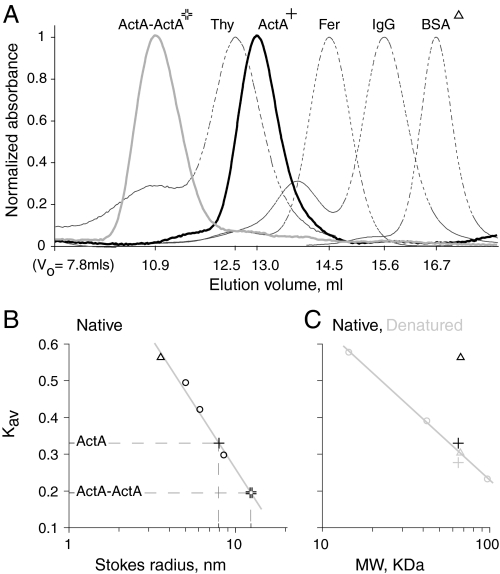
Characterization of ActA monomer and dimer by size exclusion chromatography provides insights into the structure of ActA. The expected Stokes radius for a 65-kDa globular protein is ∼3.5 nm. Our size exclusion data shows that monomeric ActA has a Stokes radius of 8.0 nm (Fig. 2), in agreement with previously published results (32, 34). A Stokes radius of 8.0 nm corresponds to a globular protein of 710 kDa. Clearly, monomeric ActA is not a conventional compact globular protein.
FIGURE 2.
Analysis of ActA monomer and dimer by gel filtration. A, native monomeric (cross) and dimeric (double cross) ActA are well separated by gel filtration over a Superose 6 column. The horizontal axis is labeled with the elution volumes corresponding to their respective peaks. The standards with their respective Stokes radii are thyroglobulin (8.5 nm), ferritin (6.1 nm), IgG (5.0 nm), and bovine serum albumin (BSA, triangle, 3.6 nm). B, labels are as in A. Plot of Kav versus the Stokes radius (on a logarithmic axis) for native proteins shows that the elution volume of native ActA monomer and dimer correspond to Stokes radii of 8.0 and 12.5 nm, respectively. C, labels are as in A. Plot of Kav versus the molecular weight (on a logarithmic axis) for denatured proteins (gray markers). For reference, the Kav values of native (black markers) bovine serum albumin (BSA; triangle) and ActA monomer (cross) are included. The protein standards, open shapes, fall on a line, as would be expected for proteins with no secondary structure. Note that Native ActA is only slightly more compact than what would be expected for a completely unfolded protein, whereas unfolded ActA is only slightly more extended than expected.
If ActA could form a dimer, then cross-linking the ActA at its C terminus would be consistent with its orientation on the surface of L. monocytogenes. Moreover, if being in close proximity to one another favors the formation of lateral ActA-ActA associations, then we would expect the Stokes radius of the dimer to be comparable with that of the monomer. This would be especially true if the two molecules are cross-linked at the C terminus and dimerize by associations at the N terminus, as has been previously proposed (35). However, gel filtration of the ActA dimer showed that it has a Stokes radius of 12.5 nm (Fig. 2). This size is consistent with two models; 1) a simple geometrical model that the dimerized ActA proteins are non-interacting extended rods that are freely hinged at the base, with an average angle between them of ∼100°, or 2) a model that assumes ActA is largely a natively unfolded protein as a dimer would be expected to be about 21/2 times as large as the monomer or 11.3 nm in radius (52).
Gel filtration under denaturing conditions suggests that ActA is a natively unfolded protein (random coil). We compared the retention time of monomeric ActA to a set of protein standards that have been rendered random coil by denaturation in 6 m guanidine hydrochloride at low pH and covalent alkylation of the cysteines. Native ActA migrates over gel filtration as a slightly more compact molecule than the model unfolded protein standards and, when denatured in guanidine, is only slightly more extended than the model random coil standards. In contrast, bovine serum albumin (BSA) shows a significant size shift on denaturation and fully denatured BSA migrates with a retention time comparable with native ActA, which is similar in molecular weight (Fig. 2). It is clear that the ActA native structure is mostly random coil. The monomeric ActA slightly overextended structure in guanidine could be due to the polyproline rich domains in ActA, which can retain structure even in guanidine (53).
Coupled to the anomalously large Stokes radius of ActA in physiological buffers is its low relative SDS-PAGE mobility, which corresponds to an apparent molecular mass of 97 kDa compared with its true molecular mass of 65 kDa. This large discrepancy in relative mobility to actual molecular mass has been at least partially attributed to a polyproline-rich central domain which is expected to be resistant to denaturation by SDS (54). However, ActA continues to migrate with an abnormally high relative mobility even when this polyproline rich domain is deleted (14). There are numerous reports of natively unfolded proteins traveling with an abnormally low relative mobility on SDS-PAGE (55, 56). The mechanisms by which proteins bind to SDS are only partially understood. In general, proteins bind to SDS with a fairly constant ratio of 1.4 g of SDS/g of protein (57). However, this binding is mostly hydrophobic in nature (57) and can even result in an increase in secondary structure especially in amphiphilic regions (54, 58). It would not be surprising that natively unfolded proteins, being very extended and hydrophilic, do not form compact helices with SDS micelles and, therefore, migrate with a low relative mobility on SDS-PAGE.
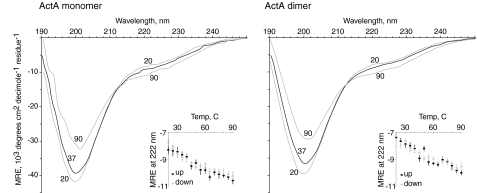
Circular Dichroism Spectroscopy of ActA Indicates That It Is Natively Unfolded—CD spectroscopy is used to estimate the relative amounts of α-helix, β-sheet, and random coil in proteins. We performed our CD analysis by collecting spectra from 190 to 260 nm taken over a range of temperatures. The spectrum of soluble monomeric ActA is characterized by a large dip centered at 200 nm (Fig. 3). Upon heating the ActA monomer, the CD spectrum shifts by increasing the signal at 200 nm and decreasing at 222 nm. Unusually, these spectra exhibit absolutely no hysteresis upon cooling, which suggests that heating ActA does not induce irreversible changes in protein structure. Moreover, there is also no evidence for a cooperative unfolding transition on heating.
FIGURE 3.
Circular dichroism of monomeric and dimeric ActA. The circular dichroism spectra of ActA monomer and dimer are virtually identical. Wavelength scans were made consecutively at 20, 37, 90, 37, and 20 °C. The scans at the same temperature showed values for mean residue ellipticity (MRE) that were identical to within the error of the instrument, about 2% between 200 and 230 nm, increasing to 5% at 195 and 238 nm. Insets, the forward and reverse melts at 222 nm show no hysteresis and demonstrate that ActA undergoes a reversible transition upon heating with no obvious cooperativity. Error bars are the S.D. taken for each data point during the measurement. The wavelength scans and temperature melts are evidence that ActA is a natively unfolded protein.
The characteristic signatures of β-sheet and α-helix are not a prominent part of the CD spectrum for ActA. Rather, when taken alone, the shape of the spectrum obtained is more indicative of either of two other types of structure, random coil and polyproline II (PII) helix. In addition, the decrease in signal at 222 nm upon heating is further evidence that the spectrum is not dominated by α-helix. The 222-nm CD signal of an α-helix should increase (rather than decrease) as the temperature is raised due to unfolding. This decrease in dichroism at 222 nm as the temperature is increased is observed with PII type helices (59, 60), and it is obvious that ActA contains regions rich in polyproline so it is likely for ActA to contain some PII helix. Although PII helices will not melt out at elevated temperatures, the lack of overall shape change upon heating is probably due to the fact that ActA is mostly random coil, and therefore, there is very little ordered structure to melt out. In addition, the overall shape of the CD spectrum is virtually identical to the CD spectra previously reported for two other natively unfolded proteins (55, 56). Analysis of the amino acid sequence of ActA using PONDR (Predictor of Natural Disordered Regions; access provided by Molecular Kinetics (copyright 2004)) predicts ActA to be 74, 100, 89, and 100% disordered using the VLXT (copyright 1999 by the Washington State University Research Foundation (61)), VSL1 (62), XL1_XT (63), and VL3 (64) algorithms, respectively. Using these estimates we predict ActA to be >90% random coil. The remainder is PII helix, and there might be a small amount of β-sheet, but there is clearly very little or no α-helix.
To see whether forced dimerization could affect the ActA overall structure, we compared the CD spectra of ActA monomer and synthetic dimer and found no significant difference (Fig. 3). These results cannot rule out an ActA-ActA interaction but certainly argue against a strong protein-protein interaction or one with a large contact area (65, 66). Our results are most consistent with the hypothesis that ActA is truly a monomeric protein even when the local concentration is extremely high (>30 mm) as rendered by forced tethering.
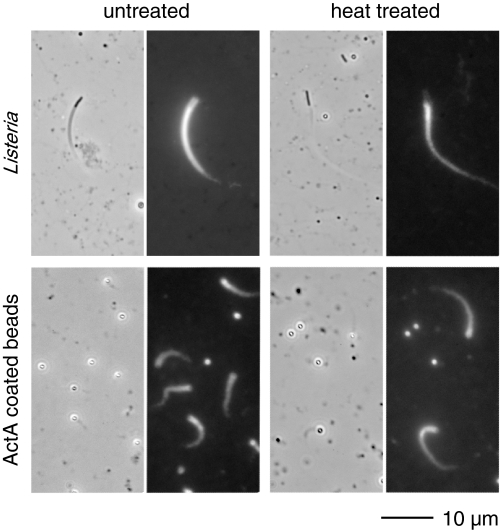
Heat-treated L. monocytogenes and ActA Support Motility— We were intrigued by the CD observation that indicated ActA might be heat-stable. We tested this hypothesis by heating L. monocytogenes to 90 °C for 3 min and then examining their ability to perform actin-based motility. Heat-treated bacteria are capable of forming actin-rich clouds and comet tails when added to Xenopus egg extract (Fig. 4). We also tested the ability of purified soluble ActA to support motility when immobilized onto latex beads after heat treatment. Soluble ActA at 1 and 10 mg/ml was heated to 90 °C for 10 min before being immobilized onto 1-m latex microspheres (11). Visually, the actin tails formed on the beads with heat-treated ActA were indistinguishable from those of unheated ActA (Fig. 4). The analysis of data gathered blind indicated that the speed of beads with heat-treated ActA are comparable with those for unheated ActA.
FIGURE 4.
ActA retains its activity after heat treatment. Motility assays with fixed L. monocytogenes and beads coated with ActA monomer that was left untreated or heat-treated showed robust motility and actin comet tail formation when placed into Xenopus extract doped with rhodamine-actin. The speeds of heated and unheated samples were comparable.
Immobilized ActA Dimers Enhance Actin Nucleation in Xenopus Egg Extracts—We wished to examine whether the artificial dimerization of ActA had any effect on its biochemical activity. To measure quantitatively the kinetics and extent of actin network formation by monomeric versus dimeric ActA, we adapted the standard in vitro motility assay. We saturated the surface of 0.7 and 1-μm latex microspheres separately with both monomeric and dimeric ActA. The total quantity of monomeric and dimeric ActA, in micrograms per unit area, that we were able to immobilize onto the beads were identical. The beads were blindly mixed in all possible combinations, placed into the Xenopus egg cytoplasmic extract motility system to record the formation of actin clouds around the microspheres, and imaged using fluorescently labeled actin. Phase/fluorescent image pairs were collected using only the phase image to locate beads for imaging. We also included fixed Listeria in our bead preparations as internal controls and to assist in measuring variability. After all the data collection and quantitation was complete, the data were unblinded, yielding four separate sets of data for monomeric as well as dimeric ActA. We normalized all the data to that from the 1-μm spheres by utilizing the fluorescence ratio from the 1-0.7-μm spheres in the trials where both bead sizes were exclusively monomer or dimer.
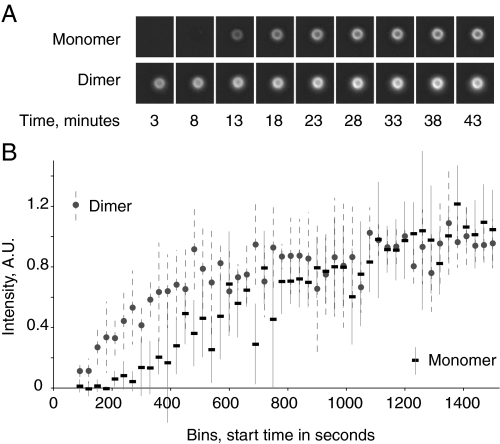
Our results show a subtle yet statistically significant difference between the monomeric and dimeric forms of immobilized ActA (Fig. 5). The final cloud size around the ActA-coated beads (as judged by accumulation of rhodamine-actin around the beads) was the same for monomeric and dimeric ActA, indicating that both forms of ActA were capable of catalyzing the assembly of equivalent filamentous actin networks. The only measurable difference between the monomeric and dimeric ActA was the actin cloud formation lag time. Monomeric ActA takes about twice as long as dimeric ActA to reach the plateau. The curve describing the accumulation of F-actin with monomeric ActA has a clear lag phase that persists until 200 s, whereas for dimeric ActA we see no visible lag phase. The lag phase for ActA dimer is either not measurable due to experimental limitations or does not exist. L. monocytogenes included in these assays had actin cloud intensities covering the entire fluorescence intensity range or well formed actin comet tails with no measurable time delay for formation of these structures. Thus, the kinetics of actin cloud assembly for bacteria more closely resembled the dimer-coated beads, with well formed actin clouds as early as 200 s, than the monomer-coated beads. However, individual-to-individual variability was much greater for bacteria than beads and for this reason the population numbers cannot be compared.
FIGURE 5.
Nucleation of F-actin cloud formation by monomeric and dimeric ActA. Beads coated with ActA dimer have a shorter lag time for cloud formation than beads with ActA monomer. A, time course of actin cloud formation around two 1-μm latex beads. The bead coated with monomeric ActA has no detectable fluorescence at 8 min, whereas the bead coated with dimeric ActA shows detectable accumulation of actin before 3 min. B, time course with data compiled from 4 assays totaling 580 beads, each observed for one time point. The curve for ActA monomer has a clear lag phase that is lacking for the ActA dimer. The lag phase for ActA dimer-coated beads is either nonexistent or too short to measure with this assay. Fixed L. monocytogenes, which were included in the assay but not shown in this figure, had clouds, tails, or no associated actin at all time points. Error bars are the S.D. of the points in each 30 s bin with a median number of 8 beads per bin. A.U., arbitrary units.
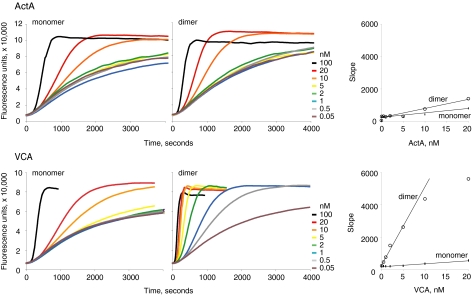
Soluble ActA Dimers Enhance Arp2/3-mediated Actin Nucleation Activity—In principle, the kinetic difference we observed for actin cloud accumulation by beads coated with ActA dimers versus monomers could be due to 1) an intrinsic difference in biochemical activity of the monomeric versus dimeric proteins at the level of activation of the Arp2/3 complex, 2) an intrinsic difference in their interaction with proteins other than Arp2/3, for example VASP (which is a native tetramer (67)), 3) a direct or indirect difference in their ability to recruit actin bundling or cross-linking proteins present in the complex Xenopus egg cytoplasmic extract, or 4) a surface-dependent effect such as a difference in their ability to capture or maintain growing actin filaments during cloud accumulation on the bead surface. To resolve this issue we examined the intrinsic ability of the ActA monomer and synthetic dimer to activate Arp2/3 actin nucleation activity in an in vitro assay containing only purified ActA, Arp2/3, and pyrene-labeled actin (25). We monitored actin filament assembly upon activation of the Arp2/3 complex by observing the increase in pyrene fluorescence signal. At equivalent concentrations of total ActA units, synthetic ActA dimer is ∼2.5-fold more potent in stimulating Arp2/3 nucleation than is ActA monomer (Fig. 6). Thus, simple physical proximity of two ActA molecules is sufficient to enhance their biochemical activity, and this effect alone can explain the more rapid kinetics of actin cloud accumulation for beads coated with ActA dimer. In addition, these experiments allow us to conclude that the increased activation of the ActA dimer need not be due to other cellular factors present in the Xenopus egg extract cloud formation assays nor due to being on a surface. Rather, in these experiments it is reasonable to conclude that the physical proximity alone is responsible for the increased Arp2/3 activation activity of the ActA dimer compared with monomer.
FIGURE 6.
Activation of Arp2/3 complex by monomeric and dimeric activators. Pyrene-actin assays were used to monitor the rate of F-actin polymerization with 2 μm actin and 20 nm Arp2/3 complex. Monomeric ActA and VCA are less efficient at catalyzing filament formation than their respective dimeric forms. The right-most graph in both rows displays the slope of the linear phase of the two left graphs with respect to activator concentration. Dimeric ActA and VCA activate the Arp2/3 complex by 2.5- and 20-fold over their respective monomeric forms.
If cooperation between two ActA molecules is due only to physical proximity, then a similar effect should be seen for any dimeric actin nucleation-promoting factor. When the VCA domain is isolated from the remainder of neuronal Wiskott-Aldrich syndrome protein, it will constitutively activate Arp2/3 complex (22, 39, 43). We made VCA fused to GST separated by a factor Xa protease site. Because GST is a parallel homodimer, this construct allowed us to explore the Arp2/3 complex activation of dimeric as well as monomeric VCA. Dimeric GST-VCA enhances Arp2/3 nucleation activity more than 20-fold compared with monomeric VCA (Fig. 6). Clearly, using dimeric VCA or ActA leads to increased activation of the Arp2/3 complex when compared with the monomer forms of either activator. This is in agreement with the enhancement of Arp2/3 nucleation seen by dimeric GST-WA (68) where the increase in activity over the monomeric form was reported to be almost 100-fold.
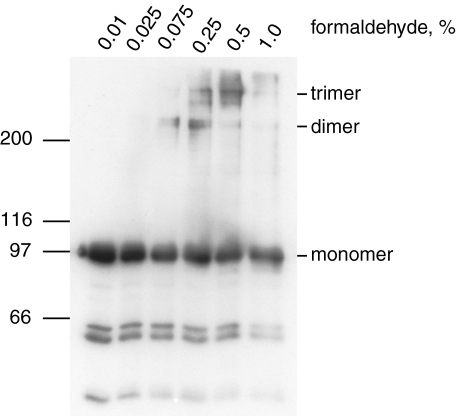
ActA Molecules Are Closely Packed on the Surface of L. monocytogenes— To better understand whether this proximity effect might influence the kinetics of actin assembly during bacterial infection, we sought to measure the density of ActA molecules on the surface of L. monocytogenes. Because ActA expression levels can vary depending on growth conditions, we performed all quantitation experiments with a single bacterial culture that we confirmed as having robust actin-based motility in the in vitro assay. Because previous studies have reported that ActA is dense enough on the surface of L. monocytogenes to be chemically cross-linked as dimers but not to higher order multimers (35), we repeated the chemical cross-linking experiments to establish the lower limit for the ActA spacing we should expect to see using other types of analysis. We used the non-reducible amine-reactive cross-linker formaldehyde to probe ActA spacing on the surface of the Mackaness strain of L. monocytogenes. After cross-linking, ActA was extracted with SDS gel loading buffer and analyzed by Western blot using a polyclonal antibody against full-length ActA. We detected the native ActA monomer, which travels with a relative mobility of ∼97 kDa as well as larger bands indicative of higher order multimers (Fig. 7). The ∼220-kDa band corresponds to where we would expect a dimer, and the unmistakable presence of even larger bands indicates that we generated a complex of more than two ActA molecules. Large amounts of cross-linker lead to a reduction in the quantity of multimers presumably due to saturation of all reactive amines or the formation of large aggregates which fail to enter the gel and migrate by SDS-PAGE. As with the previously published experiments, it is obvious that the total amount of cross-linked ActA represents only a fraction of the total, i.e. most protein remained monomeric under all conditions. Given that it is hard to predict how an unfolded protein will behave during cross-linking, we can only make an imperfect prediction about protein density using these data. A crude calculation assuming three rigid spheres in a square box four times the Stokes radius would imply 3 ActA molecules within 1024 nm2 or 0.29 ActA molecules per 100 nm2.
FIGURE 7.
Cross-linking of ActA on the bacterial surface. When L. monocytogenes is incubated with increasing amounts of formaldehyde, ActA forms higher order aggregates. ActA dimers begin to appear at 0.075% formaldehyde, and trimers are the predominant species at 0.5%. The cross-linked multimers clearly represent a minor percentage of the ActA in the samples.
More accurate measurements of ActA protein-protein spacing can be achieved by quantitating the amount of ActA per bacterium and then using the distribution data to determine the protein-protein spacing on the surface of L. monocytogenes. This is particularly relevant because ActA is not uniformly distributed on the bacterial surface but is instead concentrated at the old pole (8, 69). We used colony counts to determine that our bacterial culture had 1.11 × 109 cells/ml. To achieve quantitative removal of ActA from the surface of L. monocytogenes, we compared the amount of ActA that was extracted with SDS alone to the amount extracted after digestion of the cell wall with the Listeria-specific endolysin Hpl511 (44) followed by extraction with SDS. Removing the bacterial cell wall with Hpl511 did not change the quantity of ActA we were able to extract. Using quantitative Western analysis we determined that the amount of ActA in 1 ml of bacterial culture was 0.69 μg. This yields 5800 as the average number of ActA molecules on the surface of a single bacterium.
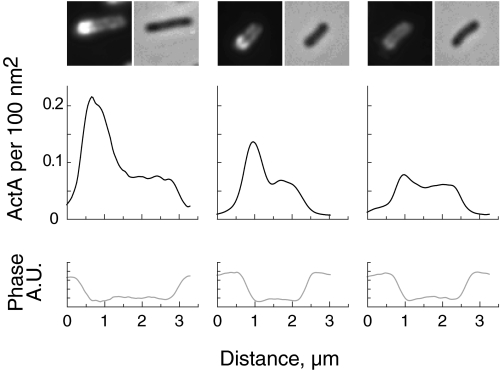
To determine the spatial distribution of these 5800 ActA molecules, we analyzed more than 350 L. monocytogenes cells labeled with an antibody against ActA to correlate fluorescence intensity with ActA surface density and cell shape (Fig. 8). The bacteria were fixed and labeled for ActA using standard immunofluorescence techniques. Phase/fluorescence image pairs were collected at random using only the phase image to locate bacteria that were not obviously in the process of completing cell division. Our fluorescence intensity profiles using indirect immunofluorescence on fixed cells matched those previously published on live cells using a red fluorescent protein-tagged ActA (70). By taking the average net fluorescence intensity per bacterium and setting this value equal to the average number of ActA molecules per bacterium, we were able to convert fluorescence intensity units into molecules of ActA per unit area (these bacteria averaged 2.2 μm long by 0.8 μm wide). The number of ActA molecules per 100 nm2 ranges from 0 to 0.27 (essentially 0–0.3). The maximum value is close to the estimate from the cross-linking data, 0.29 molecules/100 nm2.
FIGURE 8.
ActA density on L. monocytogenes. 355 L. monocytogenes were used to determine the surface density of ActA. Representative images of three bacteria showing immunofluorescently labeled ActA and the corresponding phase image are shown directly above their respective line scans. Line scans measure pixel intensities through the long axis of the bacteria. The upper line scans (black) are calibrated in ActA molecules per 100 nm2 using the fluorescence image data. The lower line scans (gray), obtained from the phase image, were used for distance/size calibrations. ActA densities over the entire data set range from 0 to 0.3 ActA molecules per 100 nm2. A.U., arbitrary units.
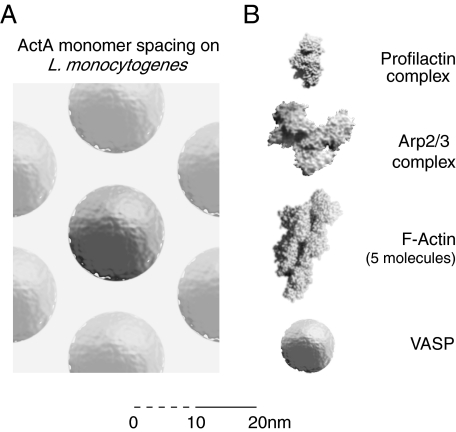
Further analysis of the fluorescence distribution data indicates that 20% of the ActA on the surface of Listeria is above the density of 0.09 molecules per 100 nm2, the minimum density for cross-linking two molecules assuming a zero-length cross-linker and a Stokes radius of 8 nm. The maximum packing density of 0.3 ActA molecules per 100 nm2 (equal to hexagonal packing with 11.4-nm sides) is not unreasonable considering that ActA has a diameter of 16 nm, and its cofactor, Arp2/3 complex, is on the order of 10 nm in size. Thus, the spacing of ActA on the surface of L. monocytogenes is almost dense enough in areas to “saturate” a two-dimensional array of ActA along with its cofactors, enabling the proximity-based cooperation that we have measured for the isolated protein (Fig. 9).
FIGURE 9.
Comparison of the physical sizes of the molecules for actin-based motility initiation. To scale (±5%) schematic of some of the molecules involved in ActA-activated actin-based motility. A, top view of a field of ActA molecules as it might appear on the surface of L. monocytogenes at their closest packing density of 0.27 ActA molecules per 100 nm2 or 1 ActA molecule per 370 nm2. Note that the actual packing may not be hexagonal, but the density will be equivalent. B, components of the ActA-mediated actin-based motility machine at the same scale as in A. The Arp2/3 complex and the profilactin complex images are based on the published structures (74, 75), and the F-actin image is based on a theoretical model (76). Visualizations were made with QuteMol (77). The entire structure for VASP has not been published, so monomeric VASP is represented as a sphere based on a published Stokes radius of 4.5 nm (67). Note that the in vitro biochemistry of ActA utilizes a minimum of 1 VASP tetramer, up to 4 profilactin complexes per VASP tetramer, one Arp2/3 complex, an elongating actin filament, and many other actin-associated cofactors. The surface environment is very crowded indeed. Scale bar in 1-nm increments for first 10 nm.
DISCUSSION
In this investigation we have confirmed previous work indicating that ActA is a monomeric protein and extended our knowledge of the state of ActA on the surface of L. monocytogenes. We have also elucidated aspects of the structure of ActA heretofore undescribed and present a model of surface-bound ActA that is consistent with all previously published results and the constraints imposed by its binding partners in the formation of filamentous actin.
Soluble ActA Is a Natively Unfolded Monomeric Protein—All of the available data are most consistent with the hypothesis that the soluble form of ActA is a purely monomeric protein. Our analytical ultracentrifugation data and that from others (20) arrive at the same conclusion, that ActA exists as a monomer in solution, at least at concentrations below 30 μm. Above this concentration the behavior of ActA becomes non-ideal. An earlier publication reporting higher order aggregates of ActA using analytical ultracentrifugation (32) may have suffered from non-ideality at the high concentrations used. Our data from gel filtration is perfectly consistent with our analytical ultracentrifugation data. Monomeric ActA migrates as a molecule with a Stokes radius of 8.0 nm with no trace of dimer, whereas a synthetic dimer migrates as a molecule with a Stokes radius of 12.5 nm. Our circular dichroism experiments, where we compare monomeric to dimeric ActA, give us additional evidence that ActA does not self-associate in a way that affects its structure even when forced into a dimer. Thus, we conclude that ActA is natively a monomer and exhibits no tendency to interact with itself even when two molecules are tethered together.
The data from gel filtration and CD are unambiguous in identifying ActA as a natively unfolded (largely random coil) protein. The Stokes radius of native ActA, 8 nm, is very close to what we would expect for a random coil of 65 kDa, and when two monomers are linked at their C terminus the size increase can be explained as a corresponding increase in the size of a random coil. Monitoring CD during temperature melts of ActA indicate that there is very little structure which can be melted out. Furthermore, the presence of PII helix can be inferred from the sequence data alone, and the shifts seen with elevated temperature would be consistent with ActA containing some PII helix. The shape of the CD spectra and behavior during thermal melting argue against ActA containing any α-helix. Moreover, the CD spectrum of ActA closely resembles that of other natively unfolded proteins (55, 56). Its behavior on SDS-PAGE is also consistent with being a hydrophilic unfolded protein. Additionally, for in vitro motility assays with ActA, it is necessary to use carboxylate-modified latex as ActA fails to bind to hydrophobic latex.
It is interesting to note that one recent publication detailing the binding of phosphoinositides to ActA presents CD spectra before and after the addition of phosphoinositides (32) that are identical to the spectra of ActA at 25 and 90 °C, respectively. This study reported a decrease in dichroism at 222 nm that was attributed to an increase in the α-helix content, but our data indicate that such a change may not be indicative of an increase in α-helix. In any case, if it does represent an increase in α-helical content, there is no reason to expect it to be a specific effect since even SDS micelles can increase the helix content of proteins (58, 71), and no biological or biochemical role for phosphoinositide binding to ActA has been demonstrated.
Forced Dimerization Enhances the Activity of Arp2/3-activating Proteins Simply by Proximity Effects—Our results demonstrate that one method of increasing the activity of ActA and other Arp2/3 activators as well is to synthetically dimerize the molecules. The net effect of dimerization of ActA is a mild increase in the activation of the Arp2/3 complex. By comparison, the increase in activation by dimeric VCA is 20-fold higher than the monomeric form. The substantial difference between the dimeric ActA and dimeric VCA is not surprising given that VCA has a significant amount of secondary structure (72). Although neither of these proteins present any evidence of being dimeric in vivo, one can explain the increase in the activity of Arp2/3 complex by at least two non-exclusive mechanisms. Having a dimer provides multiple binding sites which may increase the avidity of the interaction between Arp2/3 complex and its activator. Another possible explanation is that two nearby activated Arp2/3 complexes can cooperate with F-actin assembly because the second Arp2/3 complex may bind to the side of the first nucleated filament (73). It seems reasonable to speculate that the differences between the activities of the VCA dimer and ActA dimer might in part be due to the relatively few degrees of freedom of flexibility and parallel orientation of the VCA-GST dimer in relation to the flexible linker and wall-spanning region present in the ActA dimers. Although these subtleties are interesting from a biochemical standpoint, they are probably not relevant to L. monocytogenes motility where the asymmetric distribution of ActA on the surface is sufficient to ensure the polar formation of actin-rich comet tails and, thus, a successful infection cycle.
In addition it should be noted that our studies explore the parallel dimer and not an antiparallel dimer. Formally, it would be possible for ActA to dimerize in an antiparallel fashion. This would, presumably, be initiated by the N terminus (35). If ActA were to form an antiparallel homodimer via the N terminus alone then the ActA molecules would exist as hoops on the surface of L. monocytogenes. Given that ActA is natively disordered, our cysteine dimer could in fact still form such homodimers. One observable consequence of such N-terminal antiparallel dimerization between two disordered proteins connected at the C terminus is that it would be expected to significantly affect the Stokes radius. With our synthetic ActA dimers this is clearly not the case. Antiparallel dimerization along the entire length of the ActA would not be possible with our synthetic dimer, but such dimerization would also imply a strong association which is clearly not the case as demonstrated by size exclusion chromatography and analytical ultracentrifugation. Although such dimerization remains a theoretical possibility, it is very unlikely.
We present the first work that quantitates the distribution of ActA on the surface of L. monocytogenes in terms of actual spacing of the molecules on the surface. This study and prior studies (35) used cross-linking experiments to establish the minimum density of ActA on the bacterial cell surface. Because the flexibility of the ActA on the surface of L. monocytogenes is unknown, the packing density that we established by cross-linking is only the lower limit of what the closest packing may be. The highest density of ActA estimated by both cross-linking and calibrated immunofluorescence was determined to be 0.3 molecules per 100 nm2, equal to the spacing of the ActA molecule in a hexagonal array with 11.4-nm sides. Given the size of ActA and its binding partners, this is probably the tightest spacing possible (Fig. 9). The crystal structure of the Arp2/3 complex (74) suggests that there would need to be a space of about 10 nm around the ActA molecule for the Arp2/3 complex to easily bind. Additional factors needed for actin filament formation, VASP tetramers and profilactin, would need space to bind to ActA in addition to that occupied by the Arp2/3 complex. Indeed, if there were any more ActA molecules on the surface of the “tail end” of L. monocytogenes, there would be no room for cofactors. Any model of ActA-activated actin-based motility must take this crowded and restrictive environment on the surface of L. monocytogenes into account.
Acknowledgments
We are grateful to Robert L. Baldwin for many insightful and enjoyable discussions and Mark A. Tsuchida for helpful comments on the manuscript. We thank Mitchell C. Sanders and W. Barrett Simms for experimental contributions to the earliest phases of this project.
This work was supported, in whole or in part, by National Institutes of Health Grant R01-AI36929. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: VASP, vasodilator-stimulated phosphoprotein; GST, glutathione S-transferase; PII, polyproline II.
References
- 1.Farber, J. M., and Peterkin, P. I. (1991) Microbiol. Rev. 55 476-511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lecuit, M., Vandormael-Pournin, S., Lefort, J., Huerre, M., Gounon, P., Dupuy, C., Babinet, C., and Cossart, P. (2001) Science 292 1722-1725 [DOI] [PubMed] [Google Scholar]
- 3.Pentecost, M., Otto, G., Theriot, J. A., and Amieva, M. R. (2006) PLoS Pathog. 2 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shetron-Rama, L. M., Marquis, H., Bouwer, H. G., and Freitag, N. E. (2002) Infect. Immun. 70 1087-1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tilney, L. G., and Portnoy, D. A. (1989) J. Cell Biol. 109 1597-1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Portnoy, D. A., Auerbuch, V., and Glomski, I. J. (2002) J. Cell Biol. 158 409-414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kocks, C., Gouin, E., Tabouret, M., Berche, P., Ohayon, H., and Cossart, P. (1992) Cell 68 521-531 [DOI] [PubMed] [Google Scholar]
- 8.Kocks, C., Hellio, R., Gounon, P., Ohayon, H., and Cossart, P. (1993) J. Cell Sci. 105 699-710 [DOI] [PubMed] [Google Scholar]
- 9.Kocks, C., Marchand, J. B., Gouin, E., d'Hauteville, H., Sansonetti, P. J., Carlier, M. F., and Cossart, P. (1995) Mol. Microbiol. 18 413-423 [DOI] [PubMed] [Google Scholar]
- 10.Smith, G. A., Portnoy, D. A., and Theriot, J. A. (1995) Mol. Microbiol. 17 945-951 [DOI] [PubMed] [Google Scholar]
- 11.Cameron, L. A., Footer, M. J., van Oudenaarden, A., and Theriot, J. A. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 4908-4913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brundage, R. A., Smith, G. A., Camilli, A., Theriot, J. A., and Portnoy, D. A. (1993) Proc. Natl. Acad. Sci. U. S. A. 90 11890-11894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pistor, S., Chakraborty, T., Walter, U., and Wehland, J. (1995) Curr. Biol. 5 517-525 [DOI] [PubMed] [Google Scholar]
- 14.Smith, G. A., Theriot, J. A., and Portnoy, D. A. (1996) J. Cell Biol. 135 647-660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lauer, P., Theriot, J. A., Skoble, J., Welch, M. D., and Portnoy, D. A. (2001) Mol. Microbiol. 42 1163-1177 [DOI] [PubMed] [Google Scholar]
- 16.Pistor, S., Chakraborty, T., Niebuhr, K., Domann, E., and Wehland, J. (1994) EMBO J. 13 758-763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pistor, S., Grobe, L., Sechi, A. S., Domann, E., Gerstel, B., Machesky, L. M., Chakraborty, T., and Wehland, J. (2000) J. Cell Sci. 113 3277-3287 [DOI] [PubMed] [Google Scholar]
- 18.Welch, M. D., Iwamatsu, A., and Mitchison, T. J. (1997) Nature 385 265-269 [DOI] [PubMed] [Google Scholar]
- 19.Skoble, J., Portnoy, D. A., and Welch, M. D. (2000) J. Cell Biol. 150 527-538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Machner, M. P., Urbanke, C., Barzik, M., Otten, S., Sechi, A. S., Wehland, J., and Heinz, D. W. (2001) J. Biol. Chem. 276 40096-40103 [DOI] [PubMed] [Google Scholar]
- 21.Zalevsky, J., Grigorova, I., and Mullins, R. D. (2001) J. Biol. Chem. 276 3468-3475 [DOI] [PubMed] [Google Scholar]
- 22.Bernheim-Groswasser, A., Wiesner, S., Golsteyn, R. M., Carlier, M. F., and Sykes, C. (2002) Nature 417 308-311 [DOI] [PubMed] [Google Scholar]
- 23.Samarin, S., Romero, S., Kocks, C., Didry, D., Pantaloni, D., and Carlier, M. F. (2003) J. Cell Biol. 163 131-142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cameron, L. A., Robbins, J. R., Footer, M. J., and Theriot, J. A. (2004) Mol. Biol. Cell 15 2312-2323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Welch, M. D., Rosenblatt, J., Skoble, J., Portnoy, D. A., and Mitchison, T. J. (1998) Science 281 105-108 [DOI] [PubMed] [Google Scholar]
- 26.Chakraborty, T., Ebel, F., Domann, E., Niebuhr, K., Gerstel, B., Pistor, S., Temm-Grove, C. J., Jockusch, B. M., Reinhard, M., Walter, U., and Wehland, J. (1995) EMBO J. 14 1314-1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gertler, F. B., Niebuhr, K., Reinhard, M., Wehland, J., and Soriano, P. (1996) Cell 87 227-239 [DOI] [PubMed] [Google Scholar]
- 28.Auerbuch, V., Loureiro, J. J., Gertler, F. B., Theriot, J. A., and Portnoy, D. A. (2003) Mol. Microbiol. 49 1361-1375 [DOI] [PubMed] [Google Scholar]
- 29.Marienfeld, S., Uhlemann, E. M., Schmid, R., Kramer, R., and Burkovski, A. (1997) Antonie Van Leeuwenhoek 72 291-297 [DOI] [PubMed] [Google Scholar]
- 30.Matias, V. R., and Beveridge, T. J. (2006) J. Bacteriol. 188 1011-1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matias, V. R., and Beveridge, T. J. (2005) Mol. Microbiol. 56 240-251 [DOI] [PubMed] [Google Scholar]
- 32.Cicchetti, G., Maurer, P., Wagener, P., and Kocks, C. (1999) J. Biol. Chem. 274 33616-33626 [DOI] [PubMed] [Google Scholar]
- 33.Steffen, P., Schafer, D. A., David, V., Gouin, E., Cooper, J. A., and Cossart, P. (2000) Cell Motil. Cytoskeleton 45 58-66 [DOI] [PubMed] [Google Scholar]
- 34.Belyi, Y. F., Tartakovskii, I. S., and Prosorovskii, S. V. (1992) Med. Microbiol. Immunol. 181 283-291 [DOI] [PubMed] [Google Scholar]
- 35.Mourrain, P., Lasa, I., Gautreau, A., Gouin, E., Pugsley, A., and Cossart, P. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 10034-10039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laemmli, U. K. (1970) Nature 227 680-685 [DOI] [PubMed] [Google Scholar]
- 37.Upadhyaya, A., Chabot, J. R., Andreeva, A., Samadani, A., and van Oudenaarden, A. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 4521-4526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Higgs, H. N., Blanchoin, L., and Pollard, T. D. (1999) Biochemistry 38 15212-15222 [DOI] [PubMed] [Google Scholar]
- 39.Egile, C., Loisel, T. P., Laurent, V., Li, R., Pantaloni, D., Sansonetti, P. J., and Carlier, M. F. (1999) J. Cell Biol. 146 1319-1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spudich, J. A., and Watt, S. (1971) J. Biol. Chem. 246 4866-4871 [PubMed] [Google Scholar]
- 41.Edelhoch, H. (1967) Biochemistry 6 1948-1954 [DOI] [PubMed] [Google Scholar]
- 42.Miki, H., Miura, K., and Takenawa, T. (1996) EMBO J. 15 5326-5335 [PMC free article] [PubMed] [Google Scholar]
- 43.Hufner, K., Higgs, H. N., Pollard, T. D., Jacobi, C., Aepfelbacher, M., and Linder, S. (2001) J. Biol. Chem. 276 35761-35767 [DOI] [PubMed] [Google Scholar]
- 44.Loessner, M. J., Schneider, A., and Scherer, S. (1996) Appl. Environ. Microbiol. 62 3057-3060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Correia, I., Chu, D., Chou, Y. H., Goldman, R. D., and Matsudaira, P. (1999) J. Cell Biol. 146 831-842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Theriot, J. A., and Fung, D. C. (1998) Methods Enzymol. 298 114-122 [DOI] [PubMed] [Google Scholar]
- 47.Cohn, E. J., and Edsall, J. T. (1943) Proteins, Amino Acids, and Peptides as Ions and Dipolar Ions, pp. 157, Reinhold Publishing Corporation, New York
- 48.Lide, D. R. (2008) CRC Handbook of Chemistry and Physics, 88th Ed., pp. 4, CRC Press, Boca Raton, FL
- 49.Laue, T. M., Shah, B. D., Rideway, T. M., and Pelletier, S. L. (1992) in Analytical Ultracentrifugation in Biochemistry and Polymer Science (Harding, S. E., Rowe, A. J., and Horton, J. C., eds) pp. 90-175, Royal Society of Chemistry, Cambridge, UK
- 50.Durchschlag, H. (1986) in Thermodynamic Data for Biochemistry and Biotechnology (Hinz, H.-J., ed) pp. 45, Springer-Verlag New York Inc., New York
- 51.Adler, A. J., Greenfield, N. J., and Fasman, G. D. (1973) Methods Enzymol. 27 675-735 [DOI] [PubMed] [Google Scholar]
- 52.Creighton, T. E. (1993) Proteins: Structures and Molecular Properties, 2nd Ed., pp. 177, W. H. Freeman and Co., New York
- 53.Tiffany, M. L., and Krimm, S. (1973) Biopolymers 12 575-587 [Google Scholar]
- 54.Ruzza, P., Calderan, A., Guiotto, A., Osler, A., and Borin, G. (2004) J. Pept. Sci. 10 423-426 [DOI] [PubMed] [Google Scholar]
- 55.Werten, M. W., Wisselink, W. H., Jansen-van den Bosch, T. J., de Bruin, E. C., and de Wolf, F. A. (2001) Protein Eng. 14 447-454 [DOI] [PubMed] [Google Scholar]
- 56.Murray, C. L., Marcotrigiano, J., and Rice, C. M. (2008) J. Virol. 82 1294-1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reynolds, J. A., and Tanford, C. (1970) Proc. Natl. Acad. Sci. U. S. A. 66 1002-1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Parker, W., and Song, P. S. (1992) Biophys. J. 61 1435-1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Makarov, A. A., Adzhubei, I. A., Protasevich, I. I., Lobachov, V. M., and Fasman, G. D. (1994) Biopolymers 34 1123-1124 [DOI] [PubMed] [Google Scholar]
- 60.Rucker, A. L., and Creamer, T. P. (2002) Protein Sci. 11 980-985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Romero, P., Obradovic, Z., Li, X., Garner, E. C., Brown, C. J., and Dunker, A. K. (2001) Proteins 42 38-48 [DOI] [PubMed] [Google Scholar]
- 62.Obradovic, Z., Peng, K., Vucetic, S., Radivojac, P., and Dunker, A. K. (2005) Proteins 61 Suppl. 7, 176-182 [DOI] [PubMed] [Google Scholar]
- 63.Romero, P., Obradovic, Z., Kissinger, C. R., Villafranca, J. E., and Dunker, A. K. (1997) in Proc. I.E.E.E. International Conference on Neural Networks, Houston, TX, June 9–12, 1997, I.E.E.E., ACM
- 64.Radivojac, P., Obradovic, Z., Brown, C. J., and Dunker, A. K. (2008) Pac. Symp. Biocomput. 8 216-227 [PubMed] [Google Scholar]
- 65.Lo Conte, L., Chothia, C., and Janin, J. (1999) J. Mol. Biol. 285 2177-2198 [DOI] [PubMed] [Google Scholar]
- 66.Nooren, I. M., and Thornton, J. M. (2003) J. Mol. Biol. 325 991-1018 [DOI] [PubMed] [Google Scholar]
- 67.Bachmann, C., Fischer, L., Walter, U., and Reinhard, M. (1999) J. Biol. Chem. 274 23549-23557 [DOI] [PubMed] [Google Scholar]
- 68.Higgs, H. N., and Pollard, T. D. (2000) J. Cell Biol. 150 1311-1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rafelski, S. M., and Theriot, J. A. (2006) Mol. Microbiol. 59 1262-1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rafelski, S. M., and Theriot, J. A. (2005) Biophys. J. 89 2146-2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nielsen, M. M., Andersen, K. K., Westh, P., and Otzen, D. E. (2007) Biophys. J. 92 3674-3685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Volkman, B. F., Prehoda, K. E., Scott, J. A., Peterson, F. C., and Lim, W. A. (2002) Cell 111 565-576 [DOI] [PubMed] [Google Scholar]
- 73.Bailly, M., Ichetovkin, I., Grant, W., Zebda, N., Machesky, L. M., Segall, J. E., and Condeelis, J. (2001) Curr. Biol. 11 620-625 [DOI] [PubMed] [Google Scholar]
- 74.Robinson, R. C., Turbedsky, K., Kaiser, D. A., Marchand, J. B., Higgs, H. N., Choe, S., and Pollard, T. D. (2001) Science 294 1679-1684 [DOI] [PubMed] [Google Scholar]
- 75.Schutt, C. E., Myslik, J. C., Rozycki, M. D., Goonesekere, N. C., and Lindberg, U. (1993) Nature 365 810-816 [DOI] [PubMed] [Google Scholar]
- 76.Mendelson, R., and Morris, E. P. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 8533-8538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tarini, M., Cignoni, P., and Montani, C. (2006) IEEE Trans. Vis. Comput. Graph. 12 1237-1244 [DOI] [PubMed] [Google Scholar]