Abstract
The functions of quinone reductase 2 have eluded researchers for decades even though a genetic polymorphism is associated with various neurological disorders. Employing enzymatic studies using adrenochrome as a substrate, we show that quinone reductase 2 is specific for the reduction of adrenochrome, whereas quinone reductase 1 shows no activity. We also solved the crystal structure of quinone reductase 2 in complexes with dopamine and adrenochrome, two compounds that are structurally related to catecholamine quinones. Detailed structural analyses delineate the mechanism of quinone reductase 2 specificity toward catechol quinones in comparison with quinone reductase 1; a side-chain rotational difference between quinone reductase 1 and quinone reductase 2 of a single residue, phenylalanine 106, determines the specificity of enzymatic activities. These results infer functional differences between two homologous enzymes and indicate that quinone reductase 2 could play important roles in the regulation of catecholamine oxidation processes that may be involved in the etiology of Parkinson disease.
Cytosolic quinone reductases consist of two enzymes termed quinone reductase 1 (also referred to as QR1,2 NQO1, DT-diaphorase) and quinone reductase 2 (also referred to as QR2, NQO2) which catalyze the two-electron reduction of quinones without the formation of reactive intermediates (1). They play important roles against oxidative stress imposed by quinones. Quinone reductase 2 was first described in 1961 (2), although its biological function has eluded scientists for decades until recently (3–5). However, quinone reductase 1 has been very well studied (6). In particular, conclusive evidence points to QR1 having a protective function for cells against the toxicity of electrophiles and reactive forms of oxygen. In addition, its induction protects cells against carcinogenesis. Therefore, QR1 is acknowledged as belonging to the group of enzymes classified as phase 2 detoxification enzymes.
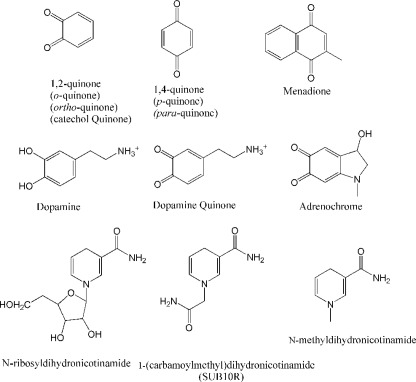
There are two major classes of quinones: 1,4-quinones (para-quinones or p-quinones, see Fig. 1) and 1,2-quinones (ortho-quinones, catechol quinones or o-quinones). Both classes of quinones can be derived from oxidation of xenobiotics as well as endogenous molecules. 1,4-Quinones include vitamin K analogues, such as VK3 (menadione), whereas catechol quinones include the oxidation products of catecholamines, amino acid tyrosine as well as estradiols (7, 8).
FIGURE 1.
Chemical structures of relevant molecules.
Although QR2 and QR1 have high sequence and structural similarities, they possess significantly different catalytic actions (9). Both QR1 and QR2 can catalyze the two-electron reduction of p-quinones such as menadione (vitamin K3), only QR1 uses NADH and NADPH as electron donors. Instead, QR2 can use N-ribosyldihydronicotinamide (NRH) or a variety of NRH analogues as electron donors in the reduction of quinones in vitro (see Fig. 1) (10). While NADH and NADPH are electron donors for a variety of enzymes in various reactions and their biological metabolism and concentrations are very well characterized, very little is known about NRH and its related analogues. Thus, it is still not clear whether NRH is the biological electron donor for QR2, and the true biological functions of QR2 require further investigation despite the significant progresses made (4, 5, 11–13).
In addition to differences in the preference of electron donors for catalysis, QR1 and QR2 have very different inhibitors. The well characterized inhibitors for QR1, such as dicumarol and Cibacron blue, do not significantly inhibit QR2 activity (3), whereas many natural polyphenols with chemopreventive properties (14) as well as some polyphenol analogs are strong inhibitors of QR2 activity (2, 15, 16). Another significant difference between the functions of QR1 and QR2 is that, even though animals with gene disruption of either protein exhibit similar phenotypes, they exhibit opposite properties with regard to menadione toxicity. Animals with QR2 gene disruption exhibit increased resistance to p-quinone toxicity, whereas animals with QR1 gene disruption show increased sensitivity to p-quinone toxicity (4). Similar differences are observed in cell lines when menadione is used as well (16). This is an intriguing paradox, as both proteins exhibit similar catalytic properties toward menadione reduction in vitro.
Epidemiological studies also demonstrate significant biological differences between these enzymes. Genetic polymorphisms of the QR1 gene, resulting in the loss or reduction of QR1 function, are associated with increased susceptibility to a variety of cancers, chemical toxicity, and drug toxicity (17–23). In contrast, genetic polymorphisms of the QR2 gene are associated with various neurological disorders, such as Parkinson disease (24, 25), schizophrenia (26), alcohol withdrawal syndrome (27), clozapine-induced agranulocytosis (28), and methamphetamine psychosis (29). Two of these studies indicated that homozygous deletion of the 29-bp sequence in the promoter region of the QR2 gene resulted in significantly reduced QR2 mRNA levels in patients (26, 27). Other genetic polymorphism resulting in the significant reduction of messenger RNA levels of QR2 is also associated with clozapine-induced agranulocytosis in schizophrenia patients (28). However, further investigation is warranted, as an in vitro transcription assay of the deletion of the 29-bp sequence in the promoter region of the QR2 gene showed the opposite effect (30), and another study conducted in the United States failed to support the association of QR2 genetic polymorphism with PD (31). Nevertheless, all these conditions are associated with neurological disorders, which led to our hypothesis that one aspect of QR2 function could be the detoxification of oxidative metabolites of neurotransmitters, specifically catechol quinones. To test this hypothesis we conducted enzymatic and structural studies to compare specificity differences between QR1 and QR2.
EXPERIMENTAL PROCEDURES
Protein Methods—Purification of QR2 was reported previously (16). The protocol of QR1 purification was similar to that of rat QR1 (32). Specifically, coding regions of human QR1 cDNA was cloned into pET23d expression vector (Novagen). Escherichia coli (Bl21) harboring the construct was cultured in LB media and induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside. Homogenous recombinant QR1 protein was purified sequentially by Cibacron blue affinity and ion exchange (Mono Q, GE Healthcare) column chromatographic methods. The QR2 and dopamine complex crystals were obtained by cocrystallization of QR2 (20 mg/ml) and 2 mm dopamine (1.6 m ammonium sulfate, 100 mm HEPES, pH 7.5). QR2-adrenochrome complex crystals were obtained by soaking QR2 crystals with 1 mm adrenochrome for 2 h in crystal freezing solution (1.2 m ammonium sulfate, 30% glycerol) (16).
Crystallographic Analysis—Crystals were frozen in solid propane in the presence of 30% glycerol as cryoprotectant. Diffraction data were collected at beamline 19BM (Argonne National Laboratory) and processed with HKL2000 (33). Crystals belonged to the space group P212121. Initial phases were determined by molecular replacement methods with CNS (34) using the dimeric QR2 structure (Protein Data Bank code 1QR2) (9). Dopamine and adrenochrome sites were identified after Fourier-difference transformation. The crystallographic analysis statistics are shown in supplemental Table 1. The final models have 95.8% residues in the most favored region, 4% in the additional allowed region and a proline residue outside the allowed region. The coordinates and structure factors were deposited at the Research Collaboratory for Structural Bioinformatics Protein Data base under the accession codes 2QMY and 2QMZ.
Enzyme Assays—The enzyme activity of QR2 in comparison with QR1 was determined spectrophotometrically at room temperature in pH 7.6 buffer (100 mm phosphate buffer, 1 μm FAD, 0.3% Tween 20) with adrenochrome as substrate. Freshly prepared adrenochrome solution in 5 mm HCl solution was used. The co-substrates used were N-methyldihydronicotinamide (NMN), SUB10R (1-(carbamoyl)dihydronicotinamide), or NADH. Because the reduced product of adrenochrome is extremely unstable and is oxidized back to adrenochrome when the assay is conducted in the presence of oxygen (35), we measured the consumption of the electron-donors NMN, SUB10R, and NADH by monitoring the reductions of UV absorbance at 360, 350, and 340 nm, respectively. Reactions were initiated by the addition of 20–30 ng of QR2 or QR1, and catalysis was monitored with a Unicam UV-visible spectrophotometer for 15 min. For the saturation curve of adrenochrome toward QR2, the reduction of SUB10R (200 μm) was measured in triplicate after 4 min with different concentrations of adrenochrome. In all assays the UV absorbance of each sample containing all the chemicals in the absence of enzyme was regarded as the background. This is especially important as reduced nicotinamide analogs can reduce adrenochrome at moderate rates (36).
RESULTS
Identification of QR2 as a Novel Catechol Quinone Reductase—QR2 is a member of the FAD-dependent oxidoreductases. It is a flavoprotein that shares extensive amino acid identity (50%) with QR1 (9, 37). QR1 has been extensively studied and has a well established role in detoxification as a member of phase II detoxification enzymes and consequently plays an important role in carcinogenesis (38). QR1 and QR2 possess near identical three-dimensional structures that extend to their catalytic active sites (supplemental Fig. 1) (9, 16). Both enzymes can efficiently catalyze the reduction of menadione (p-quinone). However, they differ in the preference of electron donors. QR1 uses the universal electron donor NADH, whereas QR2 uses reduced nicotinamide analogues with small 1-substitution groups. Its in vivo electron donor has not been characterized and has been assumed to be reduced N-ribosylnicotinamide or N-methylnicotinamide. However, one common property of these enzymes is important in that both catalyze a two-electron reduction of quinones without the formation of reactive intermediates (38).
We previously identified QR2 in a search for the cellular binding protein for resveratrol and showed that resveratrol was a potent inhibitor of QR2 activity with a dissociation constant of 35 nm (16). Because of an association of a QR2 genetic polymorphism with neurological disorders, we hypothesized that QR2 might catalyze the reduction of catechol quinones, the oxidation products of catecholamines that include the important neurotransmitters dopamine, norepinephrine, and epinephrine. This had neither been tested nor consideration given to the possibility that QR2 might be a catechol quinone reductase.
Catechol quinones are extremely unstable, making enzymatic studies technically difficult to conduct. However, the commercially available adrenochrome, the oxidation product of epinephrine (Fig. 1 for structures), is a cyclized catechol quinone that is homologous to catecholamine quinones. The quinone moiety of adrenochrome can be reduced to the dihydroxyl form. However, the reduced form is spontaneously re-oxidized back to adrenochrome in the presence of oxygen (39). In the process, electron donors are consumed, providing a basis for a sensitive assay that allows rigorous kinetic analyses. Therefore, we analyzed the enzymatic activities of both QR1 and QR2 toward adrenochrome as a representative catechol quinone. We also used a number of known QR2 electron donors to conduct the reduction of adrenochrome (see Fig. 1 for structures of electron donors used).
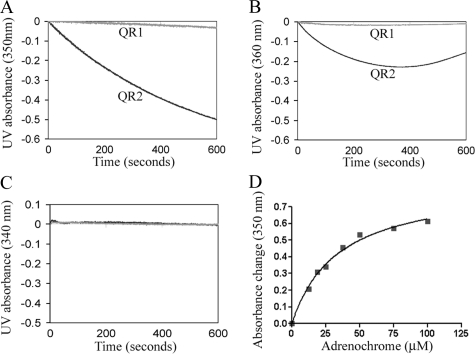
As shown in Fig. 2, A, B, and D, we demonstrate that QR2 is extraordinarily efficient in the reduction of adrenochrome with a Km value of 34 μm and a catalytic efficiency near diffusion control limits (kcat/Km = 108 m–1s–1) when QR2 electron donors such as 1-methyldihydronicotinamide or SUB10R are used. When NADH was used as the electron donor, QR2 possessed no activity for the reduction of adrenochrome (Fig. 2C). In contrast, QR1 showed no catalytic activity toward adrenochrome with any of the electron donors (Fig. 2). Fig. 2B is complicated by the fact that 1-methyldihydronicotinamide is not stable at pH below 8.0 (3). These findings argue for different cellular functions of these enzymes despite their structural similarities. The difference in electron donor is also significant, as it separates QR2 from NADH that is involved in the general cellular redox state.
FIGURE 2.
Comparisons of QR2 and QR1 activities in the reduction of adrenochrome. A, reduction of adrenochrome using SUB10R as the electron donor. Progression of the reaction was determined by monitoring the consumption of electron donors, which corresponds to UV light absorbance decrease at wavelengths around 340 nm. B, NMN was used as the electron donor. C, when NADH was used as the electron donor, neither QR1 nor QR2 exhibited any activity. D, saturation curve of the reduction of adrenochrome by QR2 using SUB10R as the electron donor.
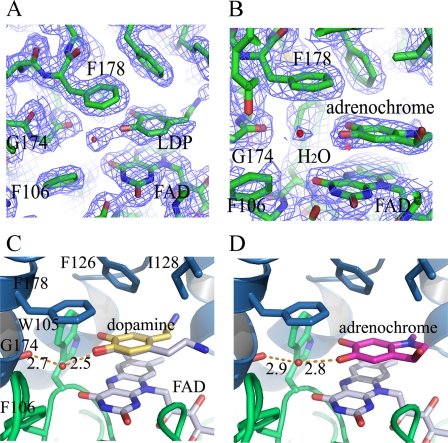
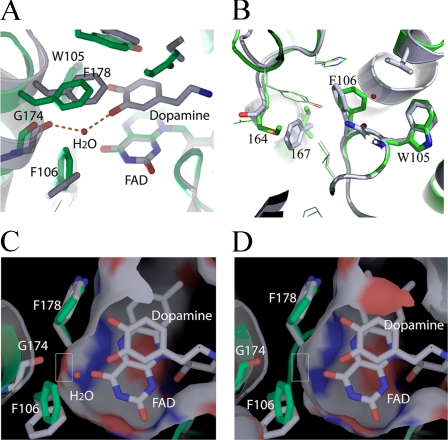
Structures of QR2 in Complexes with Dopamine and Adrenochrome—To elucidate a structural basis for the activity differences between QR1 and QR2, we crystallized QR2 in complexes with dopamine and adrenochrome. QR2-dopamine crystals were obtained by co-crystallization of QR2 in the presence of 2 mm dopamine. QR2-adrenochrome complex crystals were obtained by soaking QR2 crystals with 1 mm adrenochrome in the QR2 crystallization solution. Both compounds structurally resemble catecholamine quinones, and both molecules are similar in structure to dopamine quinone (Fig. 1). The three-dimensional structures were solved by molecular replacement methods and refined to 2.1 Å for the QR2-dopamine complex and 2.5 Å for the QR2-adrenochrome complex with CNS (34) (supplemental Table 1 for refinement statistics and Fig. 3, A and B, for electron density maps for both ligands). Both dopamine and adrenochrome bind to QR2 active sites, which are formed at the dimer interface (supplemental Fig. 2, A and B). Both molecules show well defined electron densities in their respective complexes and have average B-factors of 35 and 44 Å2 for dopamine and adrenochrome, respectively. The aromatic planes of dopamine and adrenochrome are parallel to the plane of the isoalloxazine ring of co-factor FAD molecules (Fig. 3, C and D). On the other side of the aromatic rings are hydrophobic residues, which include Phe-178, Phe-126, and Ile-128. The hydrophobic nature of the active sites of both QR1 and QR2 are well defined in the literature (9, 40).
FIGURE 3.
Structures of QR2 in complexes with dopamine and adrenochrome. A, electron density map at the active sites of QR2-dopamine complex; LDP indicates dopamine (2fo – fc, 1σ cutoff). B, electron density map at the active sites of QR2-adrenochrome complex. Adrenochrome molecule is labeled (2fo – fc, 1σ cutoff). C, detailed interaction of dopamine with QR2 at the active site. Dopamine can adopt two conformations (gray and yellow) at the active site without change in the orientation of the two phenolate groups. One phenolate group of dopamine forms a strong hydrogen bond with the main-chain carbonyl of Gly-174 through a water molecule (red dot). The distances indicated are in Å. D, adrenochrome (green) forms a strong hydrogen bond with the carbonyl group of Gly-174 through a water molecule.
The relatively narrow nature of the QR2 active sites require planar substrates and inhibitors (9, 16), which would preclude aliphatic (1,2)-diketones as substrates. The aromatic nature of QR2 active site (Trp-105, Phe-106, Phe-178, and Phe-126) also implies a preference for molecules with multiple conjugated double bonds, which would add additional π -electron interactions; catecholamine quinones belong to this class.
Both dopamine and adrenochrome form conserved strong hydrogen bonds through a water molecule with the main-chain carbonyl of Gly-174, and the hydrogen-bond lengths are all less that 3 Å. Even though dopamine displays two possible conformations at the active sites (Fig. 3C), the positions of the two hydroxyls (phenols) do not change. One of the two phenol groups forms a strong hydrogen bond with the main-chain carbonyl of Gly-174 through a water molecule (Fig. 3C). The water molecule is further held by the formation of another hydrogen bond with co-factor FAD molecule, rendering the water molecule in the QR2-dopamine complex well ordered with a B-factor of 26 Å2. Therefore, a network of hydrogen bonds among dopamine, co-factor FAD, the water molecule, and the carbonyl of Gly-174 help to orient the substrates at the active sites. Furthermore, the hydrogen bond formed between dopamine and the carbonyl group of Gly-174 through a water molecule implies that the same hydrogen bond can be formed between dopamine quinone and Gly-174 even though the ketone oxygen on quinones lacks hydrogen donors, as water molecule can act as both hydrogen bond donors and acceptors. The adrenochrome occupies a position very similar to dopamine (Fig. 3D), with one of the two ketone oxygens in adrenochrome forming a hydrogen bond with the carbonyl of Gly-174 through a water molecule. The two ketone oxygen groups in adrenochrome are positioned in a manner similar to the two hydroxyls in dopamine at the QR2 active sites.
In comparison with the QR2-menadione complex structure, both dopamine (yellow) and adrenochrome (red) occupy similar positions at the QR2 active site as menadione (green) (Fig. 4B), another QR2 substrate. For catalysis to occur, one of the ketone oxygens has to be positioned on top of the hydride donor, N5 of the isoalloxazine ring of FAD (40), even though alternative mechanism was also proposed with other substrates (41). In the QR2-dopamine complex, the distance between one of the hydroxyl oxygen is 3.5 Å from N5 of FAD molecule (Fig. 4A), whereas in QR2-adrenochrome complex, one of the ketone oxygen is 3.4 Å from N5 of FAD. In the QR2-menadione complex, one of the ketone oxygens in menadione is also 3.5 Å away from N5 of FAD molecule. All three molecules occupy similar positions at the QR2 active sites (dopamine in yellow, adrenochrome in red, and menadione in green). All these data are consistent with the catalytic mechanism proposed by Li et al. (40). Comparatively, QR2 may prefer catecholamine quinones as substrates, as there is a potential ionic interaction between the protonated amino groups in catecholamine quinones with the negatively charged carboxylate side chain of Glu-193 in QR2 (Fig. 4A) even though the distance between these two groups in the structure we solved is greater than optimum for the interaction. However, in our crystallization conditions we used high concentrations of ammonium sulfate (1.5–2 m) that could potentially disrupt the existing interactions under low ionic strength conditions. Interestingly, the carbonyl group of Gly-174 also plays a critical role in the binding of resveratrol to QR2 (16), contributing to the differentiation of QR1 and QR2 polyphenol inhibitors. In comparison with the QR2 inhibitor resveratrol (16), the substrates take up about half of the active site cavity, whereas the inhibitor occupies the entire active site (Fig. 4, C and D). An arrow indicates the exposed carbonyl oxygen from Gly-174, which forms hydrogen bonds with both substrates and inhibitors.
FIGURE 4.
Comparisons of dopamine binding with other QR2 substrates and inhibitors. A, the hydrogen bond between dopamine and the carbonyl group of Gly-174 is critical for holding its neighboring hydroxyl group situated on top of the isoalloxazine ring of co-factor FAD. The distance between one of the oxygen atoms in dopamine and N5 of FAD molecules is 3.5 Å, properly positioned for any quinone groups to accept the potential hydride transfer from FADH2. The amino group of dopamine is 3.9 Å from the carboxylate group of Glu-193. B, dopamine (yellow), adrenochrome (red), and menadione (green) are similarly positioned at the active sites of QR2. C, surface presentation of the active site of QR2. The red color indicates exposed oxygen atoms, the blue color indicates exposed nitrogen atoms, and the white (or gray) indicates carbon atoms on the surface. Dopamine occupies about half of the active site cavity, a deep hydrophobic pocket formed by the hydrophobic QR2 amino acid side chains and FAD. An arrow (yellow) indicates the exposed carbonyl group of Gly-174. D, resveratrol (blue), a strong QR2 inhibitor, occupies the entire active site. The exposed carbonyl of Gly-174 is also critical for the binding of resveratrol.
To determine the substrate specificity differences between QR1 (Protein Data Bank code 1h69) and QR2, we aligned the three-dimensional structures of QR1 with QR2 to optimize the overlap of the cofactor FADs from both proteins. Most of the active site residues are conserved between the two enzymes, which would explain their similar activities toward the reduction of menadione (12). However, for the reduction of catechol quinones, the critical interaction in the QR2-adrenochrome and QR2-dopamine complex is the hydrogen bond between the substrates and the carbonyl of Gly-174 through a water molecule. This hydrogen bond maintains the substrate in an optimum position for its neighboring ketone group (in catechol quinones) to accept the transfer of a hydride from reduced FAD (Fig. 4A).
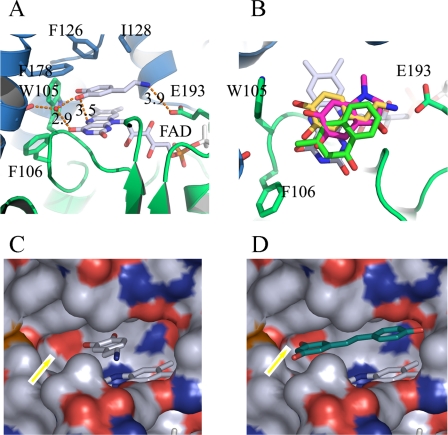
In QR2, the critical water molecule is held in position by hydrogen bonds in a space between the side chains of Phe-106 and Phe-178. In the QR1 structure, however, the side chain of amino acid Phe-106 is in a more vertical rotation (Fig. 5A, QR2 in gray and QR1 in green), making contact with the side chain of Phe-178 and, thus, forming a continuous hydrophobic area that renders the Gly-174 carbonyl group buried under a hydrophobic surface in QR1. Therefore, the water molecule that is critical for substrate (catechol quinone) orientation cannot be accommodated in the QR1 active site. This can be further illustrated by the surface presentation of the active sites. In QR2 the Gly-174 main chain carbonyl is exposed for the potential formation of hydrogen bonds with ligands (Fig. 5, C and D). In the QR1 structure the Gly-174 carbonyl is buried under a hydrophobic surface, making the formation of the hydrogen bond with any potential ligands impossible. In fact, the hydrophobic surface formed by the side chains of Phe-106 and Phe-178 in QR1 would make it energetically formidable for the potential catechol quinone substrates to adopt orientations for catalysis to occur, as that would require the polar ketone group to make contact with the hydrophobic surface. The different rotations adopted by the side-chain of Phe-106 between QR1 and QR2 are the results of different residues adjacent to them (Fig. 5B and supplemental Fig. 3). In QR2, residue 164 is serine and 167 is phenylalanine, whereas in QR1 they are methionine and isoleucine, respectively. The side chains of residue 167 are adjacent to the side chains of 106 in both structures. The steric hindrance of Ile-167 in QR1 forces Phe-106 to adopt a different rotation from that of QR2. Another residue difference, amino acid 161, an Asn in QR2 and His in QR1, accounted for the activity differences for the activation of cancer prodrug CB1954 (42). In conclusion, subtle structural differences at the active sites of two very homologous proteins account for their differences of substrate preference and, hence, differences in biological function.
FIGURE 5.
Comparisons of QR1 and QR2 structural specificities. A, structures of QR1 (green) and QR2 (gray) are aligned so that the FAD molecules overlap. The essential water molecule is in a space formed between side chains of Phe-178 and Phe-106. The hydrogen bond between dopamine and the carbonyl group of Gly-174 through a water molecule in QR2 would not be possible in QR1 due to the conformation of Phe-106 site chain. B, the different rotations of the side chains of Phe-106 between QR1 and QR2 are the results of nonconservations of neighboring residues between these two proteins, especially residues 167 and 164. The steric hindrance of Ile-167 in QR1 (phenylalanine in QR2) forces Phe-106 to adopt a different rotation in QR1. C, surface presentation of the QR2 active site according to the exposed elements with oxygen in red, nitrogen in blue, and carbon in white. The rectangle highlights the exposed carbonyl oxygen of Gly-174. D, surface presentation of the QR1 active site at the same angle as in C relative to dopamine. The carbonyl oxygen of Gly-174 is buried, as contrasted with Fig. 3C, highlighted by the rectangle, and therefore is unavailable for hydrogen bond formation.
DISCUSSION
Dopamine oxidation plays a significant role in the etiology of Parkinson disease, as the oxidation products, catechol quinones, are toxic to neurons (43–45). The detoxification of catecholamine quinones by two-electron reduction was proposed to be QR1 (46). However, various genetic studies on QR1 association with PD are inconclusive (25, 31, 47, 48). Here we established that QR2 is a catechol quinone reductase, whereas QR1 is not active when adrenochrome is employed as the substrate. Earlier studies indicated that QR1 was active in the reduction of aminochrome by monitoring oxygen consumption; however, the catalytic efficacy is very limited with a turnover rate less than 5 s–1 (36). In contrast, this study demonstrates QR2-catalyzed reduction of adrenochrome is near diffusion-controlled limits, whereas QR1 showed no detectible activity when catalytic amount is used. The relatively low Km (35 μm) value of adrenochrome toward QR2 is also a good indication that QR2 has high affinity toward catechol quinones.
The specificity of QR2 toward potential catechol quinones in comparison with QR1 is the result of structural differences at the active sites of both enzymes. The different rotations adopted by the side chains of Phe-106 are mainly the results of nonconserved neighboring residues between QR1 and QR2 (supplemental Fig. 3), especially residue 167. However, the differences extend to a number of other hydrophobic residues near 167 that include 167, 164, 146, and 169 (supplemental Fig. 3C). These residues are important in the packing of the hydrophobic core of their respective structures. Therefore, any mutational attempt to convert QR1 into QR2 in the reduction of catechol quinones most likely will not be productive.
The catechol quinone structure includes many potential substrates from normal cellular metabolites, including oxidation products of catecholamines and catechol estrogens. These molecules play important roles in many biological processes; catecholamines are important neurotransmitters, and estrogen is a reproductive hormone. Catecholamine oxidation results in the formation of catecholamine quinones (49, 50), including dopamine quinone and norepinephrine quinone. These quinones are strong electrophiles that can react with sulfhydryl-containing molecules. Dopamine is especially sensitive to oxidation, and the process is accelerated by the presence of metal ions, especially iron, manganese, and copper (51). The oxidative process can also be catalyzed by a number of enzymes, such as tyrosinase (52) and prostaglandin H synthase (53). Dopamine quinone has been reported to modify many sulfhydryl-containing proteins. To date, the target proteins identified include tyrosine hydroxylase (52, 54), dopamine transporter (55), tryptophan hydroxylase (56), and Parkin (57). The modification renders these enzymes inactive. The reactive quinone species are responsible for the toxicity of dopamine both in cell lines (58) and animal models (43, 59), leading to cell apoptosis through the induction of p53 and caspase-3 activation (58).
The implication that QR2 is involved in the detoxification of catechol quinones is also consistent with genetic studies. Genetic polymorphisms of QR2 gene are associated with various neurological disorders, which include Parkinson disease (24, 25), schizophrenia (26, 28), drug abuse (29), and others even though a study conducted in a United States population could not confirm the same association with PD (31). On the other hand, conclusive evidence demonstrates polymorphisms of QR1 gene are associated with increased susceptibility to various cancers (38, 60), whereas the association with PD is inclusive (25, 31, 47, 48). Evidently, more research on these two proteins is needed on their neurological functions.
The electron donor preferences for QR1 and QR2 also have important implications. The availability of NADH implies that QR1 will be always active, and thus, the reduction of its potential substrates will be governed by the enzymatic kinetic parameters of a particular substrate. In contrast, the electron donor for QR2 has yet to be identified even though it has been suggested to be NRH (61). However, the availability and concentration of N-ribosyldihydronicotinamide in vivo are unknown even though ribosylnicotinamide has been established as an essential intermediate in the nicotinamide-salvage pathway in yeast (62). Another excellent electron-donor in vitro for QR2 is N-methyldihydronicotinamide, the reduced form of N-methylnicotinamide that is formed from nicotinamide in vivo by nicotinamide N-methyltransferase (63). Interestingly, all PD patients have significantly elevated levels of nicotinamide N-methyltransferase (64, 65). However, whether the reduced form of N-methylnicotinamide exists in vivo is not known.
In summary, our results established that QR2 is a catechol quinone reductase when adrenochrome is used as a model substrate and, thus, could play important roles in the regulation of the catecholamine oxidation process. Additional studies on the protein expression levels of QR2 on patients with neurological disorders could further elucidate the function of this protein in neurobiochemistry.
Supplementary Material
Acknowledgments
Diffraction data were collected at beam line 19BM (Argonne National Laboratory). Use of the Advanced Photon Source was supported by the United States Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract DE-AC02-06CH11357. We also thank Dr. Ernest Lee and Dr. Joseph Schlessinger for critical readings of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 NS051548 (NINDS). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The atomic coordinates and structure factors (code 2QMZ and 2QMY) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3 and Table 1.
Footnotes
The abbreviations used are: QR2, quinone reductase 2; QR1, quinone reductase 1; PD, Parkinson disease; NRH, N-ribosyldihydronicotinamide.
References
- 1.Chen, S., Wu, K., and Knox, R. (2000) Free Radic Biol. Med. 29 276–284 [DOI] [PubMed] [Google Scholar]
- 2.Liao, S., and Williams-Ashman, H. G. (1961) Biochem. Biophys. Res. Commun. 4 208–213 [DOI] [PubMed] [Google Scholar]
- 3.Zhao, Q., Yang, X. L., Holtzclaw, W. D., and Talalay, P. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 1669–1674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Long, D. J., Jr., Iskander, K., Gaikwad, A., Arin, M., Roop, D. R., Knox, R., Barrios, R., and Jaiswal, A. K. (2002) J. Biol. Chem. 277 46131–46139 [DOI] [PubMed] [Google Scholar]
- 5.Bianchet, M. A., Erdemli, S. B., and Amzel, L. M. (2008) Vitam. Horm. 78 63–84 [DOI] [PubMed] [Google Scholar]
- 6.Dinkova-Kostova, A. T., and Talalay, P. (2000) Free Radic. Biol. Med. 29 231–240 [DOI] [PubMed] [Google Scholar]
- 7.Smythies, J., and Galzigna, L. (1998) Biochim. Biophys. Acta 1380 159–162 [DOI] [PubMed] [Google Scholar]
- 8.Cavalieri, E. L., and Rogan, E. G. (2004) Ann. N. Y. Acad. Sci. 1028 247–257 [DOI] [PubMed] [Google Scholar]
- 9.Foster, C. E., Bianchet, M. A., Talalay, P., Zhao, Q., and Amzel, L. M. (1999) Biochemistry 38 9881–9886 [DOI] [PubMed] [Google Scholar]
- 10.Knox, R. J., Jenkins, T. C., Hobbs, S. M., Chen, S., Melton, R. G., and Burke, P. J. (2000) Cancer Res. 60 4179–4186 [PubMed] [Google Scholar]
- 11.Calamini, B., Santarsiero, B. D., Boutin, J. A., and Mesecar, A. D. (2008) Biochem. J. 413 81–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foster, C. E., Bianchet, M. A., Talalay, P., Faig, M., and Amzel, L. M. (2000) Free Radic. Biol. Med. 29 241–245 [DOI] [PubMed] [Google Scholar]
- 13.Iskander, K., Li, J., Han, S., Zheng, B., and Jaiswal, A. K. (2006) J. Biol. Chem. 281 30917–30924 [DOI] [PubMed] [Google Scholar]
- 14.Kang, Y. H., and Pezzuto, J. M. (2004) Methods Enzymol. 382 380–414 [DOI] [PubMed] [Google Scholar]
- 15.Liao, S., Dulaney, J. T., and Williams-Ashman, H. G. (1962) J. Biol. Chem. 237 2981–2987 [PubMed] [Google Scholar]
- 16.Buryanovskyy, L., Fu, Y., Boyd, M., Ma, Y., Hsieh, T. C., Wu, J. M., and Zhang, Z. (2004) Biochemistry 43 11417–11426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bergamaschi, E., De Palma, G., Mozzoni, P., Vanni, S., Vettori, M. V., Broeckaert, F., Bernard, A., and Mutti, A. (2001) Am. J. Respir. Crit. Care Med. 163 1426–1431 [DOI] [PubMed] [Google Scholar]
- 18.Clairmont, A., Sies, H., Ramachandran, S., Lear, J. T., Smith, A. G., Bowers, B., Jones, P. W., Fryer, A. A., and Strange, R. C. (1999) Carcinogenesis 20 1235–1240 [DOI] [PubMed] [Google Scholar]
- 19.Fleming, R. A., Drees, J., Loggie, B. W., Russell, G. B., Geisinger, K. R., Morris, R. T., Sachs, D., and McQuellon, R. P. (2002) Pharmacogenetics 12 31–37 [DOI] [PubMed] [Google Scholar]
- 20.Krajinovic, M., Sinnett, H., Richer, C., Labuda, D., and Sinnett, D. (2002) Int. J. Cancer 97 230–236 [DOI] [PubMed] [Google Scholar]
- 21.Smith, M. T. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 7624–7626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takagi, S., Kinouchi, Y., Hiwatashi, N., Hirai, M., Suzuki, S., Takahashi, S., Negoro, K., Obana, N., and Shimosegawa, T. (2002) Anticancer Res. 22 2749–2752 [PubMed] [Google Scholar]
- 23.Zhang, J. H., Li, Y., Wang, R., Geddert, H., Guo, W., Wen, D. G., Chen, Z. F., Wei, L. Z., Kuang, G., He, M., Zhang, L. W., Wu, M. L., and Wang, S. J. (2003) World J. Gastroenterol. 9 1390–1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang, W., Le, W. D., Pan, T., Stringer, J. L., and Jaiswal, A. K. (2008) J. Gerontol. A Biol. Sci. Med. Sci. 63 127–134 [DOI] [PubMed] [Google Scholar]
- 25.Harada, S., Fujii, C., Hayashi, A., and Ohkoshi, N. (2001) Biochem. Biophys. Res. Commun. 288 887–892 [DOI] [PubMed] [Google Scholar]
- 26.Harada, S., Tachikawa, H., and Kawanishi, Y. (2003) Psychiatr. Genet. 13 205–209 [DOI] [PubMed] [Google Scholar]
- 27.Okubo, T., Harada, S., Higuchi, S., and Matsushita, S. (2003) Alcohol Clin. Exp. Res. 27 68–71 [DOI] [PubMed] [Google Scholar]
- 28.Ostrousky, O., Meged, S., Loewenthal, R., Valevski, A., Weizman, A., Carp, H., and Gazit, E. (2003) Tissue Antigens 62 483–491 [DOI] [PubMed] [Google Scholar]
- 29.Ohgake, S., Hashimoto, K., Shimizu, E., Koizumi, H., Okamura, N., Koike, K., Matsuzawa, D., Sekine, Y., Inada, T., Ozaki, N., Iwata, N., Harano, M., Komiyama, T., Yamada, M., Sora, I., Ujike, H., Shirayama, Y., and Iyo, M. (2005) Addict. Biol. 10 145–148 [DOI] [PubMed] [Google Scholar]
- 30.Wang, W., and Jaiswal, A. K. (2004) Free Radic Biol. Med. 37 1231–1243 [DOI] [PubMed] [Google Scholar]
- 31.Okada, S., Farin, F. M., Stapleton, P., Viernes, H., Quigley, S. D., Powers, K. M., Smith-Weller, T., Franklin, G. M., Longstreth, W. T., Swanson, P. D., and Checkoway, H. (2005) Neurosci. Lett. 375 178–180 [DOI] [PubMed] [Google Scholar]
- 32.Chen, H. H., Ma, J. X., Forrest, G. L., Deng, P. S., Martino, P. A., Lee, T. D., and Chen, S. (1992) Biochem. J. 284 855–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Otwinowsky, Z., and Minor, W. (1997) Methods Enzymol. 276 307–326 [DOI] [PubMed] [Google Scholar]
- 34.Brunger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T., and Warren, G. L. (1998) Acta Crystallogr. D Biol. Crystallogr. 54 905–921 [DOI] [PubMed] [Google Scholar]
- 35.Baez, S., and Segura-Aguilar, J. (1995) Biochem. Mol. Med. 56 37–44 [DOI] [PubMed] [Google Scholar]
- 36.Zafar, K. S., Siegel, D., and Ross, D. (2006) Mol. Pharmacol. 70 1079–1086 [DOI] [PubMed] [Google Scholar]
- 37.Jaiswal, A. K., Burnett, P., Adesnik, M., and McBride, O. W. (1990) Biochemistry 29 1899–1906 [DOI] [PubMed] [Google Scholar]
- 38.Ross, D., and Siegel, D. (2004) Methods Enzymol. 382 115–144 [DOI] [PubMed] [Google Scholar]
- 39.Baez, S., Segura-Aguilar, J., Widersten, M., Johansson, A. S., and Mannervik, B. (1997) Biochem. J. 324 25–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li, R., Bianchet, M. A., Talalay, P., and Amzel, L. M. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 8846–8850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Faig, M., Bianchet, M. A., Talalay, P., Chen, S., Winski, S., Ross, D., and Amzel, L. M. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 3177–3182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fu, Y., Buryanovskyy, L., and Zhang, Z. (2005) Biochem. Biophys. Res. Commun. 336 332–338 [DOI] [PubMed] [Google Scholar]
- 43.Hastings, T. G., Lewis, D. A., and Zigmond, M. J. (1996) Adv. Exp. Med. Biol. 387 97–106 [DOI] [PubMed] [Google Scholar]
- 44.Spencer, J. P., Jenner, P., Daniel, S. E., Lees, A. J., Marsden, D. C., and Halliwell, B. (1998) J. Neurochem. 71 2112–2122 [DOI] [PubMed] [Google Scholar]
- 45.Asanuma, M., and Miyazaki, I. (2006) Expert. Rev. Neurother. 6 1313–1325 [DOI] [PubMed] [Google Scholar]
- 46.van Muiswinkel, F. L., de Vos, R. A., Bol, J. G., Andringa, G., Jansen Steur, E. N., Ross, D., Siegel, D., and Drukarch, B. (2004) Neurobiol. Aging 25 1253–1262 [DOI] [PubMed] [Google Scholar]
- 47.Jiang, X. H., Yang, H., Yang, J. F., Wang, H. T., Xu, Q. Y., and Chen, B. (2004) Zhonghua Yi Xue Yi Chuan Xue Za Zhi 21 120–123 [PubMed] [Google Scholar]
- 48.Shao, M., Liu, Z., Tao, E., and Chen, B. (2001) Zhonghua Yi Xue Yi Chuan Xue Za Zhi 18 122–124 [PubMed] [Google Scholar]
- 49.Graham, D. G. (1978) Mol. Pharmacol. 14 633–643 [PubMed] [Google Scholar]
- 50.Bacq, Z. M., and Fischer, P. (1950) Arch. Int. Physiol. 57 271–276 [DOI] [PubMed] [Google Scholar]
- 51.Sulzer, D., and Zecca, L. (2000) Neurotox. Res. 1 181–195 [DOI] [PubMed] [Google Scholar]
- 52.Xu, Y., Stokes, A. H., Roskoski, R., Jr., and Vrana, K. E. (1998) J. Neurosci. Res. 54 691–697 [DOI] [PubMed] [Google Scholar]
- 53.Hastings, T. G. (1995) J. Neurochem. 64 919–924 [DOI] [PubMed] [Google Scholar]
- 54.Kuhn, D. M., Arthur, R. E., Jr., Thomas, D. M., and Elferink, L. A. (1999) J. Neurochem. 73 1309–1317 [DOI] [PubMed] [Google Scholar]
- 55.Whitehead, R. E., Ferrer, J. V., Javitch, J. A., and Justice, J. B. (2001) J. Neurochem. 76 1242–1251 [DOI] [PubMed] [Google Scholar]
- 56.Kuhn, D. M., and Arthur, R., Jr. (1998) J. Neurosci. 18 7111–7117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.LaVoie, M. J., Ostaszewski, B. L., Weihofen, A., Schlossmacher, M. G., and Selkoe, D. J. (2005) Nat. Med. 11 1214–1221 [DOI] [PubMed] [Google Scholar]
- 58.Emdadul Haque, M., Asanuma, M., Higashi, Y., Miyazaki, I., Tanaka, K., and Ogawa, N. (2003) Biochim. Biophys. Acta 1619 39–52 [DOI] [PubMed] [Google Scholar]
- 59.Hastings, T. G., Lewis, D. A., and Zigmond, M. J. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 1956–1961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kiyohara, C., Yoshimasu, K., Takayama, K., and Nakanishi, Y. (2005) Genet. Med. 7 463–478 [DOI] [PubMed] [Google Scholar]
- 61.Wu, K., Knox, R., Sun, X. Z., Joseph, P., Jaiswal, A. K., Zhang, D., Deng, P. S., and Chen, S. (1997) Arch. Biochem. Biophys. 347 221–228 [DOI] [PubMed] [Google Scholar]
- 62.Bieganowski, P., and Brenner, C. (2004) Cell 117 495–502 [DOI] [PubMed] [Google Scholar]
- 63.Aksoy, S., Szumlanski, C. L., and Weinshilboum, R. M. (1994) J. Biol. Chem. 269 14835–14840 [PubMed] [Google Scholar]
- 64.Parsons, R. B., Smith, S. W., Waring, R. H., Williams, A. C., and Ramsden, D. B. (2003) Neurosci. Lett. 342 13–16 [DOI] [PubMed] [Google Scholar]
- 65.Aoyama, K., Matsubara, K., Kondo, M., Murakawa, Y., Suno, M., Yamashita, K., Yamaguchi, S., and Kobayashi, S. (2001) Neurosci. Lett. 298 78–80 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.