Abstract
Nuclear mitochondrial pseudogenes (numts) are nonfunctional copies of mtDNA in the nucleus that have been found in major clades of eukaryotic organisms. They can be easily coamplified with orthologous mtDNA by using conserved universal primers; however, this is especially problematic for DNA barcoding, which attempts to characterize all living organisms by using a short fragment of the mitochondrial cytochrome c oxidase I (COI) gene. Here, we study the effect of numts on DNA barcoding based on phylogenetic and barcoding analyses of numt and mtDNA sequences in two divergent lineages of arthropods: grasshoppers and crayfish. Single individuals from both organisms have numts of the COI gene, many of which are highly divergent from orthologous mtDNA sequences, and DNA barcoding analysis incorrectly overestimates the number of unique species based on the standard metric of 3% sequence divergence. Removal of numts based on a careful examination of sequence characteristics, including indels, in-frame stop codons, and nucleotide composition, drastically reduces the incorrect inferences of the number of unique species, but even such rigorous quality control measures fail to identify certain numts. We also show that the distribution of numts is lineage-specific and the presence of numts cannot be known a priori. Whereas DNA barcoding strives for rapid and inexpensive generation of molecular species tags, we demonstrate that the presence of COI numts makes this goal difficult to achieve when numts are prevalent and can introduce serious ambiguity into DNA barcoding.
Keywords: cytochrome c oxidase I, Decapoda, Orthoptera
The orthology of characters is one of the fundamental and implicit assumptions in the use of DNA sequence data to reconstruct phylogeny or to establish “barcodes” for species. If the orthology assumption is violated, that is, whether paralogous sequences are unknowingly treated as orthologs, incorrect inferences are inevitable (1). This is especially true for the DNA barcoding initiative, which relies on the premise that all organisms have a unique and identifiable molecular tag, namely, a short region of mitochondrial cytochrome c oxidase subunit 1 (COI) amplified by universal primers, and that one is comparing only orthologs among species when formulating barcodes (2). As such, DNA barcoding relies on the assumption that the COI fragments generated by PCR from genomic DNA represent orthologous copies of mitochondrial DNA (mtDNA). Increasing empirical evidence suggests; however, that this assumption does not always hold true and that there are a number of molecular evolutionary processes that can hinder correct amplification and identification of the orthologs (3), including (i) duplication of the gene of interest within the mitochondrial genome (4), (ii) heteroplasmy (5), (iii) bacterial infection biasing mtDNA variation (6), and (iv) nuclear integration of mtDNA (7, 8). If a portion of COI was duplicated in a given species, conventional PCR might amplify both the correct and duplicated COI fragments, thus introducing ambiguity into the barcoding whether the paralogous copy had diverged since duplication. Heteroplasmy is the presence of a mixture of more than one type of mitochondrial genome within a single individual, and the coamplification of divergent heteroplasmic copies of mtDNA would lead to an overestimation of the number of unique species under barcoding (3). Maternally inherited symbionts, such as Wolbachia, can cause linkage disequilibrium with mtDNA and, whether a population becomes infected with such symbionts, the mtDNA associated with the initial infection will spread throughout the population and result in the homogenization of mtDNA haplotypes (6). Among closely related species, these symbionts can break through the species barrier by hybridization followed by selective sweep, resulting in identical mtDNA sequences among different species, which would cause the underestimation of the number of unique species under barcoding (9). Whereas these three processes may be relatively uncommon and limited to a small number of organisms, a fourth process, the nuclear integration of mtDNA that gives rise to nuclear mitochondrial pseudogenes (numts), is a widespread phenomenon that has been reported in many eukaryotic clades (8, 10). The effect of numts on DNA barcoding, however, has not been systematically studied to date.
The first case of numts in Metazoa was reported in the grasshopper Locusta migratoria (11), in which a copy of a mitochondrial ribosomal RNA gene was found in the nuclear genome. Lopez et al. (12) found that nearly half of the mitochondrial genome (7.9 kb) was transferred to the nuclear genome in the domestic cat and coined the term “numts.” Since then, >82 eukaryotes have been reported to have numts (8). A BLAST search of mitochondrial sequences in the published nuclear genomes suggests that nearly 99% of the mitochondrial sequences were transferred to different parts of the nucleus in both human and mouse (10). Pamilo et al. (13) reported >2,000 possible numts in the honey bee genome and found a similarly large number of numt copies in the flour beetle genome. These findings collectively indicate that numts are extremely pervasive in nature and that there may be a large number of species with unrealized numts of the COI gene in the nucleus.
The possible existence of COI numts poses a serious challenge to DNA barcoding. The fact that the COI gene can be amplified from diverse taxa by using a limited set of primers is heralded as one of the attractive features of this marker (14). It is true that relatively conserved regions within mtDNA allow the design of “universal” primers, which can amplify mitochondrial fragments from an unknown species (15). However, conserved primers can be a double-edged sword when numts are present because they can coamplify numts in addition to the target mtDNA (7, 8). If the nuclear integration of numts was an ancient and sufficient sequence divergence accumulated in the orthologous mtDNA, the conserved primers would be more likely to amplify numts in preference to mtDNA, which could possibly result in unambiguous, paralogous sequences (8). Despite this serious problem, numts have been dismissed as a minor concern for DNA barcoding (16) and the issue of numts has not been adequately addressed.
In this study, we investigate the effect of including numts in DNA barcoding in two divergent lineages of arthropods, insects, and crustaceans, which are known to have especially large numbers of numts (8, 17–19). We also examine the effect of numts at different levels of divergence: subfamily-level (grasshoppers) and species- and population-level (crayfish). Herein, we show that both grasshopper and crayfish species included in the study have numts of the COI gene and barcoding methods would incorrectly infer that single individuals belong to multiple, unique species. The prevalence of numts appears to be both species-specific and population-specific and the pattern of numt distribution is considerably different between lower-level and higher-level divergence among taxa. Finally, we demonstrate the importance of data exploration in DNA barcoding practice by examining sequence characteristics of numts.
Results and Discussion
Coamplification of Numts with Orthologous mtDNA.
Our results strongly suggest that a large number of paralogous haplotypes of various divergences are coamplified with the orthologous mtDNA sequences when conserved primers are used in both grasshoppers and crayfish, which can be identified by the presence of indels, point mutations, and in-frame stop codons [supporting information (SI) Table S1]. The majority of the coamplified paralogs can be easily considered nonfunctional numt haplotypes because of the presence of in-frame stop codons, which is especially evident in our crayfish data in which 97.3% of paralogs have stop codons. A large number of these numts have unusually high numbers of point mutations (mean = 65.52, n = 110), suggesting that nuclear integration of mtDNA would result in random accumulation of nucleotide changes. In the grasshopper data, however, there are many paralogs that cannot readily be categorized as numts because they lack in-frame stop codons and differ from the orthologous mtDNA sequences by one or two nucleotides. If the same haplotype that appears to be functional other than the ortholog is repeatedly found from a single individual, one can suspect heteroplasmy (5). Indeed, heteroplasmy seems to explain the presence of certain paralogous haplotypes in Schistocerca americana. However, there are many paralogs represented by single haplotypes with small nucleotide differences in all four grasshopper species. Although, it is unlikely that these are Taq polymerase errors during PCR because of the high fidelity polymerase we used (0.015% error rate or 0.0732 bp per reaction); we cannot rule out the possibility of PCR error amplified by additional cloning (5, 18). Also, heteroplasmy might in fact be a plausible explanation for these haplotypes because we limited our study to only 30 clones per grasshopper species, thus not exploring the full extent of heteroplasmic diversity.
If the proportion of numts is high compared with the orthologous mtDNA fragments in a given PCR product, it is possible to generate unambiguous paralogous sequences (8). This is exacerbated when the conserved primers preferentially amplify numts because of relatively ancestral sequence similarity of numts or divergence within the primer regions of the orthologous mtDNA. In this case, typical indicators of different PCR products, such as multiple bands on gels and double peaks, background noise, and ambiguity in sequence chromatograms, will not be present; hence, paralogous sequences can be mistaken as orthologous mtDNA. In fact, this exact phenomenon was observed in 18 crayfish individuals of Orconectes barri and Orconectes australis from which numts were amplified and cleanly sequenced without cloning.
Phylogenetic Analyses and Distribution of Numts.
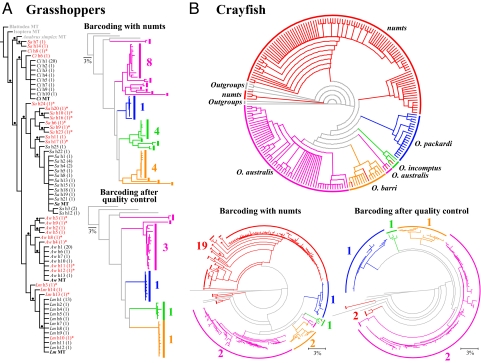
For the grasshopper data, the parsimony and the Bayesian analyses both recovered the monophyly of the orthologous mtDNA and haplotypes for three of four species (Fig. 1A); although, the topology was different in the placement of S. americana and Calliptamus italicus clades between the two methods. In each species, the largest clade was the polytomous clade consisting of the mtDNA ortholog and several similar haplotypes. The remaining haplotypes formed highly structured clades within each species, and this pattern was especially evident in Acrida willemsei and S. americana. For crayfish data, the Bayesian analysis recovered a topology mostly congruent with the parsimony analysis (Fig. 1B) and both analyses found a large clade of numt haplotypes (84 in parsimony and 82 in Bayesian) and a small clade of numts (18 in both analyses), distinctly divergent from the clearly defined clades of the orthologous mtDNA of four species. These numt clades consisted of the haplotypes from O. australis, O. barri, and Orconectes packardi, which were not necessarily grouped either by the species or the populations. Only four numt haplotypes were placed among orthologs (one in Orconectes incomptus, one in O. australis, and two in O. barri). A clade consisting of three numt haplotypes of O. barri was robustly placed near the root of trees in both analyses. Among the orthologous mtDNA clades, three of four species formed monophyletic clades, with the exception of O. australis that had one large clade sister to O. barri and a small clade basal to the australis + barri clade. Based on the phylogenetic analyses, number of indels, point mutations, in-frame stop codons, and sequence divergence, it is possible to conclude: among grasshoppers, Locusta migratoria has three numts, A. willemsei has six, C. italicus has two, and S. americana has at least 11 numts; among crayfish, O. australis has 60, O. barri has 46, O. incomptus has one, and O. packardi has four numts. On average, 32.54% and 41.88% of haplotypes generated from grasshoppers and crayfish, respectively, were numts (Table S1). It is important to note that this is a conservative estimate of the number of numts per species because it is limited by the number of individuals and clones we generated.
Fig. 1.
Phylogenetic and barcoding analyses based on orthologous mtDNA COI and paralogous numt haplotypes from grasshoppers and crayfish. (A) Grasshoppers: the cladogram on the left is a strict consensus of 41 MPTs (L = 1002; CI = 0.54; RI = 0.85). Dots above branch indicate the nodes with the bootstrap value of >75 and posterior probability of >95%. Orthologous mtDNA is indicated in bold and putative numts are indicated as red terminals. Number in parenthesis represents the number of identical copies for a particular haplotypes (h) and asterisk indicates ones with in-frame stop codons. When DNA barcoding analysis (NJ analysis based on K2P distances) is performed on the complete dataset, the number of unique species inferred based on 3% sequence divergence (colored numbers next to the vertical bars) is overestimated (barcoding with numts). After the removal of the haplotypes with indels and in-frame stop codons (barcoding after quality control), the number of unique species inferred under DNA barcoding is drastically reduced. Purple, Schistocerca americana (Sa); blue, Calliptamus italicus (Ci); green, Acrida willemsei (Aw); orange, Locusta migratoria (Lm); and gray, outgroups. (B) Crayfish: the circular cladogram on top is the strict consensus of 94 MPTs (L = 1064; CI = 0.39; RI = 0.91). Terminals are colored to indicate species. Purple, Orconectes australis; orange, O. barri; green, O. incomptus; blue, O. packardi; and gray, outgroups. All numt haplotypes are indicated as red terminals. Similarly, DNA barcoding overestimates the number of unique species when numts are included, but the removal of numts reduces the inferred number of species. Notice that even after rigorous quality control, the inferred number of unique species is actually higher than the actual number of species, suggesting that some numts are difficult to identify.
Both the grasshopper and crayfish data suggest that there can be multiple types of numts present within single individuals that vary considerably in nucleotide composition, suggesting multiple independent transfer events from the mitochondrial genome to the nucleus (8, 17). Moreover, our data suggest that these independent nuclear integration events can give rise to a family of numts that can diverge at different substitution rates. For example, we found that the relationships among the numt haplotypes of S. americana are highly structured and a similar pattern was observed in other grasshopper and crayfish species. Not only can independent transfer occur multiple times, but it can occur at very different phylogenetic levels. Whereas many numts of S. americana are closely related to the orthologous mtDNA, two numt haplotypes form a strong clade with the mtDNA ortholog of Anabrus simplex, which belongs to a different suborder within Orthoptera. Similarly, three numt haplotypes of O. barri were placed near outgroups belonging to different crayfish genera. These findings collectively suggest that there could have been an ancient nuclear integration event and that enough time has passed for these numts to have accumulated substantial sequence divergence from the orthologous mtDNA.
Our two datasets differ at the level of divergence among the ingroup species along with the distribution of numts deduced from the phylogenetic analyses. Except for two numt haplotypes of S. americana that grouped within an outgroup, all grasshopper numt haplotypes strongly grouped with their orthologous mtDNA. However, this pattern is not observed in the four closely related crayfish species. Only a small portion of numt haplotypes grouped with their orthologs, whereas the majority formed clades among themselves with no apparent population or species-specific groupings. In other words, numt haplotypes sequenced from different crayfish individuals from different populations and different species form monophyletic groups. This finding implies that the closely related crayfish species share similar types of numts that must predate the speciation events. Both patterns have been reported from other studies looking at various mitochondrial genes in diverse metazoan lineages at different phylogenetic levels (18, 20). We conclude that the distribution pattern of numts within a given group of organisms cannot be predicted a priori, but depends on the timing and the frequency of nuclear integration, which can clearly predate and postdate speciation events.
The distribution of numts in both datasets suggests that their prevalence may be lineage-specific. For example, we sequenced numts from individuals collected from 11 of 56 cave crayfish populations (Tables S2 and S3). Among the four species, numts were sequenced for 7 of 34 localities in O. australis (southernmost region of range, primarily caves in Alabama), 2 of 7 in O. barri (southernmost region of range, caves in Tennessee), 1 of 3 in O. incomptus, and 1 of 12 in O. packardi. This observed pattern does not necessarily mean that the remaining 45 populations are free of numts but it does mean that there is a nonrandom population-specific variation in the level of numt prevalence. Nuclear integration of mtDNA happens at the level of the individual (8) and a large population size can effectively dilute the amount of numts in a given population. In this case, the proportion of numts is much smaller than that of the orthologous mtDNA in a given individual, rendering the numt coamplification less likely, even with a possibility of the presence of plesiomorphic numts in the nucleus. However, whether a population experiences genetic drift because of an extreme bottleneck, numts can be fixed in a few founders, resulting in a disproportionately high level of numts. Another intrinsic factor, nuclear genome size, might also play a role that results in uneven distribution of numts. Whereas all grasshopper species have numts, A. willemsei and S. americana have especially high numbers that are divergent from each other. Bensasson et al. (21) suggested that a positive correlation between the number of numts and nuclear genome size might exist and it is possible that these two species might have larger nuclear genomes than the others. Alternatively, inherent species-specific differences in the frequency of DNA transfer from mitochondria to the nucleus and in the rate of loss of numts in the nucleus have also been suggested as possible explanations for lineage-specific numt variation (10), which may explain our observed pattern. It is also possible that the conserved primers we used were simply more efficient in amplifying numts of some individuals than others.
DNA Barcoding Overestimates the Number of Species with Coamplification of Numts.
If numts of the COI region are unknowingly coamplified with the orthologous mtDNA and used in DNA barcoding without careful exploration of sequence characteristics, the number of unique species inferred from the analysis would certainly be overestimated. For the grasshopper data, the barcoding analysis finds that the haplotypes generated from individuals from each of four species form several distinct clusters, which can be considered unique species based on the 3% sequence divergence threshold typically used in the barcoding studies. Based on the clustering pattern, one would conclude that there are a total of 17 unique species, suggesting the discovery of 13 additional cryptic species (Fig. 1A). For the crayfish data, our analysis finds that some numt haplotypes nested among the orthologous mtDNA are not divergent enough to consider them unique species under DNA barcoding. However, among the highly divergent numts, we find numerous distinct clusters that had >3% sequence divergence among each other. O. australis and O. barri each had two ortholog clusters that could be considered unique species. The barcoding analysis would thus infer a total of 25 unique species, suggesting the discovery of 21 additional cryptic species (Fig. 1B).
A careful examination of sequence characteristics before barcoding analyses drastically reduces the possibility of incorrect inferences. It is possible to identify numts on the basis of in-frame stop codons and indels and to remove these haplotypes from the original datasets. For grasshopper data, for example, the barcoding analysis recovers a total of six clusters that can be considered unique species after the removal of such obvious numt haplotypes, implying two additional cryptic species (Fig. 1A). These two clusters are formed by three haplotypes of S. americana, which have neither indels nor stop codons. When we remove a total of 108 obvious numt haplotypes from the crayfish data, the barcoding analysis finds 7 unique clusters consisting of three crayfish species, two differently sized clusters of O. australis, and two clusters formed by the numts with no stop codons (Fig. 1B). In other words, the removal of numts considerably reduces the inferred number of unique species in both grasshoppers (17 before, 6 after) and crayfish (25 before, 7 after). However, even the most careful sequence examination does not eliminate all incorrect inferences because some numts can be of the expected length without any in-frame stop codons and are not readily distinguishable from the orthologous mtDNA. An examination of nucleotide composition may serve as an effective filter for identifying highly divergent numts because of different compositional bias between mtDNA and nuclear DNA (8). In fact, two paralogous haplotypes of S. americana with no stop codons have a significantly lower AT% compared with the orthologous mtDNA, suggesting that they are numts and the clade formed by these haplotypes does not represent a cryptic species. However, even this approach cannot totally eliminate incorrect inferences because some numts (1 haplotype of S. americana, 1 haplotype of O. australis, and 2 haplotypes of O. barri) have no indels, no in-frame stop codons, and a highly similar AT% to the orthologs. It is possible that in-frame stop codons and indels lie downstream beyond the region amplified by the Folmer primers. Typical DNA barcoding studies would inevitably conclude the presence of cryptic species in such cases.
Rigorous Quality Control Against Numts Is Necessary in DNA Barcoding.
According to the standard DNA barcoding protocol published by the Consortium for the Barcode of Life (CBOL, http://barcoding.si.edu), there appears to be a minimum amount of quality control involved in generating COI sequences. By using simple protocols, it is easy to generate molecular species tags rapidly and inexpensively, which is one of the main goals of DNA barcoding initiative (22). However, molecular evolutionary processes such as the nuclear integration of mtDNA present challenges to every step of this standard protocol, and without specific quality control measures in place, the integrity of DNA barcode sequences is seriously compromised (3). Despite obvious shortcomings because of numts, the proponents of DNA barcoding have argued that “numts have proven a minor limitation to using mitochondrial barcode in groups studied so far.” Hebert et al. (16) also suggested that the taxonomic implication of numts is small. Our study clearly demonstrates that coamplification of numts with the mtDNA orthologs is not only a major limitation of DNA barcoding, but also has significant taxonomic implications. Reported cases of numts are ever increasing and numts should not be treated as mere nuisance any more. In a showcase study of DNA barcoding, Hebert et al. (16) found that 13 individuals of skipper butterfly Astraptes fulgerator had heterozygous sequences and concluded that they were numts because the nonmitochondrial sequences of these 13 individuals (obtained by subtracting “typical sequences” from the ambiguous region) were highly similar among each other. The rationale behind this conclusion was that numts would be conserved among individuals because numts represent an ancient molecular event whereas the corresponding mtDNA sequences would be more variable. However, empirical studies of numts suggest that substitution occurs at different rates once mtDNA has been integrated into nucleus (23). Brower (24) argued against the claim by Hebert et al. (16) and suggested that the 13 haplotypes that shared heterozygosity were unlikely to be numts, but likely to be heteroplasmic mtDNA at best. We downloaded these questionable sequences from GenBank (AY666889, AY666943, AY666968, and AY667044) and looked for any sign of compositional bias and other indicators of numts. In comparison with other DNA barcodes Hebert et al. (16) used, these presumed numts had identical nucleotide composition and translated amino acid sequences as the orthologous mtDNA sequences and had no indels or in-frame stop codons. In other words, these presumed numt sequences were fully functional copies of mtDNA and are likely heteroplasmic mtDNA. With careful examination of the sequences in question, Hebert and colleagues could have easily avoided their incorrect inference.
How to Control for Numts.
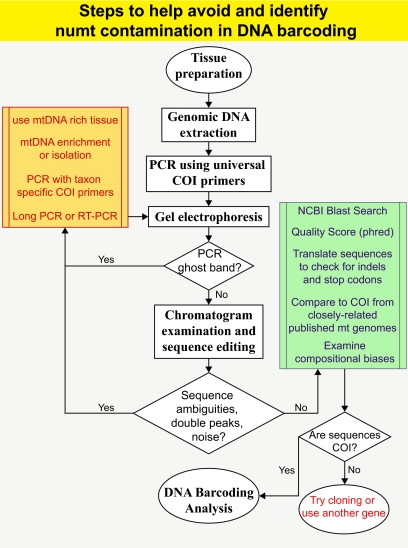
Among the few barcoding studies that did attempt to control for numts (25), researchers extracted DNA from tissues known to be rich in mitochondria and amplified slightly longer fragments (750 bp) based on the idea that numts are shorter than the barcode amplicon (26). However, muscle tissues still do contain nuclear DNA that can harbor numts and the size of numts can be highly variable and not necessarily smaller than the typical 700-bp size of DNA barcodes (10). Several methods have been suggested as means to avoid numt coamplification, including RT-PCR, long PCR, and mtDNA enrichment (8). However, these methods are often tedious, time-consuming and expensive, and their efficiency is often not high enough to totally avoid numts (27). Recent studies have questioned the universal utility of the Folmer region in DNA barcoding (28), and especially when a large number of numts are suspected in this region, it would be worthwhile to analyze additional markers other than COI gene. We recommend a number of steps that researchers should employ when using mtDNA for barcoding studies (Fig. 2). However, our suggested quality control measures against numts are neither simple nor rapid, which is at odds with the goal of DNA barcoding initiatives. DNA barcoding is a tool to aid rapid biological identification, which should be used in conjunction with other information including morphology, behavior, and ecology, and the use of other information will help reduce incorrect molecular inferences.
Fig. 2.
Suggested steps to help avoid and identify numts in DNA barcoding analysis. Whereas these steps will help reduce the chance of sequencing numts instead of the target COI, they are not guaranteed to remove all numts. Each resulting sequence must be examined as part of quality control protocols. If numts are rampant, then the isolation of COI sequences becomes difficult and it may be best to use other genes. When interpreting the results from DNA barcoding analysis, it is important to survey congruence with other molecular markers, morphology, ecology, and behavior.
Concluding Remarks.
The possible coamplification of numts is clearly a major impediment to DNA barcoding. To be fair, this is a problem that all PCR-based studies face, including phylogeography and phylogenetic studies using mtDNA. This is why both fields have largely rejected sole reliance on a single marker and emphasized congruence among multiple markers. The problem is exacerbated because the variation in the prevalence of numts appears to be a widespread phenomenon. Richly and Leister (10) surveyed numts in sequenced eukaryotic genomes and found that the number of numts ranges from none in the mosquito Anopheles gambiae to >500 in human. Although there are little or no reported cases of numts in groups such as flies (10), chicken (26), and fishes (10), a large number of eukaryotic clades including plants (29), birds (30), nonavian reptiles (31), mammals (12, 20), and arthropods (8, 11, 13, 17–19) were shown to have numts. In our study, the variation is not only clade-specific, but also species-specific and population-specific. From a barcoding perspective, the presence of numts can be disastrous. Because the DNA barcoding initiative attempts to barcode all life forms, including both organisms with known numts and other organisms that potentially have numts, this issue cannot simply be ignored. Otherwise, the number of single individuals that are inferred to be multiple species because of numt contamination may become the legacy of the DNA barcode movement.
Materials and Methods
Taxon Sampling.
To study the evolution and distribution of numts at higher-level divergence, we included four grasshopper species belonging to four different subfamilies of Acrididae. To establish the orthology of mtDNA, we used the taxa whose partial or complete mitochondrial genomes have been sequenced: Acrida willemsei (Acridinae, EU589053), Calliptamus italicus (Calliptaminae, EU589054), Locusta migratoria (Oedipodinae, EU589051), and Schistocerca americana (Cyrtacanthacridinae, EU589055). We generated numts from single individuals per species and used the same individuals that the complete mitochondrial genomes were sequenced from with an exception of L. migratoria. As outgroups, we used the COI regions of a Mormon cricket Anabrus simplex (EU589052), a cockroach Gromphadorhina portentosa (EU589049), and a termite Mastotermes darwinensis (EU589050). To study the numts at population- and species-level divergence, we included a total of 119 individuals of four closely related species belonging to the cave crayfish genus Orconectes, collected from 56 localities along the Cumberland Plateau of the Southern Appalachians: O. australis, O. barri, O. incomptus, and O. packardi. As outgroups, we included O. limosus (AF517105), Procambarus simulans (EU583575), and three species of the genus Cambarus (C. gentryi [DQ411785], C. tenebrosus [EU583576], and C. bartonii [EU583574]). COI from the complete mtDNA genome of Cherax destructor (NC_011243) was used for reference. GenBank accession numbers for the haplotypes are EU589057–EU589148 (grasshoppers) and EU583504–EU583573, EU583577–EU583678, EF207161–EF207162, and EF207165–EF207168 (crayfish). Details about numt amplification can be found in SI Methods.
Characterization of Numts.
To ensure the quality and identity, each haplotype was blasted by using MegaBLAST option against the nucleotide collection (nr/nt) as implemented in the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov/blask/Blast.cgi). Only the haplotypes that had high similarity to the COI sequence were used for the further analyses. For example, the blast search revealed that eight cloned sequences (3 from L. migratoria and 5 from A. willemsei) were of either bacterial or unknown origins and these sequences were treated as cloning error and removed from further analyses. We characterized the haplotypes in Sequencher 4.6 for length, nucleotide composition, and number of in-frame stop codons. Putative indels and point mutations were estimated by comparing the haplotype sequences against the mtDNA orthologs. The number of unique haplotypes was determined and the sequence divergence from the orthologous mtDNA sequence for each species was calculated under Kimura 2-parameter (K2P) model in MEGA 3.1 (32), as routinely used in barcoding studies, despite this model being under fit relative to the data (see below).
Data Analysis.
To study divergence pattern of numts, we performed phylogenetic analyses in both parsimony and Bayesian frameworks. For both grasshopper and crayfish datasets, the unique haplotypes and the mtDNA orthologs were aligned in MUSCLE (33) by using default parameters. For grasshopper data, we created a matrix of 69 terminals (7 mtDNA orthologs and 62 unique haplotypes) and 475 aligned nucleotides. For crayfish data, we created a matrix of 215 terminals (5 outgroups and 210 unique haplotypes of four species) and 663 aligned nucleotides. Within the parsimony framework, the aligned sequence data were analyzed with gaps treated as missing, by using search algorithms implemented in TNT (www.zmuc.dk/public/phylogeny). To assess support, we calculated standard bootstrap values based on 1,000 replicates (100 random-addition TBR replicates each) and Bremer support values, both in TNT. Within the Bayesian framework, we analyzed the datasets by using the program MrBayes 3.1 (34) after selecting best-fit models of nucleotide evolution under the AIC criteria by using MrModeltest 2.2 (program distributed by J.A.A. Nylander, Evolutionary Biology Centre, Uppsala University). The analyses consisted of running four simultaneous chains for 20 million generations for grasshopper data (GTR+G) and six simultaneous chains for 30 million generations for crayfish data (HKY+I+G), both sampling every 1,000 generations. Four independent identical Bayesian runs were performed to ensure convergence on similar results and the nodal support was assessed by using the posterior probability generated from a consensus tree of the sampled trees past burn-in determined by using Tracer 1.4 (http://beast.bio.ed.ac.uk).
To study how the presence of numts might influence the inferences from DNA barcoding, we performed a neighbor-joining (NJ) analysis under K2P model and calculated the sequence divergence among haplotypes on each dataset in MEGA 3.1 as typically used in barcoding studies (2, 16). From the NJ analyses and sequence divergence data, we then determined the number of clusters that would be considered unique species under the DNA barcoding standard (3% or higher sequence divergence). Numts are known to accumulate in-frame stop codons because they become nonfunctional after nuclear integration and are no longer under selective pressure to conserve an ORF (8). Based on the presence of in-frame stop codons, it would be possible to identify numts from the data and remove them from the analyses. We applied this method to the grasshopper and crayfish datasets and performed barcoding analyses on the reduced datasets to test whether the correct inferences would be made after such a correction.
Supplementary Material
Acknowledgments.
We thank C. Shepherd and N. Sheffield for technical assistance; H. Park for illustration; J.W. Sites for comments on the manuscript; two anonymous reviewers for improving the quality of the manuscript. This work was supported by National Science Foundation (NSF) Grants EF-0531665 to MFW and EF-0531762 to KAC.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. EU589049–EU589148, EU583504–EU583573, EU583577–EU583678, EF207161–EF207162, and EF207165–EF207168).
This article contains supporting information online at www.pnas.org/cgi/content/full/0803076105/DCSupplemental.
References
- 1.Funk DJ, Omland KE. Species-level paraphyly and polyphyly: Frequency, causes, and consequences, with insights from animal mitochondrial DNA. Annu Rev Ecol Evol Syst. 2003;34:397–423. [Google Scholar]
- 2.Hebert PDN, Cywinska A, Ball SL, deWaard JR. Biological identifications through DNA barcodes. Proc R Soc London Ser B. 2003;270:313–322. doi: 10.1098/rspb.2002.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rubinoff D, Cameron S, Will K. A genomic perspective on the shortcomings of mitochondrial DNA for “barcoding” identification. J Hered. 2006;97:581–594. doi: 10.1093/jhered/esl036. [DOI] [PubMed] [Google Scholar]
- 4.Campbell NJH, Barker SC. The novel mitochondrial gene arrangement of the cattle tick, Boophilus microplus: Fivefold tandem repetition of a coding region. Mol Biol Evol. 1999;16:732–740. doi: 10.1093/oxfordjournals.molbev.a026158. [DOI] [PubMed] [Google Scholar]
- 5.Frey JE, Frey B. Origin of intra-individual variation in PCR-amplified mitochondrial cytochrome oxidase I of Thrips tabaci (Thysanoptera: Thripidae): Mitochondrial heteroplasmy or nuclear integration? Hereditas. 2004;140:92–98. doi: 10.1111/j.1601-5223.2004.01748.x. [DOI] [PubMed] [Google Scholar]
- 6.Hurst GDD, Jiggins FM. Problems with mitochondrial DNA as a marker in population, phylogeographic and phylogenetic studies: the effects of inherited symbionts. Proc R Soc London Ser B. 2005;272:1525–1534. doi: 10.1098/rspb.2005.3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang D-X, Hewitt GM. Nuclear integrations: Challenge for mitochondrial DNA markers. Trends Ecol Evol. 1996;11:247–251. doi: 10.1016/0169-5347(96)10031-8. [DOI] [PubMed] [Google Scholar]
- 8.Bensasson D, Zhang D-X, Hartl DL, Hewitt GM. Mitochondrial pseudogenes: Evolution's misplaced witnesses. Trends Ecol Evol. 2001;16:314–321. doi: 10.1016/s0169-5347(01)02151-6. [DOI] [PubMed] [Google Scholar]
- 9.Whitworth TL, Dawson RD, Magalon H, Baudry E. DNA barcoding cannot reliably identify species of the blowfly genus Protocalliphora (Diptera: Calliphoridae) Proc R Soc London Ser B. 2007;274:1731–1739. doi: 10.1098/rspb.2007.0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Richly E, Leister D. NUMTs in sequenced eukaryotic genomes. Mol Biol Evol. 2004;21:1081–1084. doi: 10.1093/molbev/msh110. [DOI] [PubMed] [Google Scholar]
- 11.Gellissen G, Bradfield JY, White BN, Wyatt GR. Mitochondrial DNA sequences in the nuclear genome of a locust. Nature. 1983;301:631–634. doi: 10.1038/301631a0. [DOI] [PubMed] [Google Scholar]
- 12.Lopez JV, Yuhki N, Masuda R, Modi W, O'Brien SJ. Numt, a recent transfer and tandem amplification of mitochondrial DNA to the nuclear genome of the domestic cat. J Mol Evol. 1994;39:174–190. doi: 10.1007/BF00163806. [DOI] [PubMed] [Google Scholar]
- 13.Pamilo P, Viljakainen L, Vihavainen A. Exceptionally high density of NUMTs in the honeybee genome. Mol Biol Evol. 2007;24:1340–1346. doi: 10.1093/molbev/msm055. [DOI] [PubMed] [Google Scholar]
- 14.Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 1994;3:294–299. [PubMed] [Google Scholar]
- 15.Simon C, et al. Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Ann Entomol Soc Am. 1994;87:651–701. [Google Scholar]
- 16.Hebert PDN, Penton EH, Burns JM, Janzen DH, Hallwachs W. Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator. Proc Natl Acad Sci USA. 2004;101:14812–14817. doi: 10.1073/pnas.0406166101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bensasson D, Zhang D-X, Hewitt GM. Frequent assimilation of mitochondrial DNA by grasshopper nuclear genomes. Mol Biol Evol. 2000;17:406–415. doi: 10.1093/oxfordjournals.molbev.a026320. [DOI] [PubMed] [Google Scholar]
- 18.Williams ST, Knowlton N. Mitochondrial pseudogenes are pervasive and often insidious in the snapping shrimp genus Alpheus. Mol Biol Evol. 2001;18:1484–1493. doi: 10.1093/oxfordjournals.molbev.a003934. [DOI] [PubMed] [Google Scholar]
- 19.Nguyen TTT, Murphy NP, Austin CM. Amplification of multiple copies of mitochondrial cytochrome b gene fragments in the Australian freshwater crayfish, Cherax destructor Clark (Parastacidae; Decapoda) Anim Genet. 2002;33:304–308. doi: 10.1046/j.1365-2052.2002.00867.x. [DOI] [PubMed] [Google Scholar]
- 20.Mirol PM, Mascheretti S, Searle JB. Multiple nuclear pseudogenes of mitochondrial cytochrome b in Ctenomys (Caviomorpha, Rodentia) with either great similarity to or high divergence from the true mitochondrial sequence. Heredity. 2000;84:538–547. doi: 10.1046/j.1365-2540.2000.00689.x. [DOI] [PubMed] [Google Scholar]
- 21.Bensasson D, Petrov DA, Zhang D-X, Hartl DL, Hewitt GM. Genomic gigantism: DNA loss is slow in mountain grasshoppers. Mol Biol Evol. 2001;18:246–253. doi: 10.1093/oxfordjournals.molbev.a003798. [DOI] [PubMed] [Google Scholar]
- 22.Hebert PDN, Gregory TR. The promise of DNA barcoding for taxonomy. Syst Biol. 2005;54:852–859. doi: 10.1080/10635150500354886. [DOI] [PubMed] [Google Scholar]
- 23.Lopez JV, Culver M, Stephens JC, Johnson WE, O'Brien SJ. Rates of nuclear and cytoplasmic mitochondrial DNA sequence divergence in mammals. Mol Biol Evol. 1997;14:277–286. doi: 10.1093/oxfordjournals.molbev.a025763. [DOI] [PubMed] [Google Scholar]
- 24.Brower AVZ. Problems with DNA barcodes for species delimitation: ‘Ten species’ of Astraptes fulgerator reassessed (Lepidoptera: Hesperiidae) Syst Biodivers. 2006;4:127–132. [Google Scholar]
- 25.Hebert PDN, Stoeckle MY, Zemlak TS, Francis CM. Identification of birds through DNA barcodes. PLoS Biol. 2004;2:e312. doi: 10.1371/journal.pbio.0020312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pereira SL, Baker AJ. Low number of mitochondrial pseudogenes in the chicken (Gallus gallus) nuclear genome: Implications for molecular inference of population history and phylogenetics. BMC Evol Biol. 2004;4:17. doi: 10.1186/1471-2148-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thalmann O, Hebler J, Poinar HN, Pääbo S, Vigilant L. Unreliable mtDNA data due to nuclear insertions: A cautionary tale from analysis of humans and other great apes. Mol Ecol. 2004;13:321–335. doi: 10.1046/j.1365-294x.2003.02070.x. [DOI] [PubMed] [Google Scholar]
- 28.Burns JM, Janzen DH, Hajibabaei M, Hallwachs W, Hebert PDN. DNA barcodes of closely related (but morphologically and ecologically distinct) species of skipper butterflies (Hesperiidae) can differ by only one to three nucleotides. J Lepid Soc. 2007;61:138–153. [Google Scholar]
- 29.Sun CW, Callis J. Recent stable insertion of mitochondrial DNA into an Arabidopsis polyubiquitin gene by nonhomologous recombination. Plant Cell. 1993;5:97–107. doi: 10.1105/tpc.5.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sorenson MD, Quinn TW. Numts: A challenge for avian systematics and population biology. Auk. 1998;115:214–221. [Google Scholar]
- 31.Podnar M, Haring E, Pinsker W, Mayer W. Unusual origin of a nuclear pseudogene in the Italian wall lizard: Intergenomic and interspecific transfer of a large section of the mitochondrial genome in the genus Podacris (Lacertidae) J Mol Evol. 2007;64:308–320. doi: 10.1007/s00239-005-0259-0. [DOI] [PubMed] [Google Scholar]
- 32.Kumar S, Tamura K, Nei M. MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- 33.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ronquist F, Huelsenbeck JP. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.