Abstract
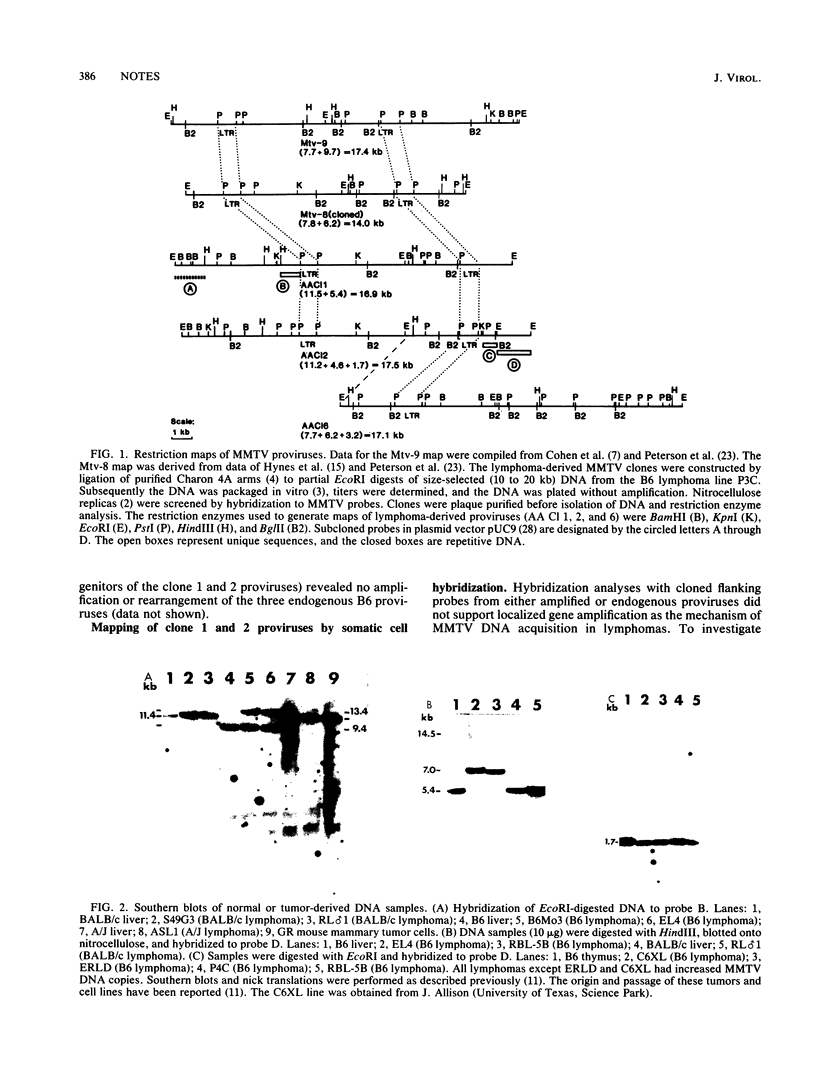
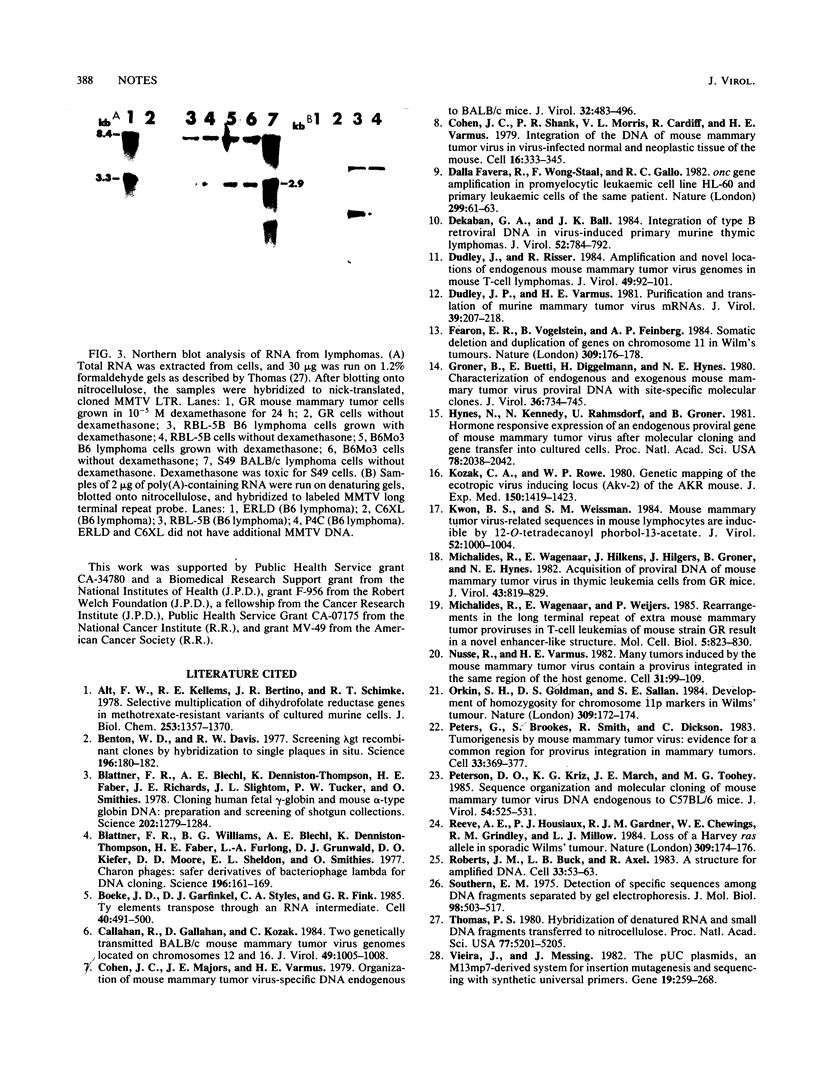
Five mouse mammary tumor virus proviruses and their flanking cellular DNA sequences have been cloned from a transplanted C57BL/6 (B6) T-cell lymphoma containing additional copies of mouse mammary tumor virus DNA. Characterization of these proviruses and their flanking DNA indicates that B6 lymphomas contain many newly integrated mouse mammary tumor virus copies synthesized by a mechanism(s) which generates polymorphism or deletions or both.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alt F. W., Kellems R. E., Bertino J. R., Schimke R. T. Selective multiplication of dihydrofolate reductase genes in methotrexate-resistant variants of cultured murine cells. J Biol Chem. 1978 Mar 10;253(5):1357–1370. [PubMed] [Google Scholar]
- Benton W. D., Davis R. W. Screening lambdagt recombinant clones by hybridization to single plaques in situ. Science. 1977 Apr 8;196(4286):180–182. doi: 10.1126/science.322279. [DOI] [PubMed] [Google Scholar]
- Blattner F. R., Blechl A. E., Denniston-Thompson K., Faber H. E., Richards J. E., Slightom J. L., Tucker P. W., Smithies O. Cloning human fetal gamma globin and mouse alpha-type globin DNA: preparation and screening of shotgun collections. Science. 1978 Dec 22;202(4374):1279–1284. doi: 10.1126/science.725603. [DOI] [PubMed] [Google Scholar]
- Blattner F. R., Williams B. G., Blechl A. E., Denniston-Thompson K., Faber H. E., Furlong L., Grunwald D. J., Kiefer D. O., Moore D. D., Schumm J. W. Charon phages: safer derivatives of bacteriophage lambda for DNA cloning. Science. 1977 Apr 8;196(4286):161–169. doi: 10.1126/science.847462. [DOI] [PubMed] [Google Scholar]
- Boeke J. D., Garfinkel D. J., Styles C. A., Fink G. R. Ty elements transpose through an RNA intermediate. Cell. 1985 Mar;40(3):491–500. doi: 10.1016/0092-8674(85)90197-7. [DOI] [PubMed] [Google Scholar]
- Callahan R., Gallahan D., Kozak C. Two genetically transmitted BALB/c mouse mammary tumor virus genomes located on chromosomes 12 and 16. J Virol. 1984 Mar;49(3):1005–1008. doi: 10.1128/jvi.49.3.1005-1008.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J. C., Majors J. E., Varmus H. E. Organization of mouse mammary tumor virus-specific DNA endogenous to BALB/c mice. J Virol. 1979 Nov;32(2):483–496. doi: 10.1128/jvi.32.2.483-496.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J. C., Shank P. R., Morris V. L., Cardiff R., Varmus H. E. Integration of the DNA of mouse mammary tumor virus in virus-infected normal and neoplastic tissue of the mouse. Cell. 1979 Feb;16(2):333–345. doi: 10.1016/0092-8674(79)90010-2. [DOI] [PubMed] [Google Scholar]
- Dalla-Favera R., Wong-Staal F., Gallo R. C. Onc gene amplification in promyelocytic leukaemia cell line HL-60 and primary leukaemic cells of the same patient. Nature. 1982 Sep 2;299(5878):61–63. doi: 10.1038/299061a0. [DOI] [PubMed] [Google Scholar]
- Dekaban G. A., Ball J. K. Integration of type B retroviral DNA in virus-induced primary murine thymic lymphomas. J Virol. 1984 Dec;52(3):784–792. doi: 10.1128/jvi.52.3.784-792.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley J. P., Varmus H. E. Purification and translation of murine mammary tumor virus mRNA's. J Virol. 1981 Jul;39(1):207–218. doi: 10.1128/jvi.39.1.207-218.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley J., Risser R. Amplification and novel locations of endogenous mouse mammary tumor virus genomes in mouse T-cell lymphomas. J Virol. 1984 Jan;49(1):92–101. doi: 10.1128/jvi.49.1.92-101.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fearon E. R., Vogelstein B., Feinberg A. P. Somatic deletion and duplication of genes on chromosome 11 in Wilms' tumours. Nature. 1984 May 10;309(5964):176–178. doi: 10.1038/309176a0. [DOI] [PubMed] [Google Scholar]
- Groner B., Buetti E., Diggelmann H., Hynes N. E. Characterization of endogenous and exogenous mouse mammary tumor virus proviral DNA with site-specific molecular clones. J Virol. 1980 Dec;36(3):734–745. doi: 10.1128/jvi.36.3.734-745.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes N. E., Kennedy N., Rahmsdorf U., Groner B. Hormone-responsive expression of an endogenous proviral gene of mouse mammary tumor virus after molecular cloning and gene transfer into cultured cells. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2038–2042. doi: 10.1073/pnas.78.4.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak C. A., Rowe W. P. Genetic mapping of the ecotropic virus-inducing locus Akv-2 of the AKR mouse. J Exp Med. 1980 Nov 1;152(5):1419–1423. doi: 10.1084/jem.152.5.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon B. S., Weissman S. M. Mouse mammary tumor virus-related sequences in mouse lymphocytes are inducible by 12-O-tetradecanoyl phorbol-13-acetate. J Virol. 1984 Dec;52(3):1000–1004. doi: 10.1128/jvi.52.3.1000-1004.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalides R., Wagenaar E., Hilkens J., Hilgers J., Groner B., Hynes N. E. Acquisition of proviral DNA of mouse mammary tumor virus in thymic leukemia cells from GR mice. J Virol. 1982 Sep;43(3):819–829. doi: 10.1128/jvi.43.3.819-829.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalides R., Wagenaar E., Weijers P. Rearrangements in the long terminal repeat of extra mouse mammary tumor proviruses in T-cell leukemias of mouse strain GR result in a novel enhancer-like structure. Mol Cell Biol. 1985 Apr;5(4):823–830. doi: 10.1128/mcb.5.4.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusse R., Varmus H. E. Many tumors induced by the mouse mammary tumor virus contain a provirus integrated in the same region of the host genome. Cell. 1982 Nov;31(1):99–109. doi: 10.1016/0092-8674(82)90409-3. [DOI] [PubMed] [Google Scholar]
- Orkin S. H., Goldman D. S., Sallan S. E. Development of homozygosity for chromosome 11p markers in Wilms' tumour. Nature. 1984 May 10;309(5964):172–174. doi: 10.1038/309172a0. [DOI] [PubMed] [Google Scholar]
- Peters G., Brookes S., Smith R., Dickson C. Tumorigenesis by mouse mammary tumor virus: evidence for a common region for provirus integration in mammary tumors. Cell. 1983 Jun;33(2):369–377. doi: 10.1016/0092-8674(83)90418-x. [DOI] [PubMed] [Google Scholar]
- Peterson D. O., Kriz K. G., Marich J. E., Toohey M. G. Sequence organization and molecular cloning of mouse mammary tumor virus DNA endogenous to C57BL/6 mice. J Virol. 1985 May;54(2):525–531. doi: 10.1128/jvi.54.2.525-531.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeve A. E., Housiaux P. J., Gardner R. J., Chewings W. E., Grindley R. M., Millow L. J. Loss of a Harvey ras allele in sporadic Wilms' tumour. Nature. 1984 May 10;309(5964):174–176. doi: 10.1038/309174a0. [DOI] [PubMed] [Google Scholar]
- Roberts J. M., Buck L. B., Axel R. A structure for amplified DNA. Cell. 1983 May;33(1):53–63. doi: 10.1016/0092-8674(83)90334-3. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Thomas P. S. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5201–5205. doi: 10.1073/pnas.77.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]