Abstract
The use of luminescent colloidal quantum dots in biological investigations has increased dramatically over the past several years due to their unique size-dependent optical properties and recent advances in biofunctionalization. In this review, we describe the methods for generating high-quality nanocrystals and report on current and potential uses of these versatile materials. Numerous examples are provided in several key areas including cell labeling, biosensing, in vivo imaging, bimodal magnetic-luminescent imaging, and diagnostics. We also explore toxicity issues surrounding these materials and speculate about the future uses of quantum dots in a clinical setting.
Keywords: quantum dot, nanoparticle, biosensor, fluorescence, imaging, immunoassay, toxicity, FRET
Introduction
Colloidal quantum dots (QDs) have completed the transition from a once curious demonstration of quantum confinement in semiconductors to ubiquitous fluorophores providing unique insights in biological investigations (Alivisatos 2004; Medintz et al 2005b; Michalet et al 2005). With well-established inorganic synthetic techniques now available for generating high quality QDs in solution, many experimentalists have focused on applications using these novel materials that exploit the numerous unique physical and optical properties of these luminescent nanoparticles. In 1998, two seminal papers appearing consecutively in an issue of Science provided the first glimpse of the vast potential of quantum dots as probes for studying biology as well as a strong impetus for their rapid growth in such applications (Bruchez et al 1998; Chan and Nie 1998). As we approach a decade since those initial reports, the current literature is rife with examples of quantum dots used in specific biological applications. This trend is directly attributable to the unique characteristics exhibited by these nanocrystals and related to considerable improvements in synthetic techniques. Although they are often portrayed simply as alternatives to organic dyes, QDs have many important distinguishing characteristics that are unlike those of molecular fluorophores (Murray et al 2000). In particular, resistance to photobleaching is unparalleled in semiconductor nanoparticles allowing them to retain their luminescent properties for remarkably long periods under continuous illumination. Perhaps most widely known is the variation of QD optical properties with particle size (Figure 1a) which permits tunable absorption and emission spectra. QDs can also be synthesized to be effectively monodispersed, leading to exceptionally narrow emission spectra with desirable symmetric profiles (cf. Figure 1b). While these principal advantages of colloidal QDs have been known for many years, only recently has the field matured to the extent that there is significant practical use of these materials.
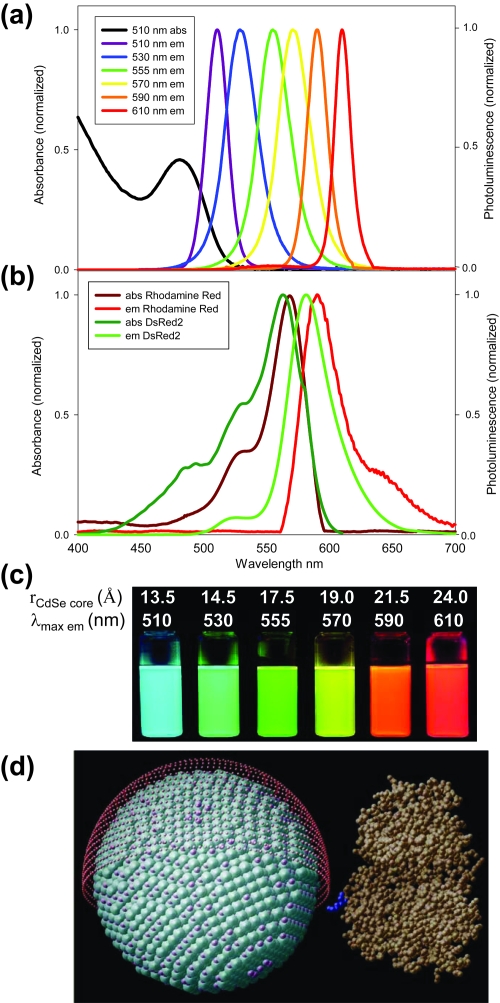
Figure 1.
Comparison of Rhodamine Red (RR) and DsRed2 spectra to those of representative samples of QDs. Multiple QD emission spectra fit within the same spectral window as that of an organic or genetically encoded dye. (a) Absorption and emission of xis6 different QD populations. The black line shows the absorption of the 510 nm emitting QDs. (b) Absorption and emission of RR, a common organic dye, and genetically encoded DsRed2 protein (Baird et al 2000). (c) Color photo demonstrating the size-tunable fluorescence properties and spectral range of the six QD dispersions plotted in (a) versus average CdSe core size. All samples were excited at 365 nm with a UV lamp. For the 610 nm emitting QDs, this demonstrates an effective Stokes shift of about 250 nm. (d) Comparison of QD size to a maltose binding protein (MBP) molecule. 555 nm emitting CdSe-ZnS core-shell QDs, diameter ~60 Å, surface functionalized with dihydrolipoic acid (red shell ~9–11 Å) has a diameter ~78–82 Å. Diagram depicts the homogeneous orientation MBP assumes relative to the QD (Copyright © 2004 National Academy of Sciences USA) (Medintz et al 2004). MBP is a midsize protein (Mr ~44 kDa) with dimensions of 30 × 40 × 65 Å (Medintz et al 2003). Source: Medintz IL, Uyeda HT, et al 2005. Quantum dot bioconjugates for imaging, labeling, and sensing. Nature Materials 4:435–46. Reproduced with permission.
Biological applications require luminescent probes that remain stably dispersed in aqueous media over a wide range of pH and ionic strengths, and accordingly there has been substantial effort devoted to generating water-soluble nanocrystals. Early studies considered aqueous phase synthesis using inverse micelles which naturally produce water-soluble QDs, however these methods have generally produced lower quality materials than methods using organic coordinating solvents (Kortan et al 1990). Fortunately, there are now numerous effective methods available for creating hydrophilic QDs post-synthesis which can be divided into two main categories: complete cap exchange or native surface modification. The former involves the displacement of hydrophobic ligands with hydrophilic moieties that coordinate with surface atoms in the outermost shell layer. In the case of CdSe-ZnS QDs, this usually entails replacing alkyl amines, phosphines, and other surface active ligands with thiols covalently linked to hydrophilic groups (Figure 2). Mono and dithiol groups have been shown to stably bind QDs with ZnS shells (Chan and Nie 1998; Mattoussi et al 2000). In select cases, these modified materials are stable in aqueous media for months to a year (or more) without significant aggregation or loss of function (Uyeda et al 2005). The latter method retains the native hydrophobic capping ligands, however an amphiphilic molecule (such as a phospholipid) is introduced that favorably associates with the hydrophobic alkyl chains while simultaneously exposing a hydrophilic group to the aqueous solvent (Dubertret et al 2002). This has the advantage of retaining the exceptional passivating features of the native hydrophobic ligands (eg, alkyl phosphines) and has to this point been the preferred procedure for commercially-produced biocompatible QDs.
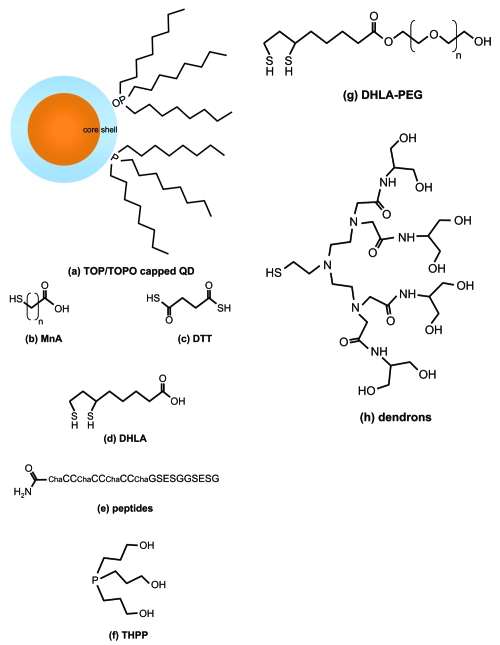
Figure 2.
Examples of surface ligands used to produce water soluble nanocrystals. Following synthesis, the hydrophobic coordinating ligands in (a) can be replaced with hydrophilic moieties through a cap exchange process. Some possibilities include: (b) mercapto n-alkyl acids (MnA; eg, mercaptoacetic acid, MAA) (Chan and Nie 1998), (c) dithiothreitol (DTT) (Pathak et al 2001), (d) dihydrolipoic acid (DHLA) (Mattoussi et al 2000), (e) peptides containing appropriate high affinity residues (e.g, Cys – C and His – H) (Pinaud et al 2004), (f) trishydroxypropyl phosphine (THPP) (Kim and Bawendi 2003), (g) dihydrolipoic acid-polyethylene oxide (DHLA-PEG) (Uyeda et al 2005), (h) dendrons (Guo et al 2003). Many of these can be processed further to crosslink the ligands, add new functional groups, and/or attach biomolecules (QD/ligands not drawn to scale).
QD bioconjugates merge the specific biological function of molecules such as DNA, proteins, and peptides with the novel photophysical properties of luminescent inorganic nanocrystals (Figure 1d). These hybrid nanomaterials are possible largely due to the solubilization methods described previously which allow biomolecules to remain stable and functional following assembly to nanoparticles. Traditional fluorescence methods (using organic dyes or rare earth metal chelates) have revolutionized many clinical methods and procedures such as diagnostic assays. Similarly, QDs have considerable potential to further improve existing methods and inspire entirely new diagnostic strategies due to their extraordinary photophysical properties. This review discusses some of the present clinical applications of luminescent quantum dots and explores their future prospects in this area.
Cell labeling
One of the broadest uses of fluorescent probes in biology is the labeling of cellular structures. Naturally, the earliest demonstrated uses of QDs in biology were to label cells with a new class of bright and stable fluorophores. Multicolor labeling of cells is a powerful technique for visualizing many of these structures simultaneously, such as cytoskeletal proteins or organelles, and to elucidate intracellular processes. Although cell labeling with organic dyes has been commonplace for decades, using multiple labels simultaneously remains a cumbersome procedure due to the narrow absorption profiles of most dyes. Effective multicolor labeling requires an assortment of filters to properly excite and collect fluorescence from specific dye molecules. Moreover, if laser excitation is used, multiple sources are typically required to excite all of the dyes labeling the cell which can be expensive and requires a complex microscopy arrangement specific to the experiment. The continuous excitation of dyes inevitably results in significant photobleaching that quenches the luminescence over short time scales (seconds to minutes). This severely limits the practical observation time for a sample, even with the addition of various anti-bleaching chemical agents (Lakowicz 1999).
By contrast, QDs are excellent fluorescent probes for long-term multicolor cell labeling. Due to their exceptionally broad absorption profiles (Figure 1a), they can be efficiently excited at any wavelength smaller than their initial band edge absorption. As the excitation photon energy increases (and wavelength decreases), the QD absorption likewise increases suggesting that extremely large effective Stokes shifts (spectral intervals between excitation and emission maxima) are possible (Leatherdale et al 2002). The effective brightness per probe particle is also superior with quantum dots as evidenced by their large molar absorption cross-sections which are a consequence of their nanometer size and composition. Perhaps the most impressive feature of QDs is their resistance to photobleaching which is quantitatively orders of magnitude superior to common organic dyes. Occasionally, QDs can even demonstrate photo-induced brightening where the luminescence quantum yield (QY) noticeably increases under continuous excitation possibly due to annealing of their surfaces. Multiplexed emission is also more spectrally isolated with QDs due to narrow and symmetric fluorescence profiles (routinely <40 nm full-width at half-maximum) which reduces signal crosstalk and increases the number of labels than can be used simultaneously in a single system (Dabbousi et al 1997). This is especially useful in quantitative fluorescence studies where spectral deconvolution is required to monitor individual signal channels (De Rosa et al 2001).
Beginning with the first successful demonstrations of QD cell labeling by the Alivisatos and Nie groups, QDs have been widely used as luminescent cell markers that identify molecular structures. However, overcoming the challenge of creating stable, water-soluble materials that could be delivered within cells was formidable. Two unique strategies were ultimately used to achieve a similar goal. Bruchez and colleagues (1998) used phalloidin to specifically target actin filaments, followed by exposure to streptavidin and biotinylated QDs to form a sandwich structure. Electrostatically-charged QDs bearing trimethoxysilylpropyl urea and acetate groups were shown to label the nucleus. Alternatively, Chan and Nie (1998) demonstrated transferrin-mediated endocytic uptake of QDs within HeLa cells which resulted in efficient and preferential delivery of transferrin-labeled QDs versus unlabeled QDs. These early studies proved that QDs could perform similar to organic dye fluorophores within cells and provided initial guidance regarding successful strategies for efficient cell delivery.
Many examples have since followed where QDs have been used to label cellular structures both within and external to the cell membrane. Using conventional methods, QDs have been delivered inside cells via receptor-mediated pathways where specific ligands were attached to QDs to induce cellular uptake, as well as nonspecific endocytosis (ie, pinocytosis) where cells were incubated with a concentrated QD solution (Jaiswal et al 2003). Delivery of QDs via endocytosis has inherent benefits because it invokes native cellular mechanisms to transfer nanoparticles across the cell membrane. It is consequently thought to be the least disruptive delivery method as well as the most versatile. QDs capped with compact carboxylated ligands have shown reasonable rates of nonspecific endocytosis (Jaiswal et al 2003), however receptor-mediated uptake has demonstrated better efficiency by specifically targeting receptors displayed on the cell surface (Chan and Nie 1998; Lidke et al 2004; Bharali et al 2005). As expected, ligands previously identified to cue and facilitate receptor-mediated endocytosis continue to function effectively when attached to QD surfaces resulting in efficient delivery of the modified nanoparticle assembly within cells, but with the potential drawback of the cargoes remaining largely confined inside the endosomes (Delehanty et al 2006). If the intended destination of a QD bioconjugate is a specific organelle or general release into the cytosol, this perpetual confinement presents a significant obstacle.
Alternative delivery methods that introduce microscopic mechanical defects in the cell membrane are also options for inserting nanoparticle cargoes, but with an elevated risk of traumatizing the cell which may compromise long term viability and perturb normal function. Electroporation and microcapillary injection have been used to introduce nanoparticles into live cells with varying success (Dubertret et al 2002; Derfus et al 2004a; Uyeda et al 2005). In particular, microcapillary injection is suitable for introducing well-dispersed QD bioconjugates directly into the cytosol which could be desirable for certain sensing applications. However, the serial nature of this method is time-consuming and therefore considerably limits the number of cells that can be labeled in a practical period and subsequently isolated. Other methods including ballistic delivery and scrape loading are also possibilities, but have not been widely reported.
With the availability of commercial labeling kits, the prevalence of QDs in cell labeling applications has increased dramatically. Despite their popularity and success, commercial materials currently have somewhat limited potential due to the use of specific proprietary coatings and surface ligands to passivate and stabilize the nanoparticles. Similar to the challenges encountered in gene therapy applications, given the complexity of targeted delivery and function within cells it is likely that more flexible and customized QD systems will be required for long-term intracellular imaging applications.
Biosensing and energy transfer
Due to their unique physical and optical properties, colloidal QDs have been used to develop new methods of biosensing. By attaching biomolecules to the QD surface, it is possible to generate complex bioconjugates that merge biological specificity and function with the desirable optical characteristics of QDs. In many cases the nanometer size of QDs allows the nanocrystal to become a central structural component that can accommodate numerous copies of a particular biomolecule (eg, protein or DNA) or several different biomolecules simultaneously; as a result, these bioconjugates are sometimes referred to as “nanosensors.” As robust fluorophores, QDs are compatible with conventional biosensing techniques that implement fluorescence to produce a large measurable signal. Fluorescence resonance energy transfer (FRET) in particular has been a popular method of signal transduction due to its sensitivity to molecular scale interactions (Lakowicz 1999; Clapp et al 2004; Medintz et al 2004). An early FRET-based technique was demonstrated by Patolsky and colleagues (2003) for the study of telomerization and DNA replication dynamics. Thiolated DNA was attached to the surface of water-soluble CdSe-ZnS QDs. In the presence of appropriate enzymes (telomerase or Klenow fragment) and dye-labeled nucleotides, the emission spectra showed a time dependent red-shift indicating efficient energy transfer from QD to dye specifically due to telomerization or replication, respectively. The telomerization results were corroborated with AFM images showing growth of DNA extending from the QD surface. The same group later looked at QD-based FRET for detecting DNA hybridization and cleavage (Gill et al 2005). They noted that full recovery of the fluorescence emission following the addition of enzyme was not possible due to nonspecific interactions between the dyes and nanoparticles. This could be a significant limitation in this particular arrangement, however nonspecific interactions might be reduced through unique surface passivation methods.
Medintz and colleagues (2003) used a related approach to develop a prototype FRET-based QD biosensor capable of detecting the nutrient sugar maltose in solution. Maltose binding protein (MBP), pre-bound to an analog sugar-dye complex, was conjugated to water-soluble QDs resulting in many MBP attached to each QD. In this initial state, fluorescence from the QD was significantly quenched by the nonemissive dye. As maltose is added to solution, the analog sugar-dye complex is progressively displaced resulting in a substantial increase in fluorescence signal with increasing concentration. A reagentless implementation of a maltose biosensor was also demonstrated where the QD transfers energy to a Cy3 dye covalently attached near the binding pocket of MBP (Medintz et al 2005a). Sensing is achieved by monitoring the Cy3 fluorescence as a function of maltose concentration where the quantum yield of the dye varies due to differences in solvent interaction with the dye between the bound and unbound states.
In another demonstration of FRET-based biosensing, Medintz and colleagues (2006) developed QD-peptide bioconjugates designed to measure rates of enzymatic digestion. Peptides were synthesized to have domains specific for QD attachment, structural rigidity, protease recognition, and dye labeling. In a manner similar to the maltose biosensor, the intact QD bioconjugates exhibit quenched fluorescence due to the proximity of the dye labels. However, as an appropriate enzyme is added to solution, some of the peptides are cleaved allowing the dyes to diffuse away from the QD surface resulting in an increased fluorescence signal. By altering the recognition sequence (consisting of 2–4 carefully chosen amino acid residues), the biosensor can be tuned to respond to a specific enzyme. More recently, Shi and colleagues (2006) demonstrated a QD FRET-based probe for enzymatic activity using compact dye-labeled tetra-peptide ligands. The probe was able to detect increased metalloproteinase activity in the extracellular matrix surrounding cancerous breast cells. A very recent communication by Gill and colleagues (2006) reports a multifunctional enzymatic probe for tyrosinase and thrombin. The former is observed following the oxidation of tyrosine near the surface of a QD, and the latter by specific scission of a linker peptide which interrupts energy transfer.
An analogous signal transduction method using charge to modulate QD fluorescence was demonstrated by Sandros and colleagues (2005) to detect maltose in solution. Similar to the FRET-based maltose biosensor described above, this method utilizes MBP to specifically recognize the disaccharide. However, in this system a Ru complex transfers charge to the nearby QD thus quenching fluorescence when the protein is in an unbound/open conformation. As MBP binds maltose in solution, the hinge-bending movement of the protein positions the Ru complex sufficiently far away from the QD surface to disrupt charge transfer and enable fluorescence emission from the exciton. In a slightly different configuration, the same group developed a nanosensor selective for the fatty acid palmitate using QDs bound to intestinal fatty acid binding protein (Aryal and Benson 2006). In this arrangement, it is thought that a solvent occupancy effect, rather than a protein conformational change, is responsible for modulating the QD fluorescence. In both systems where a Ru complex is conjugated to QDs via a protein, the sensing method appears to be general for many protein-ligand pairs where charge transfer is responsible for signal transduction; however, the exact mechanism has yet to be fully elucidated.
Collectively, the preceding methods demonstrate the versatility and functionality of QDs in fluorescence biosensors. Energy transfer in these systems allows sensitive measurements of events such as binding, digestion, or conformational changes occurring on the molecular scale. Many of the benefits of QDs are unique and allow development of nanosensors that can retain their function over long periods and potentially in living tissue. Most of the reports in the literature are preliminary demonstrations, yet they offer a tantalizing preview of what is possible using QDs.
In vivo imaging
The ability to visualize native processes occurring in living organisms is invaluable for clinical diagnostic applications, yet remains elusive in practice due to conventional imaging limitations and the availability of suitable fluorescence markers. Regarding the latter, many organic dyes have very short lifetimes (~1 ns), are susceptible to photodegradation, and show inadequate fluorescence brightness (Lakowicz 1999). Additionally, tissue autofluorescence can exhibit similar spectroscopic characteristics making it difficult to resolve the desired signal from unwanted background. Due to their unique photophysical properties, QDs are promising fluorophores for in vivo fluorescence imaging and can overcome many of the usual limitations of dyes.
An early study by Akerman and colleagues (2002) examined targeted delivery of QDs to specific locations within live mice, however fluorescence imaging was performed after the animal was sacrificed and the tissue sectioned. Noninvasive, real-time in vivo fluorescence imaging requires exciting fluorophores and detecting their emission through tissue which is invariably hindered by scattering and absorption of both the excitation and emission wavelengths. Although CdSe-ZnS QDs are by far the most common choice for high quantum yield nanocrystals, their visible fluorescence (restricted within a range from 470 to 650 nm) is not well-suited for imaging through tissue. Nonetheless, Gao and colleagues (2004) showed that CdSe-ZnS QDs could be used to image human tumors implanted in mice by implementing an image processing algorithm to isolate signal contributions from QDs and autofluorescence. However, their method was limited to imaging structures very near the surface due to the limited penetration depth of visible wavelengths. A similar study was reported by Ballou and colleagues (2004) where they used several imaging methods, including transmission electron microscopy, to explore long term QD partitioning in tissue.
Using a different semiconductor core material and specifying an appropriate nanocrystal size, QDs can be excited and observed in the near-infrared (near-IR or NIR) which coincides with a range of optical transparency for living tissue (Figure 3a) (Lim et al 2003). Due to their size and composition, QDs exhibit an unusually large molar extinction coefficient which leads to improved absorption of photo-excitation and effective brightness. Size tuning can further optimize the peak emission wavelength for maximum fluorescence penetration through relatively thick tissue (>1 cm). This concept was first demonstrated by Kim and colleagues (2004) using so-called “Type II” QDs composed of core-shells with offset bandgaps. In this study, near-IR QDs were used to identify sentinel lymph nodes (directly invaded by metastatic cancer cells) in mice. Using a filtered halogen source and an IR camera, the collection of QDs within tissue was monitored in real-time to identify a region for surgical resection (Figure 4). The size of these nanocrystals (~15–20 nm diameter) was shown to be ideal for their efficient collection and retention in lymph nodes. This study demonstrates the feasibility of near-IR imaging for real-time surgical assistance, however the toxicity issues (to be discussed) associated with most QD materials remain a formidable hurdle to use in humans.
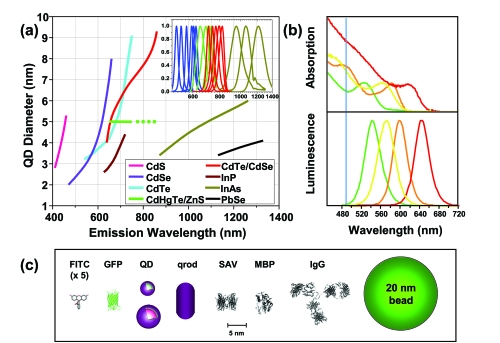
Figure 3.
(a) Emission maxima and sizes of quantum dots of different composition. Quantum dots can be synthesized from various types of semiconductor materials (II-VI: CdS, CdSe, CdTe; III-V: InP, InAs.; IV-VI: PbSe) characterized by different bulk band gap energies. The curves represent experimental data from the literature on the dependence of peak emission wavelength on QD diameter. The range of emission wavelength is 400 to 1350 nm, with size varying from 2.0 to 9.5 nm (excluding the organic passivation/solubilization layer). All spectra are typically around 30 to 50 nm (full width at half maximum). Inset: Representative emission spectra for some materials. Data for CdHgTe-ZnS have been extrapolated to the maximum emission wavelength obtained in the Weiss group. (b) Absorption (upper curves) and emission (lower curves) spectra of four CdSe-ZnS QD samples. The blue vertical line indicates the 488 nm line of an argon-ion laser, which can be used to efficiently excite all four types of QD simultaneously. (c) Size comparison of QDs and comparable probes/biomolecules. FITC, fluorescein isothiocyanate; GFP, green fluorescent protein; QD, green (4 nm, top) and red (6.5 nm, bottom) CdSe-ZnS QD; qrod, rod-shaped QD (sizes reported on Invitrogen’s web site). Three proteins – streptavidin (SAV), maltose binding protein (MBP), and immunoglobulin G (IgG) – have been used for further functionalization of QDs and add to the final size of the QD, in conjunction with the solubilization chemistry. Copyright © 2005 AAAS. Michalet X, Pinaud FF, et al 2005. Quantum dots for live cells, in vivo imaging, and diagnostics. Science, 307:538–44. Reproduced with permission.
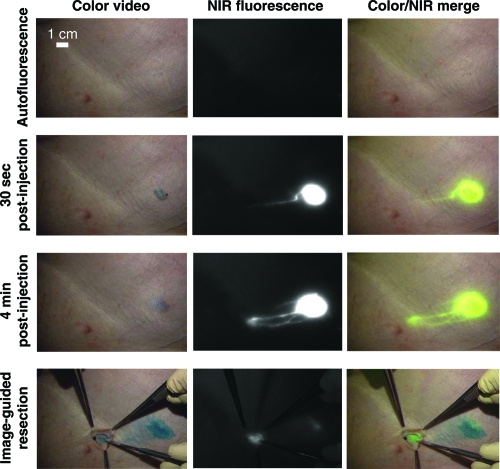
Figure 4.
NIR QD imaging in vivo. Images showing the surgical field of a pig injected intradermally with 400 pmol of NIR QDs in the right groin. Four time points are shown from top to bottom: before injection (autofluorescence), 30 s after injection, 4 min after injection and during image-guided resection. For each time point, color video (left), NIR fluorescence (middle) and color-NIR merge (right) images are shown. Note the lymphatic vessel draining to the sentinel node from the injection site. Copyright © 2004 Nature Publishing Group. Kim S, Lim YT, Soltesz EG, et al 2004. Near-infrared fluorescent type II quantum dots for sentinel lymph node mapping. Nature Biotechnology 22:93–7. Reproduced with permission.
In vivo fluorescence imaging has undoubtedly benefited from the development of QD fluorophores where their unique properties allow more flexible imaging strategies. For example, the expanded use of multi-photon microscopy can provide a more efficient method of exciting QDs in vivo. This is due to the exceptional multi-photon absorption cross-section of semiconductor nanocrystals (Larson et al 2003). In certain cases, the effective brightness of QD fluorophores is two or more orders of magnitude higher than organic dyes using multi-photon excitation. By preferentially exciting fluorophores very near the highly focused excitation beam, high-resolution fluorescence images can be generated while eliminating out-of-focus and autofluorescence contributions. This method has yet to be fully realized, however. Other in vivo imaging modalities such as magnetic resonance imaging (MRI) and positron emission tomography (PET) could also benefit from QDs (Michalet et al 2005).
Diagnostics
One medical area where QDs may have significant impact is in diagnostics and clinical assays. The unique properties of QDs have been investigated almost exclusively for two techniques that require the use of diagnostic fluorophores: immunolabeling and nucleic acid detection. As QDs are a relatively new class of materials, the focus has been primarily on proof-of-concept demonstrations with various bioconjugation approaches being tested.
Immunolabeling
For the purposes of this review, we define immunolabeling loosely as the use of antibody-driven specific binding to a target protein or biomolecule and the visualization of this event with some form of QD labeling. Table 1 presents a semi-comprehensive list where QDs and immunolabeling have been used for mostly proof-of-concept diagnostic purposes.
Table 1.
Diagnostic related uses of quantum dots
| Target/biomarker | Tissue/cell type | Labeling configuration | Comment | Ref |
|---|---|---|---|---|
| Respiratory syncytial virus | HEp-2 cells | Commercial streptavidin-coated QDs | Track intracellular viral proteins | (Bentzen et al 2005) |
| Red blood cell antigens | Erythrocytes | Glutaraldehyde functionalized QDs bound to monoclonal anti-A | Monitor A-antigens | (de Farias et al 2005) |
| c-Abl proteins | K-562 leukemia cells | Uncapped CdSe QDs; broad white emission | Western blot and flow cytometry analysis | (Zhelev et al 2006) |
| Biotinylated cholera toxin | Plasma membrane of neurons | Avidin-coated QDs | Indirect staining of GM1 gangliosides | (Jaiswal et al 2004) |
| B- and T-cell antigens | Fixed lymph nodes | Commercial IgG-coated QDs | Simultaneous detection | (Zahavy et al 2005) |
| p-glycoprotein | Breast adenocarcinoma cells | QDs immobilized within polymeric beads | Monitor multidrug resistance phenotype | (Stsiapura et al 2004) |
| CA-125 tumor marker | Ovarian cells | Commercial streptavidin-coated QDs | Monitor ovarian carcinoma | (Wang et al 2004) |
| PMP70 peroxisomal membrane proteins | Liver cells | Commercial streptavidin-coated QDs | Monitor hypolipidemic drug therapy | (Colton et al 2004) |
| Metastatic potential | Human mammary epithelial tumor cells | SiO2-capped CdSe QDs | Correlated uptake of QDs during cellular migration with cells metastatic potential | (Parak et al 2002) |
| Band 3 protein | Human erythrocytes | Commercial antibody-coated QDs | Could monitor invasion of a malaria parasite | (Tokumasu and Dvorak 2003) |
| Mortalin | Human fetal fibroblasts | Commercial streptavidin-coated QDs | Monitor differential staining in cancer | (Kaul et al 2003) |
| p-glycoprotein/cytokeratin | Breast adenocarcinoma cells/skin basal carcinoma | Polyallylamine coated QDs; Antibody attached via EDC chemistry | Demonstrated superior photostability for QDs | (Sukhanova et al 2004) |
| HER2 cancer marker | Breast cancer cells | Commercial streptavidin and IgG linked QDs | Labeled in both live and fixed cells | (Wu et al 2003) |
| Epidermal growth factor (EGF) receptor | Breast cancer cells | CdTe QD-coated magnetic polystyrene nanospheres coated with EGF | Labeling and magnetic separation | (Chu et al 2006a) |
| Carcinoembryonic antigen | Human colon carcinoma cells | PEG modified CdTe QDs | Antibodies noncovalently associated with the QDs | (Hu et al 2006) |
| Prostate specific membrane antigen (PSMA) | Prostate tumor cells | Commercial streptavidin-coated QDs with biotinylated aptamers specific for PSMA | PSMA detected in both fixed and live cells | (Chu et al 2006b) |
| CD36 | Human monocytic cells and atherosclerotic tissue sections | Commercial QDs | Flow cytometry and confocal microscopic analysis | (Kahn et al 2006) |
| CD4/CD8 ratio | Peripheral blood | Commercial QDs | Automated microscopy with four-color labeling | (Bocsi et al 2006) |
Abbreviations: EDC, 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride; PEG, polyethylene glycol; QD, quantum dots.
Many of the reported examples target the detection of various cancer markers within cells including c-Abl, CA-125, p-glycoprotein, mortalin, cytokeratin, prostate specific membrane antigen, and carcinoembryonic antigen (see Table 1). These markers are associated with transformation or metastasis and are usually indicative of both the cancer and its progression and are thus of pathological interest. Other uses include intracellular viral monitoring, blood cell antigen typing, visualizing drug therapy effects on cellular metabolism, and monitoring of cell markers. The only prominent nonimmunological exception is an example where the metastatic potential of cell lines was shown to correlate well with their ability to take up QDs from culture wells during migration (Parak et al 2002). However, this is more of a scientific curiosity and will likely never find use as a prognostic indicator.
For almost half of the examples cited in Table 1, commercially available QDs (Quantum Dot Corporation/Invitrogen) have been used which are either purchased pre-coated with streptavidin (for conjugation with biotinylated antibodies) or some type of species-specific IgG. This suggests that for many of these immunolabeling uses, commercial QD materials may be adequate and thus obviate the need to synthesize, solubilize, and functionalize custom materials. The greatest potential of QDs in this area is “multiplexing” or the simultaneous detection of multiple targets, since multiple colors of QDs can be excited with a single wavelength far removed from their respective emissions (Figure 3b). Indeed, examples have been reported of simultaneous cellular labeling with up to five QD colors (Medintz et al 2005b). In terms of pathology and diagnosis, this would enable the colocalization of many markers within the same cells/tissues and allow determination of their spatial distribution. Based on the typical emission spectra of QDs, it is reasonable to anticipate ten or more colors of QDs (or QDs and dyes) could be used simultaneously. Furthermore, if a UV excitation source is used in combination with more red-emitting QDs (approaching the near-IR), brighter signals can be achieved as the molar extinction coefficients of these materials increases significantly towards the UV. However, the limiting barrier is bioconjugation. Streptavidin-coated QDs will bind any biotinylated molecule indiscriminately and thus each “color” or antibody-labeled QD must either be prepared separately or a different and specific attachment scheme must be found for each combination of antibody and QD. For example the use of anti-mouse IgG coated QDs (targeting mouse-derived antibodies) in combination with anti-goat IgG coated QDs (targeting goat-derived antibodies) and streptavidin coated QDs, etc. This is not easily accomplished as there are currently a limited number of such QD coatings available, and this will be further complicated by the choice of specific antibody for a particular experiment. Although antibody cross-reactivity is a serious issue that can further limit any potential “multiplexed” immunological assays, Goldman and colleagues (2004) were still able to demonstrate four-color simultaneous small molecule toxin detection using a commercial plate reader and one excitation wavelength. Although not in vivo diagnostics per se, this did demonstrate the strong potential of QD usage in combination with antibodies.
Nucleic acid detection
From a diagnostic standpoint, QDs have been used in this area primarily as a visualization tool for nucleic acid array detection, in homogenous mutation assays, or as the fluorophore in fluorescence in situ hybridization (FISH). Again, this work has been largely “proof of concept” with several different bioconjugation strategies tested. For nucleic acid arrays, the majority of studies have attempted to combine the ability to multiplex with the increased sensitivity derived from QD photostability to improve detection and reduce both the sample and probe concentrations required. Gerion and coworkers demonstrated single nucleotide polymorphism (SNP) of p53 tumor suppressor gene mutations and multiallele detection of hepatitis B and C virus genes with a commercial scanner and two colors of QDs (Gerion et al 2003). In a strong demonstration of QD potential for multiplexed analysis, Shepard accomplished a simultaneous “eight-plex” array consisting of both QDs and commercial cyanine dyes for co-detection of Bacillus anthracis genes (Shepard 2006). A number of other groups have also used Qbeads or QDs embedded in microspheres to simplify the sample handling, expand the number of available colors by using codes consisting of QD mixtures in each bead, and increase the relative number of QDs hybridized per spot and hence the signal. Xu and colleagues (2003) focused on using such Qbeads for SNP typing while Eastman and colleagues (2006) demonstrated improved sensitivity in gene expression arrays.
Homogenous mutation assays based on two-color fluorescence coincidence analysis have also been reported (Zhang et al 2005; Yeh et al 2006; Zhang and Johnson 2006). This method labels the target and the probe with different QDs, or QD-dye combinations and then detects the presence of both by their coincidence in a small focal volume. FRET configurations of QDs and dyes have also been used in this type of analysis (Zhang et al 2005). The principal benefit of this format is that it can have single molecule sensitivity obviating the need for sample or signal amplification. In QD-based FISH analysis, the focus has been to show that QDs could be superior to conventional fluorophores due to increased photostability and the ability to multiplex or both. Xiao and Barker demonstrated increased photostability of QD based probes versus conventional dyes for the probing of human metaphase chromosomes (see Figure 5). Chan and colleagues, and Bentolila and Weiss also demonstrated increased sensitivity and the ability to multiplex in these assay formats (Chan et al 2005; Bentolila and Weiss 2006). These results indicate that the relatively large QD-DNA probe complexes can still penetrate the tight cellular and chromosomal structures while retaining hybridization specificity. In contrast to the bioconjugation issues mentioned above for immunolabeling, the use of streptavidin QDs coupled to biotinylated DNA appears to suffice for this type of analysis. However, several groups have commented on the need for long linkers between the DNA probe portion and the QD (Bakalova et al 2005; Chan et al 2005). It is not unreasonable to expect more studies using FISH analysis with QDs in the near future as researchers become increasingly interested in multiplexing simplified optical detection schemes.
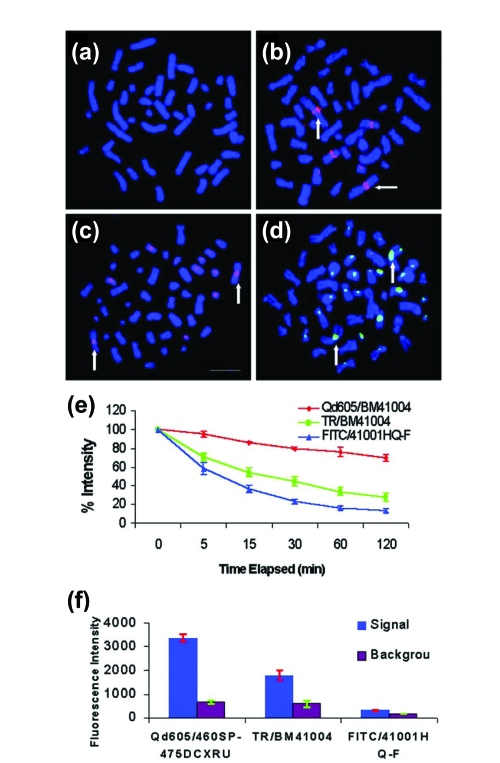
Figure 5.
Specificity of fluorophore – streptavidin detection of biotinylated total human DNA probe in metaphase chromosomes and photostability (Mulder et al 2006). (a) Control (no fluorophore – streptavidin conjugate); (b) streptavidin – Qdot 605 detection of chromosome 1q12 region (vertical and horizontal arrows); (c) Texas Red – streptavidin detection of biotinylated DNA hybridized to 1q12 (vertical arrows) and (d) FITC – streptavidin detection of 1q12 sites (vertical arrows). Bar in panel (c) is 10 μm. (e) Signal decay upon continuous illumination with fluorescence microscope/mercury illumination in metaphase chromosome band 1q12 during 2 h continuous illumination. Red is Qdot 605, green is Texas Red, and blue is FITC. (f) Total intensity of whole interphase nuclei during 120 ms illumination (blue bars) and background (red bars). N = 3 cells in each. Copyright © 2004 Oxford University Press. Xiao Y, Barker PE. 2004. Semiconductor nanocrystal probes for human metaphase chromosomes. Nucleic Acids Research, 32:e28. Reproduced with permission.
Toxicity
The issue of QD toxicity presents perhaps the most serious obstacle to a full exploration of their in vivo usage in biomedical imaging. The toxicity arises primarily from two sources: 1. the semiconductor materials that commonly constitute the QD core (and sometimes the overcoating shell) which can leach under certain circumstances (Derfus et al 2004b), and 2. the generation of reactive and free radical species during excitation (Bakalova et al 2004a, 2004b). The toxicity of metals such as Cd and Se is well documented (Colvin 2003; Hoet et al 2004; Hardman 2006; Nel et al 2006). These heavy metals can cross the blood-brain barrier, can accumulate in adipose tissue with biological excretion half-lives greater than ten years, are primarily toxic to the liver and kidneys, and are considered possible teratogens and probable carcinogens (Colvin 2003; Hoet et al 2004; Hardman 2006; Nel et al 2006). In addition to this direct toxicity source, the unique QD nanoscale structure presents a complex set of physiochemical characteristics that further compounds any simple studies or conclusions in this area. As detailed previously, the nanocrystalline cores can be constituted from different combinations of binary semiconductors such as CdSe, CdTe, CdS, and InP. Further, the cores are commonly encapsulated with a secondary semiconductor material and are then functionalized with a variety of surface coating ligands including small thiolated molecules or larger amphiphilic polymers for aqueous compatibility (Medintz et al 2005b; Michalet et al 2005). Additionally, even though the physical dimensions are on the nanoscale, the QDs can have a wide range of sizes with diameters ranging from <2 nm to greater than 10 nm; the latter is beyond the size limit that is passively excreted by the kidneys (Hoet et al 2004; Michalet et al 2005; Hardman 2006; Nel et al 2006). For biological use, QDs can be further modified with either proteins, such as NeutrAvidin, or other biomolecules such as DNA. Cumulatively, this combination of materials and physical properties serves to confound any systematic study of toxicity, even before issues such as dosage or exposure time are formally addressed. Nevertheless, a body of literature is slowly accumulating that is contributing to our understanding of this important issue.
Although much of the early research in this area was collected piecemeal, a number of excellent review articles have been published that summarize much of what is known in this area. Hoet and colleagues (2004) reviewed most of the data available up until 2004 on the health effects of nanoparticles. The authors point out that upon casual exposure, nanoparticles would enter the body mainly via the lungs and intestines and to a lesser extent the skin. These routes are primarily attributable to the small size of the materials. Hardman (2006) has recently authored one of the most extensive reviews on the toxicity of semiconductor QDs in which the cumulative results from almost all previous in vivo studies (both cellular and small animal) are summarized. This study pays particular attention to the effects of different core-shell materials and their solubilizing ligands. The results that are summarized in these reviews reflect publications that are primarily observational where the authors have reported the effects of a given QD material on a particular cell line or animal, at some specified concentration(s) for some exposure time (Hoet et al 2004; Hardman 2006). In general, the results were mixed with some reporting no visible toxicity (Jaiswal et al 2003) while others reported high cytotoxicity (Hoshino et al 2004). This can be interpreted to reflect what is posited above: the choice of core-shell material and solubilizing cap in conjunction with dosage/exposure time will obscure any simple assessments of toxicity. It is also notable that most of the cell lines used in these studies were transformed (immortal and primarily derived from different cancers) and thus might not reflect either true toxicity or even a normal cellular response.
Recently, several reports have appeared that have focused exclusively on QD toxicity with more systematic approaches. Chan’s group has reported on the distribution, sequestration, and clearance of mercaptoundecanoic acid (MUA) functionalized QDs in rats (Fischer et al 2006). They found that QDs coated with bovine serum albumin (BSA) are cleared more rapidly from serum than those coated with crosslinked lysine. Further, most of the BSA coated QDs (99%) are found in the liver after 90 minutes as compared to the lysine QDs (40%). This result reflects, perhaps, one of the primary metabolic roles of endogenous serum albumin. Interestingly, these QDs were not excreted from the rats even after 10 days. In another study, Chen’s group undertook a global molecular and cellular response investigation of skin and lung fibroblast cell lines when exposed to QDs coated with crosslinked silane and polyethylene glycol (PEG) (Zhang et al 2006). The authors found negligible toxicity in these model cells and attribute this result to appropriate sequestration of the core-shell material and the biocompatibility of the surface-bound PEG. Mattoussi’s group has reported on the dosage effects of QDs on different cell lines (Delehanty et al 2006). They found that acute (1 hour) versus chronic (continuous) exposure and labeling of different concentrations of QDs to two cell lines resulted in a significantly different impact on subsequent cellular viability. No toxicity was noted for acute exposure at all concentrations while chronic exposure manifested significant toxicity and loss of viability with increasing concentration of materials.
More generally, it is important to reiterate that almost all synthetic precursor materials used to make semiconductor QDs are toxic. Some can be physically dangerous since they are pyrophoric, while others are only toxic upon ingestion or prolonged exposure (Medintz et al 2005b). Hardman provides some fascinating perspective on synthetic scales and dosage issues by pointing out that a 2 g synthesis of 100 nm diameter particles contains enough material to dose every human on earth with around 300,000 particles (Hardman 2006). This highlights the importance of proper laboratory procedures during synthesis and appropriate disposal practices as more QD materials are synthesized worldwide. Researchers interested in determining the toxicity of a particular QD (or other nanoparticle) are not without resources. Foreseeing a need in this area, the National Cancer Institute (NCI) in conjunction with several other US health agencies have established the Nanotechnology Characterization Laboratory (NCL) to perform preclinical efficacy and toxicity testing of nanoparticles (http://ncl.cancer.gov/). As part of its services, the NCL will characterize nanoparticles’ physical attributes, their in vitro biological properties, and their in vivo compatibility. The time required to characterize a particular nanomaterial is anticipated to be around 18 months. The Center for Biological and Environmental Nanotechnology (CBEN), a National Science Foundation (NSF) funded Nanoscale Science and Engineering Center at Rice University, provides information and focuses research on the environmental impact and toxicity of a variety of nanomaterials including QDs (http://cohesion.rice.edu/centersandinst/cben/index.cfm). The International Council on Nanotechnology (ICON), CBEN’s multi-stakeholder initiative, provides another valuable resource by maintaining a nanomaterial-specific Environmental, Health, and Safety Database that can be accessed by the public ( http://icon.rice.edu/research.cfm). The continuing debate in this area mirrors that of the general debate on the long-term environmental and health effects of all nanoscale materials (Colvin 2003; Nel et al 2006).
Bimodal luminescent-magnetic quantum dots for imaging
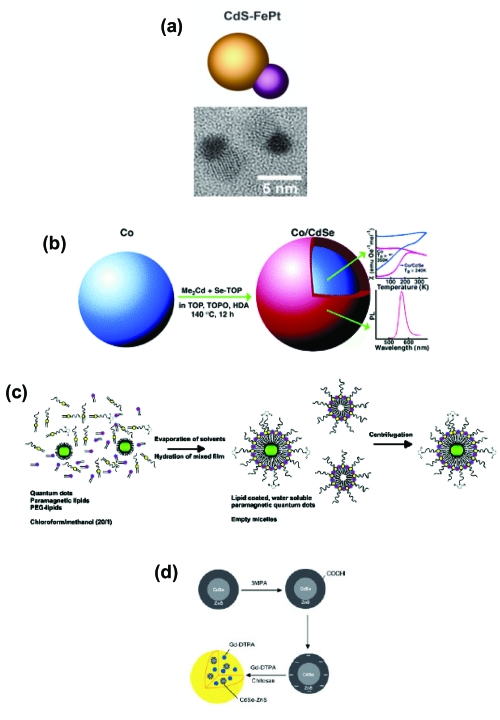
As noted throughout, QDs are excellent deep tissue and in vivo imaging/contrast agents due to several key properties. Although this field is still in its infancy, an application based on combining QD fluorescent imaging with magnetic resonance imaging (MRI) to deliver superior contrast and possibly detailed three-dimensional in vivo images has already emerged. The bulk of the research in this area has focused on synthesis and characterization. A variety of strategies exist for creating such bimodal materials; these are highlighted in Figure 4. In a one example, Gu and colleagues were able to generate a nanocrystalline luminescent/magnetic CdS/FePt heterodimer in a stepwise manner (Figure 6a; Gu et al 2004). Klimov’s group took a slightly different approach by growing a CdSe shell over a Co core (Figure 6b; Kim et al 2005). Rather than using a direct synthesis route, Mulder and colleagues (2006) coated CdSe-ZnS core-shell QDs with a PEGylated phospholipid and a gadolinium diethylene triamine pentaacetate-bisstearylamide (Gd-DTPA-BSA) paramagnetic lipid (Figure 6c). Also bypassing a direct synthesis, Tan and Zhang (2005) encapsulated CdSe-ZnS core-shell QDs and paramagnetic Gd-DTPA chelates into chitosan nanobeads (Figure 6d). Most of these materials have only undergone initial testing to demonstrate that they still retain the parent materials’ native luminescent and magnetic properties as synthesized and after intracellular delivery. Some remaining issues before wide-spread implementation include low QD quantum yield and appropriate relaxivity. Despite this, we can expect the great interest in this area to drive the synthesis of a variety of related materials and will shortly begin to see reports of increasingly more detailed in vivo images that have been generated with these bimodal materials. Of particular interest will be the ability to combine these bimodal probes with tumor specific antibodies for in vivo delivery and targeting.
Figure 6.
Strategies for creating bifunctional luminescent-magnetic QD materials. (a) Schematic and TEM image of heterodimeric luminescent/magnetic CdS/FePt nanoparticles. Copyright © 2004 American Chemical Society. Gu H, Zheng R, et al 2004. Facile one-pot synthesis of bifunctional heterodimers of nanoparticles: A conjugate of quantum dot and mangetic nanoparticles. J Am Chem Soc 126:5664–5. Reproduced with permission. (b) Schematic of the synthesis and properties of Co/CdSe core/shell nanocomposite nanocrystals. Copyright © 2005 American Chemical Society. Kim H, Achermann M, et al 2005. Synthesis and characterization of Co/CdSe core/shell nanocomposites: Bifunctional magnetic-optical nanocrystals. J Am Chem Soc 127:544–6. Reproduced with permission. (c) Schematic representation of the preparation of QDs with a paramagnetic micellar coating. Copyright © 2006 American Chemical Society. Mulder WJM., Koole R, et al 2006. Quantum dots with a paramagnetic coating as a bimodal molecular imaging probe. Nano Letters 6:1–6. Reproduced with permission. (d) Schematic drawing of the synthesis of chitosan nanobeads that encapsulate both semiconductor QDs and paramagnetic Gd-DTPA. Copyright © 2005 Wiley-VCH. Tan WB, Zhang Y. 2005. Multifunctional quantum-dot-based magnetic chitosan nanobeads. Adv Mater, 17:2375–80. Reproduced with permission.
Future outlook
What can we expect for the future clinical applications of quantum dots? QDs are already having an impact in molecular pathology. Ventana Medical Systems (http://www.ventanadiscovery.com) has recently started marketing the QD Map family of products. These are immunohistochemistry reagent kits for automated slide processing and fluorescent detection of fixed specimens. The selling point of this product family mirrors the list of unique QD photophysical properties, namely photostability, single source excitation, narrow emission, multiplexing capabilities and high quantum yield. However, the lack of consistent and reproducible methods to conjugate many different biomolecules such as antibodies, protein markers, DNA and RNA to QDs in a systematic manner with control over their ratio, orientation, and avidity continues to hamper their further use in clinical diagnostics. As this ability to couple biological recognition agents to QDs slowly improves, we can expect more commercial products incorporating QDs for clinical, diagnostic, and research purposes. The ability to combine QD-based analysis into a multiplexed format, be it immunohistochemistry or in situ nucleic acid hybridization, will be particularly significant and should dramatically increase throughput and engender deep multi-marker studies that analyze correlations between RNA-protein expression and disease state. For biomedical imaging, the unanswered questions and complexity of the QD toxicity issue means that they will almost exclusively be used for research purposes in cellular and animal models.
References
- Akerman ME, Chan WCW, Laakkonen P, et al. Nanocrystal targeting in vivo. Proc Natl Acad Sci. 2002;99:12617–21. doi: 10.1073/pnas.152463399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alivisatos AP. The use of nanocrystals in biological detection. Nature Biotechnol. 2004;22:47–52. doi: 10.1038/nbt927. [DOI] [PubMed] [Google Scholar]
- Aryal BP, Benson DE. Electron donor solvent effects provide biosensing with quantum dots. J Am Chem Soc. 2006;128:15986–7. doi: 10.1021/ja066658f. [DOI] [PubMed] [Google Scholar]
- Baird GS, Zacharias DA, Tsien RY. Biochemistry, mutagenesis, and oligomerization of DsRed, a red fluorescent protein from coral. Proc Natl Acad Sci. 2000;97:11984–9. doi: 10.1073/pnas.97.22.11984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakalova R, Ohba H, Zhelev Z, et al. Quantum dots as photosensitizers. Nature Biotechnol. 2004a;22:1360–1. doi: 10.1038/nbt1104-1360. [DOI] [PubMed] [Google Scholar]
- Bakalova R, Ohba H, Zhelev Z, et al. Quantum dot anti-CD conjugates: Are they potential photosensitizers or potentiators of classical photosensitizing agents in photodynamic therapy of cancer? Nano Letters. 2004b;4:1567–73. [Google Scholar]
- Bakalova R, Zhelev Z, Ohba H, et al. Quantum dot-conjugated hybridization probes for preliminary screening of siRNA sequences. J Am Chem Soc. 2005;127:11328–35. doi: 10.1021/ja051089h. [DOI] [PubMed] [Google Scholar]
- Ballou B, Lagerholm BC, Ernst LA, et al. Noninvasive imaging of quantum dots in mice. Bioconjug Chem. 2004;15:79–86. doi: 10.1021/bc034153y. [DOI] [PubMed] [Google Scholar]
- Bentolila LA, Weiss S. Single-step multicolor fluorescence in situ hybridization using semiconductor quantum dot-DNA conjugates. Cell Biochem Biophys. 2006;45:59–70. doi: 10.1385/CBB:45:1:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentzen EL, House F, Utley TJ, et al. Progression of respiratory syncytial virus infection monitored by fluorescent quantum dot probes. Nano Letters. 2005;5:591–5. doi: 10.1021/nl048073u. [DOI] [PubMed] [Google Scholar]
- Bharali D, Lucey D, Harishankar J, et al. folate-receptor-mediated delivery of InP quantum dots for bioimaging using confocal and two-photon microscopy. J Am Chem Soc. 2005;127:11364–71. doi: 10.1021/ja051455x. [DOI] [PubMed] [Google Scholar]
- Bocsi J, Lenz D, Mittag A, et al. Automated four-color analysis of leukocytes by scanning fluorescence microscopy using quantum dots. Cytometry Part A. 2006;69A:131–4. doi: 10.1002/cyto.a.20217. [DOI] [PubMed] [Google Scholar]
- Bruchez M, Jr, Moronne M, Gin P, et al. Semiconductor nanocrystals as fluorescent biological labels. Science. 1998;281:2013–16. doi: 10.1126/science.281.5385.2013. [DOI] [PubMed] [Google Scholar]
- Chan PM, Yuen T, Ruf F, et al. Method for multiplex cellular detection of mRNAs using quantum dot fluorescent in situ hybridization. Nucleic Acids Res. 2005;33:e161. doi: 10.1093/nar/gni162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan WCW, Nie S. Quantum dot bioconjugates for ultrasensitive nonisotopic detection. Science. 1998;281:2016–8. doi: 10.1126/science.281.5385.2016. [DOI] [PubMed] [Google Scholar]
- Chu MQ, Song X, Cheng D, et al. Preparation of quantum dot-coated magnetic polystyrene nanospheres for cancer cell labelling and separation. Nanotechnology. 2006a;17:3268–73. [Google Scholar]
- Chu TC, Shieh F, Lavery LA, et al. Labeling tumor cells with fluorescent nanocrystal-aptamer bioconjugates. Biosens Bioelectron. 2006b;21:1859–66. doi: 10.1016/j.bios.2005.12.015. [DOI] [PubMed] [Google Scholar]
- Clapp AR, Medintz IL, Mauro JM, et al. Fluorescence resonance energy transfer between quantum dot donors and dye-labeled protein acceptors. J Am Chemical Society. 2004;126:301–10. doi: 10.1021/ja037088b. [DOI] [PubMed] [Google Scholar]
- Colton HM, Falls JG, Ni H, et al. Visualization and quantitation of peroxisomes using fluorescent nanocrystals: Treatment of rats and monkeys with fibrates and detection in the liver. Toxicol Sci. 2004;80:183–92. doi: 10.1093/toxsci/kfh144. [DOI] [PubMed] [Google Scholar]
- Colvin VL. The potential environmental impact of engineered nanomaterials. Nature Biotechnol. 2003;21:1166–70. doi: 10.1038/nbt875. [DOI] [PubMed] [Google Scholar]
- Dabbousi BO, Rodriguez-Viejo J, Mikulec FV, et al. (CdSe)ZnS core-shell quantum dots: synthesis and optical and structural characterization of a size series of highly luminescent materials. J Phys Chem B. 1997;101:9463–75. [Google Scholar]
- de Farias PMA, Santos BS, de Menezes FD, et al. Investigation of red blood cell antigens with highly fluorescent and stable semiconductor quantum dots. J Biomed Optics. 2005;10:044023. doi: 10.1117/1.1993257. [DOI] [PubMed] [Google Scholar]
- De Rosa SC, Herzenberg LA, Herzenberg LA, et al. 11-color, 13-parameter flow cytometry: Identification of human naive T cells by phenotype, function, and T-cell receptor diversity. Nat Med. 2001;7:245–8. doi: 10.1038/84701. [DOI] [PubMed] [Google Scholar]
- Delehanty JB, Medintz IL, Pons T, et al. Self-assembled quantum dot-peptide bioconjugates for selective intracellular delivery. Bioconjug Chem. 2006;17:920–7. doi: 10.1021/bc060044i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derfus AM, Chan WCW, Bhatia SN. Intracellular delivery of quantum dots for live cell labeling and organelle tracking. Adv Mater. 2004a;16:961–6. [Google Scholar]
- Derfus AM, Chan WCW, Bhatia SN. Probing the Cytotoxicity of Semiconductor Quantum Dots. Nano Letters. 2004b;4:11–18. doi: 10.1021/nl0347334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubertret B, Skourides P, Norris DJ, et al. In vivo imaging of quantum dots encapsulated in phospholipid micelles. Science. 2002;298:1759–62. doi: 10.1126/science.1077194. [DOI] [PubMed] [Google Scholar]
- Eastman PS, Ruan WM, Doctolero M, et al. Qdot nanobarcodes for multiplexed gene expression analysis. Nano Letters. 2006;6:1059–64. doi: 10.1021/nl060795t. [DOI] [PubMed] [Google Scholar]
- Fischer HC, Lichuan L, Pang KS, et al. Pharmacokinetics of Nanoscale Quantum Vivo Distribution, Sequestration, and Clearance in the Rat. Advanced Functional Materials. 2006;16:1299–305. [Google Scholar]
- Gao X, Cui Y, Levenson RM, et al. In vivo cancer targeting and imaging with semiconductor quantum dots. Nature Biotechnol. 2004;22:969–76. doi: 10.1038/nbt994. [DOI] [PubMed] [Google Scholar]
- Gerion D, Chen F, Kannan B, et al. Room-Temperature Single-Nucleotide Polymorphism and Multiallele DNA Detection Using Fluorescent Nanocrystals and Microarrays. Anal Chem. 2003;75:4766–72. doi: 10.1021/ac034482j. [DOI] [PubMed] [Google Scholar]
- Gill R, Freeman R, Xu JP, et al. Probing biocatalytic transformations with CdSe-ZnS QDs. J Am Chem Soc. 2006;128:15376–7. doi: 10.1021/ja066636t. [DOI] [PubMed] [Google Scholar]
- Gill R, Willner I, Shweky I, et al. Fluorescence resonance energy transfer in CdSe/ZnS-DNA conjugates: Probing hybridization and DNA cleavage. J Phys Chem B. 2005;109:23715–9. doi: 10.1021/jp054874p. [DOI] [PubMed] [Google Scholar]
- Goldman ER, Clapp AR, Anderson GP, et al. Multiplexed Toxin Analysis Using Four Colors of Quantum Dot Fluororeagents. Anal Chem. 2004;76:684–8. doi: 10.1021/ac035083r. [DOI] [PubMed] [Google Scholar]
- Gu H, Zheng R, Zhang X, et al. Facile one-pot synthesis of bifunctional heterodimers of nanoparticles: A conjugate of quantum dot and magnetic nanoparticles. J Am Chem Soc. 2004;126:5664–5. doi: 10.1021/ja0496423. [DOI] [PubMed] [Google Scholar]
- Guo W, Li JJ, Wang YA, et al. Conjugation chemistry and bioapplications of semiconductor box nanocrystals prepared via dendrimer bridging. Chem Mater. 2003;15:3125–33. [Google Scholar]
- Hardman R. A toxicological review of quantum dots:toxicity depends on physicochemical and environmental factors. Environ Health Perspect. 2006;114:165–72. doi: 10.1289/ehp.8284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoet PH, Bruske-Hohlfeld I, Salata OV. Nanoparticles - known and unknown health risks. J Nanobiotechnol. 2004;2:2–12. doi: 10.1186/1477-3155-2-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino A, Hanaki K, Suzuki K, et al. Applications of T-lymphoma labeled with fluorescent quantum dots to cell tracing markers in mouse body. Biochem Biophys Res Comm. 2004;314:46–53. doi: 10.1016/j.bbrc.2003.11.185. [DOI] [PubMed] [Google Scholar]
- Hu FQ, Ran YL, Zhou ZA, et al. Preparation of bioconjugates of CdTe nanocrystals for cancer marker detection. Nanotechnology. 2006;17:2972–7. [Google Scholar]
- Jaiswal JK, Goldman ER, Mattoussi H, et al. Use of quantum dots for live cell imaging. Nature Meth. 2004;1:73–8. doi: 10.1038/nmeth1004-73. [DOI] [PubMed] [Google Scholar]
- Jaiswal JK, Mattoussi H, Mauro JM, et al. Long-term multiple color imaging of live cells using quantum dot bioconjugates. Nature Biotechnol. 2003;21:47–51. doi: 10.1038/nbt767. [DOI] [PubMed] [Google Scholar]
- Kahn E, Vejux A, Menetrier F, et al. Analysis of CD36 expression on human monocytic cells and atherosclerotic tissue sections with quantum dots – Investigation by flow cytometry and spectral imaging microscopy. Anal Quant Cytol Histol. 2006;28:14–26. [PubMed] [Google Scholar]
- Kaul Z, Yaguchi T, Kaul SC, et al. Mortalin imaging in normal and cancer cells with quantum dot immuno-conjugates. Cell Res. 2003;13:503–7. doi: 10.1038/sj.cr.7290194. [DOI] [PubMed] [Google Scholar]
- Kim H, Achermann M, Balet LP, et al. Synthesis and characterization of Co/CdSe core/shell nanocomposites: Bifunctional magnetic-optical nanocrystals. J Am Chem Soc. 2005;127:544–6. doi: 10.1021/ja047107x. [DOI] [PubMed] [Google Scholar]
- Kim S, Bawendi MG. Oligomeric Ligands for Luminescent and Stable Nanocrystal Quantum Dots. J Am Chem Soc. 2003;125:14652–3. doi: 10.1021/ja0368094. [DOI] [PubMed] [Google Scholar]
- Kim S, Lim YT, Soltesz EG, et al. Near-infrared fluorescent type II quantum dots for sentinel lymph node mapping. Nature Biotechnol. 2004;22:93–7. doi: 10.1038/nbt920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortan AR, Hull R, Opila RL, et al. Nucleation and growth of cadmium selendie on zinc sulfide quantum crystallite seeds, and vice versa, in inverse micelle media. J Am Chem Soc. 1990;112:1327–32. [Google Scholar]
- Lakowicz JR. Principles of Fluorescence Spectroscopy. Kluwer Academic/Plenum Publishers; 1999. [Google Scholar]
- Larson DR, Zipfel WR, Williams RM, et al. Water-soluble quantum dots for multiphoton fluorescence imaging in vivo. Science. 2003;300:1434–7. doi: 10.1126/science.1083780. [DOI] [PubMed] [Google Scholar]
- Leatherdale CA, Woo WK, Mikulec FV, et al. On the absorption cross section of CdSe nanocrystal quantum dots. J Phys Chem B. 2002;106:7619–22. [Google Scholar]
- Lidke DS, Nagy P, Heintzmann R, et al. Quantum dot ligands provide new insights into erbB/HER receptor-mediated signal transduction. Nature Biotechnol. 2004;22:198–203. doi: 10.1038/nbt929. [DOI] [PubMed] [Google Scholar]
- Lim YT, Kim S, Nakayama A, et al. Selection of quantum dot wavelengths for biomedical assays and imaging. Mol Imag. 2003;2:50–64. doi: 10.1162/15353500200302163. [DOI] [PubMed] [Google Scholar]
- Mattoussi H, Mauro JM, Goldman ER, et al. Self-assembly of Cdse-zns quantum dot bioconjugates using an engineered recombinant protein. J Am Chem Soc. 2000;122:12142–50. [Google Scholar]
- Medintz IL, Clapp AR, Brunel FM, et al. Proteolytic activity monitored by fluorescence resonance energy transfer through quantum-dot-peptide conjugates. Nature Mater. 2006;5:581–9. doi: 10.1038/nmat1676. [DOI] [PubMed] [Google Scholar]
- Medintz IL, Clapp AR, Mattoussi H, et al. Self-assembled nanoscale biosensors based on quantum dot FRET donors. Nature Mater. 2003;2:630–8. doi: 10.1038/nmat961. [DOI] [PubMed] [Google Scholar]
- Medintz IL, Clapp AR, Melinger JS, et al. A reagentless biosensing assembly based on quantum dot donor förster resonance energy transfer. Adv Mater. 2005a;17:2450–5. [Google Scholar]
- Medintz IL, Konnert JH, Clapp AR, et al. A fluorescence resonance energy transfer derived structure of a quantum dot-protein bioconjugate nanoassembly. Proc Natl Acad Sci. 2004;101:9612–17. doi: 10.1073/pnas.0403343101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medintz IL, Uyeda HT, Goldman ER, et al. Quantum dot bioconjugates for imaging, labeling and sensing. Nature Mater. 2005b;4:435–46. doi: 10.1038/nmat1390. [DOI] [PubMed] [Google Scholar]
- Michalet X, Pinaud FF, Bentolila LA, et al. Quantum dots for live cells, in vivo imaging, and diagnostics. Science. 2005;307:538–44. doi: 10.1126/science.1104274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder WJM, Koole R, Brandwijk RJ, et al. Quantum dots with a paramagnetic coating as a bimodal molecular imaging probe. Nano Letters. 2006;6:1–6. doi: 10.1021/nl051935m. [DOI] [PubMed] [Google Scholar]
- Murray CB, Kagan CR, Bawendi MG. Synthesis and characterization of monodisperse nanocrystals and close-packed nanocrystal assemblies. Ann Rev Mater Sci. 2000;30:545–610. [Google Scholar]
- Nel A, Xia T, Madler L, et al. Toxic potential of materials at the nanolevel. Science. 2006;311:622–7. doi: 10.1126/science.1114397. [DOI] [PubMed] [Google Scholar]
- Parak WJ, Boudreau R, Le Gros M, et al. Cell motility and metastatic potential studies based on quantum dot imaging of phagokinetic tracks. Adv Mater. 2002;14:882–5. [Google Scholar]
- Pathak S, Choi SK, Arnheim N, et al. Hydroxylated quantum dots as luminescent probes for in situ hybridization. J Am Chem Soc. 2001;123:4103–4. doi: 10.1021/ja0058334. [DOI] [PubMed] [Google Scholar]
- Patolsky F, Gill R, Weizmann Y, et al. Lighting-up the dynamics of telomerization and DNA replication by CdSe-ZnS quantum dots. J Am Chem Soc. 2003;125:13918–19. doi: 10.1021/ja035848c. [DOI] [PubMed] [Google Scholar]
- Pinaud F, King D, Moore H-P, et al. Bioactivation and cell targeting of semiconductor CdSe/ZnS nanocrystals with phytochelatin-related peptides. J Am Chem Soc. 2004;126:6115–23. doi: 10.1021/ja031691c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandros MG, Gao D, Benson DE. A modular nanoparticle-based system for reagentless small molecule biosensing. J Am Chem Soc. 2005;127:12198–9. doi: 10.1021/ja054166h. [DOI] [PubMed] [Google Scholar]
- Shepard JRE. Polychromatic microarrays: Simultaneous multicolor array hybridization of eight samples. Anal Chem. 2006;78:2478–86. doi: 10.1021/ac060011w. [DOI] [PubMed] [Google Scholar]
- Shi L, DePaoli V, Rosenzweig N, et al. Synthesis and application of quantum dots FRET-based protease sensors. J Am Chem Soc. 2006;128:10378–9. doi: 10.1021/ja063509o. [DOI] [PubMed] [Google Scholar]
- Stsiapura V, Sukhanova A, Artemyev M, et al. Functionalized nanocrystal-tagged fluorescent polymer beads: synthesis, physicochemical characterization and immunolabeling application. Anal Biochem. 2004;334:257–65. doi: 10.1016/j.ab.2004.07.006. [DOI] [PubMed] [Google Scholar]
- Sukhanova A, Devy M, Venteo L, et al. Biocompatible fluorescent nanocrystals for immunolabeling of membrane proteins and cells. Anal Biochem. 2004;324:60–7. doi: 10.1016/j.ab.2003.09.031. [DOI] [PubMed] [Google Scholar]
- Tan WB, Zhang Y. Multifunctional quantum-dot-based magnetic chitosan nanobeads. Adv Mater. 2005;17:2375–80. [Google Scholar]
- Tokumasu F, Dvorak J. Development and application of quantum dots for immunocytochemistry of human erythrocytes. J Microsc. 2003;211:256–61. doi: 10.1046/j.1365-2818.2003.01219.x. [DOI] [PubMed] [Google Scholar]
- Uyeda HT, Medintz IL, Jaiswal JK, et al. Synthesis of compact multidentate ligands to prepare stable hydrophilic quantum dot fluorophores. J Am Chem Soc. 2005;127:3870–8. doi: 10.1021/ja044031w. [DOI] [PubMed] [Google Scholar]
- Wang HZ, Wang HY, Liang RQ, et al. Detection of tumor marker CA125 in ovarian carcinoma using quantum dots. Acta Biochim Biophys Sin. 2004;36:681–6. doi: 10.1093/abbs/36.10.681. [DOI] [PubMed] [Google Scholar]
- Wu X, Liu H, Liu J, et al. Corrigendum: Immunofluorescent labeling of cancer marker Her2 and other cellular targets with semiconductor quantum dots. Nature Biotechnol. 2003;21:452. doi: 10.1038/nbt764. [DOI] [PubMed] [Google Scholar]
- Xu HX, Sha MY, Wong EY, et al. Multiplexed SNP genotyping using the Qbead (TM) system: a quantum dot-encoded microsphere-based assay. Nucleic Acids Res. 2003;31:e43. doi: 10.1093/nar/gng043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh HC, Ho YP, Shih IM, et al. Homogeneous point mutation detection by quantum dot-mediated two-color fluorescence coincidence analysis. Nucleic Acids Res. 2006:34. doi: 10.1093/nar/gkl021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahavy E, Freeman E, Lustig S, et al. Double labeling and simultaneous detection of B- and T cells using fluorescent nano-crystal (q-dots) in paraffin-embedded tissues. J Fluoresc. 2005;15:661–5. doi: 10.1007/s10895-005-2972-x. [DOI] [PubMed] [Google Scholar]
- Zhang C-Y, Johnson LW. Homogeneous rapid detection of nucleic acids using two-color quantum dots. The Analyst. 2006;131:484–8. doi: 10.1039/b514309h. [DOI] [PubMed] [Google Scholar]
- Zhang CY, Yeh HC, Kuroki MT, et al. Single-quantum-dot-based DNA nanosensor. Nature Mater. 2005;4:826–31. doi: 10.1038/nmat1508. [DOI] [PubMed] [Google Scholar]
- Zhang TT, Stilwell JL, Gerion D, et al. Cellular effect of high doses of silica-coated quantum dot profiled with high throughput gene expression analysis and high content cellomics measurements. Nano Letters. 2006;6:800–8. doi: 10.1021/nl0603350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhelev Z, Bakalova R, Ohba H, et al. Uncoated, broad fluorescent, and size–homogeneous CdSe quantum dots for bioanalyses. Anal Chem. 2006;78:321–30. doi: 10.1021/ac0511896. [DOI] [PubMed] [Google Scholar]