Abstract
The neutral mutation rate is equal to the base substitution rate when the latter is not affected by natural selection. Differences between these rates may reveal that factors such as natural selection, linkage, or a mutator locus are affecting a given sequence. We examined the neutral base substitution rate by measuring the sequence divergence of ∼30,000 pairs of inactive orthologous L1 retrotransposon sequences interspersed throughout the human and chimpanzee genomes. In contrast to other studies, we related ortholog divergence to the time (age) that the L1 sequences resided in the genome prior to the chimpanzee and human speciation. As expected, the younger orthologs contained more hypermutable CpGs than the older ones because of their conversion to TpGs (and CpAs). Consequently, the younger orthologs accumulated more CpG mutations than the older ones during the ∼5 million years since the human and chimpanzee lineages separated. But during this same time, the younger orthologs also accumulated more non-CpG mutations than the older ones. In fact, non-CpG and CpG mutations showed an almost perfect (R2 = 0.98) correlation for ∼97% of the ortholog pairs. The correlation is independent of G + C content, recombination rate, and chromosomal location. Therefore, it likely reflects an intrinsic effect of CpGs, or mutations thereof, on non-CpG DNA rather than the joint manifestation of the chromosomal environment. The CpG effect is not uniform for all regions of non-CpG DNA. Therefore, the mutation rate of non-CpG DNA is contingent to varying extents on local CpG content. Aside from their implications for mutational mechanisms, these results indicate that a precise determination of a uniform genome-wide neutral mutation rate may not be attainable.
The divergence rate (number of base substitutions over time) between orthologous sequences not under natural selection is usually taken as the neutral mutation rate (Nachman and Crowell 2000). Deviations from the neutral mutation rate may provide presumptive evidence that a sequence is under selection and thus of functional importance. Such inferences as well as the applicability of molecular clocks for phylogenetic analysis depend on an accurate determination of neutral base substitution rates. However, a number of factors can affect mutation rates. One is DNA replication (i.e., the number of germ line cell divisions [Ellegren 2007]), because DNA synthesis is not error-free.
Additionally, mutation rates reportedly differ dramatically both within and between chromosomes. These differences have been correlated with such factors as recombination, G + C content, gene content, and the presence of CpG dinucleotides (e.g., Lercher et al. 2001; Ebersberger et al. 2002; Hardison et al. 2003; Malcom et al. 2003; Hellmann et al. 2005; Taylor et al. 2006). Because the C of CpG is a preferred site of methylation, and methyl-C is prone to spontaneous deamination to T, CpGs are hypermutable (Ehrlich and Wang 1981). Therefore, a positive correlation between CpGs and mutation rate would be expected. However, the hypermutablity of methylated CpGs does not fully explain the correlation, as it persists even after excluding CpG mutations from the calculations of mutation rate (The Chimpanzee Sequencing and Analysis Consortium 2005; Hellmann et al. 2005). Thus, the correlation between CpGs and mutation rate might be a joint manifestation of some higher-order factor (The Chimpanzee Sequencing and Analysis Consortium 2005; Hellmann et al. 2005).
We addressed the above issues by comparing the divergence of ancestral L1 (long interspersed nuclear element 1 [LINE-1]) retrotransposon DNA between humans and chimpanzees. L1 DNA is uniquely suited for this purpose: Essentially all ancestral L1 inserts are assuredly nonfunctional DNA fossils that evolved without selection (i.e., neutrally). Therefore, measurements of their divergence cannot be biased by the inadvertent inclusion of conserved nongenic sequences (e.g., Dermitzakis et al. 2003; Thomas et al. 2003). Additionally, as L1 inserts are highly similar in both sequence and distribution throughout the genome, they provide a common substrate for mutation whatever their chromosomal location (Furano 2000; Khan et al. 2006). Furthermore, because fixed L1 inserts rarely participate in ectopic homologous recombination (Cooper et al. 1998; McNaughton et al. 1998; Richard et al. 1994), they are rarely subject to homogenization by gene conversion. Thus, their divergence provides a reliable record of base substitutions.
However, here we exploited an additional important property of L1 DNA that results from its unique evolutionary dynamics in mammals. L1 evolution repeatedly generated distinct, but closely related, L1 families that were active at different times during the evolution of their hosts, and then went extinct (Furano 2000). Consequently, the members of these highly similar families should differ in their CpG content, because they will have resided in the ancestral genome for different times. As the CpGs of L1 inserts would likely have been fully methylated (Orend et al. 1995; Yoder et al. 1997; Remus et al. 1999), they should have been converted to TpGs (and CpAs) to an extent proportional to their time in the genome (Hwang and Green 2004). Therefore, everything else being equal, the mutation rates of L1 sequences that differ in CpG content should indicate whether the relationship between CpG content and non-CpG divergence is an intrinsic property of the compared sequences, or is dependent on extrinsic factors particular to their chromosomal environments.
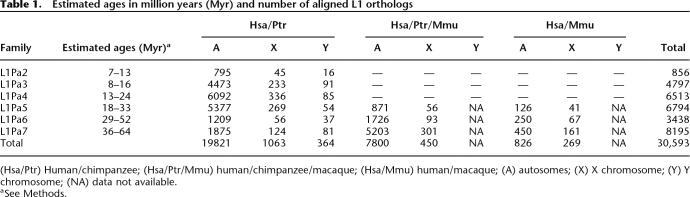
Accordingly, we determined the divergence of ∼30,000 pairs of L1 orthologs in humans and chimpanzees. These belonged to six closely related L1 families that were active at different times in the chimpanzee/human ancestor 6–53 million years ago (Mya) (Fig. 1) (Furano et al. 2004; Khan et al. 2006; Giordano et al. 2007). Because orthologous (syntenic) sequences, regardless of age, should be identical by descent, their divergence should reflect the mutations that accumulated since humans and chimpanzees diverged. The issue here then is whether the different families of L1 orthologs diverged at the same rate during this time.
Figure 1.
The relationship between the age of L1 families and the phylogeny of humans, chimpanzees, and macaques. The age range of the six different L1 families (gray rectangles) was estimated from their divergence relative to the divergence time of humans (H), chimpanzees (P), and macaques (M) as described in the Methods. The extended limb of the gray rectangle for the L1Pa2 family indicates that this L1 family is still active in chimpanzees but went extinct in humans sometime after the chimpanzee/human divergence. The 4–7 Myr range of the estimate for this divergence (see Methods) is shown on the phylogenetic tree. (Double-headed arrows, T2–T7) Times between the mean age of each L1 family and the mean of the time of the chimpanzee/human divergence (see Methods). (Red arrow, Tall) Time from the mean of the chimpanzee/human divergence to the present. The divergences between the chimpanzee and human ortholog pairs include only the nucleotide changes that occurred during the Tall interval.
As expected, the younger L1 ortholog pairs contained significantly more CpGs, and, consequently, more CpG mutations, than the older ones. However, the younger L1 orthologs also accumulated significantly more non-CpG mutations than the older ones. In fact, the non-CpG and CpG mutation rates show a near perfect (R2 = 0.98) correlation for 97% of the examined orthologs. G + C content, recombination rate, or chromosomal location do not account for the different mutation rates of older and younger L1 family orthologs. Thus, it appears as if CpG content per se is intimately related to the non-CpG mutation rate. We discuss possible mechanisms whereby CpGs, or mutations at CpGs, could affect the mutation of neighboring non-CpG DNA.
Results
The use of L1 retrotransposon DNA for estimating mammalian mutation rates
Mammalian L1 DNA is particularly well suited for comparing the extent of nucleotide differences (divergence) between neutrally evolving orthologous (syntenic) sequences in the species of interest. L1 retrotransposons reside in the host genome but can replicate autonomously by copying (retrotransposing) their RNA transcripts into genomic DNA. L1 copies are inserted throughout the genome, but most are defective (e.g., 5′-truncated) and therefore evolve without selection as nonfunctional DNA (for review, see Furano 2000). Replication-competent copies are also produced, some of which are variants. Eventually, a given variant gives rise to a novel L1 family that supplants the preexisting active family, which eventually ceases to replicate.
This process has occurred repeatedly during mammalian evolution. As fixed L1 inserts are rarely excised or homogenized by gene conversion (e.g., International Human Genome Sequencing Consortium 2001; Mouse Genome Sequencing Consortium 2002; The Chimpanzee Sequencing and Analysis Consortium 2005; Han et al. 2007), 15%–20% of the mammalian genomes studied to date consists of fossils of no longer active L1 families (Fig. 1; Smit et al. 1995; Furano 2000). Consequently, given extant species share thousands of orthologous L1 insertions inherited from their common ancestor. Because distinct L1 families generated these orthologs, they resided in the genome for different times (Fig. 1). As orthologous L1 elements are identical by descent, they should differ only by the number of substitutions accumulated since the species diverged (plus any surviving ancestral polymorphisms).
Recovery and distribution of aligned orthologous L1 sequences
We aligned 30,593 L1 DNA inserts from the human genome with their orthologous counterparts from the chimpanzee and macaque genomes and the corresponding L1 family-specific consensus sequence (Table 1). The orthologs that belong to the L1Pa2–L1Pa4 families (∼70% of the total) are unique to human and chimpanzee (Hsa and Ptr), as these L1 families emerged after apes and Old World monkeys diverged (Fig. 1). Orthologs from the older L1Pa5–L1Pa7 families amplified before Old World monkeys and apes diverged were recovered from all three species. Some human/macaque (Mmu) orthologs were not recovered from the chimpanzee, reflecting differences between the completeness of the genome databases. As the macaque Y chromosome sequence is not yet available, these orthologs are missing from our data set.
Table 1.
Estimated ages in million years (Myr) and number of aligned L1 orthologs
(Hsa/Ptr) Human/chimpanzee; (Hsa/Ptr/Mmu) human/chimpanzee/macaque; (Hsa/Mmu) human/macaque; (A) autosomes; (X) X chromosome; (Y) Y chromosome; (NA) data not available.
aSee Methods.
We recovered 856 orthologs for the youngest L1Pa2 family. These represent ∼22% of the estimated genomic copy number of this family in humans and <3% of the total nucleotides in our data set (see Methods; Supplemental Fig. S1; Supplemental Table S1). We expected a low yield of L1Pa2 orthologs because this family arose close to the split of the human and chimpanzee lineages (Fig. 1). It remains active in chimpanzee (Lee et al. 2007) but went extinct in the human lineage, giving way to the L1Pa1 (Ta) family (Boissinot et al. 2000). Although the low yield of L1Pa2 orthologs may not be as representative of genomic regions as the other orthologs, we included them as they represent the most recent L1 family shared between humans and chimpanzees.
The L1Pa3 orthologs represent ∼58% of the genomic copies of this family in humans, somewhat lower than the recovery for the remaining older families (Supplemental Fig. S1). Some evidence suggests that the activity of L1Pa3 persisted after the human and chimpanzee lineages diverged (Mills et al. 2006; but also see Lee et al. 2007). About 73% (SE ± 3.7%) of the genomic members of the L1Pa4–L1Pa7 families (∼68% of L1Pa4 to ∼79% of L1Pa7) were represented in our ortholog data set. However, the yield of orthologs from the sex chromosomes was somewhat less: 43% for X and 50% for Y (for possible explanations, see Supplemental Data).
Except for L1Pa2, the relative recovery of orthologs across each chromosome mimics the chromosomal densities of each L1 family, which are fairly similar for each family (Supplemental Fig. S1). Also, except for L1Pa2, the L1 orthologs of each family are similarly distributed within each chromosome (Supplemental Fig. S2). And, finally, the size distribution of the orthologs from both the autosomes and sex chromosomes generally recapitulated that reported for their genomic copies and includes both full-length and truncated L1 elements (Boissinot et al. 2001; Song and Boissinot 2007; Supplemental Fig. S3). Thus, the L1 orthologs (with the exception of L1Pa2) comprise a representative sample of the genomic inserts of each L1 family, which in turn represent similar genomic environments.
CpGs of L1 sequences are converted to TpGs and CpAs over time
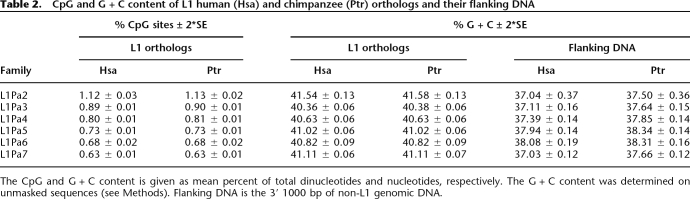
Because the L1 families resided in the ancestral genome for different times prior to the speciation of humans and chimpanzees (Fig. 1), their CpG contents should decrease with time as this dinucleotide mutates to TpG (or CpA) in a clock-like fashion (Hwang and Green 2004). This occurs because most CpGs in mammalian genomes, especially in retrotransposons, are methylated on the C residue, which undergoes spontaneous deamination to T (Coulondre et al. 1978; Bird 1980; Ehrlich et al. 1982; Orend et al. 1995; Yoder et al. 1997; Remus et al. 1999). Table 2 shows that the orthologs of the older L1 families contain fewer CpGs than the younger ones. Thus, the L1Pa7 sequences contain only ∼60% as many CpGs as do the L1Pa2 orthologs, regardless of species.
Table 2.
CpG and G + C content of L1 human (Hsa) and chimpanzee (Ptr) orthologs and their flanking DNA
The CpG and G + C content is given as mean percent of total dinucleotides and nucleotides, respectively. The G + C content was determined on unmasked sequences (see Methods). Flanking DNA is the 3′ 1000 bp of non-L1 genomic DNA.
Because CpGs account for only ∼1% of the total G + C in L1 families, their G + C content should hardly change with time. In addition, the minimal differences between the G + C content of either the orthologs or their genomic environment (Table 2) would not be expected to affect mutation rate (see Fig. 4 in Hellmann et al. 2005). The similarity in G + C content of their genomic environment would be expected, as the genomic distribution of these families is basically the same (Supplemental Fig. S2). To determine if the lower recovery of CpGs from the older L1 families was due to their mutation to TpGs (or CpAs), we determined the frequency of these dinucleotides at positions in the older families that should correspond to CpGs.
We aligned the corresponding nucleotide positions of full-length members of each L1 family extracted from the human database (Supplemental Table S4). We put these sequences in the same sequence register by aligning them to a reference L1 element, L1.3 (Dombroski et al. 1993), an active member of the currently active human Ta1 (L1Pa1) family (Boissinot et al. 2000). Aside from some minor insertions and deletions, and the diagnostic nucleotide differences that distinguish the L1 families, these sequences are highly similar (e.g., Supplemental Fig. S5; Boissinot et al. 2000; Khan et al. 2006). Therefore, we could unambiguously align all of the corresponding nucleotide positions and then determine whether CpGs of younger families corresponded to TpGs (or CpAs) of older families.
Figure 2 shows the percentage of corresponding dinucleotides that were either CpG or (TpG + CpA) for each family. The results are as expected in that the decrease in CpG dinucleotides was compensated by an approximately comparable increase in (TpG + CpA) dinucleotides at the corresponding position. We also analyzed the frequency of bases at the first (C) and second (G) position of these corresponding dinucleotide positions (Supplemental Table S9). These results showed that ∼85% of the CpGs missing from the older L1 families were compensated by a gain in TpGs plus CpAs (i.e., 42.6% gain in [TpG + CpA] ÷ 50.4% loss of CpGs for full-length L1 elements; Supplemental Table S9].
Figure 2.
Relative percentage of CpG and TpG (CpA) at corresponding nucleotide positions of various L1 families. This determination was made on full-length members aligned as described in the Methods. The ordinate gives the relative percentage of CpG and (TpG + CpA) in full-length members of the various L1 families at positions corresponding to a CpG in the relevant L1 family-specific ancestral consensus sequence. The sums of the [CpG + (TpG + CpA)] percentages range from ∼83% in the three oldest families (L1Pa5–L1Pa7) to ∼92% in the three younger families (L1Pa2–L1Pa4). Also see Supplemental Table S9.
Divergence of L1 orthologs as a function of their age in the genome
Almost without exception, each ortholog pair is the result of a unique historical L1 insertion and thus ideally would be identical at the time of speciation (Tall; Fig. 1). Divergence then would simply equal the number of times corresponding nucleotide positions differed between the chimpanzee and human orthologs divided by the total number of ortholog pairs compared. However, humans and chimpanzees descended from an ancestral population, not a single individual. Thus, base differences (ancestral polymorphisms) were undoubtedly present in the ancestral population, including L1 orthologs. On the other hand, recent studies indicate that, in the absence of selection, ancestral polymorphisms would not likely persist for the 5–7 million years (Myr) since speciation, and thus would not contribute to present-day divergence (Asthana et al. 2005; Kehrer-Sawatzki and Cooper 2007). In any event, any persistent ancestral polymorphisms should affect the divergence of all the ortholog pairs and thus not affect comparisons between the L1 families.
We compared the divergence of the L1Pa2–L1Pa7 orthologs over the entire length of the L1 elements. Figure 3 shows the base-wise divergence at CpG and non-CpG sites for the 3′ 1200 bp of ORF2 of the autosomal orthologs. Two major results, which are typical of the rest of the element, are apparent: First, every CpG site (red arrowheads, L1Pa2) is a mutational hot spot (defined as a nucleotide position where ≥0.1 of the human and chimpanzee orthologs differ from each other; see legend to Fig. 3). These hotspots eventually disappear in the older families because their corresponding CpGs had mutated to TpGs (or CpAs) (Fig. 2; Supplemental Table S9). However, their mutation rates vary, as some CpG hotspots (e.g., 1 and 2) disappear faster than others over time (e.g., 7 and 8). At sites 7 and 8, CpGs have been retained in enough members of older families to still be scored as mutational hotspots. Residual CpG hotspot activity also explains the presence of some mutational hotspots in some families that do not correspond to CpG sites in other families (downward pointing arrows in Fig. 3). Reconstruction of the ancestral consensus sequences for each L1 family (see Methods) showed that some such sites (e.g., ancestral CpGs 2 and 4; Fig. 3) correspond to CpGs in the older families that were not conserved during L1 evolution.
Figure 3.
Divergence of L1 orthologs at single base pair resolution. The region shown corresponds to the 3′ 1200 bp of ORF2. The ordinate gives the fraction of the number of changes between the chimpanzee/human orthologs at each position. (Red triangles) CpGs present in L1Pa2, (lettered inverted green arrowheads) ancestral CpGs (ancCG_1, ancCG_2, etc.), (magenta lettered peaks) some non-CpG hot spots; only hotspot “d” corresponds to a CpT dinucleotide in L1Pa2 and L1Pa3 (see text). CpG hot spots are defined as a divergence >0.1 (solid line), and non-CpG hot spots as divergence >0.05. Also note that some CpG hot spots in the younger families persist as hot spots in the older families even after the frequency of CpGs in the older orthologs has fallen below the threshold value to appear as a CpG in the current consensus sequence (see Methods for definition of current and ancestral consensus sequences). An example of the data underlying this plot is shown for the 3′ 186 bp in Supplemental Figure S7.
Second, Figure 3 shows that non-CpG nucleotide positions of the younger L1 orthologs are also more divergent than the corresponding positions in the older orthologs. Thus, the non-CpG mutation rate is positively correlated with the CpG mutation rate, which in turn reflects CpG content. Although some of the non-CpG divergent positions are mutational hot spots (defined as ≥0.05 divergence, some of which are labeled with letters), most are not. In addition, Figure 3 also shows that the distribution of non-CpG divergence at particular positions is not random between families. Thus, positions of higher or lower divergence in one family are more likely to have a similar divergence in another family (also evident in regions other than that shown in Figure 3, Kendall’s correlation P < 0.001).
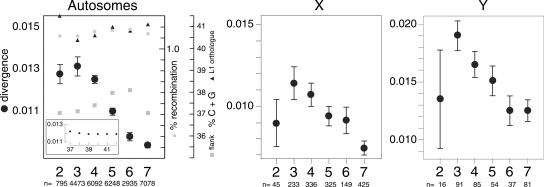
Figure 4 shows the median non-CpG divergences for the autosomes and sex chromosomes. Except for L1Pa2, the ortholog pairs from the older families diverged (mutated) at a significantly lower rate than those of the younger families on the autosomes and sex chromosomes. The means for these divergences are shown in Supplemental Table S2. The disparate results with the L1Pa2 orthologs, especially for the sex chromosomes, likely result from their low copy number. Figure 4 also shows that the minimal differences between the G + C content of the different L1 ortholog families, or their flanking DNA sequences (<1.2% in both cases; Table 2), are not sufficient to account for the differences in the non-CpG mutation rates. This conclusion is based on data like that presented in the inset in Figure 4. These results (extracted from Fig. 4 in Hellmann et al. 2005) show that the divergence between syntenic regions of the chimpanzee and human genomes (Y-axis) is essentially invariant over the range of G + C content (X-axis) considered here.
Figure 4.
Divergence of autosomal and sex chromosomal members of different L1 families. The median (black circles) and confidence intervals around the median (Strelen 2004) of the non-CpG ortholog divergences for the L1Pa2–L1Pa7 families were determined as described in the Methods. The means of the recombination rates (gray circles) are shown for each L1 family (see Methods). Mean % G + C content for the human orthologs (filled triangles) and their flanking DNA from Table 2 (gray squares) are also given. The number (n) of ortholog pairs for each L1 family (2–7) is shown for the data used for the divergence measurements. We were able to assign recombination rates to ≥98% of the L1 orthologs. (Inset) Portion of the curve fit extracted from Figure 4 published by (Hellmann et al. 2005). The Y-axis shows the divergence between syntenic regions of the chimpanzee and human genomes as a function of G + C content (X-axis).
We also determined the recombination rates associated with each L1 ortholog family (see Methods). The mean values (±SE) of these recombination rates were 1.12 (±0.03), 1.12 (±0.01), 1.15 (±0.01), 1.16 (±0.01), 1.17 (±0.02), and 1.13 (±0.01), respectively, for the L1Pa2, L1Pa3, L1Pa4, L1Pa5, L1Pa6, and L1Pa7 families. These means are also shown in Figure 4. Although the slight differences between the recombination rates are statistically significant (ANOVA, F = 2.86, P = 0.014), they too are insufficient to account for the lower non-CpG divergence of the old L1 orthologs compared with the younger ones (Fig. 4). However, Figure 5 shows that for 97% of the examined orthologs, non-CpG mutations are nearly perfectly (R2 = 0.98) correlated with CpG mutations.
Figure 5.
The relationship between the percentage of non-CpG and CpG mutations. Both classes of mutations for the L1 orthologs were determined as described in the Methods. We calculated the correlation coefficient both with (dashed line) and without (solid line) the L1Pa2 orthologs because the orthologs for this family were recovered at a far lower frequency than the older families (see text). As a result they provided only ∼3% as much DNA sequence (in Mb) of the total base pairs in our data set. The reduced data set for the L1Pa2 family could partly explain why the relationship between CpG content and divergence of this family is different from that expected from the other families. It may also explain why the divergences of the L1Pa2 orthologs differed far more between the chromosomes than that of the other families (Supplemental Fig. S6).
Divergence of L1 orthologs on different chromosomes
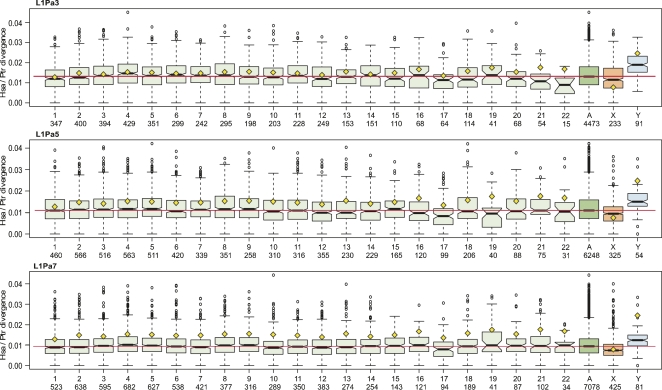
One surprising outcome from large-scale interspecies genomic comparisons was the report of statistically significant, autosome-wide differences in sequence divergence (Lercher et al. 2001; Ebersberger et al. 2002; Hardison et al. 2003; Malcom et al. 2003; The Chimpanzee Sequencing and Analysis Consortium 2005). Figure 6 shows that the divergence of the L1Pa3 orthologs fairly well recapitulates that reported by the chimpanzee genome consortium (The Chimpanzee Sequencing and Analysis Consortium 2005). The yellow diamonds in Figure 6 indicate the median values of whole-genome divergence between the syntenic regions of the chimpanzee and human genomes from Figure 1b in reference The Chimpanzee Sequencing and Analysis Consortium (2005). With the possible exception of chromosomes 21 and 22, both the median divergence values and pattern of autosomal divergences found for the L1Pa3 orthologs track fairly well with the published whole-genome comparisons (The Chimpanzee Sequencing and Analysis Consortium 2005).
Figure 6.
Box blots of the distribution of chromosomal divergence values. The median divergence with a 95% confidence interval (notches) is given for the chimpanzee/human orthologs from the L1Pa3, L1Pa5, and L1Pa7 families. The number below each chromosome is the number of ortholog pairs compared. (A) Combined divergence for all the autosomes, (red line) median value of this divergence, (open circles) indicate outliers. (Box plots for all of the families are shown in Supplemental Fig. S6). (Yellow diamonds) Median value of whole-genome chromosomal divergences between syntenic regions of the chimpanzee and human genomes from Figure 1b in reference The Chimpanzee Sequencing and Analysis Consortium (2005).
Although the reported divergence values (The Chimpanzee Sequencing and Analysis Consortium 2005) included mutations at CpG sites, these mutations account for only ∼4% of the total variance between the autosomes (The Chimpanzee Sequencing and Analysis Consortium 2005). Thus, the similarity between the L1Pa3 divergences (which did not include CpG mutations) and the whole-genome divergences indicates that for most autosomes the divergence of the putatively neutral L1Pa3 orthologs is a reasonable proxy for whole-genome divergence. The consortium investigators concluded that the differences between autosomal divergences were statistically significant (Kruskal–Wallis test, P < 3 × 10−15 [The Chimpanzee Sequencing and Analysis Consortium 2005]). The smallest P-value that we obtained with this test was 1.58 × 10−7 for L1Pa3. The values returned by this test for the other chromosomes ranged from nonsignificant (L1Pa6) to 1.16 × 10−5 (L1Pa7) (Supplemental Table S3).
One difficulty in interpreting the above results is that just a few deviant median values can lead to low P-values in the Kruskal–Wallis test. For example, except for chromosomes 21 and 22, only the confidence intervals (notches on the box plot) of the L1Pa3 divergences for chromosomes 4 and 17 did not overlap the median of the total autosomal divergence (red line, Fig. 6). Therefore, we carried out pairwise comparisons between the divergences of each of the autosomes (except 21 and 22) and tested for statistical significance using the Wilcoxon rank sum test with Bonferroni correction for multiple comparisons (see Methods; Supplemental Table S4). Only five pairwise differences had P-values < 0.05: chromosome 4 versus 1, 12, and 17; chromosome 6 versus 1 and 17. Applying the Wilcoxon test to the pairwise comparisons of the divergences of the other families produced the following chromosome pairs with P-values < 0.05: L1Pa2, 10 versus 18; L1Pa4, none; L1Pa5, 17 versus 3–9; L1Pa6, none; L1pa7, 4 versus 1, 7, 10, 12, 17 (Supplemental Fig. S4).
Thus, of the 1140 (190 × 6) possible pairwise comparisons, only 18 (1.6%) were statistically significant, and of these only two did not involve chromosomes 4 or 17. And even these chromosomes did not show notably consistent statistically significant differences with any other chromosomes except each other. Taken together, these results provide little statistical support for general autosome-wide differences using the divergence of the assuredly neutral orthologous L1 sequences. However, as mentioned above, the divergences of the L1Pa3 orthologs are congruent with the median values of the genome-wide divergences (The Chimpanzee Sequencing and Analysis Consortium 2005). Therefore, pairwise comparisons between the published values might well yield results similar to those found here.
Figure 6 also shows that the extent of divergence for each autosome was less when measured with the L1Pa7 orthologs than with the younger (e.g., L1Pa3) orthologs (for all of the families, see Supplemental Fig. S6). Furthermore, the extent of the decrease in divergence (D) is the same for each autosome. We found this result when we normalized the ratios of chromosomal divergences for L1Pa3 to those obtained for L1Pa7; i.e., [(D-L1Pa3chr1/D-L1Pa3chr2) / (D-L1Pa7chr1/D-L1Pa7chr2)]. A value of 1 indicates that the decreased divergence measured by L1Pa7 relative to L1Pa3 was the same for chromosomes 1 and 2; the mean (range) of this ratio for the pairwise comparisons between all the autosomes (except 21 and 22) is 1.03 (0.87–1.23). A one-way ANOVA analysis showed no significant difference between the pairwise comparisons except for chromosomes 21 and 22.
Determining the extent of male bias in mutation rate
Autosomes and sex chromosomes spend different lengths of time in males and females. Therefore, the ratios of their divergences will reflect a male bias (α) in mutation rate to the extent that it is affected by errors in DNA replication. This is because the number of germline cell divisions (i.e., DNA replications) differs between males and females (Miyata et al. 1987). Figure 6 (and Supplemental Fig. S6) shows that the least variance in the divergence of both the autosomes and sex chromosomes was obtained with the L1Pa7 (and L1Pa6) orthologs. This result and the fact these orthologs are the least divergent may indicate that the mutation rates of the L1Pa6 and L1Pa7 orthologs, being least affected by CpGs, more faithfully reflect the effect of DNA replication than the divergence of the younger orthologs. Therefore, we used the mean divergences of these families (Supplemental Table S2) to calculate a mean α (95% confidence interval) for the different ratios of autosomal and sex chromosome divergences (i.e., α(A/X), α(Y/A), α(Y/X)), respectively, of 1.74 (1.06–2.92), 1.67 (1.28–2.35), 1.69 (1.32–2.19), for L1Pa6; and 4.31 (2.87–7.13), 1.82 (1.46–2.35), 2.38 (1.93–2.94), for L1Pa7. If only the number of germline differences determines chromosomal divergence ratios, then the value of α should be the same regardless of the ratio used. The mean α from these determinations is 2.3.
The above calculations did not include any correction for ancestral polymorphisms (Makova and Li 2002), for, as mentioned above, this correction seems unwarranted (Asthana et al. 2005; Kehrer-Sawatzki and Cooper 2007). Our uncorrected mean value for α of 2.3 is consistent with those published by some (e.g., Bohossian et al. 2000; Patterson et al. 2006) of 1.7 and 1.9, respectively, but not those calculated by others (e.g., Makova and Li 2002; Taylor et al. 2006). The latter investigators applied corrections for persistent ancestral polymorphisms and found a value for α of 5.2. Thus, we also calculated α applying the values for ancestral polymorphisms (Taylor et al. 2006) and present these results along with the calculations of α using the divergences of all the families in Supplemental Table S5. We also did not include any correction for modern polymorphisms, as these are a function of the mutation rate, which is what we are trying to measure. Furthermore, as the recovery of any given polymorphism in any individual will depend on its allele frequency and the population size, there seems little justification for applying a general correction for them.
Discussion
A major conclusion from this work is that CpG content is positively correlated with non-CpG mutations in orthologous pairs of L1 sequences from humans and chimpanzees. It had already been shown that G + C content and recombination rate are positively correlated with mutation rate (e.g., Hardison et al. 2003; Hellmann et al. 2005). The minimal differences in both parameters between the age classes of L1 orthologs (Table 2; Methods) are not sufficient to explain the different divergence rates of the older and younger orthologs (Table 2; Fig. 4). In contrast, Figure 5 shows there is a near perfect correlation (R2 = 0.98) between CpG and non-CpG mutations for 97% of the ∼30,000 ortholog pairs (Table 1). Both the non-CpG divergence (Figs. 3, 4; Supplemental Table S2) and the CpG content (Table 2; Fig. 2; Supplemental Table S9) of the orthologs were inversely proportional to the time that they had resided in the common ancestor of chimpanzees and humans prior to their speciation (Fig. 1). Therefore, the CpG content of the orthologs of the different L1 families would have differed when the chimpanzee and human lineages separated (Hwang and Green 2004), which was the starting point for our measurements of sequence divergence.
Others had also noted a positive correlation between CpG content and divergence at non-CpG sites upon comparisons of human and chimpanzee syntenic DNA (The Chimpanzee Sequencing and Analysis Consortium 2005; Hellmann et al. 2005). But because total genomic DNA was compared in these studies, it was reasonably proposed that the correlation might be a joint manifestation of some higher-order factor (The Chimpanzee Sequencing and Analysis Consortium 2005; Hellmann et al. 2005). However, this consideration does not likely apply to our results. First, the different families of L1 orthologs have an overlapping distribution throughout the genome (Table 2; Fig. 4; Supplemental Fig. S2). Thus, their mutation rates should be similarly subjected to any higher-order genomic environmental factors that might affect mutation rate. Second, except for CpG content, the L1 orthologs have highly similar DNA sequences (Supplemental Fig. S5). Therefore, they provide a common substrate for mutation wherever they are located. Thus, the different mutation rates of the older and younger L1 orthologs seem to result from an intrinsic effect of their CpG content.
The possibility that the correlation between CpGs and non-CpG mutations is merely coincidental, rather than causal, seems most unlikely. A coincidental relationship would imply that the longer a neutrally evolving sequence resides in the genome the less susceptible it becomes to mutation, and at the same rate for both CpG and non-CpG sites. However, only the methyl-C in CpG sites mutates with a clock-like rate, whereas the mutation rates of Cs in other contexts and of all other nucleotides are affected by factors other than time (Hwang and Green 2004). The C of CpG is a preferred site of methylation in mammals (Ehrlich et al. 1982), and methyl-C is intrinsically hypermutable because of spontaneous deamination to T, thereby producing a T/G mismatch (Coulondre et al. 1978; Bird 1980). Despite mismatch repair processes, methylated CpGs that are not under selection are converted with time to TpGs (or CpAs) (Hwang and Green 2004). Consequently, hypermutable CpGs are converted to dinucleotides that have a “normal” mutation rate.
The issue then is whether the non-CpG sites in the younger orthologs are also being converted with time from an intrinsically high mutable state to a less mutable state, and at the same pace as the CpG sites. Casane et al. (1997) suggested that the release from natural selection, as would be the case for newly derived pseudogene copies of protein-encoding genes, would lead to an increase in mutation rate. This mechanism should apply only to nucleotide positions that are intrinsically highly mutable (e.g., methyl-Cs of methylated CpGs) but were under selection in their former context. Otherwise, the mutation rate of a pseudogene should be governed just by the factors that affect the neutral rate of its new chromosomal location. Nonetheless, we compared the divergence of the three different codon positions for ORF1 and ORF2. As the third codon position is normally under far less selective pressure than the first two positions, release from selection would have resulted in a higher divergence of positions one and two compared with position three. However, the divergences of positions one and two were not significantly higher than that of position three (Supplemental Table S6).
Additionally, there is no evidence that the mutability of neutral non-CpG sequences might change with time. No other nucleotide in mammals is as intrinsically mutable as methyl-C (Hwang and Green 2004), and no dinucleotide other than CpG is underrepresented in mammalian DNA (Duret and Galtier 2000). However, the C of both CpA and CpT (at about one-fourth the CpA rate) can be methylated, at least in embryo-derived cells (Woodcock et al. 1997; Ramsahoye et al. 2000). Together, these dinucleotides were methylated 10%–20% of the extent of CpG methylation in such cells. Although these sites are potentially prone to mutation due to deamination of the methyl-C, these dinucleotides are no more mutable than Cs in other sequence contexts (Hwang and Green 2004).
Furthermore, the distributions of CpAs and CpTs in the L1 orthologs were not correlated with regions of higher and lower non-CpG divergence (Supplemental Table S7). And finally, only one of the six non-CpG hot spots (d) highlighted in Figure 3 contains a CpT. Although it is conceivable that any random mutational hot spot can mutate to a less mutable sequence, there is no a priori reason why random mutations could not convert sites of “normal” mutation rate to ones of increased mutability. Therefore, we would not expect that such random fluctuations in mutability should show an age-dependence toward lower mutability, which is what we found for the non-CpG mutation rate of the L1 orthologs. Considering all of the above, we think it most unlikely that the near perfect correlation between CpG and non-CpG mutations is merely coincidental. As the lower CpG content of the older L1 orthologs is almost certainly a function of their methylation, then we need to consider how methyl-CpG, or mutations thereof, might affect the non-CpG mutation rate.
Methyl-CpG might affect the non-CpG mutation rate of DNA by enhancing its conversion to silent (closed) chromatin (e.g., Jaenisch and Bird 2003; Pennings et al. 2005), which has a higher mutation rate than open chromatin (Prendergast et al. 2007). These results were found by comparing the divergence of orthologous sequences between chimpanzees and humans, and included intergenic and intronic DNA as well as “ancient” repeats (i.e., those shared by murine rodents and primates). Accounting for our results solely by this mechanism would mean that the older, lower-divergence, CpG-poor L1 orthologs are preferentially located in open (low mutation rate) chromatin, while the younger, higher-divergence, CpG-rich L1 orthologs are in closed (high mutation rate) chromatin. However, there are several problems with this explanation. First, ancient repeats, far older than any analyzed here and likely more CpG-poor than any analyzed here, are not excluded from closed chromatin (Prendergast et al. 2007). Therefore, recruitment of a given sequence into silent chromatin may not be a direct function of its CpG content. That CpG methylation can occur subsequent to chromatin silencing supports this idea (Bird 2002). Second, young and old L1 orthologs are intermingled over much shorter ranges than the ∼100-kb stretches over which closed and open chromatin extend (Gilbert et al. 2004). Thus, unless open and closed chromatin can alternate at the kilobase range, differential chromatin silencing would not likely explain the differences in ortholog divergence.
One mechanism that could directly couple non-CpG and CpG mutations is if repair of the latter (i.e., T/G mismatches resulting from deamination of methyl-C) produces mutations at non-CpG sites. Eukaryotes contain T/G-specific mismatch repair systems that repair this mismatch at ∼90% efficiency (Walsh and Xu 2006). One involves excision of the thymine by a glycosylase and subsequent repair of the abasic site by the base excision repair mechanism (Walsh and Xu 2006). Recruitment of an error-prone Y family DNA polymerase to effect this repair could produce mutations at non-CpG sites (for reviews, see Goodman 2002; Rattray and Strathern 2003). In fact, one of the more error-prone Y family polymerases, pol iota, may have evolved specifically to correct T/G mismatches (Vaisman and Woodgate 2001; Vaisman et al. 2001). This enzyme has error rates on undamaged DNA of 10−4 to 10−2 and prefers to insert a G, rather than an A, opposite a T. Thus, if the strand bearing the T at a T/G mismatch is the template during mismatch repair, the G will be restored, increasing the probability of preserving the original C/G base pair. However, if, after incorporating the G, pol iota is not immediately replaced at the replication site by a high-fidelity DNA polymerase, mismatched bases could result.
Switching between high- and low-fidelity DNA polymerases during DNA replication has been demonstrated in vitro for prokaryotic DNA replication. The Escherichia coli error-prone Y family DNA polymerase pol IV can coexist in a DNA replication complex with the high-fidelity pol III polymerase (Indiani et al. 2005). These studies showed that pol IV seamlessly preempts DNA replication by pol III when the equivalent of a DNA lesion is encountered. However, pol III remains bound to the replication complex, and after the lesion is passed pol III resumes replication, although pol IV remains in the replication complex. Therefore, if the resumption of DNA synthesis by the high-fidelity pol III is delayed, mismatches could be introduced by continued replication by pol IV.
Our proposal that error-prone DNA repair of T/G mismatches might explain the correlation between CpG and non-CpG mutations has certain merits: It straightforwardly explains how CpG mutations increase non-CpG mutations, and could account for the unexpectedly high level of transversions (in addition to the expected preponderance of transitions) at CpG sites (Ebersberger et al. 2002; Taylor et al. 2006), which we also observed (data not shown). Furthermore, our proposed mechanism should be experimentally testable in vivo. Experimental support for this idea would imply that methylatable CpGs might even be considered mutagenic. Thus, the mutation rate of a region of DNA could be increased by any event that increases its content of methylatable CpGs: e.g., expansions of CpG-containing repeats (Usdin and Grabczyk 2000), transposon inserts (Yoder et al. 1997; Kazazian 2004), or integration of foreign sequences including viral DNA (Orend et al. 1995; Remus et al. 1999).
Whatever the explanation, the close correspondence that we found between CpG content and the mutation rate at non-CpG sites means that eliminating CpG mutations from mutation rate measurements will not necessarily compensate for their effect. Thus, conclusions about the selective forces on sequences based on deviations of their divergence from an assumed neutral rate could be problematic. In addition, CpG content did not uniformly affect the mutation of non-CpG sites in the L1 orthologs (Fig. 3; Supplemental Table S7). Thus, even if all other factors are equal, the divergence of neutral sequences may be variably contingent on CpG content. Thus, it might not be so surprising why determining the effect of factors such as DNA replication on the neutral mutation rate has been so problematic (Ellegren 2007).
Methods
Isolation of L1 orthologs
Sequence and annotation data were retrieved from the University of California, Santa Cruz download site (http://genome.ucsc.edu/). The following assemblies were used here: human genome-freeze March 2006 (UCSC hg18, NCBI Build 36.1); chimpanzee-freeze March 2006 (UCSC panTro2); and macaque-freeze January 2006 (UCSC rheMac2). Repeat Masker track files based on the Repeat Masker program and RepBase library were used to obtain L1 family information and genome coordinates (ftp://hgdownload.cse.ucsc.edu/goldenPath/). Based on the track file information, sequences of L1 elements in the human genome were retrieved from each chromosome and stored in FASTA files. The following L1 sequences were excluded from our database: L1 sequences shorter than 100 bp and/or missing 3′ UTR regions. As most L1 elements are 5′-truncated (Voliva et al. 1983; Furano 2000), L1 elements with a 3′ UTR most likely correspond to a single insertion event. Application of these two filters removed ∼20% of the elements from the RepeatMasker output. We also removed records with ambiguous information (e.g., insertions that could not be precisely located on a chromosome) and sequences >10 kb, as a typical full-length L1 element is ∼6-kb long. These filters removed an additional 1%–2% of the RepeatMasker output. The remaining L1 elements were taken as the genomic copy numbers for each family.
We found orthologous (syntenic) insertions, (i.e., those identical by descent) between human and chimpanzee or macaque genome by converting the human genome coordinates for L1 insertions between assemblies using the command line tool liftOver (version 134 for Mac OSX, http://genome.ucsc.edu/). The following parameters were used: the minimum ratio of bases that must remap was set to 0.85 (–minMatch), and multiple output regions were not allowed. The program and the appropriate chain files (e.g., human–chimpanzee and human–macaque) can be downloaded from the UCSC website (http://genome.ucsc.edu/). The converted L1 element insertions were compared with the RepeatMasker track files for the target species’ L1 families (e.g., chimpanzee and macaque). Records that did not correspond to the query L1 family, L1 sequences >10 kb, and multiple hits with overlapping regions or ambiguous coordinate information in the target genome were removed from the data set.
Determination of the current consensus sequences of L1 families
Full-length L1 elements were isolated from the human, chimpanzee, and macaque databases to build what we refer to as current L1-family-specific consensus sequences for each species. Current, because they are derived from L1 sequences in the modern (current) genomes of the species examined. The current consensus sequences are distinguished from ancestral consensus sequences in which the ancestral state of the CpGs has been restored (see next section). We aligned the full-length elements with the multiple sequence alignment application MUSCLE (Edgar 2004). The alignments were checked by hand using the multiple sequence alignment editor SEAVIEW (Galtier et al. 1996). Consensus sequences with 60% similarity threshold were built for each species and each L1 family studied (L1Pa2, L1Pa3, L1Pa4, L1Pa5, L1Pa6, L1Pa7). Based on these 60% threshold consensus sequences, multi-species and family-specific L1 consensus sequences were obtained using a 100% similarity threshold. The number of full-length elements used for each consensus sequence is given in Supplemental Table S10.
Determining the ancestral L1 family-specific consensus sequences
We reconstructed the ancestral L1 family-specific consensus sequences by restoring CpG sites to the family-specific current consensus sequences. We put all of the consensus sequences in the same register by alignment to the modern L1.3 element. Supplemental Figure S5 shows that these sequences are highly similar (also see Boissinot et al. 2000; Khan et al. 2006). We also aligned all of the orthologs for each family to their respective multi-species L1 family-specific consensus sequences and thereby determined the base frequency in the ortholog population at every position in the consensus sequence. As we could unambiguously align all of the corresponding nucleotide positions, we could determine whether CpGs of younger families corresponded to TpGs (or CpAs) of older families. In addition, by using the sequence information from the aligned orthologs, we could: (1) in many cases confirm that the TpGs (CpAs) in older families, which correspond to CpGs in younger families, were actually CpGs because some of the orthologs of the older families still retained CpGs at these sites; (2) infer the ancestral sequence of CpN or NpA sites; (3) confirm the presence of ancestral CpG sites, i.e., CpGs that were present only in ancestral L1 families. An example of (2) is shown in Supplemental Table S8. Several examples of (3) are pointed out in Figure 3. Deriving ancestral consensus L1 sequences as described above is quite straightforward and has been used successfully before; e.g., to resuscitate an active 5′ UTR L1 promoter sequence from current inactive ancestral mouse L1 5′ UTR sequences (Adey et al. 1994) and, in our laboratory, to construct an active ancestral L1Pa5 ORF1p protein from current defective ancestral versions of this sequence (A.V.F. and J.-C.W., unpubl.).
Sequence alignment and other DNA sequence manipulations
We used MUSCLE to align the members of each ortholog pair using the family-specific ancestral L1 consensus as a reference sequence. Non-L1 DNA sequences were masked and excluded from the alignments, as were ambiguous regions in the L1 sequences, i.e., the start point of the 5′ UTR and the poly-A tail. We used EMBOSS (European Molecular Biology Open Software Suite; Rice et al. 2000) for general sequence handling and sequence comparisons and generated custom UNIX, Perl, and Python scripts as necessary.
Divergence calculations
Base-by-base divergence
A site was considered if it contained a nonambiguous nucleotide (e.g., A, T, C, or G) present in both lineages (i.e., human, chimpanzee) and any nucleotide (A, T, C, G, or N) present in the consensus sequence; thereby, gaps and insertions were ignored. Divergence at each base was calculated by dividing the number of times a given site was different between humans and the chimpanzee by the total number of ortholog pairs compared for that particular site. This method was used to obtain data like that shown in Figure 3.
Overall non-CpG ortholog divergence
We masked and excluded all known mutational hot spots: the G-rich polypurine tract (GRPPT) in the 3′ UTR, all the dinucleotide sites corresponding to CpG sites in the relevant ancestral consensus sequence, and any other CpG present in a particular ortholog alignment. Pairwise divergence was determined by dividing the number of nucleotide sites different between a pair of orthologous L1 sequences by the total number of aligned sites (Ebersberger et al. 2002) from these fully masked orthologous sequences alignments. The same criteria as above for the base-wise divergence was applied: i.e., only nonambiguous nucleotides for both human and chimpanzee that corresponded to a base present in the consensus were considered, and gaps and insertions were ignored. Divergence values were not corrected for superimposed or back mutations as the number of substitutions was small. This method was used to obtain data like that shown in Figure 4.
Overall CpG and non-CpG mutation rates
For calculating these rates we only considered sites where at least one species (human or chimpanzee) is identical to the family-specific species-wide consensus sequence. Ambiguous nucleotides and gaps were ignored. We also eliminated sites directly flanked by an insertion or a deletion in any one of the three sequences (i.e., either member of the ortholog pair or the consensus). This restriction assures that arbitrary placement of nucleotides on either side of a gap during alignments does not skew the divergence measurements (Khelifi et al. 2006). We also excluded the hypermutable GRPPT in the 3′ UTR (see above).
We divided the sequences into CpG and non-CpG sites. CpG sites were defined as described above in the sections on “Determining the ancestral L1 family-specific consensus sequences.” For the determination of non-CpG divergence we also excluded CCG and CGG sites (Meunier and Duret 2004). To determine the numbers of non-CpG and CpG mutations, we compared all of the chimpanzee and human orthologs to the species-wide consensus sequences and counted those instances where either of the two orthologs differed from the consensus as a mutation. This method was used to obtain data like that shown in Figure 5.
Estimation of L1 family ages
We estimated the age of the L1 families using the median non-CpG divergence, D, of the orthologs for each family, and estimated times, t, of the human (Hsa)/chimpanzee (Ptr) divergence of 4–7 Mya, and of the Hsa/macaque (Mmu) divergence of 23–28 Mya. These species divergence times were the consensus of those reported in several studies (Goodman et al. 1998; Chen and Li 2001; Glazko and Nei 2003; Pilbeam and Young 2004; Patterson et al. 2006). Thus, the range of the above divergence times, tHP and tHM, were used in the following formulae: DHsa/Con / (DHsa/Ptr * 2 tHP) or DPtr/Con / (DHsa/Ptr * 2tHP); DHsa/Con / (DHsa/Mmu * 2 tHP) or DPtr/Con / (DPtr/Mmu * 2tHP). The 2 in the denominator assumes that the divergence of any pair orthologs from each other will be twice that of either member of the pair from the family consensus (Con); i.e., equal rates of base substitution in each branch. The calculations also assume that similar neutral substitution rates governed the divergence of the ortholog pairs along all the branches of the relevant primate lineages. Although the latter assumption might not be strictly true, the correspondence between the recovery of the various L1 family orthologs from the various species, the estimated ages of the L1 families, and the times of divergence of Hsa, Ptr, and Mmu indicate that the assumption is reasonable enough for ordering the L1 families by age. In addition, our ordering of the L1 families agreed completely with a recent analysis based on an entirely different method for determining the relative age of L1 families in primates (Giordano et al. 2007).
Recombination rates
We extracted the deCODE recombination rates (1-Mb intervals) (Kong et al. 2002) from the recombRate table associated with the hg18 build on the UCSC genome browser. We mapped the average female/male recombination rates (i.e., the decodeAvg) onto each of the human orthologs for each L1 family.
Statistical analysis
A χ2 goodness-of-fit test showed that the CpG dinucleotide and G + C content (Table 2) were normally distributed. Therefore, the SE of these determinations was calculated on this basis. Except as indicated, all statistical calculations were performed with the R project for statistical computing (www.R-project.org; R_Development_Core_Team 2007). One-way ANOVA was carried out with Holms correction for multiple comparisons using the statistical tools in KaleidaGraph (www.synergy.com).
Acknowledgments
We thank Deborah M. Hinton and Natalia Wesolowska for their valuable suggestions during the preparation of the manuscript. We are grateful to the UCSC Genome Browser Database group for providing various sequences files and programs. Furthermore, we thank Marcos Antezana (U. Chicago) for stimulating discussions on mutational aspects of the response to foreign DNA in eukaryotes. The Intramural Research Program of the NIH, NIDDK, funded A.V.F. and J.-C.W., and the National Museum of Natural History, France funded L.P.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.076455.108.
References
- Adey N.B., Tollefsbol T.O., Sparks A.B., Edgell M.H., Hutchison C.A.I., Tollefsbol T.O., Sparks A.B., Edgell M.H., Hutchison C.A.I., Sparks A.B., Edgell M.H., Hutchison C.A.I., Edgell M.H., Hutchison C.A.I., Hutchison C.A.I. Molecular resurrection of an extinct ancestral promoter for mouse L1. Proc. Natl. Acad. Sci. 1994;91:1569–1573. doi: 10.1073/pnas.91.4.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asthana S., Schmidt S., Sunyaev S., Schmidt S., Sunyaev S., Sunyaev S. A limited role for balancing selection. Trends Genet. 2005;21:30–32. doi: 10.1016/j.tig.2004.11.001. [DOI] [PubMed] [Google Scholar]
- Bird A.P. DNA methylation and the frequency of CpG in animal DNA. Nucleic Acids Res. 1980;8:1499–1504. doi: 10.1093/nar/8.7.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. DNA methylation patterns and epigenetic memory. Genes & Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- Bohossian H.B., Skaletsky H., Page D.C., Skaletsky H., Page D.C., Page D.C. Unexpected similar rates of nucleotide substitution found in male and female hominids. Nature. 2000;406:622–625. doi: 10.1038/35020557. [DOI] [PubMed] [Google Scholar]
- Boissinot S., Chevret P., Furano A.V., Chevret P., Furano A.V., Furano A.V. L1 (LINE-1) retrotransposon evolution and amplification in recent human history. Mol. Biol. Evol. 2000;17:915–928. doi: 10.1093/oxfordjournals.molbev.a026372. [DOI] [PubMed] [Google Scholar]
- Boissinot S., Entezam A., Furano A.V., Entezam A., Furano A.V., Furano A.V. Selection against deleterious LINE-1-containing loci in the human lineage. Mol. Biol. Evol. 2001;18:926–935. doi: 10.1093/oxfordjournals.molbev.a003893. [DOI] [PubMed] [Google Scholar]
- Casane D., Boissinot S., Chang B.H., Shimmin L.C., Li W., Boissinot S., Chang B.H., Shimmin L.C., Li W., Chang B.H., Shimmin L.C., Li W., Shimmin L.C., Li W., Li W. Mutation pattern variation among regions of the primate genome. J. Mol. Evol. 1997;45:216–226. doi: 10.1007/pl00006223. [DOI] [PubMed] [Google Scholar]
- Chen F.C., Li W.H., Li W.H. Genomic divergences between humans and other hominoids and the effective population size of the common ancestor of humans and chimpanzees. Am. J. Hum. Genet. 2001;68:444–456. doi: 10.1086/318206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Chimpanzee Sequencing and Analysis Consortium Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- Cooper D.M., Schimenti K.J., Schimenti J.C., Schimenti K.J., Schimenti J.C., Schimenti J.C. Factors affecting ectopic gene conversion in mice. Mamm. Genome. 1998;9:355–360. doi: 10.1007/s003359900769. [DOI] [PubMed] [Google Scholar]
- Coulondre C., Miller J.H., Farabaugh P.J., Gilbert W., Miller J.H., Farabaugh P.J., Gilbert W., Farabaugh P.J., Gilbert W., Gilbert W. Molecular basis of base substitution hotspots in Escherichia coli. Nature. 1978;274:775–780. doi: 10.1038/274775a0. [DOI] [PubMed] [Google Scholar]
- Dermitzakis E.T., Reymond A., Scamuffa N., Ucla C., Kirkness E., Rossier C., Antonarakis S.E., Reymond A., Scamuffa N., Ucla C., Kirkness E., Rossier C., Antonarakis S.E., Scamuffa N., Ucla C., Kirkness E., Rossier C., Antonarakis S.E., Ucla C., Kirkness E., Rossier C., Antonarakis S.E., Kirkness E., Rossier C., Antonarakis S.E., Rossier C., Antonarakis S.E., Antonarakis S.E. Evolutionary discrimination of mammalian conserved non-genic sequences (CNGs) Science. 2003;302:1033–1035. doi: 10.1126/science.1087047. [DOI] [PubMed] [Google Scholar]
- Dombroski B.A., Scott A.F., Kazazian H.H.J., Scott A.F., Kazazian H.H.J., Kazazian H.H.J. Two additional potential retrotransposons isolated from a human L1 subfamily that contains an active retrotransposable element. Proc. Natl. Acad. Sci. 1993;90:6513–6517. doi: 10.1073/pnas.90.14.6513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L., Galtier N., Galtier N. The covariation between TpA deficiency, CpG deficiency, and G+C content of human isochores is due to a mathematical artifact. Mol. Biol. Evol. 2000;17:1620–1625. doi: 10.1093/oxfordjournals.molbev.a026261. [DOI] [PubMed] [Google Scholar]
- Ebersberger I., Metzler D., Schwarz C., Paabo S., Metzler D., Schwarz C., Paabo S., Schwarz C., Paabo S., Paabo S. Genomewide comparison of DNA sequences between humans and chimpanzees. Am. J. Hum. Genet. 2002;70:1490–1497. doi: 10.1086/340787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich M., Wang R.Y., Wang R.Y. 5-Methylcytosine in eukaryotic DNA. Science. 1981;212:1350–1357. doi: 10.1126/science.6262918. [DOI] [PubMed] [Google Scholar]
- Ehrlich M., Gama-Sosa M.A., Huang L.H., Midgett R.M., Kuo K.C., McCune R.A., Gehrke C., Gama-Sosa M.A., Huang L.H., Midgett R.M., Kuo K.C., McCune R.A., Gehrke C., Huang L.H., Midgett R.M., Kuo K.C., McCune R.A., Gehrke C., Midgett R.M., Kuo K.C., McCune R.A., Gehrke C., Kuo K.C., McCune R.A., Gehrke C., McCune R.A., Gehrke C., Gehrke C. Amount and distribution of 5-methylcytosine in human DNA from different types of tissues of cells. Nucleic Acids Res. 1982;10:2709–2721. doi: 10.1093/nar/10.8.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegren H. Characteristics, causes and evolutionary consequences of male-biased mutation. Proc. Biol. Sci. 2007;274:1–10. doi: 10.1098/rspb.2006.3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furano A.V. The biological properties and evolutionary dynamics of mammalian LINE-1 retrotransposons. Prog. Nucleic Acid Res. Mol. Biol. 2000;64:255–294. doi: 10.1016/s0079-6603(00)64007-2. [DOI] [PubMed] [Google Scholar]
- Furano A.V., Duvernell D.D., Boissinot S., Duvernell D.D., Boissinot S., Boissinot S. L1 (LINE-1) retrotransposon diversity differs dramatically between mammals and fish. Trends Genet. 2004;20:9–14. doi: 10.1016/j.tig.2003.11.006. [DOI] [PubMed] [Google Scholar]
- Galtier N., Gouy M., Gautier C., Gouy M., Gautier C., Gautier C. SEAVIEW and PHYLO_WIN: Two graphic tools for sequence alignment and molecular phylogeny. Comput. Appl. Biosci. 1996;12:543–548. doi: 10.1093/bioinformatics/12.6.543. [DOI] [PubMed] [Google Scholar]
- Gilbert N., Boyle S., Fiegler H., Woodfine K., Carter N.P., Bickmore W.A., Boyle S., Fiegler H., Woodfine K., Carter N.P., Bickmore W.A., Fiegler H., Woodfine K., Carter N.P., Bickmore W.A., Woodfine K., Carter N.P., Bickmore W.A., Carter N.P., Bickmore W.A., Bickmore W.A. Chromatin architecture of the human genome: Gene-rich domains are enriched in open chromatin fibers. Cell. 2004;118:555–566. doi: 10.1016/j.cell.2004.08.011. [DOI] [PubMed] [Google Scholar]
- Giordano J., Ge Y., Gelfand Y., Abrusan G., Benson G., Warburton P.E., Ge Y., Gelfand Y., Abrusan G., Benson G., Warburton P.E., Gelfand Y., Abrusan G., Benson G., Warburton P.E., Abrusan G., Benson G., Warburton P.E., Benson G., Warburton P.E., Warburton P.E. Evolutionary history of mammalian transposons determined by genome-wide defragmentation. PLoS Comput. Biol. 2007;3:e137. doi: 10.1371/journal.pcbi.0030137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazko G.V., Nei M., Nei M. Estimation of divergence times for major lineages of primate species. Mol. Biol. Evol. 2003;20:424–434. doi: 10.1093/molbev/msg050. [DOI] [PubMed] [Google Scholar]
- Goodman M.F. Error-prone repair DNA polymerases in prokaryotes and eukaryotes. Annu. Rev. Biochem. 2002;71:17–50. doi: 10.1146/annurev.biochem.71.083101.124707. [DOI] [PubMed] [Google Scholar]
- Goodman M., Porter C.A., Czelusniak J., Page S.L., Schneider H., Shoshani J., Gunnell G., Groves C.P., Porter C.A., Czelusniak J., Page S.L., Schneider H., Shoshani J., Gunnell G., Groves C.P., Czelusniak J., Page S.L., Schneider H., Shoshani J., Gunnell G., Groves C.P., Page S.L., Schneider H., Shoshani J., Gunnell G., Groves C.P., Schneider H., Shoshani J., Gunnell G., Groves C.P., Shoshani J., Gunnell G., Groves C.P., Gunnell G., Groves C.P., Groves C.P. Toward a phylogenetic classification of primates based on DNA evidence complemented by fossil evidence. Mol. Phylogenet. Evol. 1998;9:585–598. doi: 10.1006/mpev.1998.0495. [DOI] [PubMed] [Google Scholar]
- Han K., Konkel M.K., Xing J., Wang H., Lee J., Meyer T.J., Huang C.T., Sandifer E., Hebert K., Barnes E.W., Konkel M.K., Xing J., Wang H., Lee J., Meyer T.J., Huang C.T., Sandifer E., Hebert K., Barnes E.W., Xing J., Wang H., Lee J., Meyer T.J., Huang C.T., Sandifer E., Hebert K., Barnes E.W., Wang H., Lee J., Meyer T.J., Huang C.T., Sandifer E., Hebert K., Barnes E.W., Lee J., Meyer T.J., Huang C.T., Sandifer E., Hebert K., Barnes E.W., Meyer T.J., Huang C.T., Sandifer E., Hebert K., Barnes E.W., Huang C.T., Sandifer E., Hebert K., Barnes E.W., Sandifer E., Hebert K., Barnes E.W., Hebert K., Barnes E.W., Barnes E.W., et al. Mobile DNA in Old World monkeys: A glimpse through the rhesus macaque genome. Science. 2007;316:238–240. doi: 10.1126/science.1139462. [DOI] [PubMed] [Google Scholar]
- Hardison R.C., Roskin K.M., Yang S., Diekhans M., Kent W.J., Weber R., Elnitski L., Li J., O'Connor M., Kolbe D., Roskin K.M., Yang S., Diekhans M., Kent W.J., Weber R., Elnitski L., Li J., O'Connor M., Kolbe D., Yang S., Diekhans M., Kent W.J., Weber R., Elnitski L., Li J., O'Connor M., Kolbe D., Diekhans M., Kent W.J., Weber R., Elnitski L., Li J., O'Connor M., Kolbe D., Kent W.J., Weber R., Elnitski L., Li J., O'Connor M., Kolbe D., Weber R., Elnitski L., Li J., O'Connor M., Kolbe D., Elnitski L., Li J., O'Connor M., Kolbe D., Li J., O'Connor M., Kolbe D., O'Connor M., Kolbe D., Kolbe D., et al. Covariation in frequencies of substitution, deletion, transposition, and recombination during eutherian evolution. Genome Res. 2003;13:13–26. doi: 10.1101/gr.844103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmann I., Prufer K., Ji H., Zody M.C., Paabo S., Ptak S.E., Prufer K., Ji H., Zody M.C., Paabo S., Ptak S.E., Ji H., Zody M.C., Paabo S., Ptak S.E., Zody M.C., Paabo S., Ptak S.E., Paabo S., Ptak S.E., Ptak S.E. Why do human diversity levels vary at a megabase scale? Genome Res. 2005;15:1222–1231. doi: 10.1101/gr.3461105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang D.G., Green P., Green P. Bayesian Markov chain Monte Carlo sequence analysis reveals varying neutral substitution patterns in mammalian evolution. Proc. Natl. Acad. Sci. 2004;101:13994–14001. doi: 10.1073/pnas.0404142101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indiani C., McInerney P., Georgescu R., Goodman M.F., O’Donnell M., McInerney P., Georgescu R., Goodman M.F., O’Donnell M., Georgescu R., Goodman M.F., O’Donnell M., Goodman M.F., O’Donnell M., O’Donnell M. A sliding-clamp toolbelt binds high- and low-fidelity DNA polymerases simultaneously. Mol. Cell. Biol. 2005;19:805–815. doi: 10.1016/j.molcel.2005.08.011. [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Jaenisch R., Bird A., Bird A. Epigenetic regulation of gene expression: How the genome integrates intrinsic and environmental signals. Nat. Genet. 2003;33(Suppl.):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- Kazazian H.H. Mobile elements: Drivers of genome evolution. Science. 2004;303:1626–1632. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- Kehrer-Sawatzki H., Cooper D.N., Cooper D.N. Structural divergence between the human and chimpanzee genomes. Hum. Genet. 2007;120:759–778. doi: 10.1007/s00439-006-0270-6. [DOI] [PubMed] [Google Scholar]
- Khan H., Smit A., Boissinot S., Smit A., Boissinot S., Boissinot S. Molecular evolution and tempo of amplification of human LINE-1 retrotransposons since the origin of primates. Genome Res. 2006;16:78–87. doi: 10.1101/gr.4001406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khelifi A., Meunier J., Duret L., Mouchiroud D., Meunier J., Duret L., Mouchiroud D., Duret L., Mouchiroud D., Mouchiroud D. GC content evolution of the human and mouse genomes: Insights from the study of processed pseudogenes in regions of different recombination rates. J. Mol. Evol. 2006;62:745–752. doi: 10.1007/s00239-005-0186-0. [DOI] [PubMed] [Google Scholar]
- Kong A., Gudbjartsson D.F., Sainz J., Jonsdottir G.M., Gudjonsson S.A., Richardsson B., Sigurdardottir S., Barnard J., Hallbeck B., Masson G., Gudbjartsson D.F., Sainz J., Jonsdottir G.M., Gudjonsson S.A., Richardsson B., Sigurdardottir S., Barnard J., Hallbeck B., Masson G., Sainz J., Jonsdottir G.M., Gudjonsson S.A., Richardsson B., Sigurdardottir S., Barnard J., Hallbeck B., Masson G., Jonsdottir G.M., Gudjonsson S.A., Richardsson B., Sigurdardottir S., Barnard J., Hallbeck B., Masson G., Gudjonsson S.A., Richardsson B., Sigurdardottir S., Barnard J., Hallbeck B., Masson G., Richardsson B., Sigurdardottir S., Barnard J., Hallbeck B., Masson G., Sigurdardottir S., Barnard J., Hallbeck B., Masson G., Barnard J., Hallbeck B., Masson G., Hallbeck B., Masson G., Masson G., et al. A high-resolution recombination map of the human genome. Nat. Genet. 2002;31:241–247. doi: 10.1038/ng917. [DOI] [PubMed] [Google Scholar]
- Lee J., Cordaux R., Han K., Wang J., Hedges D.J., Liang P., Batzer M.A., Cordaux R., Han K., Wang J., Hedges D.J., Liang P., Batzer M.A., Han K., Wang J., Hedges D.J., Liang P., Batzer M.A., Wang J., Hedges D.J., Liang P., Batzer M.A., Hedges D.J., Liang P., Batzer M.A., Liang P., Batzer M.A., Batzer M.A. Different evolutionary fates of recently integrated human and chimpanzee LINE-1 retrotransposons. Gene. 2007;390:18–27. doi: 10.1016/j.gene.2006.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lercher M.J., Williams E.J., Hurst L.D., Williams E.J., Hurst L.D., Hurst L.D. Local similarity in evolutionary rates extends over whole chromosomes in human-rodent and mouse-rat comparisons: Implications for understanding the mechanistic basis of the male mutation bias. Mol. Biol. Evol. 2001;18:2032–2039. doi: 10.1093/oxfordjournals.molbev.a003744. [DOI] [PubMed] [Google Scholar]
- Makova K.D., Li W.-H., Li W.-H. Strong male-driven evolution of DNA sequences in humans and apes. Nature. 2002;416:624–626. doi: 10.1038/416624a. [DOI] [PubMed] [Google Scholar]
- Malcom C.M., Wyckoff G.J., Lahn B.T., Wyckoff G.J., Lahn B.T., Lahn B.T. Genic mutation rates in mammals: Local similarity, chromosomal heterogeneity, and X-versus-autosome disparity. Mol. Biol. Evol. 2003;20:1633–1641. doi: 10.1093/molbev/msg178. [DOI] [PubMed] [Google Scholar]
- McNaughton J.C., Cockburn D.J., Hughes G., Jones W.A., Laing N.G., Ray P.N., Stockwell P.A., Petersen G.B., Cockburn D.J., Hughes G., Jones W.A., Laing N.G., Ray P.N., Stockwell P.A., Petersen G.B., Hughes G., Jones W.A., Laing N.G., Ray P.N., Stockwell P.A., Petersen G.B., Jones W.A., Laing N.G., Ray P.N., Stockwell P.A., Petersen G.B., Laing N.G., Ray P.N., Stockwell P.A., Petersen G.B., Ray P.N., Stockwell P.A., Petersen G.B., Stockwell P.A., Petersen G.B., Petersen G.B. Is gene deletion in eukaryotes sequence-dependent? A study of nine deletion junctions and nineteen other deletion breakpoints in intron 7 of the human dystrophin gene. Gene. 1998;222:41–51. doi: 10.1016/s0378-1119(98)00466-1. [DOI] [PubMed] [Google Scholar]
- Meunier J., Duret L., Duret L. Recombination drives the evolution of GC-content in the human genome. Mol. Biol. Evol. 2004;21:984–990. doi: 10.1093/molbev/msh070. [DOI] [PubMed] [Google Scholar]
- Mills R.E., Bennett E.A., Iskow R.C., Luttig C.T., Tsui C., Pittard W.S., Devine S.E., Bennett E.A., Iskow R.C., Luttig C.T., Tsui C., Pittard W.S., Devine S.E., Iskow R.C., Luttig C.T., Tsui C., Pittard W.S., Devine S.E., Luttig C.T., Tsui C., Pittard W.S., Devine S.E., Tsui C., Pittard W.S., Devine S.E., Pittard W.S., Devine S.E., Devine S.E. Recently mobilized transposons in the human and chimpanzee genomes. Am. J. Hum. Genet. 2006;78:671–679. doi: 10.1086/501028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata T., Hayashida H., Kuma K., Mitsuyasu K., Yasunaga T., Hayashida H., Kuma K., Mitsuyasu K., Yasunaga T., Kuma K., Mitsuyasu K., Yasunaga T., Mitsuyasu K., Yasunaga T., Yasunaga T. Male-driven molecular evolution: A model and nucleotide sequence analysis. Cold Spring Harb. Symp. Quant. Biol. 1987;52:863–867. doi: 10.1101/sqb.1987.052.01.094. [DOI] [PubMed] [Google Scholar]
- Mouse Genome Sequencing Consortium Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Nachman M.W., Crowell S.L., Crowell S.L. Estimate of the mutation rate per nucleotide in humans. Genetics. 2000;156:297–304. doi: 10.1093/genetics/156.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orend G., Knoblauch M., Kammer C., Tjia S.T., Schmitz B., Linkwitz A., Meyer G., Maas J., Doerfler W., Knoblauch M., Kammer C., Tjia S.T., Schmitz B., Linkwitz A., Meyer G., Maas J., Doerfler W., Kammer C., Tjia S.T., Schmitz B., Linkwitz A., Meyer G., Maas J., Doerfler W., Tjia S.T., Schmitz B., Linkwitz A., Meyer G., Maas J., Doerfler W., Schmitz B., Linkwitz A., Meyer G., Maas J., Doerfler W., Linkwitz A., Meyer G., Maas J., Doerfler W., Meyer G., Maas J., Doerfler W., Maas J., Doerfler W., Doerfler W. The initiation of de novo methylation of foreign DNA integrated into a mammalian genome is not exclusively targeted by nucleotide sequence. J. Virol. 1995;69:1226–1242. doi: 10.1128/jvi.69.2.1226-1242.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson N., Richter D.J., Gnerre S., Lander E.S., Reich D., Richter D.J., Gnerre S., Lander E.S., Reich D., Gnerre S., Lander E.S., Reich D., Lander E.S., Reich D., Reich D. Genetic evidence for complex speciation of humans and chimpanzees. Nature. 2006;441:1103–1108. doi: 10.1038/nature04789. [DOI] [PubMed] [Google Scholar]
- Pennings S., Allan J., Davey C.S., Allan J., Davey C.S., Davey C.S. DNA methylation, nucleosome formation and positioning. Brief. Funct. Genomic. Proteomic. 2005;3:351–361. doi: 10.1093/bfgp/3.4.351. [DOI] [PubMed] [Google Scholar]
- Pilbeam D., Young N., Young N. Hominoid evolution: Synthesizing disparate data. Comptes Rendus Palevol. 2004;3:444–456. [Google Scholar]
- Prendergast J.G., Campbell H., Gilbert N., Dunlop M.G., Bickmore W.A., Semple C.A., Campbell H., Gilbert N., Dunlop M.G., Bickmore W.A., Semple C.A., Gilbert N., Dunlop M.G., Bickmore W.A., Semple C.A., Dunlop M.G., Bickmore W.A., Semple C.A., Bickmore W.A., Semple C.A., Semple C.A. Chromatin structure and evolution in the human genome. BMC Evol. Biol. 2007;7:72. doi: 10.1186/1471-2148-7-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R_Development_Core_Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna: 2007. [Google Scholar]
- Ramsahoye B.H., Biniszkiewicz D., Lyko F., Clark V., Bird A.P., Jaenisch R., Biniszkiewicz D., Lyko F., Clark V., Bird A.P., Jaenisch R., Lyko F., Clark V., Bird A.P., Jaenisch R., Clark V., Bird A.P., Jaenisch R., Bird A.P., Jaenisch R., Jaenisch R. Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc. Natl. Acad. Sci. 2000;97:5237–5242. doi: 10.1073/pnas.97.10.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattray A.J., Strathern J.N., Strathern J.N. Error-prone DNA polymerases: When making a mistake is the only way to get ahead. Annu. Rev. Genet. 2003;37:31–66. doi: 10.1146/annurev.genet.37.042203.132748. [DOI] [PubMed] [Google Scholar]
- Remus R., Kammer C., Heller H., Schmitz B., Schell G., Doerfler W., Kammer C., Heller H., Schmitz B., Schell G., Doerfler W., Heller H., Schmitz B., Schell G., Doerfler W., Schmitz B., Schell G., Doerfler W., Schell G., Doerfler W., Doerfler W. Insertion of foreign DNA into an established mammalian genome can alter the methylation of cellular DNA sequences. J. Virol. 1999;73:1010–1022. doi: 10.1128/jvi.73.2.1010-1022.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P., Longden I., Bleasby A., Longden I., Bleasby A., Bleasby A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Richard M., Belmaaza A., Gusew N., Wallenburg J.C., Chartrand P., Belmaaza A., Gusew N., Wallenburg J.C., Chartrand P., Gusew N., Wallenburg J.C., Chartrand P., Wallenburg J.C., Chartrand P., Chartrand P. Integration of a vector containing a repetitive LINE-1 element in the human genome. Mol. Cell. Biol. 1994;14:6689–6695. doi: 10.1128/mcb.14.10.6689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit A.F.A., Tóth G., Riggs A.D., Jurka J., Tóth G., Riggs A.D., Jurka J., Riggs A.D., Jurka J., Jurka J. Ancestral, mammalian-wide subfamilies of LINE-1 repetitive sequences. J. Mol. Biol. 1995;246:401–417. doi: 10.1006/jmbi.1994.0095. [DOI] [PubMed] [Google Scholar]
- Song M., Boissinot S., Boissinot S. Selection against LINE-1 retrotransposons results principally from their ability to mediate ectopic recombination. Gene. 2007;390:206–213. doi: 10.1016/j.gene.2006.09.033. [DOI] [PubMed] [Google Scholar]
- Strelen J.C. Proceedings of the 36th Conference on Winter Simulation. Winter Simulation Conference; Washington, DC: 2004. The accuracy of a new confidence interval method; pp. 654–662. [Google Scholar]
- Taylor J., Tyekucheva S., Zody M., Chiaromonte F., Makova K.D., Tyekucheva S., Zody M., Chiaromonte F., Makova K.D., Zody M., Chiaromonte F., Makova K.D., Chiaromonte F., Makova K.D., Makova K.D. Strong and weak male mutation bias at different sites in the primate genomes: Insights from the human–chimpanzee comparison. Mol. Biol. Evol. 2006;23:565–573. doi: 10.1093/molbev/msj060. [DOI] [PubMed] [Google Scholar]
- Thomas J.W., Touchman J.W., Blakesley R.W., Bouffard G.G., Beckstrom-Sternberg S.M., Margulies E.H., Blanchette M., Siepel A.C., Thomas P.J., McDowell J.C., Touchman J.W., Blakesley R.W., Bouffard G.G., Beckstrom-Sternberg S.M., Margulies E.H., Blanchette M., Siepel A.C., Thomas P.J., McDowell J.C., Blakesley R.W., Bouffard G.G., Beckstrom-Sternberg S.M., Margulies E.H., Blanchette M., Siepel A.C., Thomas P.J., McDowell J.C., Bouffard G.G., Beckstrom-Sternberg S.M., Margulies E.H., Blanchette M., Siepel A.C., Thomas P.J., McDowell J.C., Beckstrom-Sternberg S.M., Margulies E.H., Blanchette M., Siepel A.C., Thomas P.J., McDowell J.C., Margulies E.H., Blanchette M., Siepel A.C., Thomas P.J., McDowell J.C., Blanchette M., Siepel A.C., Thomas P.J., McDowell J.C., Siepel A.C., Thomas P.J., McDowell J.C., Thomas P.J., McDowell J.C., McDowell J.C., et al. Comparative analyses of multi-species sequences from targeted genomic regions. Nature. 2003;424:788–793. doi: 10.1038/nature01858. [DOI] [PubMed] [Google Scholar]
- Usdin K., Grabczyk E., Grabczyk E. DNA repeat expansions and human disease. Cell. Mol. Life Sci. 2000;57:914–931. doi: 10.1007/PL00000734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaisman A., Woodgate R., Woodgate R. Unique misinsertion specificity of poliota may decrease the mutagenic potential of deaminated cytosines. EMBO J. 2001;20:6520–6529. doi: 10.1093/emboj/20.22.6520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaisman A., Tissier A., Frank E.G., Goodman M.F., Woodgate R., Tissier A., Frank E.G., Goodman M.F., Woodgate R., Frank E.G., Goodman M.F., Woodgate R., Goodman M.F., Woodgate R., Woodgate R. Human DNA polymerase iota promiscuous mismatch extension. J. Biol. Chem. 2001;276:30615–30622. doi: 10.1074/jbc.M102694200. [DOI] [PubMed] [Google Scholar]
- Voliva C.F., Jahn C.L., Comer M.B., Hutchison C.A., Edgell M.H., Jahn C.L., Comer M.B., Hutchison C.A., Edgell M.H., Comer M.B., Hutchison C.A., Edgell M.H., Hutchison C.A., Edgell M.H., Edgell M.H. The L1Md long interspersed repeat family in the mouse: Almost all examples are truncated at one end. Nucleic Acids Res. 1983;11:8847–8859. doi: 10.1093/nar/11.24.8847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh C.P., Xu G.L., Xu G.L. Cytosine methylation and DNA repair. Curr. Top. Microbiol. Immunol. 2006;301:283–315. doi: 10.1007/3-540-31390-7_11. [DOI] [PubMed] [Google Scholar]
- Woodcock D.M., Lawler C.B., Linsenmeyer M.E., Doherty J.P., Warren W.D., Lawler C.B., Linsenmeyer M.E., Doherty J.P., Warren W.D., Linsenmeyer M.E., Doherty J.P., Warren W.D., Doherty J.P., Warren W.D., Warren W.D. Asymmetric methylation in the hypermethylated CpG promoter region of the human L1 retrotransposon. J. Biol. Chem. 1997;272:7810–7816. doi: 10.1074/jbc.272.12.7810. [DOI] [PubMed] [Google Scholar]
- Yoder J.A., Walsh C.P., Bestor T.H., Walsh C.P., Bestor T.H., Bestor T.H. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997;13:335–340. doi: 10.1016/s0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]