Abstract
Oligonucleotides containing locked nucleic acid bases (LNAs) have increased affinity for complementary DNA sequences. We hypothesized that enhanced affinity might allow LNAs to recognize chromosomal DNA inside human cells and inhibit gene expression. To test this hypothesis, we synthesized antigene LNAs (agLNAs) complementary to sequences within the promoters of progesterone receptor (PR) and androgen receptor (AR). We observed inhibition of AR and PR expression by agLNAs but not by analogous oligomers containing 2'-methoxyethyl bases or noncomplementary LNAs. Inhibition was dose dependent and exhibited IC50 values of <10 nM. Efficient inhibition depended on the length of the agLNA, the location of LNA bases, the number of LNA substitutions, and the location of the target sequence within the targeted promoter. LNAs targeting sequences at or near transcription start sites yielded better inhibition than LNAs targeting transcription factor binding sites or an inverted repeat. These results demonstrate that agLNAs can recognize chromosomal target sequences and efficiently block gene expression. agLNAs could be used, for gene silencing, as cellular probes for chromosome structure, and therapeutic applications.
The development of oligonucleotides and oligonucleotide mimics for sequence-specific recognition of chromosomal DNA inside cells confronts several challenges (1). Compounds must be able to enter cells, pass into the nucleus, and bind chromosomal DNA with high specificity. Binding must occur in spite of base-pairing at the target site and complexation of the genomic sequence with histones, transcription factors, and other DNA binding proteins. Once bound, the association of an oligomer with the chromosome must be sufficiently stable and long-lasting to affect gene expression. These challenges make recognition of DNA more complex than recognition of mRNA and overcoming them requires understanding the chemical, biophysical, and biological properties of native nucleic acids and their chemically modified analogs and mimics.
We have shown that peptide nucleic acids1 (PNAs) (Figure 1), a class of DNA/RNA mimic with an uncharged amide backbone (2,3), can target chromosomal DNA at transcription start sites and inhibit gene expression inside cells (4,5). Inhibition of gene expression by promoter-targeted antigene PNAs (agPNAs, we use the term antigene to differentiate molecules that are complementary to chromosomal DNA from antisense oligomers that are complementary to mRNA) demonstrated that synthetic molecules could access sequence information at transcription start sites and that binding was sufficient to block expression.
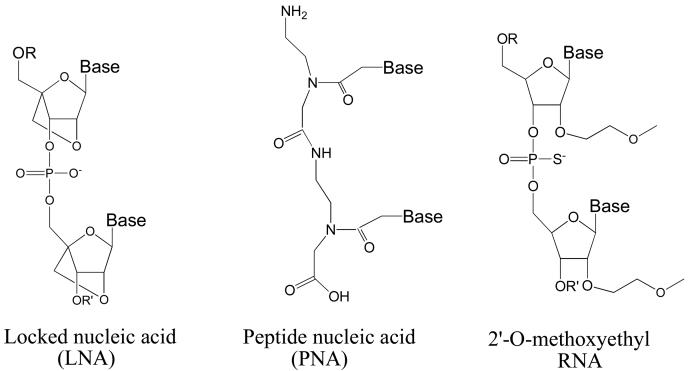
FIGURE 1.
Structures of LNA, PNA, and MOE bases.
The PNAs used in our previous studies were mixed sequences (i.e. containing all four PNA bases) and were designed to recognize their targets by Watson-Crick base-pairing and strand invasion rather than by triple helix formation (4,5). With their neutral amide backbone and propensity to invade duplex DNA, PNAs have unique structural and electronic properties that set them apart from single-stranded oligonucleotides that contain negatively charged phosphodiester backbones. Because of the special properties of PNA, we were uncertain whether other classes of oligomer could also achieve efficient Watson-Crick recognition of DNA inside cells.
We chose locked nucleic acids (LNAs) (6-9) to test whether efficient recognition of promoter sequences within chromosomal DNA could be extended to oligomers with phosphodiester backbones. LNA bases contain a methylene bridge that connects the 2'-oxygen of the ribose with the 4' carbon (Figure 1). This bridge results in a locked 3'-endo conformation, which reduces the conformational flexibility of the ribose. Reduced flexibility lowers the entropic penalty paid upon hybridization and yields a remarkable increase in hybridization affinity. Substitution of a single LNA base within a DNA or RNA oligomer can enhance melting temperature Tm values by 5 -10 °C, and multiple LNA substitutions allow Tm values to be precisely tailored for specific applications. The introduction of LNA bases also increases resistance to digestion by nucleases (10).
LNAs are an attractive option for nucleic acid recognition because it is synthetically straightforward to intersperse LNA, DNA, and RNA bases and the introduction of a relatively small number of LNA bases can dramatically enhance the affinity of an oligonucleotide for its complement. The strengths of LNA are being exploited for preclinical investigations and therapeutic development. LNAs can inhibit gene expression at the level of translation (11) or alter splicing in mouse models (12). In The mouse experiment demonstrating inhibition of translation also suggested that LNAs containing phosphorothioate substitutions can exhibit significant hepatotoxicity (11). It is possible, however, that revised LNA designs might avoid this problem. An LNA targeting Bcl-1 is being tested in Phase I/II trials for the treatment of Chronic Lymphocytic Leukemia (13,14).
Most attempts using LNAs to recognize cellular targets have focused on recognition of RNA (11,12,15-17)), but there have also been reports of recognition of DNA. Catchpole and colleagues demonstrated that LNAs could invade plasmid DNA and remain associated with plasmid after delivery into cells (18). Giovannageli and colleagues showed that triplex-forming LNAs could block gene expression from plasmids inside permeabilized cells and bind to genomic DNA in intact cells (19,20).
LNA substitutions couple the high affinity binding that characterizes recognition by PNAs with retention of the phosphodiester backbone of DNA or RNA. We hypothesized therefore, that LNAs would be ideal candidates for examining whether efficient inhibition of gene expression could be extended beyond PNAs to negatively-charged synthetic oligonucleotides.
Here, we observe that LNAs targeting transcription start sites within promoter DNA efficiently inhibit expression of progesterone receptor (PR) and androgen receptor (AR). Inhibition is dependent on target sequence, degree of LNA substitution, and LNA length. These experiments introduce LNA as an option for antigene inhibition of gene expression.
MATERIALS AND METHODS
Oligonucleotides
LNA oligonucleotides were synthesized and characterized by MALDI-TOF mass spectrometry at SIGMA-PROLIGO (Paris, France). Upon arrival each LNA was purified using Microspin™ G-25 Columns (Amersham Biosciences) according to the manufacturer's instructions. 2'-methoxyethyl oligonucleotides were synthesized at ISIS Pharmaceuticals. Concentrations were determined by UV-spectrophotometry. The absorbance for each LNA oligonucleotide at 260 nm was noted. Extinction coefficients for each LNA were provided by SIGMA-PROLIGO. Concentrations of 2'-methoxyethyl RNA oligonucleotides were calculated based on a 40 Mg/mL conversion per optical density unit at 260 nm. 20 MM stock solutions of each LNA or 2'-methoxyethyl RNA (2'-MOE) oligonucleotides were made and maintained at −20°C. Melting temperature (Tm) values were calculated using a web-based program (lna-tm.com) (42,43) assuming 0.1 M salt and a 1.5 μM concentration for each strand. For some duplexes, Tm values were experimentally determined in 0.1 M sodium monophosphate (NaH2PO4) pH 7.4 and a 1.5 μM concentration of each strand. Repeated Tm values for measurement of the same sample were within ± 2°C. Calculated and experimentally determined Tm values were within 1-3 °C.
Cell Culture
T-47D or MCF7 breast cancer cells were obtained from the American Type Culture Collection (ATCC) and maintained at 37°C and 5% CO2 in standard media: RPMI-1640 (ATCC) supplemented with 10% heat-inactivated (56°C, 1 h) fetal bovine serum (FBS, Gemini Bioproducts), 0.4 units mL−1 of bovine insulin, and 0.5% MEM nonessential amino acids (Sigma).
Lipid-mediated Transfection
T-47D cells were plated at 80,000 cells per six-well plate (Costar) two days before transfection (21). LNA or 2'-MOE oligonucleotides were transfected using Oligofectamine (Invitrogen) according to the manufacturer's instructions. Prior to transfection, LNA stocks were heated at 75°C for 6 min to dissolve any aggregates that may have formed. Per well on a 6-well plate, 25 nM (0.9 μL lipid) or 50 nM (1.8 μL lipid) single-stranded LNA or 2'-MOE oligonucleotides in Optimem (Invitrogen) were added to a final volume of 250 μL and were incubated for 20 min. Optimem was added to the LNA-lipid mixture for a final volume of 1.25 mL then added to cells. Media was exchanged 24 h later with supplemented RPMI as described above. On day 3, cells were passaged 1:3 into new six-well plates. Cells were transfected a second time on day 5. Cells were harvested on day 8.
MCF-7 cells were plated at 180,000 cells per six-well plate (Costar) two days before transfection. Transfection (Day 0) with LNAs was performed with Oligofectamine according to the manufacturer's instructions. Per well, 25 nM (0.9 μL lipid) or 50 nM (1.8 μL lipid) single-stranded LNA in Optimem were added to a final volume of 250 μL and were incubated for 20 minutes. Optimem was added to the LNA-lipid mixture for a final volume of 1.25 mL then added to cells. Media was exchanged 24 h later with supplemented RPMI as described above. On day 3, cells were passaged 1:3 into new six-well plates. Cells were transfected a second time on day 5. Cells were harvested on day 8.
Analysis of PR or AR Expression
Cells were harvested using trypsin. Cells were washed once with 1 × PBS buffer, which was then aspirated, and then the cells were treated with a trypsin solution (0.05 % Trypsin, 0.53 mM EDTA· 4Na, Invitrogen) at 37 °C for 2 min. Trypsin was inactivated using 800 μL of RPMI media. The contents of each well were transferred separately into 1.5 mL microfuge tubes and centrifuged at 3500 rpm for 15 min at 4 °C. Cells were then resuspended with 40-50 μL of ice-cold lysis buffer (120 mM Tris-base, pH 7.4, 120 mM NaCl, 1 mM Na2-EDTA, 1 mM DTT, 10 mM ß-glycerophosphate, 0.1 mM sodium fluoride, 0.1 mM sodium vanadate, 0.5 % v/v Nonidet P-40) containing Complete Protease Inhibitor Cocktail (Roche, Indianapolis, IN) and then stored at −80°C . After thawing on ice, samples were centrifuged at 12000 rpm for 15 min at 4 °C to pellet debris.
Protein concentration was determined for each sample in a 96-well plate format by the BCA method (Piece, Rockford, IL). Western analysis by SDS-PAGE was performed using standard methods. The membranes were blocked with 5% milk/PBS-Tween (Sigma) for 1 h and placed on a rocker platform with primary antibody rabbit monoclonal antibodies (1: 1000) raised against PR or AR (Cell Signaling, MA) in 5% milk/PBS-Tween overnight at 4 °C. The membranes were washed three times for 5 min each in PBS-Tween. Secondary antibody conjugate (HRP conjugate goat anti-rabbit or goat anti-mouse were diluted 1: 5000 in 5 % milk/PBS-Tween and placed on a rocker platform for 50 min at room temperature. Membranes were then washed three times with 15 min each in PBS-Tween. Each membrane was incubated for 4 min in 4 mL of Super Signal West Pico Chemiluminescent substrate (Pierce), then drained, placed in a transparent sheet protector, exposed to BioMax Light film (Eastman Kodak Company, Rochester, NY) for 1-60 s, and developed according to manufacture's recommendations. Control antibody was mouse anti-β-actin (Sigma).
RESULTS AND DISCUSSION
Design of LNAs
We chose to target sequences within the promoters for progesterone receptor (PR) (22-24) (Table 1). PR is expressed as two isoforms, PR-B and PR-A. Each isoform is encoded by its own promoter and the promoter for PR-B is upstream from the promoter for PR-A. The locations of the transcription start sites for PR had been independently determined by two laboratories (22,23) and were confirmed by 5'-RACE (25). All quantifications are based on inhibition of expression of PR-B. LNAs were complementary to either the sense or the antisense strand of the promoter. Unless otherwise noted, experiments were performed in T47D cells, a human breast cancer cell lines that expresses PR at high levels when grown in normal media.
Table 1.
LNA and 2′-MOE oligomers used in these studiesa.
| LNA | Target | (Sense/Antisense) | Sequence | Tm°C (Calc./Exp.) | |
|---|---|---|---|---|---|
| LNAs Complementary to Progesterone Receptor (PR) | |||||
| LNA | PR1 | −2/+17 | (Sense) | CCagtCCaCagCtgtCaCt | 81 |
| LNA | PR2 | −9/+10 | (Antisense) | gCTgTggaCTggCCagaCa | 88 |
| LNA | PR3 | −9/+10 | (Sense) | tGtctGGccAGtccAcAGc | 83 |
| LNA | PR4 | −47/−28 | (Sense) | TTcccTccTcccTGGaGac | 81 |
| LNA | PR5 | +126/+145 | (Sense) | tGaGctGaaGGcaaaGGGt | 84 |
| LNA | PR6 | +137/+156 | (Sense) | TCaTgaCTgagCTgaaggC | 79 |
| LNA | PR7 | −9/+7 | (Sense) | tGtctGGccAGtccAc | 75/76 |
| LNA | PR8 | −9/+4 | (Sense) | tGtctGGccAGtc | 68/67 |
| LNA | PR9 | −9/+1 | (Sense) | tGtctGGccA | 57/56 |
| LNA | PR10 | −9/+10 | (Sense) | tGtctGGccaGtccacaGc | 76/79 |
| LNA | PR11 | −9/+10 | (Sense) | tGTcTGGccAGTccAcAGc | 91 |
| LNA | PR12 | −11/+8 | (Sense) | gCTgtCTggCcagTccACa | 86 |
| LNA | PR13 | −52/−33 | (Sense) | TtTGggCGggGccTccCTa | 89 |
| LNA | PR14 | −70/−51 | (Sense) | aTtGgGgTaGggAgGggCt | 91 |
| LNA | PR15 | +90/+109 | (Sense) | gCTttCActTgtCaTtTGa | 73 |
| LNA | PR16 | +106/+125 | (Sense) | TgAgtgAaaTCtaCaaCcC | 71 |
| LNA | PR17 | −9 to +10 | (Sense) | TGTctggccagtccacAGC | 76 |
| LNA | PR18 | −9 to +10 | (Sense) | tgtctgGCCaGTCcacagc | 80 |
| LNA | PR19 | −9 to +10 | (Sense) | tgTCtgGCcaGTccACagC | 84 |
| LNA | PR20 | −9 to +10 | (Sense) | tGtcTggcCagtCcacAgc | 78/81 |
| LNAs Androgen Receptor (AR) | |||||
| LNA | AR1 | −8/ +9 | (Sense) | cACCtccCAgcgCcccCTc | 90 |
| LNA | AR2 | −13/ +6 | (Sense) | CTctcCaccTCccaGCgcC | 84 |
| LNA | AR3 | −24/ −6 | (Sense) | gTTgcATttGctctcCACc | 78 |
| Noncomplementary control LNAs | |||||
| LNAscr1 | N/A | N/A | cCacaGCtgTCcagTtGGc | ||
| LNAhcv | N/A | N/A | CTAcgaGaCctCccGggGC | ||
| LNAscr2 | N/A | N/A | cAgttGTcaGCtggCccac | ||
| 2′-Methoxyethyl RNA's Complementary to PR | |||||
| MOE | PR2 | −9/+10 | (Antisense) | GCUGUGGACUGGCCAGACACA | 82 |
| MOE | PR3 | −9/+10 | (Sense) | UGUCUGGCCAGUCCACAGCUG | 81 |
LNAs and MOEs are listed 5' to 3'. LNA-modified bases are represented as capital letters, and MOE-modified bases are represented as underlined capital letters. Unmodified bases are shown in lowercase. DNA bases are lower case. Sense LNAs and MOEs are complementary to the template strand of chromosomal DNA. Antisense LNAs and MOEs are complementary to the transcribed PR mRNA. Sense/antisense refers to the orientation of the LNA or MOE oligomer, not the orientation of the target sequence. Calculated and experimentally-determined Tm and values were obtained as described in MATERIALS AND MEHTODS (42,43).
LNA bases are linked by the same phosphate backbone as found in DNA or RNA and can be introduced at any position of an oligonucleotide during automated synthesis. We varied the location and number of LNA substitutions to investigate the effect of these variables on inhibition. LNAs were designed to be complementary to sequences at the transcription start site and to regions further upstream or downstream. Pedersen and colleagues had previously shown that LNA pentanucleotides could block transcription by E. coli polymerase in cell free assays (26), a result similar to that achieved in classic studies by Sigman and coworkers using RNA pentanucleotides (27).
Inhibition of PR Expression by LNAs
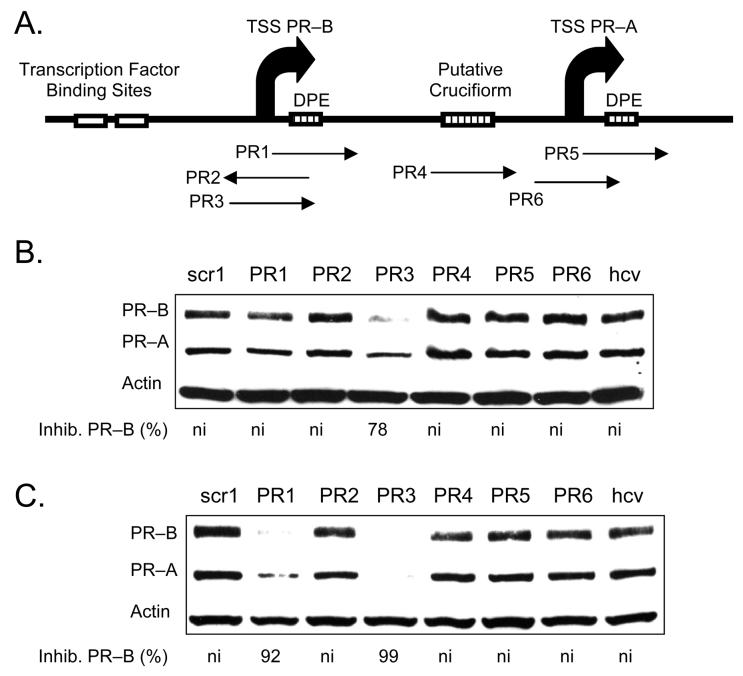
We synthesized 19 base LNAs PR1 to PR6 to be complementary to target sequences that were structurally or functionally significant within the PR gene (Figure 2 A). These initial LNAs were designed to evenly distribute LNA bases throughout the oligomer and to simplify synthesis by minimizing the number of different LNA bases used (e.g. some oligomers had only C LNA bases, others C and T only, ect.). PR1, PR2, and PR3 were complementary to the transcription start site for the PR-B isoform. PR4 targeted an inverted repeat that may form a cruciform structure and we reasoned that it might be especially accessible to binding by the LNA. PR5 targets a downstream promoter element that helps regulate PR-A expression and PR6 targets the transcription start site for the PR-A isoform.
FIGURE 2.
Western analysis of inhibition of PR protein expression by agLNAs complementary to different target sequences. Strand orientation is given in Table 1. (A) Schematic showing relative positions of target sites for agLNAs. (B) Inhibition of PR expression using agLNAs at 25 nM. (C) Inhibition of PR expression using agLNAs oligonucleotides at 50 nM. ni: no significant (<30%) inhibition. Percent inhibition of PR–B is relative to the average of noncomplementary LNAs LNAscr1 and LNAhcv. TSS: transcription start site.
To achieve inhibition, it was necessary to transfect LNAs twice. LNAs would be introduced into cells using lipid and the cells grown to confluence, typically 3-4 days. Cells were then passaged, transfected a second time, and harvested for analysis after an additional 3-4 days. We had previously observed that agPNAs that target PR also require two transfections to achieve potent inhibition of PR expression (4,5).
We introduced the LNAs into cells by standard protocols using cationic lipid (21). When added at 25 nM, PR3, which targeted the transcription start site (−9 to +10), was the only LNA to exhibit >50 % inhibition of PR expression (Figure 2 B). When LNAs were present at 50 nM, adjacent LNA PR1 (−2 to +17) also inhibited expression (Figure 2 C). LNA PR1 and LNA PR3 are complementary to the transcribed (antisense) strand of the promoter and are in the sense orientation relative to PR mRNA. LNA PR2, which is complementary to PR3 and targets the nontranscribed (sense) strand of the promoter, did not inhibit gene expression. Control LNAs that were not complementary to PR did not inhibit PR expression.
Concentration Dependence of Inhibition
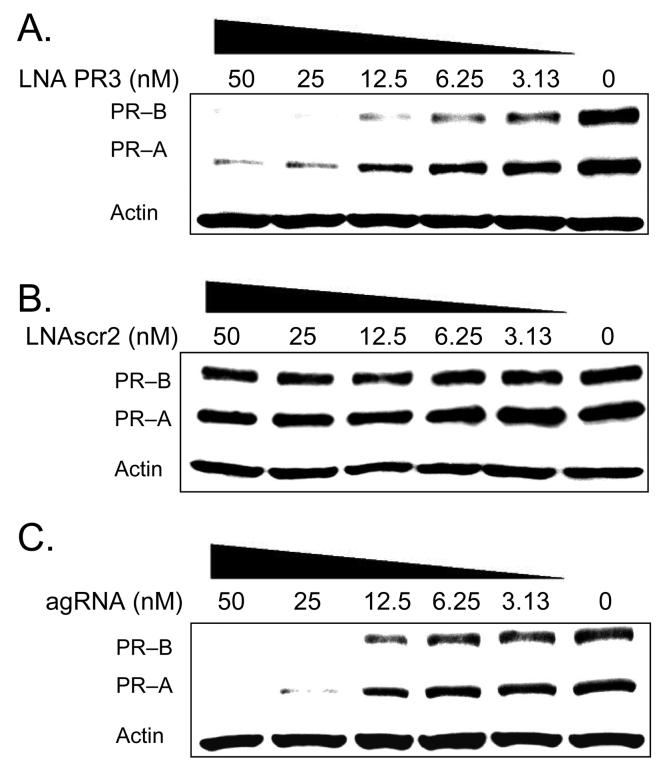
We added PR3 at varying concentrations to characterize its potency (Figure 3). Potent inhibition was observed as low as 3 nM and we calculated an IC50 value of approximately 8 nM (Figure 3 A). The scrambled control LNA did not block expression at any concentration (Figure 3 B). We compared inhibition by agLNAs with inhibition by an antigene RNA (agRNA) complementary to the sequence targeted by PR3 (Figure 3 C). We recently demonstrated that duplex agRNAs can target chromosomal DNA and efficiently block gene expression (25,28), and agRNAs provide a useful benchmark for evaluating agLNAs. We observed that agRNA-mediated inhibition of PR was similar to that achieved by agLNA PR3, with an IC50 value of approximately 15 nM (Figure 3 C). These data suggest that agLNAs can rival agRNAs for potent gene silencing.
FIGURE 3.
Western analysis comparing the potency of inhibition of PR protein expression by agLNA PR3, LNAscr2, and an agRNA. (A) Inhibition of PR expression by increasing concentrations of LNA PR3, which targets −9 to +10 relative to the PR–B transcriptional start site. (B) Lack of inhibition of PR expression by increasing concentrations of a scrambled LNA control, LNAscr2. (C) Inhibition of PR expression by increasing concentrations of an agRNA, which targets −9 to +10 relative to the PR–B transcriptional start site.
Unlike the agLNAs or agPNAs (4,5), which required two transfections for activity, agRNAs required only one transfection. The more rapid action of the agRNA relative to the agLNAs or agPNAs targeting the same sequence within PR suggests that the mechanism of action of the two antigene agents is significantly different. Inhibition by agRNAs requires expression of Argonaute1 and Argonaute2 (28,29), proteins involved in RNA interference (RNAi). It is likely that these proteins, and possibly others, facilitate recognition by duplex RNA and allow it to take place more rapidly. agLNAs and agPNAs, by contrast, are unnatural molecules that are less likely to be recognized by cellular proteins and guided to their molecular targets.
Linkage of Expression of PR-A and PR-B Isoforms
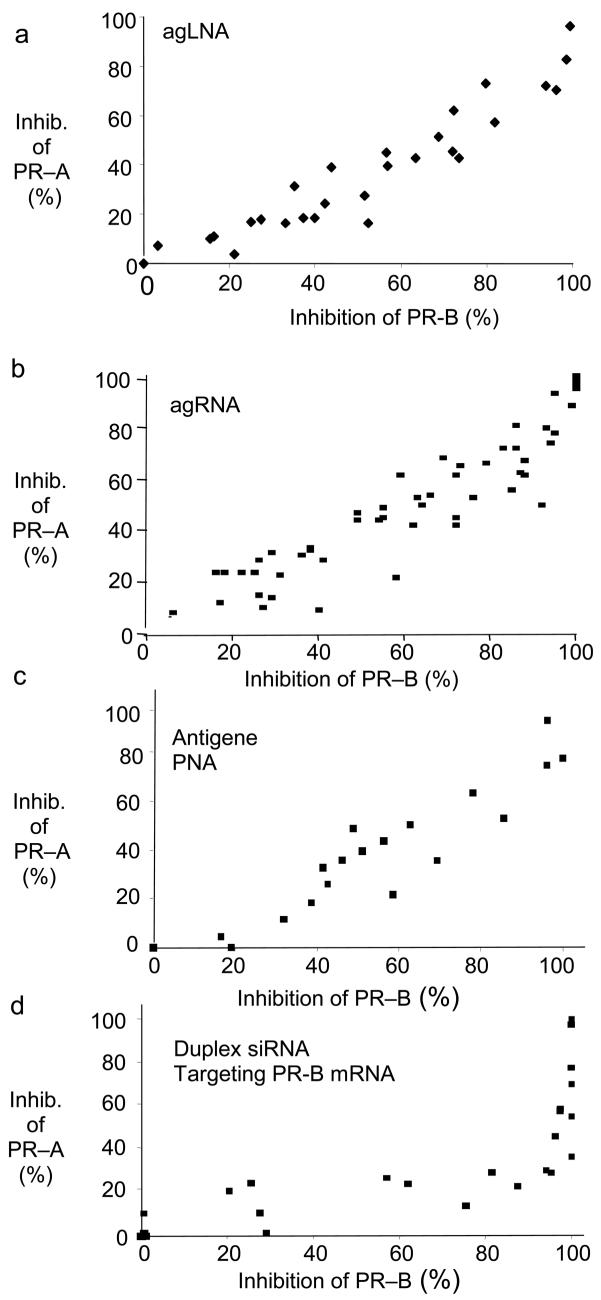
agLNAs PR3 and PR1 target the transcription start site for PR-B, a site that is approximately 800 bases distant from the transcription start site for PR-A. Nevertheless, addition of these agLNAs to cells reduces expression of both the PR-B isoform and the PR-A isoform (Figure 2 and Figure 3). To characterize the linkage between inhibition of PR-B and PR-A expression by LNAs, we quantified data from several independent experiments and plotted inhibition of PR-A as a function of inhibition of PR-B. We observed that inhibition of PR-B and PR-A varied linearly (i.e. the level of inhibition of PR-A was approximately the same as the level of inhibition of PR-B at a given concentration of LNA) (Figure 4 A).
FIGURE 4.
Comparison of the linkage of PR–B and PR–A expression levels upon treatment by A) agLNAs targeting the PR promoter, B) agRNAs targeting the PR promoter (4,25), C) agPNAs targeting the PR promoter (4,25), D) an siRNA targeting the mRNA encoding PR-B (4,25). Antisense PNAs targeting PR-B mRNA produced a linkage similar to D) (4).
We had previously shown the same result with analogous agRNAs (25) and agPNAs (4) targeting the PR-B promoter (Figure 4 B and C). Antisense PNAs or siRNAs that target PR-B mRNA, by contrast, produced an exponential linkage between inhibition of PR-B and PR-A (4), with PR-A expression becoming apparent only when high levels of PR-B expression were achieved (Figure 4 D). The common PR-A/PR-B linkage exhibited by agLNAs, agPNAs, and agRNAs underscores the fundamental difference between targeting chromosomal DNA rather than mRNA and that a specific antigene phenotype can be achieved regardless of the chemistry of the silencing approach.
One explanation for the proportional linkage exhibited by the three different antigene strategies (agLNA, agPNA, agRNA) is that recognition of the PR-B start site not only blocks the initiation of transcription of PR-B, but also prevents initiation of transcription from the PR-A start site. The PNA may reduce the ability of RNA polymerase and transcription factors to scan the chromosome and locate the promoter for PR-A. The fact that three chemically distinct antigene silencing strategies yield the same subtle phenotype supports the biological relevance of the phenotype and suggests the usefulness of comparing the results of complementary silencing strategies.
Effect of LNA Length on Inhibition of Gene Expression by agLNAs
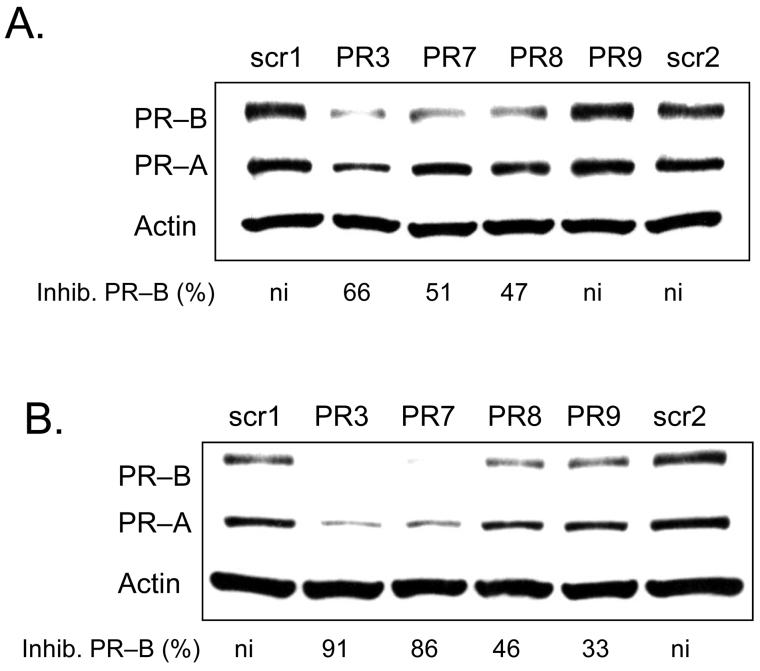
Oligonucleotide length is an important variable for gene silencing strategies, affecting cost, specificity, and potency. To test the effect of varying LNA length we compared inhibition by 19 base LNA PR3 with analogous truncated LNAs PR7 (16 bases), PR8 (13 bases), and PR9 (10 bases). When assayed at 25 nM, PR3, PR7, and PR8 blocked 47-66 % of PR expression (Figure 5 A). When assayed at 50 nM, PR3 and PR7 caused substantial inhibition (91 and 86% respectively) while shorter LNAs PR8 and PR9 inhibited less than 50 % of PR expression (Figure 5 B). These data suggest that length is an important determinant for the efficacy of promoter-targeted LNAs.
FIGURE 5.
Western analysis of inhibition of PR protein expression by agLNAs of different lengths targeting −9 to +10 relative to the transcriptional start site of PR–B. Inhibition of PR by LNA PR3 (19 bases long), LNA PR7 (16 bases long), LNA PR8 (13 bases long), and LNAPR9 (10 bases long) all at 25 nM (A) and 50 nM (B). Percent inhibition of PR–B is relative to the average of the LNAscr1 and LNAscr2. ni: no significant (<30%) inhibition.
Effect of Varying LNA Substitution
The flexibility of LNA synthesis methods permits LNA bases to be interspersed at different sites within an oligonucleotide that is mostly DNA. The location and number of LNA substitutions is likely to affect the melting temperature (Tm) and biological activity of the resulting LNA. To test the effect on inhibition of varied LNA substitutions, we synthesized a variety of LNA-DNA chimera (LNAs PR17 to PR20) with LNA bases introduced at different sites.
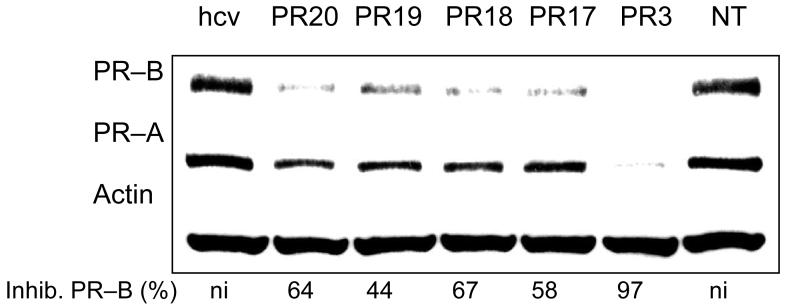
The LNAs were designed to examine different motifs for interspersing LNA bases throughout the oligomer. All LNAs were nineteen bases long and identical in sequence to inhibitory LNA PR3. LNA PR17 had six LNA bases grouped at its 3' and 5' termini in sets of three. This design resembles standard LNA gapmers used for antisense gene inhibition (15,16). LNA PR18 had six LNA bases grouped in the center. LNA PR19 had four pairs of LNA bases spread throughout the molecule (and nine total LNA bases), while LNA PR20 had five single bases scattered evenly. LNAs PR17-PR20 and LNA PR3 had similar predicted Tm values (Table 1).
We introduced these different designs into T47D cells. Experiments were performed in quadruplicate. Each of these redesigned LNAs inhibited expression of PR protein, but none was as efficient as the original design PR3 (Figure 6). These data suggest that optimal efficacy of agLNAs is sensitive to placement of LNA bases within the LNA-DNA chimera but that a broad spectrum of designs can lead to reduced protein expression. Optimal designs will need to be determined empirically.
FIGURE 6.
Western analysis of inhibition of PR protein expression by agLNAs of different designs. NT: no treatment control. All agLNAs are complementary to −9 to +10 relative to the transcriptional start site of PR–B. Percent inhibition is relative to the negative control treatment with LNAhcv
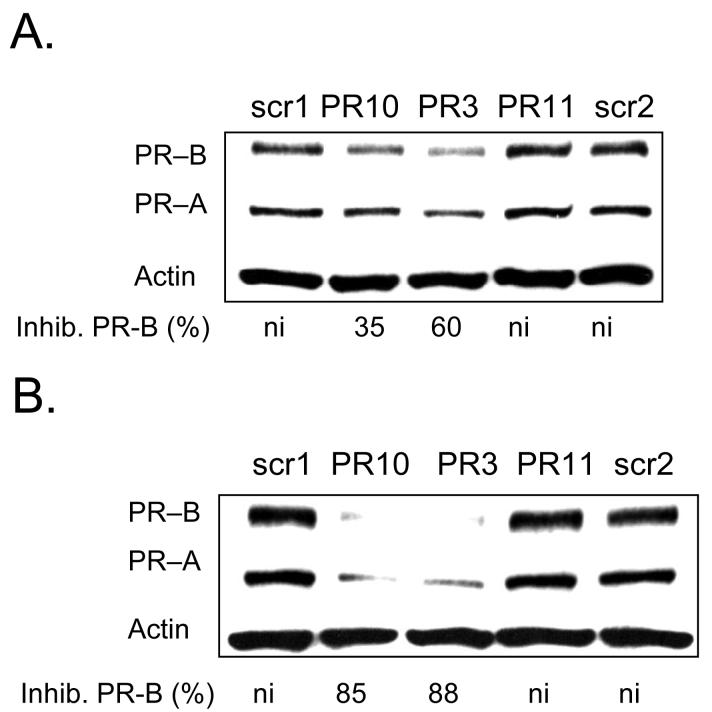
We also tested two LNAs that were analogous in length and sequence to LNA PR3 but different numbers of LNA substitutions (Figure 7). LNA PR3 contained eight LNA bases, while LNA PR10 contained five and LNA PR11 contained eleven. We observed that that LNA PR10 inhibited PR expression at levels that were similar to those achieved by LNA PR3. LNA PR11, by contrast, did not inhibit PR expression at 25 or 50 nM even though it had the highest potential for strong base-pairing with the target sequence and the highest calculated melting temperature (Table 1). It is possible that the high LNA content of LNA PR11 reduced its efficacy by promoting formation of intramolecular LNA structure, causing aggregation of LNA oligomers, or enhancing binding to non-targeted cellular nucleic acids.
FIGURE 7.
Western analysis of inhibition of PR protein expression by agLNAs with different predicted thermal stabilities. Inhibition of PR by LNA PR10 (predicted Tm of 76°C), LNA PR3 (predicted Tm of 83°C), and LNA PR11 (predicted Tm of 91°C) shown at 25 nM (A) and at 50 nM (B). Percent inhibition of PR–B is relative to the average of noncomplementary LNAs LNAscr1 and LNAscr2. All agLNAs are complementary to −9 to +10 relative to the transcriptional start site of PRB. ni: no significant (<30%) inhibition.
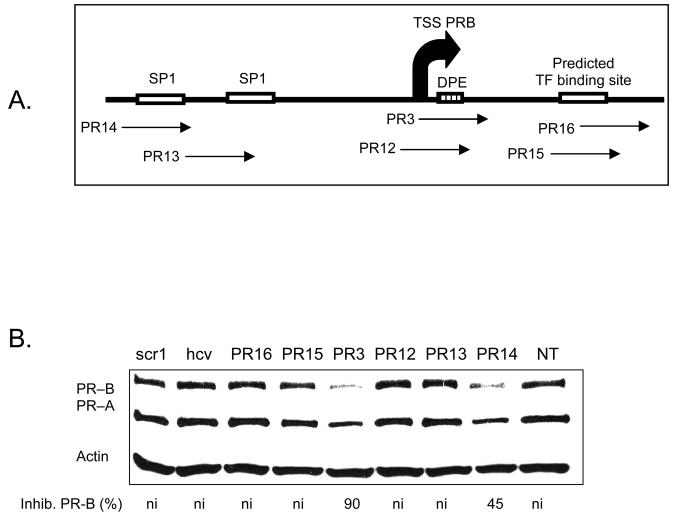
Effect of Targeting Transcription Factor Binding Sites
To gain additional insights into the potential of agLNAs for inhibiting gene expression, we designed agLNAs to be complementary to additional sequences with the PR promoter. These sequences were chosen because of their functional importance as transcription factor binding sites (Figure 8 A). Results from four independent experiments revealed that most of these agLNAs yielded little or no inhibition of gene expression (Figure 8 B). agLNA PR14 targeting an SP1 site exhibited reproducible inhibition (25-60 %, 35 % average), suggesting that it was having a modest effect. These data suggest that the transcription start site is a particularly vulnerable target for agLNAs relative to other functionally important sequences.
FIGURE 8.
(A) a schematic of PR and the relative target sites for each LNA. (B) Western analysis of inhibition of PR protein expression by agLNAs targeting putative transcription factor binding sites. Percent inhibition of PR–B is relative to the average of the LNAscr1 and LNAhcv. ni: no significant (<30%) inhibition. TSS: transcription start site. NT: no treatment.
Comparison with Analogous 2'-Methoxyethyl Oligonucleotides
2'-Methoxyethyl (2'-MOE) RNA (Figure 1) is another widely used chemical modification (30). 2'-MOE RNA substitutions increase resistance to nucleases and affinity for complementary sequences but individual MOE substitutions do not increase Tm values to the same extent as do LNA substitutions. Like LNA, oligomers that contain 2'-MOE possess a negatively charged phosphodiester backbone. 2'-MOE-containing oligomers are being used in multiple Phase 1 and Phase 2 clinical trials with apparent initial success (14), making them an important benchmark for comparisons with other oligomer chemistries.
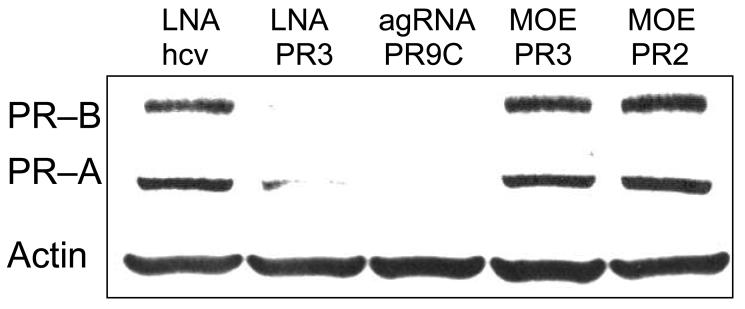
We designed twenty-one base MOE oligomers analogous in sequence to LNA PR3 (MOE PR3) (targeting the sense strand) or LNA PR2 (MOE PR2) (targeting the antisense strand). Neither MOE oligomer inhibited expression of PR when added at 50 nM (Figure 9). PR3 was tested in parallel and inhibited expression.
FIGURE 9.
Western analysis comparing inhibition of PR protein expression by 2'-MOE or LNA oligonucleotides. All oligonucleotides were present at 50 nM. agRNA PR9C was used for a positive control. Noncomplementary LNA LNAhcv was used as a negative control.
One explanation for better inhibition by LNA PR3 relative to the analogous MOE is that LNA bases yield higher affinity binding than MOE bases because of their ribose constraints. Such high affinity interactions may be critical for recognition. For example, a patch of LNA bases may be necessary to initiate and anchor base-pairing at the chromosomal target. It is also possible, however, that we have not identified an optimal MOE design and that oligonucleotides that contain mixtures of MOE and DNA bases may be more effective. We have tested only one MOE design and oligomers with other combinations of MOE and DNA bases might be more effective antigene agents.
Inhibition of AR Expression by LNAs
To begin to investigate the generality of inhibition of gene expression by promoter-targeted LNAs, we designed LNAs to be complementary to the transcription start site for androgen receptor (AR) (31) and introduced them into T47D cells. We had previously shown that agRNAs could inhibit AR expression (25,28), suggesting that the promoter was accessible and that synthetic oligomers could mediate recognition.
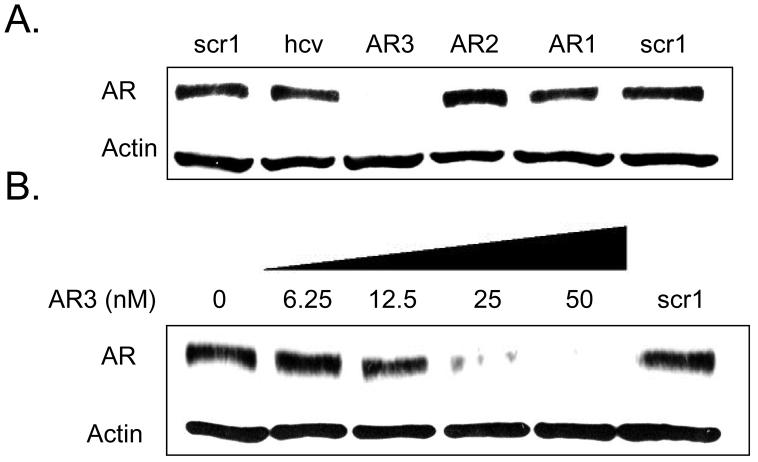
LNAs AR1, AR2, and AR3 targeted sites −8/+9, −13/+6, and −24/−6, respectively. When assayed at 50 nM AR3 was highly active (Figure 10 A). Addition of varied concentrations of AR3 showed an IC50 value of 12 nM (Figure 10 B), similar to that previously observed for LNA-mediated inhibition of PR expression (Figure 3). These data suggest that gene silencing by agLNAs may be a general phenomenon and encourage its application to other genes.
FIGURE 10.
Western analysis of inhibition of AR protein expression by agLNAs. (A) Inhibition of AR expression using agLNAs at 50 nM. (B) Inhibition of AR Expression using increasing concentrations of LNA AR3, which targets −24 to −5 relative to the transcriptional start site of AR. The negative control, LNAscr1, was present at 50 nM.
AR1, AR2, and AR3 target overlapping sequences and with much different efficacies (Figure 10 A). We have previously observed a similar phenomenon with promoter-targeted RNAs complementary to adjacent sequences (25,32). The differences in the activities of these three LNAs may be due to the composition of the LNA or to different structures/stabilities at the target sequences. Promoters are complex molecular structures subject to many unknown variables. Our data suggest that investigators interested in using promoter-targeted LNAs should select several sequences for initial testing.
Potency of PR Inhibition in MCF-7 Cells
PR expression varies substantially between cell lines and this variation is an important phenotypic marker in breast cancer. To determine whether the levels of endogenous expression of PR protein would affect the ability of LNAs to inhibit gene expression, we introduced LNAs into MCF-7 breast cancer cells. MCF-7 cells were chosen because they express much lower amounts of PR (10-fold lower levels of RNA, 30 fold levels of protein) (28) than do T47D cells.
We had previously shown that agRNAs that inhibited PR expression in T47D cells were unable to block PR expression in MCF-7 cells. Indeed, some of these agRNAs actually caused PR expression to dramatically increase (32). Conversely, agPNA-peptide conjugates were able to block gene expression in both T47D and MCF-7 cells (33) suggesting that different antigene approaches could produce divergent effects when introduced into differing cellular contexts.
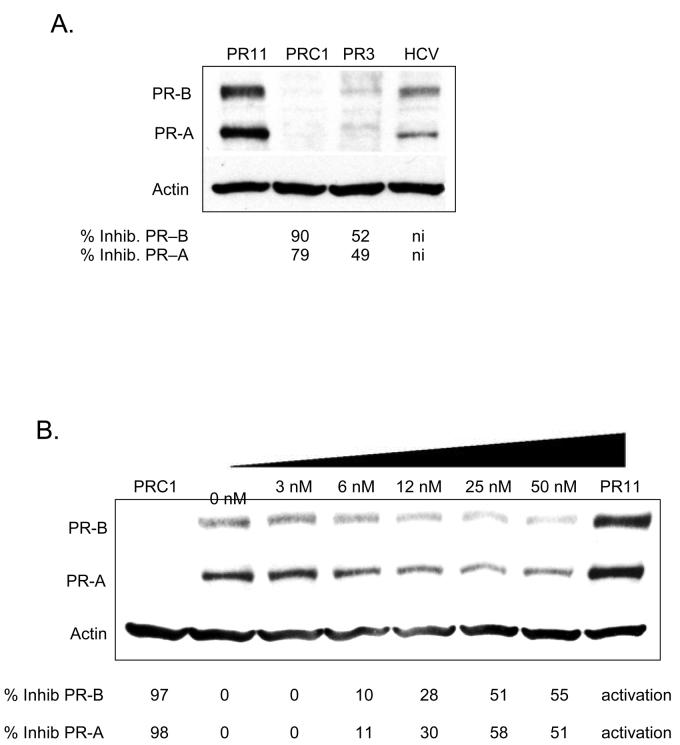
We tested benchmark agLNA PR3 for its ability to inhibit gene expression in MCF-7 cells. agLNA PR3 also inhibited gene expression, but at a lower level than had been achieved in T47D cells (Figure 11 A). Dose response data indicated an IC50 value of 20 nM with a maximal potency of efficacy of 50% inhibition (Figure 11 B). These data suggest that agLNAs can inhibit poorly expressed genes, but that achieving highly efficient gene silencing may be more difficult than in cells that highly express the targeted gene (e.g. T47D cells).
FIGURE 11.
Western analysis comparing the potency of inhibition of PR protein expression by agLNA PR3. These experiments were performed in MCF7 cells. (A) Inhibition of PR expression by LNA PR3 at 50 nM. PR11 is an activating agRNA and is a positive control, PRC1 is an siRNA targeting PR mRNA and is a positive control for inhibition. Percent inhibition is relative to noncomplementary LNAhcv. (B) Inhibition of PR expression by increasing concentrations of an LNA PR3. Percent inhibition is relative to the 0 nM treatment. PR11 is a positive control RNA that activates PR expression.
We also tested two RNA duplexes as positive controls. A duplex RNA (PRC1) that targets PR mRNA effectively blocked PR expression, demonstrating that PR expression could be efficiently blocked by transfected nucleic acids in MCF-7 cells (Figure 11 A and B). agRNA targeting −11 to +8 activated gene expression (Figure 11 A and B), replicating activation that we had observed previously (32).
Comparison of Chromosomal Recognition by PNAs, duplex RNAs, and LNAs
We have shown that a variety of different antigene oligomers can target promoter sequences within chromosomal DNA and efficiently block gene expression, including agPNAs (4,5), agPNA-peptide conjugates (33), duplex agRNAs (25,28), and now agLNAs. Our data from these studies make it clear that chromosomal sequences near transcription start sites are accessible to recognition by synthetic nucleic acids and nucleic acid mimics.
While the end result, gene inhibition, is the same, PNAs, duplex RNAs, and LNAs have substantially different properties. LNA and duplex RNA have charged phosphodiester backbones, whereas the amide PNA backbone is uncharged. PNAs and LNAs are single-stranded, whereas agRNAs are introduced into cells as a duplex. Silencing by agRNAs is known to require expression of argonaute proteins (25,28), whereas it is less likely that recognition by agLNAs or agPNAs depends on specific protein interactions because their altered ribose (LNA) or artificial amide (PNA) backbones have not evolved to optimize interactions with proteins.
Duplex agRNAs may also have a different nucleic acid target than agPNAs or agLNAs. As noted above, the activity of agRNAs requires expression of argonaute proteins (28,29). Argonaute proteins are known to promote binding to RNA (34,35), not DNA. Furthermore, it is known that many genes express antisense transcripts that overlap the promoter sequences responsible for controlling mRNA synthesis (36,37), providing potential RNA targets that extend beyond mRNA. It is possible, therefore, that agRNAs directly bind to RNA and exert indirect effects on chromosomal DNA (38).
The uncertainty about whether agRNAs bind RNA or chromosomal DNA suggests that further work will be necessary to conclusively establish that target of agPNAs and agLNAs. The nucleic acid wiring at some promoter sequences may be much more complex than had been assumed previously, and direct targeting of chromosomal DNA by PNAs and LNAs, while a leading explanation for the inhibition of expression that we observe, should not be assumed.
One observation from our work is that agRNAs require only one transfection to be effective (27,28), whereas agLNAs, agPNAs (4,5), and agPNA-peptide conjugates (33) require two transfections. This observation might be explained if 1) argonaute proteins facilitate recognition of target sequences by agRNAs, whereas agLNAs and agPNAs must find their targets without the aid of evolved helper proteins and 2) agRNAs may recognize a relatively accessible RNA target, whereas agLNAs and agPNAs may need to recognize a DNA sequence whose accessibility may be much more limited because it is hindered by bound proteins and extensive base-pairing.
The substantial chemical and mechanistic differences between these three promoter-targeted approaches are important and suggest that they will may have different strengths as probes for structure and function near transcription start sites.
Significance of agLNAs for Laboratory and Clinical Development
Single-stranded antisense oligonucleotides and double-stranded siRNAs appear to be making good progress in the laboratory and the clinic (14). Given this progress, it is necessary to consider whether agLNAs and other chromosome-targeted synthetic molecules (including agPNAs, agRNAs, triple-helix forming oligonucleotides (39), and pyrimidine polyamides (40)) merit further development. For basic science, agLNAs and other antigene approaches provide direct insights into accessibility and function at their target sequence that cannot be gained using approaches that target mRNA.
For applied science and therapeutic development, antigene approaches provide another option for addressing the difficult problem of achieving adequate potency and specificity in vivo by unlocking chromosomal DNA as a molecular target. In some cases, potency may be enhanced by targeting the chromosome in the nucleus rather than mRNA in the cytoplasm. Because antisense LNAs are already being used in clinical trials, the route to clinically relevant agLNAs might be relatively short. One caveat for clinical development, however, is the recent observation of hepatotoxicity in mouse model experiments (11). Finally, we have recently shown that duplex agRNAs can activate gene expression (32) in cells and it is known that agPNA-peptide conjugates can activate expression in cell extract (41). Gene activation cannot be acheived by siRNAs or antisense oligonucleotides and activating RNAs widen the scope of genes that can be targets for therapeutic development. More research will be needed to determine whether LNAs can also act as gene activators, but the ability to deliver LNAs to promoter DNA encourages the belief that this outcome is feasible.
Conclusion
We have demonstrated that LNAs can target sequences within promoter DNA and block endogenous gene expression of two different nuclear hormone receptors, PR and AR. LNA joins duplex RNA and PNA as an effective antigene strategy, and the three approaches provide a collection of chemically and mechanistically distinct choices for blocking gene expression at the level of the chromosome. Inhibition by agLNAs is dependent on the target sequence within the gene, LNA length, percentage of LNA bases, and the location of LNA substitution. Our ability to use agLNAs to block expression of two genes in two attempts suggests that the strategy may be general, but experimenters should expect to test several LNAs before identifying an active inhibitor.
Successful use of LNAs to target chromosomal DNA, coupled with demonstrations that LNAs can be active in animals after systemic administration (11,12), suggests that promoter-targeted LNAs are a new option for controlling gene expression for experimentation and therapeutic development.
ACKNOWLEDGEMENTS
This work was supported by the National Institutes of Health (NIGMS 60642 and NIGMS 73042) and the Robert A. Welch Foundation (I-1244).
Abbreviations
- LNA
Locked nucleic acid
- agLNA
antigene LNA
- PNA
peptide nucleic acid
- agPNA
antigene PNA
- 2'-MOE
2'-methoxyethyl
- PR
progesterone receptor
- AR
androgen receptor
REFERENCES
- 1.Kaihatsu K, Janowski BA, Corey DR. Recognition of chromosomal DNA with PNAs. Chem. Biol. 2004;11:749–758. doi: 10.1016/j.chembiol.2003.09.014. [DOI] [PubMed] [Google Scholar]
- 2.Nielsen PG, Egholm M, Berg RH, Buchardt O. Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science. 1991;254:1497–1500. doi: 10.1126/science.1962210. [DOI] [PubMed] [Google Scholar]
- 3.Nielsen PE. Gene targeting by peptide nucleic acid. Meth. Mol. Biol. 2005;288:343–358. doi: 10.1385/1-59259-823-4:343. [DOI] [PubMed] [Google Scholar]
- 4.Janowski BA, Kaihatsu K, Huffman KE, Schwartz JC, Ram R, Hardy D, Mendelson CR, Corey DR. Inhibiting transcription of chromosomal DNA with antigene peptide nucleic acids. Nature Chem. Biol. 2005;1:210–215. doi: 10.1038/nchembio724. [DOI] [PubMed] [Google Scholar]
- 5.Janowski BA, Hu J, Corey DR. Silencing gene expression by targeting chromosomal DNA with antigene peptide nucleic acids and duplex RNAs. Nature Protocols. 2006;1:436–443. doi: 10.1038/nprot.2006.64. [DOI] [PubMed] [Google Scholar]
- 6.Koshkin AA, Singh SK, Nielsen P, Rajwanshi VK, Kumar R, Meldgaard M, Olsen CE, Wengel J. LNA (locked nucleic acids): Synthesis of the adenine, cytosine, guanine, 5-methylcytosine, thyme, and uracil bicylonucleoside monomers, oligomerisation, and unprecedented nucleic acid recognition. Tetrahedon. 1998;54:3607–3630. [Google Scholar]
- 7.Obika S, Nanbu D, Hari Y, Andoh J, Morio K, Doi T, Imanishi T. Stability and structural features of the duplexes containing nucleoside analogues with a fixed N-type conformation 2'-O,4'-C-methyleneribonucleotides. Tet. Lett. 1998;39:5401–5404. [Google Scholar]
- 8.Braasch DA, Corey DR. Locked nucleic acid (LNA): fine-tuning the recognition of DNA and RNA. Chem. Biol. 2001;8:1–7. doi: 10.1016/s1074-5521(00)00058-2. [DOI] [PubMed] [Google Scholar]
- 9.Vester B, Wengel J. LNA (locked nucleic acid): high-affinity targeting of complementary RNA and DNA. Biochemistry. 2004;43:13233–13241. doi: 10.1021/bi0485732. [DOI] [PubMed] [Google Scholar]
- 10.Frieden M, Hansen HF, Koch T. Nuclease stability of LNA oligonucleotides and LNA-DNA chimera. Nucleosides Nucleotides Nucl. Acids. 2003;22:1041–1043. doi: 10.1081/NCN-120022731. [DOI] [PubMed] [Google Scholar]
- 11.Swayze EE, Siwkowski AM, Wancewicz EV, Migawa MT, Wyrzykiewicz TK, Hung G, Monia BP, Bennett AC. Antisense oligonucleotides containing locked nucleic acid improve potency but cause significant hepatotoxicity in animals. Nucl. Acids Res. 2007;35:687–700. doi: 10.1093/nar/gkl1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roberts J, Palma E, Sazani P, Orum H, Choo M, Kole R. Efficient and persistant splice switching by systemically delivered LNA oligonucleotides in mice. Mol. Ther. 2006;14:471–475. doi: 10.1016/j.ymthe.2006.05.017. [DOI] [PubMed] [Google Scholar]
- 13.Frieden M, Orum H. The application of locked nucleic acids in the treatment of cancer. IDrugs. 2006;9:706–711. [PubMed] [Google Scholar]
- 14.Corey DR. RNAi learns from antisense. Nat. Chem. Biol. 2007;3:8–11. doi: 10.1038/nchembio0107-8. [DOI] [PubMed] [Google Scholar]
- 15.Kurreck J, Wyszko E, Gillen C, Erdmann VA. Design of antisense oligonucleotides stabilized by locked nucleic acids. Nucl. Acids Res. 2002;30:1911–1918. doi: 10.1093/nar/30.9.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Braasch DB, Liu Y, Corey DR. Antisense gene inhibition by locked nucleic acids. Nucl. Acids Res. 2002;30:5160–5167. doi: 10.1093/nar/gkf651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grunweller A, Wyszko E, Bieber B, Jahnel R, Erdmann VA, Kurreck J. Comparison of different antisense strategies in mammalian cells using locked nucleic acids, 2′-O-methyl RNA, phosphorothioates and small interfering RNA. Nucl. Acids Res. 2003;31:3185–3193. doi: 10.1093/nar/gkg409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hertoghs KML, Ellis JH, Catchpole IR. Use of locked nucleic acid oligonucleotides to add functionality to plasmid DNA. Nucl. Acids Res. 2003;31:5817–5830. doi: 10.1093/nar/gkg801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brunet E, Alberti P, Perrouault L, Babu R, Wengel J, Giovannangeli C. Exploring cellular activity of locked nucleic acid-modified triplex-forming oligonucleotides and defining its molecular basis. J. Biol. Chem. 2005;280:20076–20085. doi: 10.1074/jbc.M500021200. [DOI] [PubMed] [Google Scholar]
- 20.Brunet E, Corgnali M, Cannata F, Perrouault L, Giovannangeli C. Targeting chromosomal sites using locked nucleic acid-modified triplex-forming oligonucleotides: study of efficiency dependence on DNA nuclear environment. Nucl. Acids Res. 2006;34:4546–4553. doi: 10.1093/nar/gkl630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Braasch DA, Corey DR. Cellular Effects of Locked Nucleic Acids. Current Protocols in Nucleic Acid Chemistry. 2002:4.13.1–4.13.10. doi: 10.1002/0471142700.nc0413s09. [DOI] [PubMed] [Google Scholar]
- 22.Kastner P, Krust A, Turcotte B, Stropp U, Tora L, Gronemeyer H, Chambon P. Two distinct estrogen regulated gene promoters generate transcripts encoding two functionally different human progesterone receptor forms A and B. EMBO J. 1990;9:1603–1614. doi: 10.1002/j.1460-2075.1990.tb08280.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Misrahi M, Venencie P–Y, Saugier–Veber P, Sar S, Dessen P, Milgrom E. Structure of the human progesterone receptor gene. Biochim. Biophys. Acta. 1993;1216:289–292. doi: 10.1016/0167-4781(93)90156-8. [DOI] [PubMed] [Google Scholar]
- 24.Conneely OM, Jericevic BM, Lydon JP. Progesterone receptors in mammary gland development and tumorigenesis. J. Mam. Gland Bio. Neoplasia. 2003;8:205–214. doi: 10.1023/a:1025952924864. [DOI] [PubMed] [Google Scholar]
- 25.Janowski BA, Huffman KE, Schwartz JC, Ram R, Hardy D, Shames DS, Minna JD, Corey DR. Inhibiting gene expression at transcription start sites in chromosomal DNA by antigene RNAs. Nature Chem. Biol. 2005;1:216–222. doi: 10.1038/nchembio725. [DOI] [PubMed] [Google Scholar]
- 26.Filichev VV, Vester B, Hansen LH, Abdel Aal MT, Babu BR, Wengel J, Pedersen EB. Enhanced inhibition of transcription start by targeting 2'-OMe pentaribonucleotides comprising locked nucleic acids and intercalating nucleic acids. ChemBioChem. 2005;6:1181–1184. doi: 10.1002/cbic.200400457. [DOI] [PubMed] [Google Scholar]
- 27.Milne L, Xu Y, Perrin DM, Sigman DS. An approach to gene-specific transcription inhibition using oligonucleotides complementary to the template strand of the open complex. Proc. Natl. Acad. Sci. USA. 2000;97:3136–3141. doi: 10.1073/pnas.050544597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Janowski BA, Huffman KE, Schwartz JC, Ram R, Nordsell R, Shames DS, Minna JD, Corey DR. Ago1 and Ago2 link mammalian transcriptional silencing with RNAi. Nat. Struct. Mol. Biol. 2006;13:787–792. doi: 10.1038/nsmb1140. [DOI] [PubMed] [Google Scholar]
- 29.Kim DH, Villeneuve LM, Morris KV, Rossi JJ. Argonaute-1 directs siRNA mediated transcriptional silencing in human cells. Nature Struc. Mol. Biol. 2006;13:793–797. doi: 10.1038/nsmb1142. [DOI] [PubMed] [Google Scholar]
- 30.Baker BF, Lot SS, Condon TP, Cheng-Flournoy S, Lesnik EA, Sasmor HM, Bennett CF. 2'-O-(2-methoxy)ethyl-modified anti-intercellular adhesion molecule 1 (ICAM-1) oligonucleotides selectively increase the ICAM-1 mRNA level and inhibit formation of the ICAM-1 translation initiation complex in human umbilical vein endothelial cells. J. Biol. Chem. 1997;272:11994–20000. doi: 10.1074/jbc.272.18.11994. [DOI] [PubMed] [Google Scholar]
- 31.Tilley WD, Marcelli M, McPhaul MJ. Expression of the human androgen receptor gene utilizes a common promoter in diverse human tissues and cell lines. J. Biol. Chem. 1990;265:13776–13781. [PubMed] [Google Scholar]
- 32.Janowski BA, Younger ST, Hardy DB, Ram R, Huffman KE, Corey DR. Activating gene expression in mammalian cells with promoter-targeted duplex RNAs. Nat. Chem. Biol. 2007;3:166–173. doi: 10.1038/nchembio860. [DOI] [PubMed] [Google Scholar]
- 33.Hu J, Corey DR. Inhibiting gene expression with peptide nucleic acid (PNA) peptide conjugates that target chromosomal DNA. Biochemistry. 2007 doi: 10.1021/bi700230a. in Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parker JS, Barford D. Argonaute: a scaffold for the function of short regulatory RNAs. Trends Biochem. Sci. 2006;31:622–630. doi: 10.1016/j.tibs.2006.09.010. [DOI] [PubMed] [Google Scholar]
- 35.Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 36.Chen J, Sun M, Kent WJ, Huang X, Xie H, Wang W, Zhou G, Zhang R, Rowley JD. Over 20 % of human transcripts might form sense-antisense pairs. Nucl. Acids. Res. 2004;32:4812–4820. doi: 10.1093/nar/gkh818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wahlstedt C. Natural sense and noncoding RNA transcripts as potential drug targets. Drug Discovery Today. 2006;11:503–508. doi: 10.1016/j.drudis.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 38.Corey DR. Regulating mammalian transcription with RNA. Trends Biochem. Sci. 2005;30:655–658. doi: 10.1016/j.tibs.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 39.Besch R, Giovannageli C, Schuh T, Kammerbauer C, Deglitz K. Characterization and quanification of triple helix formation in chromosomal DNA. J. Mol. Biol. 2004;341:979–989. doi: 10.1016/j.jmb.2004.05.079. [DOI] [PubMed] [Google Scholar]
- 40.Nickols NG, Jacobs CS, Farkas ME, Dervan PB. Improved nuclear localization of DNA-binding polyamides. Nucl. Acids. Res. 2007;35:363–370. doi: 10.1093/nar/gkl1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu B, Han Y, Ferdous A, Corey DR, Kodadek T. Transcription activation by a PNA-peptide chimera in a mammalian cell extract. Chem. Biol. 2003;10:909–916. doi: 10.1016/j.chembiol.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 42.McTigue PM, Peterson RJ, Kahn JD. Sequence-dependent thermodynamic parameters for locked nucleic acid LNA-DNA duplex formation. Biochemistry. 2004;43:5388–5405. doi: 10.1021/bi035976d. [DOI] [PubMed] [Google Scholar]
- 43.SantaLucia J. A unified view of polymer, dumbbell, and oligonucleotide nearest-neighbor thermodynamics. Proc. Natl. Acad. Sci. USA. 1998;95:1460–1465. doi: 10.1073/pnas.95.4.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]