Abstract
The influenza virus hemagglutinin contains four major regions that are recognized by antibodies able to neutralize viral infectivity. To investigate the effect of an antibody response directed against each of these sites on viral evolution, influenza virus A/PR/8/34 (H1N1) was grown in allantois-on-shell cultures in the presence of a mixture of monoclonal antihemagglutinin antibodies. This selection mixture contained antibodies (two or three antibodies per antigenic site) whose concentrations were adjusted to achieve equal neutralization titers against each of the four antigenic sites. By varying the ratio of input virus to selection mixture concentration, we observed that variant viruses emerged under conditions of partial neutralization. Each of the four variants characterized in detail differed from the parental virus in its interaction with cellular receptors and exhibited minimal changes in antigenicity. Thus, these variants were virtually indistinguishable from wild-type viruses, as assessed by the binding of 103 monoclonal antihemagglutinin antibodies in an indirect radioimmunoassay. Despite this, many of the same antibodies demonstrated decreased titers to the variants in hemagglutination inhibition tests. The magnitude of the differences depended on the indicator erythrocytes used (much greater differences were detected with chicken erythrocytes than with human erythrocytes). Hemagglutination mediated by the variants was more resistant to neuraminidase treatment of erythrocytes than hemagglutination mediated by the parental virus. These findings are consistent with the idea that the variants were initially selected by virtue of their increased avidity for host cell receptors. Sequencing of viral RNA revealed that each of the variants differed from the parental virus by a single amino acid alteration in its HA1 subunit. Two of the changes were close to the proposed receptor binding site on hemagglutinin and could directly alter receptor binding, while a third was located near the trimer interface and may have increased receptor binding by altering monomer-monomer interactions.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- CHOPPIN P. W., TAMM I. Studies of two kinds of virus particles which comprise influenza A2 virus strains. II. Reactivity with virus inhibitors in normal sera. J Exp Med. 1960 Nov 1;112:921–944. doi: 10.1084/jem.112.5.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- COHEN A., BIDDLE F. The effect of passage in different hosts on the inhibitor sensitivity of an Asian influenza virus strain. Virology. 1960 Jun;11:458–473. doi: 10.1016/0042-6822(60)90087-8. [DOI] [PubMed] [Google Scholar]
- Caton A. J., Brownlee G. G., Yewdell J. W., Gerhard W. The antigenic structure of the influenza virus A/PR/8/34 hemagglutinin (H1 subtype). Cell. 1982 Dec;31(2 Pt 1):417–427. doi: 10.1016/0092-8674(82)90135-0. [DOI] [PubMed] [Google Scholar]
- Crecelius D. M., Deom C. M., Schulze I. T. Biological properties of a hemagglutinin mutant of influenza virus selected by host cells. Virology. 1984 Nov;139(1):164–177. doi: 10.1016/0042-6822(84)90337-4. [DOI] [PubMed] [Google Scholar]
- Gerhard W., Yewdell J., Frankel M. E., Webster R. Antigenic structure of influenza virus haemagglutinin defined by hybridoma antibodies. Nature. 1981 Apr 23;290(5808):713–717. doi: 10.1038/290713a0. [DOI] [PubMed] [Google Scholar]
- Holland J., Spindler K., Horodyski F., Grabau E., Nichol S., VandePol S. Rapid evolution of RNA genomes. Science. 1982 Mar 26;215(4540):1577–1585. doi: 10.1126/science.7041255. [DOI] [PubMed] [Google Scholar]
- MEDILL-BROWN M., BRIODY B. A. Mutation and selection pressure during adaptation of influenza virus to mice. Virology. 1955 Sep;1(3):301–312. doi: 10.1016/0042-6822(55)90026-x. [DOI] [PubMed] [Google Scholar]
- Naeve C. W., Hinshaw V. S., Webster R. G. Mutations in the hemagglutinin receptor-binding site can change the biological properties of an influenza virus. J Virol. 1984 Aug;51(2):567–569. doi: 10.1128/jvi.51.2.567-569.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nohinek B., Gerhard W., Schulze I. T. Characterization of host cell binding variants of influenza virus by monoclonal antibodies. Virology. 1985 Jun;143(2):651–656. doi: 10.1016/0042-6822(85)90407-6. [DOI] [PubMed] [Google Scholar]
- Raymond F. L., Caton A. J., Cox N. J., Kendal A. P., Brownlee G. G. Antigenicity and evolution amongst recent influenza viruses of H1N1 subtype. Nucleic Acids Res. 1983 Oct 25;11(20):7191–7203. doi: 10.1093/nar/11.20.7191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers G. N., Paulson J. C., Daniels R. S., Skehel J. J., Wilson I. A., Wiley D. C. Single amino acid substitutions in influenza haemagglutinin change receptor binding specificity. Nature. 1983 Jul 7;304(5921):76–78. doi: 10.1038/304076a0. [DOI] [PubMed] [Google Scholar]
- Rogers G. N., Pritchett T. J., Lane J. L., Paulson J. C. Differential sensitivity of human, avian, and equine influenza A viruses to a glycoprotein inhibitor of infection: selection of receptor specific variants. Virology. 1983 Dec;131(2):394–408. doi: 10.1016/0042-6822(83)90507-x. [DOI] [PubMed] [Google Scholar]
- Schild G. C., Oxford J. S., de Jong J. C., Webster R. G. Evidence for host-cell selection of influenza virus antigenic variants. Nature. 1983 Jun 23;303(5919):706–709. doi: 10.1038/303706a0. [DOI] [PubMed] [Google Scholar]
- Ward C. W. Structure of the influenza virus hemagglutinin. Curr Top Microbiol Immunol. 1981;94-95:1–74. doi: 10.1007/978-3-642-68120-2_1. [DOI] [PubMed] [Google Scholar]
- Wiley D. C., Wilson I. A., Skehel J. J. Structural identification of the antibody-binding sites of Hong Kong influenza haemagglutinin and their involvement in antigenic variation. Nature. 1981 Jan 29;289(5796):373–378. doi: 10.1038/289373a0. [DOI] [PubMed] [Google Scholar]
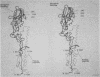
- Wilson I. A., Skehel J. J., Wiley D. C. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature. 1981 Jan 29;289(5796):366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- Yewdell J. W., Gerhard W. Antigenic characterization of viruses by monoclonal antibodies. Annu Rev Microbiol. 1981;35:185–206. doi: 10.1146/annurev.mi.35.100181.001153. [DOI] [PubMed] [Google Scholar]
- Yewdell J. W., Webster R. G., Gerhard W. U. Antigenic variation in three distinct determinants of an influenza type A haemagglutinin molecule. Nature. 1979 May 17;279(5710):246–248. doi: 10.1038/279246a0. [DOI] [PubMed] [Google Scholar]
- Yewdell J., Gerhard W. Delineation of four antigenic sites on a paramyxovirus glycoprotein via which monoclonal antibodies mediate distinct antiviral activities. J Immunol. 1982 Jun;128(6):2670–2675. [PubMed] [Google Scholar]