Abstract
Collaboration between MuA transposase and its activator protein, MuB, is essential for properly regulated transposition. MuB activates MuA catalytic activity, selects target DNA, and stimulates transposition into the selected target site. Selection of appropriate target DNA requires ATP hydrolysis by the MuB ATPase. By fusing MuB to a site-specific DNA-binding protein, the Arc repressor, we generated a MuB variant that could select target DNA independently of ATP. This Arc-MuB fusion protein allowed us to test whether ATP binding and hydrolysis by MuB are necessary for stimulation of transposition into selected DNA, a process termed target delivery. We find that with the fusion proteins, MuB-dependent target delivery occurs efficiently under conditions where ATP hydrolysis is prevented by mutation or use of ADP. In contrast, no delivery was detected in the absence of nucleotide. These data indicate that the ATP- and MuA-regulated DNA-binding activity of MuB is not essential for target delivery but that activation of MuA by MuB strictly requires nucleotide-bound MuB. Furthermore, we find that the fusion protein directs transposition to regions of the DNA within 40–750 bp of its own binding site. Taken together, these results suggest that target delivery by MuB occurs as a consequence of the ability of MuB to stimulate MuA while simultaneously tethering MuA to a selected target DNA. This tethered-activator model provides an attractive explanation for other examples of protein-stimulated control of target site selection.
Keywords: ATP–ADP switch, genetic recombination, transposition immunity, transposon target, transposition targeting
Transposable elements have been found in every species studied, and these often-considered “selfish DNAs” appear to have a tremendous impact on the evolution of their hosts (1). To comprehend how transposons have become ubiquitous and what effect they have had on evolution, we must understand the mechanisms that govern their activity and target site choice. Several transposons depend on nucleotide cofactors to regulate transposase activity or to choose appropriate target sites. For example, the bacterial Tn552 transposon appears to employ GTP for efficient transposition (2), and GTP stimulates assembly of the initial synaptic complex of the Drosophila P element transposase (3, 4). In contrast, GTP inhibits target DNA capture by the RAG recombinase, the transposase-like protein that initiates V(D)J recombination, thereby suppressing RAG-mediated transposition (5). Transposition of Mu and Tn7 uses target selection proteins that require ATP to choose the target DNA (6–10). Thus, although it is clear that nucleotide cofactors regulate a number of transposition reactions, it is largely unknown how these cofactors affect the individual steps of transposition. Here, we investigate the role of ATP in target delivery during Mu transposition.
The genome of the Mu bacteriophage is a replicative transposon. Transposition is mediated by two Mu-encoded proteins: MuA and MuB. MuA, the transposase, is a member of the transposase/retroviral integrase protein superfamily (11, 12). MuA binds specific sites at each end of the Mu genome (13). The MuA subunits at the two DNA ends assemble to form a MuA tetramer that catalyzes the cleavage and joining reactions necessary for transposition (14–17). The second protein, MuB, is an activator of MuA (17–20). Unlike MuA, MuB is an ATPase (6) and thus exists in distinct, nucleotide-controlled states. ATP-bound MuB (MuB·ATP) binds DNA tightly (Kd ≈80 nM) and with little sequence preference (20). ADP-bound MuB (MuB·ADP) binds DNA ≈10-fold more weakly (Kd ≈790 nM), as does nucleotide-free MuB (Kd ≈1,200 nM) (20). Therefore, ATP hydrolysis by MuB is coupled with dissociation of MuB from the DNA (21). As will be explained below, control of MuB ATPase activity is essential for determining the target sites during transposition.
Mu avoids transposing into or near (within ≈15 kb of) its own genome, a process termed transposition target immunity (6, 7, 22). Other transposons, including Tn7 and Tn3, also exhibit target immunity, which is considered a strategy for avoiding self-destruction (9, 23–25). During Mu transposition, target immunity is mediated by MuB and its interactions with MuA and ATP (6, 7). Steps in this process are as follows. Initially, MuB·ATP binds DNA, exhibiting a modest preference for A/T-rich sequences (26, 27). MuA, which is bound near the ends of the Mu genome, can interact with nearby MuB and stimulates MuB ATPase activity (6, 28, 29). Because MuB·ADP has a lower affinity for DNA, hydrolysis triggers MuB to dissociate from the DNA (21). As a result, MuB molecules bound to DNA near the Mu genome ends, and thus near MuA, are cleared from the DNA. Eventually, MuB accumulates on DNA far from copies of the Mu genome (7). A second interaction between MuB and MuA must also occur, in which MuB stimulates MuA to catalyze transposition into the MuB-bound DNA. In this interaction, MuB “delivers” the target DNA to MuA. Thus, MuB serves two key roles in Mu targeting: selecting distant DNA and promoting transposition into this selected DNA.
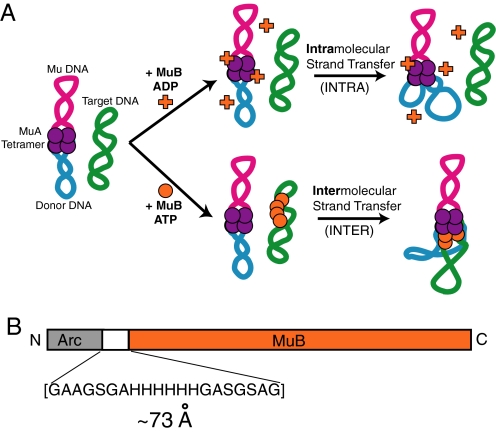
Mu target immunity can be recapitulated in vitro (6, 30). To assay immunity, a donor plasmid containing the Mu sequences necessary for transposition and a second, non-Mu, target plasmid are incubated with MuA, MuB, and ATP. Under these conditions, MuB preferentially selects the non-Mu plasmid and delivers this DNA to the transposase for recombination. We refer to this process as intermolecular transposition or INTER (Fig. 1A). In contrast, if MuB is absent, or if ADP is present in place of ATP, then target immunity fails, and transposition events occur almost exclusively into the donor plasmid's own sequence (intramolecular transposition or INTRA) (Fig. 1A). MuB can, however, contribute to intramolecular transposition, as demonstrated by the observation that INTRA is more efficient in the presence of MuB·ADP than in the absence of MuB (19, 20). Therefore, in addition to the ability of MuB to select target DNA and to deliver selected DNA to MuA, MuB can also stimulate transposition into unselected DNA.
Fig. 1.
Overview of MuB role in transposition and schematic of the MuB fusions. (A) Diagram of Mu transposition showing the pathways that give rise to intramolecular and intermolecular products and the role of ADP, ATP, and MuB. (B) Diagram of the Arc-MuB fusion protein highlighting the sequence of the inserted linker region. Note that the version of Arc used in these experiments is a monomer when denatured, folds as a dimer, and forms a tetramer when bound to its operator site on DNA.
We have described three roles of the MuB protein: (i) selection of target sites that are far from copies of the Mu genome, (ii) delivery of selected DNA to the transposase (i.e., stimulation of INTER), and (iii) activation of transposition into nonselected DNA (i.e., stimulation of INTRA). These three activities differ in their requirement for ATP hydrolysis by MuB. Specifically, target selection critically depends on MuB ATPase activity and the linkage between high-affinity DNA binding, ATP binding, and MuA control of ATP hydrolysis (6, 7). In contrast, stimulation of INTRA does not require ATP hydrolysis because this process is supported by MuB·ADP (20).
It remains unclear whether ATP-regulated DNA binding and the MuA control of ATP hydrolysis are important for delivery of selected target DNA. Previous experiments demonstrate that MuB bound to adenosine 5′-[γ-thio]triphosphate (ATPγS), a poorly hydrolyzed ATP analog, supports intermolecular transposition, suggesting that ATP hydrolysis is not necessary for target delivery (7). However, ATPγS-bound MuB makes larger oligomers than ATP·MuB and is not always as efficient at delivering target DNA as is the ATP-bound form (31). Moreover, ATPγS is not an ideal ATP mimic. ATPγS is commonly contaminated with ADP, and thus multiple conformations of a protein may be present in reactions containing ATPγS. Furthermore, some MuB mutants defective in ATP hydrolysis are also defective in INTER transposition, despite their ability to bind target DNA and to interact with the MuA tetramer (20). Therefore, contrary to studies with ATPγS, experiments with MuB ATPase mutants suggest that ATP regulation of DNA binding may play a significant role in intermolecular transposition. Thus, a clear picture of the role of ATP control of MuB activities is lacking. We sought a new approach to dissecting the mechanism of target delivery by making MuB fusion proteins that allow DNA binding and ATP binding to be controlled separately.
In this work, we probe whether or not ATP- and MuA-regulated DNA binding by MuB is important to the mechanism of target delivery. To this end, we created MuB variants that carry the full-length MuB sequence and an unrelated DNA-binding domain. These fusion proteins bind target DNA irrespective of the MuB nucleotide state. Interestingly, the fusion proteins stimulate intermolecular transposition, even under conditions that prevent ATP hydrolysis. Thus, we conclude that ATP control of DNA binding by MuB is not essential for efficient target delivery. However, nucleotide binding by MuB is still critical, although ADP can suffice, indicating that MuB activation of MuA strictly requires a nucleotide-bound state of the protein. These data support a model in which target delivery by MuB occurs as a consequence of the ability of MuB to stimulate MuA while increasing the local concentration of target DNA with respect to MuA. This model of targeting by a tethered-activator provides an attractive explanation for other examples of regulated target site choice during transposition.
Results
Engineering of MuB Fusion Proteins.
Because the ATP-bound state of MuB is coupled to its ability to bind target DNA (20), it is difficult to investigate the role of one activity without disturbing the other. Therefore, to determine whether ATP controlled DNA binding by MuB is fundamental for the mechanism of target delivery, we uncoupled the MuB DNA-binding activity from its ability to bind and hydrolyze ATP. This divorce was accomplished by creating a MuB fusion protein composed of an ATP-independent DNA-binding protein connected by a 20-aa flexible linker to the N terminus of MuB. The resulting fusion protein binds DNA independently of nucleotide, thereby separating the ability of MuB to bind target from its ATP state.
We chose the Arc repressor protein (Arc) (32) as the DNA-binding protein because it is small (6.2 kDa) and its DNA-binding activity is nucleotide-independent. Arc is a dimer (33), and two Arc dimers bind tightly to a specific Arc operator DNA sequence (half-maximal binding at 1.8 nM Arc dimer equivalents) (34). We conjectured that the Arc high affinity for Arc operator DNA and its ability to multimerize might partially mimic MuB DNA-binding characteristics. MuB·ATP binds DNA tightly, with a Kd of ≈80 nM (20), and MuB·ATP forms polymers on DNA (21, 28). As is discussed below, the Arc repressor also binds nonspecific DNA sequences, albeit much less tightly (half-maximal binding at 230 nM Arc dimer equivalents) (34).
We made two versions of the Arc-MuB fusion protein: one with Arc fused to wild-type MuB (FPWT) and one with Arc fused to MuBins101N (FPinsN) (Fig. 1B). MuBins101N contains an Asp inserted into the Walker A box of the ATP-binding motif (20, 35, 36). This previously characterized variant of MuB is defective in ATP hydrolysis and in DNA binding but is still capable of stimulating MuA to catalyze intramolecular transposition (20). Under the conditions used in our experiments, MuBins101N binds ATP, although it cannot catalyze ATP hydrolysis (20). Creating two versions of the fusion protein allowed the MuB ability to hydrolyze ATP to be blocked in distinct ways: by mutation, with FPinsN, or by using only ADP with the FPWT. Because the fusion proteins bind DNA via the Arc domain but, under these conditions, cannot hydrolyze ATP, they are excellent reagents for investigating the importance of nucleotide regulation of MuB during target delivery.
Fusion Proteins Bind Arc Sites in an ATP-Independent Manner.
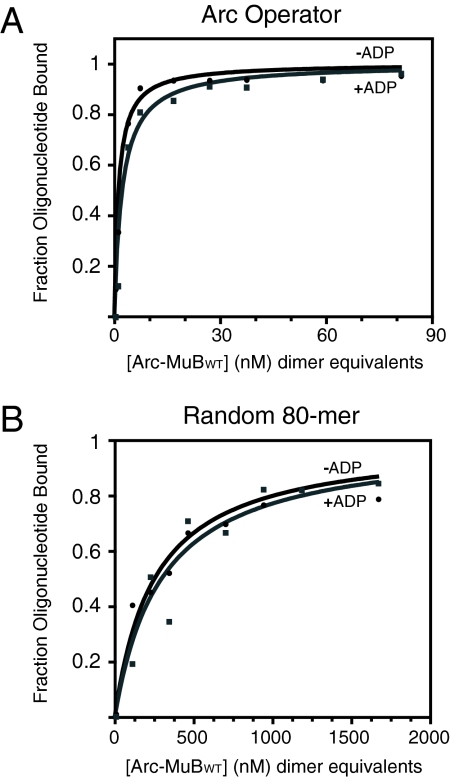
To characterize the DNA-binding activity of the MuB fusion proteins, we performed gel shift assays under the same conditions as the transposition experiments, with 80-bp DNA fragments that either did or did not contain the 21-bp Arc operator. FPWT, in the presence of ADP, bound Arc operator DNA half-maximally at a concentration of ≈1.5 nM dimer equivalents (Fig. 2A). Given that FPWT·ADP binds DNA as tightly as MuB·ATP, we successfully uncoupled the MuB nucleotide state from its ability to bind DNA. In the presence of ADP, FPWT bound non-Arc operator DNA half-maximally at a concentration of ≈300 nM dimer equivalents (Fig. 2B). Based on this clear preference of FPWT for the Arc operator site over nonspecific DNA, we conclude that the Arc region of the fusion protein is fully functional.
Fig. 2.
Characterization of the DNA-binding activity of the wild-type MuB fusion protein. (A) Gel shift assay performed with 0.1 nM DNA fragment, FPWT, and presence or absence of ADP as indicated. The 80-bp fragment contained a single copy of the Arc operator sequence. (B) Same as in A except that the DNA fragment lacked the Arc operator sequence. Binding experiments in both A and B were repeated multiple times, and the data presented are representative curves.
FPWT binds equally well to both specific and nonspecific DNA whether or not ADP is present (Fig. 2). Because MuB binds DNA ≈1.5 times more tightly in the presence of ADP than in its absence, the fact that the ability of FPWT to bind DNA is unaffected by ADP suggests that the fusion protein is binding principally via the Arc domain. In addition, FPWT binds Arc operator DNA equally tightly in the presence of ATP as without nucleotide (data not shown). It should be noted, however, that FPWT bound non-Arc operator DNA ≈10-fold more tightly in the presence of ATP than in its absence (data not shown), indicating that FPWT can bind nonspecific DNA via the MuB domain in the presence of ATP. For this reason, ADP was used in subsequent experiments performed with FPWT non-Arc operator target.
We also tested the functionality of the MuB portion of our Arc-MuB fusion proteins by assaying stimulation of intramolecular transposition in vitro. INTRA was much more efficient in the presence of the fusion proteins than in the absence of any MuB variant, indicating that the MuB portions of the proteins were indeed active (see below).
Fusion Proteins Support INTER Without ATP Regulation of DNA Binding.
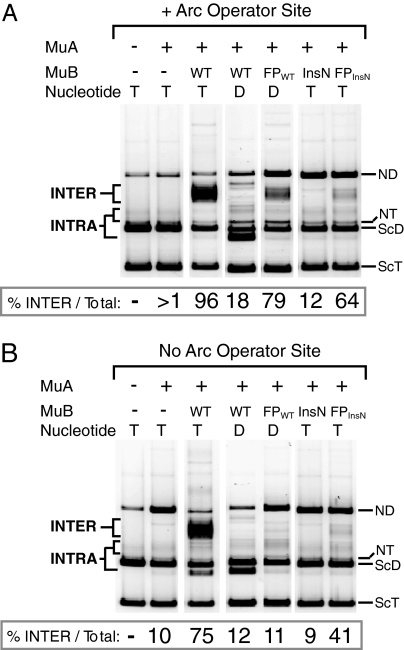
To determine whether ATP control of MuB activities is necessary for MuB-stimulated target delivery, we assayed the ability of the fusion proteins to direct transposition into foreign target DNA. We performed in vitro recombination assays, in which the fusion proteins, MuA, Mu donor DNA, and a second, non-Mu, target plasmid were incubated for 2 h at 30°C. The target plasmid either did, or did not, contain a single copy of the Arc operator. After incubation, samples were run on an agarose gel to separate the INTRA products from the INTER products. Both FPWT bound to ADP and FPinsN bound to ATP are unable to hydrolyze ATP but are able to bind target DNA via their Arc regions.
If ATP control of MuB is specifically required for target delivery, then the fusion proteins should be inefficient at INTER compared with wild-type MuB. Consequently, the majority of the recombination products are expected to be the result of INTRA. However, if ATP control of MuB is not necessary, then the fusion proteins should support recombination into bound DNA. In this case, we would expect to see a preference for INTER when the target DNA contains a copy of the Arc operator site.
Both fusion proteins supported INTER into plasmids containing the Arc operator sequence (Fig. 3A). As expected, wild-type MuB·ATP also supported efficient INTER, whereas MuBinsN·ATP and wild-type MuB·ADP failed to generate these INTER products (Fig. 3A). Moreover, when the target DNA lacked the Arc operator site, only wild-type MuB·ATP efficiently targeted recombination to intermolecular sites (Fig. 3B). Although the fusion proteins did support a modest level of INTER into non-Arc target DNA, this activity could be explained by the relatively high affinity of Arc for nonspecific DNA. Experiments like those in Fig. 3 were repeated with an unrelated pair of Arc and non-Arc target plasmids with similar results (data not shown).
Fig. 3.
The fusion proteins stimulate intermolecular transposition into DNA molecules carrying an Arc site. (A) Agarose gel of products from in vitro transposition reactions containing different variants of MuB as shown above the lanes. Reactions were incubated for 2 h and contained Arc operator target plasmid (pCS13) and either ATP (T) or ADP (D) as indicated. All of the images are lanes from the same gel with the same image contrast. The identity of INTRA and INTER products was verified by Southern hybridization with appropriate probes. The DNA species are labeled as follows: ScT, supercoiled target; ScD, supercoiled donor (mini-Mu); NT, nicked target; ND, nicked donor. (B) The reactions and electrophoresis were the same as in A except target plasmid lacked Arc operator sequence (pUC19). All of the images are lanes from the same gel with the same image contrast.
Importantly, in the absence of nucleotide, FPWT did not support INTER into Arc operator DNA. In these experiments, the amount of INTER was undetectable, as was true for the no MuB control (data not shown). These data reveal that target delivery by FPWT requires MuB to be in an nucleotide-bound state and that this nucleotide can be either ATP or ADP.
To determine whether the fusion proteins preferentially support INTER over INTRA, we calculated the percentage of total transposition products that were INTER products in each sample (see values below gels in Fig. 3 A and B). Remarkably, with both FPWT·ADP and FPinsN·ATP, more than half of the transposition products were INTER, indicating that the fusion proteins preferentially directed transposition into the intermolecular target DNA.
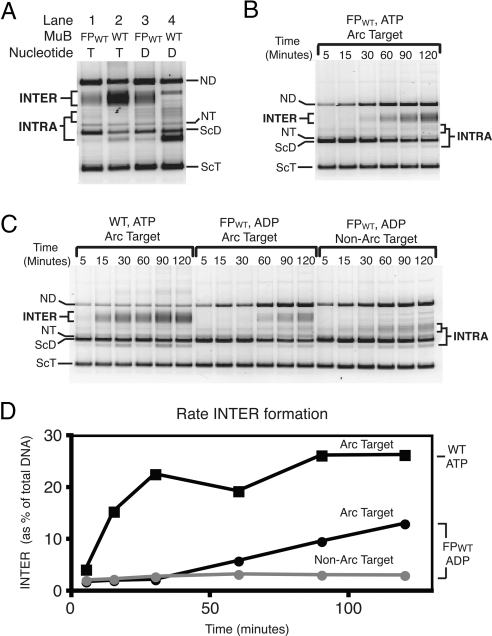
We consistently observed that FPWT·ADP and FPWT·ATP were equally efficient at stimulating INTER into Arc operator containing plasmids (Fig. 4A, lanes 1 and 3, and B and C). If the fusion proteins were binding target DNA via their MuB domains, we would expect INTER to be more efficient in the presence of ATP because wild-type MuB binds DNA ≈10 times more tightly in the presence of ATP than in the presence of ADP. Because the fusion proteins exhibit similar activity in the presence of ATP or ADP, we conclude that the fusion proteins are primarily binding target DNA via their Arc domains.
Fig. 4.
Kinetics of transposition reactions supported by MuB variants. (A) Agarose gel of products generated in the presence of MuB or FPWT in the presence of either ATP or ADP. Reactions were incubated for 2 h and contained Arc operator target plasmid (pCS13). DNA species are labeled as in Fig. 3. (B) Agarose gel of products generated in reactions containing FPWT, ATP, and the Arc operator target. Reactions were incubated for the length of time indicated. (C) Same as in B, although the MuB variant and target DNA were altered as indicated. (D) Graphical representation of the data from C.
In summary, our results indicate that FPWT·ADP and FPinsN·ATP can preferentially target transposition to intermolecular DNA and this targeting depends on the presence of an Arc operator site in the non-Mu DNA molecule. These data support the hypothesis that ATP regulation of MuB DNA interaction is not mechanistically required for the stimulation of MuA-catalyzed transposition into distant DNA.
Fusions Proteins Promoting Target Delivery with Slower Kinetics.
Although we determined that our fusion proteins support intermolecular transposition, we wanted to investigate the reaction kinetics. FPWT in the presence of either ATP or ADP formed ≈40% as much INTER product as MuB·ATP after 2 h (Fig. 4 C and D). However, product accumulation supported by the fusion protein, with either ATP or ADP, was much slower than that promoted by MuB·ATP. The MuB·ATP reaction was nearly complete by 30 min, whereas transposition promoted by the fusion protein was just detectable at 30 min and still linear after 2 h (Fig. 4D). The reaction kinetics supported by FPWT with ATP and ADP were very similar (Fig. 4 B and C). The reduced rate of FPWT compared with MuB is likely because of the difference in the number of binding sites in the target DNA for each protein. Whereas only two Arc dimers bind specifically to the Arc operator sequence present in the target DNA, wild-type MuB can bind to any site along the target plasmid. Thus, there are many more initial MuB-nucleated target sites attractive for wild-type MuB than for the fusion protein in these transposition reactions. Consistent with this hypothesis, transposition events were more clustered near the Arc operator sites when FPWT was responsible for delivery than in reaction containing wild-type MuB (see below). To reduce the discrepancy in the number of binding sites for FPWT and MuB, we attempted in vitro recombination reactions by using short DNA fragments as target DNA. Unfortunately, such short target molecules altered the dependence of the reaction on MuB, making the results impossible to compare with the results found with plasmid DNA molecules (data not shown).
Fusion Proteins Target Transposition to DNA near the Arc Operator Site.
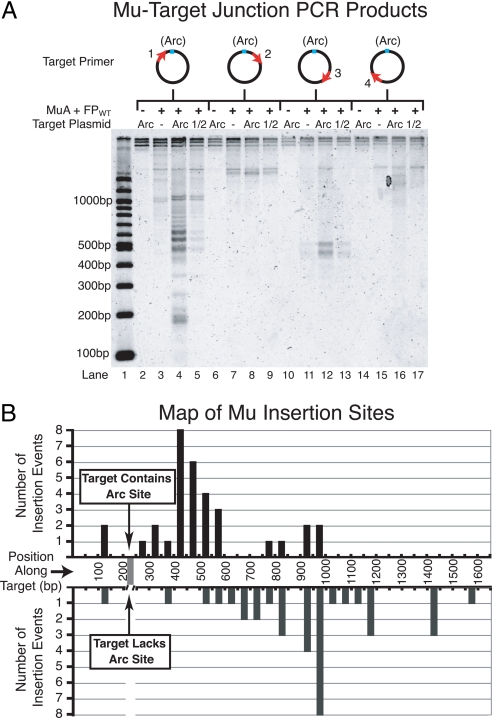
The fusion proteins clearly targeted transposition preferentially to plasmids carrying an Arc operator. However, it was uncertain where on the plasmid the recombination was occurring. Did the fusion proteins target transposition directly into the Arc operator, nearby the operator, or throughout the plasmid? To address this question, we mapped transposition events from in vitro transposition reactions containing FPWT·ADP and either Arc operator or non-Arc operator target plasmids. Mapping was accomplished by PCR-amplifying the donor–target joint DNA molecules, using one primer specific to the Mu end on the donor DNA and one primer specific to the target plasmid. PCR products were run on an agarose gel, and each resulting band on the gel corresponded to a transposition event that occurred at a specific distance from the target primer. This experiment was repeated with four different target primers, spaced evenly around the target plasmid (Fig. 5). The experiment was performed with target plasmids that either lacked a copy of the Arc operator, contained a single copy of the Arc operator, or contained a single copy of only the right half of the Arc operator (Fig. 5).
Fig. 5.
Maps of intermolecular transposition events. (A) Agarose gel of PCR amplified donor–target joints from in vitro transposition reactions containing FPWT·ADP and either pCS14 (non-Arc), pCS15 (Arc), or pCS16 (1/2 Arc) target plasmids. Diagrams above the gel indicate the position of the target primer relative to the Arc operator. Note that the samples contained different amounts of total DNA because transposition was more efficient into plasmids containing a copy of the Arc operator, and therefore, these samples contained a much larger number of donor–target joint molecules that could be amplified during PCR. This discrepancy is apparent upon inspecting the total signal per lane for Arc operator vs. non-Arc operator samples. However, because we do not expect a change in the concentration of donor–target DNA joints to cause a large PCR bias, it is informative to compare the relative intensities of bands within one lane to those within another lane. (B) Graph representing the frequency of transposition events at positions along the target DNA based on the recovery of cloned Mu–target junctions. We mapped a total of 33 events into pCS15 (Arc target) and 35 events into pCS14 (non-Arc target) from reactions containing FPWT·ADP. Individually mapped events were binned into 50-bp intervals. Donor–target joints were PCR-amplified by using primer 1 (see A).
The results of this global mapping experiment revealed a different pattern of transposition events into plasmids containing the Arc operator site compared with plasmids lacking the operator (Fig. 5). A cluster of bands corresponding to insertion events ≈40 bp to ≈340 bp 3′ of the Arc operator site was clearly present only in samples in which the target plasmid contained the Arc operator site (Fig. 5A, compare lanes 3 and 4). Another set of bands that were from insertion events ≈110 bp to ≈10 bp 5′ of the Arc operator site also appeared only when the operator was present (Fig. 5A, compare lanes 3 and 4). We did not, however, detect bands unique to the Arc operator samples at positions far from the Arc operator (Fig. 5A, lanes 6–17). Overall, the presence of an Arc operator site in the target DNA altered the profile of transposition events mediated by the fusion proteins by rendering regions of the DNA on either side of the Arc operator more susceptible to transposition.
To start to investigate the number of Arc operator-bound fusion proteins required to mediate target delivery, we created target plasmids that contained only the right half of the Arc operator. This half-site binds one Arc dimer compared with the tetramer-bound full site. Target plasmids containing the Arc half-site were substantially less efficient substrates for INTER supported by the fusion proteins (data not shown). Likewise, in mapping experiments, samples containing half operator target plasmids exhibited a banding pattern after PCR intermediate between that of samples containing non-Arc operator plasmids and that of samples containing full Arc operator plasmids (Fig. 5A). From these data, we conclude that the half Arc operator is a less robust signal than the full Arc operator for transposition targeting by the fusion proteins. Thus, binding of four Arc-MuB subunits is superior to two DNA-bound subunits at initiating events that lead to targeting of Mu transposition.
To validate the target site-selection conclusions from our global mapping experiments, we cloned the PCR-amplified donor–target DNA joints from Fig. 5. Each clone was sequenced to determine the site of the insertion event. The PCR primers allowed recovery of insertion events from ≈150 bp 5′ of the Arc operator to ≈1,400 bp 3′ of the operator. Consistent with the global mapping results, we found that FPWT·ADP targeted transposition to different locations depending on whether or not the target DNA contained the Arc operator [Fig. 5B and supporting information (SI) Fig. S1]. The regions of the target DNA into which transposition occurred most frequently in the absence of the Arc operator were also sites of transposition in the presence of the Arc operator. That these Arc-independent target sites appeared to be less pronounced when the Arc operator was present was because of the abundance of the Arc-dependent target sites overriding the signal from the Arc-independent target sites. Neither the transposition hot spots observed with the Arc operator plasmid nor those observed with the non-Arc plasmid consistently correspond to the Mu consensus sequence (5′-C-C/T-G/C-A/G-G-3′) (27, 37). It remains unclear why particular regions of the DNA are preferred target sites for Mu transposition under our reaction conditions. Perhaps, as has been suggested, the local structure of the DNA has important influence in determining the location of Mu targeting (38).
Interestingly, we did not observe a single insertion event into the Arc operator itself. The data in Fig. 5B were derived from multiple in vitro transposition reactions that were PCR-amplified and cloned separately (see Methods and Fig. S1). Thus, we conclude that the fusion proteins target transposition to locations near the Arc operator site but disfavor recombination into the operator sequence itself.
Discussion
MuB is an ATP- and MuA-regulated DNA-binding protein that controls transposition target site choice. We have created Arc-MuB fusion proteins that effectively uncouple MuB ATP binding and ATPase activity from its target DNA-binding activity. We find that these fusion proteins can target transposition to distant plasmids containing a copy of the Arc operator site. This targeting is robust even if only ADP is present or if the MuB portion of the fusion protein is mutated to prevent ATP hydrolysis. Therefore, fusion protein-mediated stimulation of INTER can occur when control of DNA binding by ATP has been disrupted. Based on these results, we can conclude that the process of target delivery does not fundamentally require the ATP-regulated DNA-binding activity of MuB. This regulation, in contrast, is essential for control of target site selection. Specifically, the process of target immunity, by which MuB accumulates on DNA far from any copies of the Mu genome, depends on MuA-stimulated ATP hydrolysis by MuB (6, 7, 22). However, our data suggest that once a biased distribution of MuB has been generated, transposition into the MuB-bound DNA can occur independently of ATP-regulated control of the DNA-binding activity of MuB. That MuB ATPase activity and its control of the DNA interaction appear solely necessary for target selection highlights the evolutionary importance of target immunity for the Mu transposon.
Our results clearly establish that, although the ATP-bound form of MuB is not needed for stimulation of INTER, nucleotide-bound MuB is essential. With the wild-type Arc-MuB fusion protein, either ATP or ADP supported INTER to an equal extent, but, in the absence of nucleotide, no activity was detected. These data suggest that ATP/ADP promotes a conformational change in MuB that is important for its ability to stimulate catalysis by the MuA transpososome. Perhaps this conformational change is stabilizing the dimeric form of MuB (31); however, our current results suggest that stabilization of a MuB dimer may not be sufficient to explain MuB dependence on nucleotide. Arc repressor is a constitutive dimer, and therefore the fusion protein would be expected to stabilize dimer contacts within MuB, perhaps making nucleotide less important for this function. Furthermore, ADP and ATP activated the fusion protein identically (within error), yet ATP promotes multimerization of MuB more effectively than does ADP (31). Thus, we favor the idea that nucleotide binding by MuB is required for MuA activation, not simply for stabilizing the dimeric state of MuB.
Our current model for MuB regulation of target choice is that MuB activates MuA to perform INTER in much the same way that it activates MuA to perform INTRA. In other words, we hypothesize that MuB stimulates INTER by activating the catalytic activity of MuA (as for INTRA), but it does so while simultaneously increasing the local concentration of the DNA molecule to which it is bound. Thus, MuB delivers target DNA by tethering that DNA to the MuA tetramer. Several other transposases select target DNA by interacting with proteins that are bound to the preferred target site. For example, Ty5 integrase interacts with heterochromatin-associated Sir4p to target Ty5 transposition to silent DNA (39), and Ty3 integrase interacts with transcription factor TFIIIB to target Ty3 transposition upstream of RNA polymerase III promoters (40). In addition, the proteins that catalyze Tn7 transposition, TnsA and TnsB, interact with two other Tn7-encoded proteins, TnsC and TnsD, to target transposition downstream of the glmS gene and to mediate target immunity in a manner similar to Mu (9, 41–43). It is attractive to consider a similar tethered-activator model for targeting other transpons that use genome-bound proteins.
One aspect of the tethered-activator model for DNA targeting that we do not fully understand is how the DNA bound by MuB is so strongly preferred. When MuB bound to target DNA interacts with the MuA tetramer, the local concentration of the MuB-bound DNA relative to the MuA tetramer increases. However, the local concentration of the flanking donor DNA relative to the MuA tetramer is also very likely to be quite high. Why, then, do the majority (75–95%) of the transposition events occur into the MuB-bound DNA? One possible answer is that the flanking donor DNA is in a particularly poor orientation to be accessed by the transposase, and thus, the DNA tethered to MuB is a favored target. Perhaps, transposase–DNA complexes have evolved to have this type of “inhibitory” conformation toward neighboring DNA to promote more large-scale element movements.
In creating our Arc-MuB fusion proteins we have engineered a version of MuB that can preferentially target transposition to a particular DNA molecule. We hope that this system will prove a useful tool for further investigation of the mechanisms underlying Mu transposition as well as inform future research aimed at redirecting the target site choice of transposons.
Methods
DNA.
The donor plasmid was pMK586 (mini-Mu) (44). Non-Arc operator target plasmid was either pUC19 or pCS14 (pJF122 with 180 bp added to the multiple cloning site). Arc operator target plasmid was either pCS13 (pUC19 with atagtagagtgcttctatcat cloned into the EcoRI site) or pCS15 (pCS14 with atagtagagtgcttctatcat cloned into the EcoRI site). Half-Arc operator target plasmid was pCS16 (pCS14 with atagtagagtgctgtattcat cloned into the EcoRI site). All plasmids were purified by a plasmid mega kit (Qiagen) followed by CsCl/ethidium bromide ultracentrifugation. Eighty-base pair oligonucleotides used in our gel shift assays were ordered from Invitrogen. The sequences are as follows: Arc operator, 5′-catcaccgaaacgtccgaggcagcaagttatgatagaagcactctactatggagtcataatgtgcctgtcattgagacga-3′; non-Arc operator, 5′-catcaccgaaacgtccgaggcagcaagtttcgcacgtccgcacagcacgtggagtcataatgtgcctgtcattgagacga-3′.
Proteins.
MuA was purified as described by Baker et al. (45). HU was purified as described by Baker and Luo (11). Wild-type MuB and MuBins101N were purified as described by Yamauchi and Baker (20).
FPWT and FPins101N were cloned into pET20b (Novagen) and expressed in bacterial strain ER2556. Cells were grown in TB to an OD600 of 0.55–0.75, induced with 0.7 mM IPTG for 2 h, centrifuged at 6,000 × g for 20 min. Cell pellets were resuspended in 50 mM Tris (pH 8), 10% sucrose, 2.5 mM DTT, 12.5 mM EDTA, plus protease inhibitor mixture (Calbiochem). Cell were French pressed, and lysate was cleared by centrifugation at 20,000 × g for 30 min. Ammonium sulfate was added to 35%, and precipitation was allowed to occur for 1 h at 4°C. Sample was centrifuged for 15 min at ≈40,000 × g, and the resulting pellet was resuspended in denaturing buffer (6 M guanidine HCl, 100 mM NaH2PO4, 10 mM Tris, 10 mM imidazole, pH 8.0). Protein was then batch bound to Ni-nitrilotriacetic acid beads (Qiagen), washed with denaturing buffer, and eluted with 6 M guanidine HCl, 20 mM NaH2PO4, 10 mM Tris, 400 mM imidazole, pH 8.0. Eluted samples were subjected to several dialysis steps: first into MuB buffer [1 M NaCl, 25 mM Hepes (pH 7.6), 0.1 mM EDTA, 20% glycerol, 2 mM DTT] + 2 M guanidine HCl, then into MuB buffer + 0.5 guanidine HCl, and finally into MuB buffer.
Gel Shift Assays.
Binding reactions contained 100 mM NaCl, 0.1 mM EDTA, 2.5 mM Hepes-KOH (pH 7.6), 3.5% glycerol, 1 mM DTT, 25 mM Tris·HCl (pH 8), 10 mM MgCl2, 0.1 nM 80-bp oligonucleotide (see DNA) that was 32P-radiolabeled at the 5′ end, and FPWT (concentration varied). Some reactions also contained 2 mM ADP. Reactions were incubated for 4 h at 30°C. Loading solution (0.8 volume) (11.25% glycerol, 2.25× loading dye) was added to each sample immediately before loading the sample onto a polyacrylamide gel. Samples were run on a 5% native polyacrylamide gel (19:1 acrylamide:bisacrylamide) in 0.5× TBE (44.5 mM Tris, 44.5 mM boric acid, 1 mM EDTA) at ≈15 V/cm. Gels were dried at 76°C and exposed on a storage phosphor screen (Amersham Biosciences) for 2–3 days. Exposed phosphor screens were viewed by using a Typhoon 9400. Bands were quantitated by using ImageQuant (Amersham Biosciences), and curves were fit by using KaleidaGraph (Synergy Software).
Transposition Assays.
Transposition reactions in Figs. 3 A and B, and 4A included: 100 mM NaCl, 3.5% glycerol, 1 mM DTT, 25 mM Tris·HCl (pH 8), 10 mM MgCl2, 2 mM ADP or ATP (as indicated), 0.1 mg/ml BSA, 10 μg/ml pMK586 (donor), 10 μg/ml pUC19 (non-Arc target) or pCS13 (Arc target) (as indicated), 130 nM HU, 40 nM MuA, 300 nM MuB or MuBins101N or FPWT or FPinsN (as indicated). Reactions were incubated for 2 h at 30°C and stopped with 0.2 volume of STOP solution (2.5% SDS, 50 mM EDTA, 30% glycerol, bromophenol blue). Reaction products were run on a 0.9% high-gelling temperature (HGT)–agarose (Cambrex) gel in 1× TAB [40 mM Tris·HCl (pH 8.0), 3.6 mM EDTA, 27 mM sodium acetate] at ≈5 V/cm for 2 h at 4°C. Gels were stained with Vistra Green (1:10,000 dilution ; Amersham Biosciences) and visualized on a Molecular Dynamics fluoroimager. Band intensities were quantitated by using ImageQuant TL (Amersham Biosciences).
Reactions in Fig. 4 B–D were performed as above, except that the MuA concentration was 30 nM, and the reactions were stopped after the indicated amount of time. Reactions for the mapping experiments in Fig. 5 were also performed as above, except that the MuA concentration was 30 nM, the reactions were incubated for 3 h, and pCS14 (non-Arc), pCS15 (Arc), or pCS16 (1/2 Arc) was used as the target plasmid.
Global Mapping Experiment.
To map globally the location of Mu transposition events, transposition reactions were performed as described above. The products of these reactions were subjected to proteinase K treatment followed by phenol:chloroform extraction and ethanol precipitation. Samples were resuspended in H2O and used as the template in subsequent PCR. PCR was performed with a Mu-specific primer (5′-cccggtttttttcgtacttcaagtgaatcaataca-3′) and one of four different target-specific primers: primer 1, 5-′cgttttttgggctaacaggaggaattaacctag-3′; primer 2, 5′-caacttcagcagcacgtaggggac-3′; primer 3, 5′-cgggtgtggtcgccatgatcg-3′; primer 4, 5′-gcatgtgtcagaggttttcaccgtcatc-3′.
PCR products were run on a 1.8% metaphore agarose (Cambrex) gel in 1× TBE at 5 V/cm at 4°C. Gels were stained with Vistra Green (1:10,000 dilution; Amersham Biosciences) and visualized on a Typhoon 4900.
Mapping Insertion Sites.
To map specifically individual insertion events, we cloned and sequenced the PCR-amplified donor–target joints generated in the global mapping experiments (above). We cloned those donor–target joints that had been primer 1-amplified from transposition reactions containing FPWT·ADP and pCS14 (non-Arc target) or pCS15 (Arc target). Donor–target joints were cloned by using the TOPO TA cloning kit (Invitrogen), and individual clones were sequence by the MIT Biopolymers Laboratory.
Supplementary Material
Acknowledgments.
We thank all members of the Baker laboratory for insightful advice relating to this project and to the writing of this manuscript. We are especially grateful to Elizabeth S. C. Oakes and Kathryn M. Lemberg for their help, respectively, in the purifications of MuA and MuB. T.A.B. is an employee of the Howard Hughes Medical Institute, and this work was supported by National Institutes of Health Grant GM-49224.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0805868105/DCSupplemental.
References
- 1.Curcio MJ, Derbyshire KM. The outs and ins of transposition: From Mu to kangaroo. Nat Rev Mol Cell Biol. 2003;4:865–877. doi: 10.1038/nrm1241. [DOI] [PubMed] [Google Scholar]
- 2.Coros AM, Twiss E, Tavakoli NP, Derbyshire KM. Genetic evidence that GTP is required for transposition of IS903 and Tn552 in Escherichia coli. J Bacteriol. 2005;187:4598–4606. doi: 10.1128/JB.187.13.4598-4606.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaufman PD, Rio DC. P element transposition in vitro proceeds by a cut-and-paste mechanism and uses GTP as a cofactor. Cell. 1992;69:27–39. doi: 10.1016/0092-8674(92)90116-t. [DOI] [PubMed] [Google Scholar]
- 4.Tang M, Cecconi C, Kim H, Bustamante C, Rio DC. Guanosine triphosphate acts as a cofactor to promote assembly of initial P-element transposase–DNA synaptic complexes. Genes Dev. 2005;19:1422–1425. doi: 10.1101/gad.1317605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsai CL, Schatz DG. Regulation of RAG1/RAG2-mediated transposition by GTP and the C-terminal region of RAG2. EMBO J. 2003;22:1922–1930. doi: 10.1093/emboj/cdg185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maxwell A, Craigie R, Mizuuchi K. B protein of bacteriophage Mu is an ATPase that preferentially stimulates intermolecular DNA strand transfer. Proc Natl Acad Sci USA. 1987;84:699–703. doi: 10.1073/pnas.84.3.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adzuma K, Mizuuchi K. Target immunity of Mu transposition reflects a differential distribution of Mu B protein. Cell. 1988;53:257–266. doi: 10.1016/0092-8674(88)90387-x. [DOI] [PubMed] [Google Scholar]
- 8.Gamas P, Craig NL. Purification and characterization of TnsC, a Tn7 transposition protein that binds ATP and DNA. Nucleic Acids Res. 1992;20:2525–2532. doi: 10.1093/nar/20.10.2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stellwagen AE, Craig NL. Avoiding self: Two Tn7-encoded proteins mediate target immunity in Tn7 transposition. EMBO J. 1997;16:6823–6834. doi: 10.1093/emboj/16.22.6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stellwagen AE, Craig NL. Mobile DNA elements: Controlling transposition with ATP-dependent molecular switches. Trends Biochem Sci. 1998;23:486–490. doi: 10.1016/s0968-0004(98)01325-5. [DOI] [PubMed] [Google Scholar]
- 11.Baker TA, Luo L. Identification of residues in the Mu transposase essential for catalysis. Proc Natl Acad Sci USA. 1994;91:6654–6658. doi: 10.1073/pnas.91.14.6654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rice P, Mizuuchi K. Structure of the bacteriophage Mu transposase core: A common structural motif for DNA transposition and retroviral integration. Cell. 1995;82:209–220. doi: 10.1016/0092-8674(95)90308-9. [DOI] [PubMed] [Google Scholar]
- 13.Craigie R, Mizuuchi M, Mizuuchi K. Site-specific recognition of the bacteriophage Mu ends by the Mu A protein. Cell. 1984;39:387–394. doi: 10.1016/0092-8674(84)90017-5. [DOI] [PubMed] [Google Scholar]
- 14.Mizuuchi K, Adzuma K. Inversion of the phosphate chirality at the target site of Mu DNA strand transfer: Evidence for a one-step transesterification mechanism. Cell. 1991;66:129–140. doi: 10.1016/0092-8674(91)90145-o. [DOI] [PubMed] [Google Scholar]
- 15.Aldaz H, Schuster E, Baker TA. The interwoven architecture of the Mu transposase couples DNA synapsis to catalysis. Cell. 1996;85:257–269. doi: 10.1016/s0092-8674(00)81102-2. [DOI] [PubMed] [Google Scholar]
- 16.Namgoong SY, Harshey RM. The same two monomers within a MuA tetramer provide the DDE domains for the strand cleavage and strand transfer steps of transposition. EMBO J. 1998;17:3775–3785. doi: 10.1093/emboj/17.13.3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Williams TL, Jackson EL, Carritte A, Baker TA. Organization and dynamics of the Mu transpososome: Recombination by communication between two active sites. Genes Dev. 1999;13:2725–2737. doi: 10.1101/gad.13.20.2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baker TA, Mizuuchi M, Mizuuchi K. MuB protein allosterically activates strand transfer by the transposase of phage Mu. Cell. 1991;65:1003–1013. doi: 10.1016/0092-8674(91)90552-a. [DOI] [PubMed] [Google Scholar]
- 19.Surette MG, Chaconas G. Stimulation of the Mu DNA strand cleavage and intramolecular strand transfer reactions by the Mu B protein is independent of stable binding of the Mu B protein to DNA. J Biol Chem. 1991;266:17306–17313. [PubMed] [Google Scholar]
- 20.Yamauchi M, Baker TA. An ATP–ADP switch in MuB controls progression of the Mu transposition pathway. EMBO J. 1998;17:5509–5518. doi: 10.1093/emboj/17.18.5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Greene EC, Mizuuchi K. Direct observation of single MuB polymers: Evidence for a DNA-dependent conformational change for generating an active target complex. Mol Cell. 2002;9:1079–1089. doi: 10.1016/s1097-2765(02)00514-2. [DOI] [PubMed] [Google Scholar]
- 22.Reyes O, Beyou A, Mignotte-Vieux C, Richaud F. Mini-Mu transduction: Cis inhibition of the insertion of Mud transposons. Plasmid. 1987;18:183–192. doi: 10.1016/0147-619x(87)90061-8. [DOI] [PubMed] [Google Scholar]
- 23.Arciszewska LK, Drake D, Craig NL. Transposon Tn7: Cis-acting sequences in transposition and transposition immunity. J Mol Biol. 1989;207:35–52. doi: 10.1016/0022-2836(89)90439-7. [DOI] [PubMed] [Google Scholar]
- 24.Lee CH, Bhagwat A, Heffron F. Identification of a transposon Tn3 sequence required for transposition immunity. Proc Natl Acad Sci USA. 1983;80:6765–6769. doi: 10.1073/pnas.80.22.6765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maekawa T, Yanagihara K, Ohtsubo E. A cell-free system of Tn3 transposition and transposition immunity. Genes Cells. 1996;1:1007–1016. doi: 10.1046/j.1365-2443.1996.d01-216.x. [DOI] [PubMed] [Google Scholar]
- 26.Manna D, Wang X, Higgins NP. Mu and IS1 transpositions exhibit strong orientation bias at the Escherichia coli bgl locus. J Bacteriol. 2001;183:3328–3335. doi: 10.1128/JB.183.11.3328-3335.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mizuuchi M, Mizuuchi K. Target site selection in transposition of phage Mu. Cold Spring Harb Symp Quant Biol. 1993;58:515–523. doi: 10.1101/sqb.1993.058.01.058. [DOI] [PubMed] [Google Scholar]
- 28.Greene EC, Mizuuchi K. Dynamics of a protein polymer: The assembly and disassembly pathways of the MuB transposition target complex. EMBO J. 2002;21:1477–1486. doi: 10.1093/emboj/21.6.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Greene EC, Mizuuchi K. Target immunity during Mu DNA transposition: Transpososome assembly and DNA looping enhance MuA-mediated disassembly of the MuB target complex. Mol Cell. 2002;10:1367–1378. doi: 10.1016/s1097-2765(02)00733-5. [DOI] [PubMed] [Google Scholar]
- 30.Mizuuchi K. In vitro transposition of bacteriophage Mu: A biochemical approach to a novel replication reaction. Cell. 1983;35:785–794. doi: 10.1016/0092-8674(83)90111-3. [DOI] [PubMed] [Google Scholar]
- 31.Adzuma K, Mizuuchi K. Steady-state kinetic analysis of ATP hydrolysis by the B protein of bacteriophage Mu: Involvement of protein oligomerization in the ATPase cycle. J Biol Chem. 1991;266:6159–6167. [PubMed] [Google Scholar]
- 32.Susskind MM. A new gene of bacteriophage P22 which regulates synthesis of antirepressor. J Mol Biol. 1980;138:685–713. doi: 10.1016/0022-2836(80)90060-1. [DOI] [PubMed] [Google Scholar]
- 33.Vershon AK, Youderian P, Susskind MM, Sauer RT. The bacteriophage P22 Arc and Mnt repressors: Overproduction, purification, and properties. J Biol Chem. 1985;260:12124–12129. [PubMed] [Google Scholar]
- 34.Robinson CR, Sauer RT. Covalent attachment of Arc repressor subunits by a peptide linker enhances affinity for operator DNA. Biochemistry. 1996;35:109–116. doi: 10.1021/bi9521194. [DOI] [PubMed] [Google Scholar]
- 35.Walker JE, Saraste M, Runswick MJ, Gay NJ. Distantly related sequences in the α- and β-subunits of ATP synthase, myosin, kinases, and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pause A, Sonenberg N. Mutational analysis of a DEAD box RNA helicase: The mammalian translation initiation factor eIF-4A. EMBO J. 1992;11:2643–2654. doi: 10.1002/j.1460-2075.1992.tb05330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haapa-Paananen S, Rita H, Savilahti H. DNA transposition of bacteriophage Mu: A quantitative analysis of target site selection in vitro. J Biol Chem. 2002;277:2843–2851. doi: 10.1074/jbc.M108044200. [DOI] [PubMed] [Google Scholar]
- 38.Manna D, Breier AM, Higgins NP. Microarray analysis of transposition targets in Escherichia coli: The impact of transcription. Proc Natl Acad Sci USA. 2004;101:9780–9785. doi: 10.1073/pnas.0400745101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xie W, et al. Targeting of the yeast Ty5 retrotransposon to silent chromatin is mediated by interactions between integrase and Sir4p. Mol Cell Biol. 2001;21:6606–6614. doi: 10.1128/MCB.21.19.6606-6614.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kirchner J, Connolly CM, Sandmeyer SB. Requirement of RNA polymerase III transcription factors for in vitro position-specific integration of a retrovirus-like element. Science. 1995;267:1488–1491. doi: 10.1126/science.7878467. [DOI] [PubMed] [Google Scholar]
- 41.Skelding Z, Queen-Baker J, Craig NL. Alternative interactions between the Tn7 transposase and the Tn7 target DNA-binding protein regulate target immunity and transposition. EMBO J. 2003;22:5904–5917. doi: 10.1093/emboj/cdg551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Waddell CS, Craig NL. Tn7 transposition: Two transposition pathways directed by five Tn7-encoded genes. Genes Dev. 1988;2:137–149. doi: 10.1101/gad.2.2.137. [DOI] [PubMed] [Google Scholar]
- 43.Waddell CS, Craig NL. Tn7 transposition: Recognition of the attTn7 target sequence. Proc Natl Acad Sci USA. 1989;86:3958–3962. doi: 10.1073/pnas.86.11.3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mizuuchi M, Baker TA, Mizuuchi K. DNase protection analysis of the stable synaptic complexes involved in Mu transposition. Proc Natl Acad Sci USA. 1991;88:9031–9035. doi: 10.1073/pnas.88.20.9031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baker TA, Mizuuchi M, Savilahti H, Mizuuchi K. Division of labor among monomers within the Mu transposase tetramer. Cell. 1993;74:723–733. doi: 10.1016/0092-8674(93)90519-v. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.