Abstract
Alfalfa is economically the most important forage legume worldwide. A recurrent challenge to alfalfa production is the significant yield loss caused by disease. Although knowledge of molecular mechanisms underlying host resistance should facilitate the genetic improvement of alfalfa, the acquisition of such knowledge is hampered by alfalfa's tetrasomic inheritance and outcrossing nature. However, alfalfa is congeneric with the reference legume Medicago truncatula, providing an opportunity to use M. truncatula as a surrogate to clone the counterparts of many agronomically important genes in alfalfa. In particular, the high degree of sequence identity and remarkably conserved genome structure and function between the two species enables M. truncatula genes to be used directly in alfalfa improvement. Here we report the map-based cloning of RCT1, a host resistance (R) gene in M. truncatula that confers resistance to multiple races of Colletotrichum trifolii, a hemibiotrophic fungal pathogen that causes anthracnose disease of alfalfa. RCT1 is a member of the Toll-interleukin-1 receptor/nucleotide-binding site/leucine-rich repeat (TIR-NBS-LRR) class of plant R genes and confers broad-spectrum anthracnose resistance when transferred into susceptible alfalfa plants. Thus, RCT1 provides a novel resource to develop anthracnose-resistant alfalfa cultivars and contributes to our understanding of host resistance against the fungal genus Colletotrichum. This work demonstrates the potential of using M. truncatula genes for genetic improvement of alfalfa.
Keywords: Colletotrichum trifolii, disease resistance, Medicago sativa
Alfalfa (Medicago sativa L.), known as the “Queen of Forages,” is the world's most important and widely grown forage legume. Alfalfa is rich in proteins, vitamins and minerals, providing highly nutritious hay and pasture for animal and dairy production. In the United States, alfalfa ranks with wheat as the third most important crop after corn and soybeans (United States Department of Agriculture Crop Values, 2005, 2006; www.nass.usda.gov). Like other legume species, alfalfa contributes to the sustainability of agricultural ecosystems because of its capacity for symbiotic nitrogen fixation. Moreover, the combination of its high biomass production, perennial growth habit, and ability to fix atmospheric nitrogen, has led to increased interest in using alfalfa as a biofuel feedstock for production of ethanol and other industrial materials.
Alfalfa production has been negatively impacted by damaging pests and pathogens. On an annual basis, ≈20% of the U.S. alfalfa hay crop is lost to disease, amounting to losses exceeding $1 billion (1). An improved understanding of genetic and molecular mechanisms underlying host defense will offer novel tools to develop resistant alfalfa cultivars, thus providing an efficient and environmentally sound strategy to control alfalfa diseases. Cultivated alfalfa is autotetraploid (2n = 4x = 32) and out-crossing, making it recalcitrant to genetic analysis, while its diploid relative Medicago truncatula is a comparatively simple genetic and genomic system, and has emerged as a reference species for the study of legume biology (2). The two species share conserved genome structure and content (3), and thus it is anticipated that M. truncatula can serve as a surrogate for cloning the counterparts of many economically important genes in alfalfa. In the case of disease resistance, the family of NBS-LRR disease resistance (R) genes has been extensively characterized at the sequence and phylogenetic levels in M. truncatula (4, 5). In parallel, the long history of cultivation of alfalfa provides numerous examples of disease phenotypes that could be mitigated, if an R gene(s) with appropriate specificities were identified. In such cases, discovery of R genes with novel specificities in M. truncatula could have direct applicability to cultivated alfalfa.
Anthracnose of alfalfa, caused by the fungal pathogen Colletotrichum trifolii, is one of the most destructive diseases of alfalfa worldwide. The same pathogen also causes anthracnose on closely related forage legumes, including annual medic species (Medicago spp.) and clovers (Trifolium spp.). Three races of C. trifolii (i.e., races 1, 2 and 4) have been described based on differential responses of alfalfa cultivars (6–8), with strain specificity in alfalfa conferred by two independent dominant resistance genes, An1 and An2 (7, 9). An1 confers resistance to race 1 and likely, race 4, whereas An2 confers resistance to races 1 and 2. It is noteworthy that the race 3 of C. trifolii was reported in 1982 (10), but this fungus was subsequently reclassified as C. destructivum (11).
Defense responses of M. truncatula against C. trifolii are similar to those observed in alfalfa and other annual Medicago species, including hypersensitive reactions in incompatible interactions and delayed induction of resistance mechanisms in compatible interactions (12–16). Alfalfa responses to C. trifolii infection also involve the production of pterocarpan and isoflavonoid phytoalexins (12, 17). In a previous report, we described the genetic and physical localization of the RCT1 (for resistance to C. trifolii race 1) locus in M. truncatula (16). Here we report the map-based cloning of RCT1. RCT1 encodes a TIR-NBS-LRR type R protein that confers broad-spectrum anthracnose resistance when transferred into the susceptible alfalfa plants. Thus, RCT1 provides a new resource to develop anthracnose-resistant alfalfa cultivars and contributes to our understanding of disease resistance mechanisms against the fungal genus Colletotrichum. This study also highlights the potential of “translational” research from M. truncatula to the forage legume alfalfa.
Results
Map-Based Cloning of RCT1.
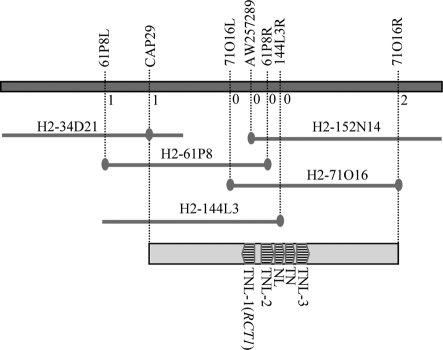
We previously mapped the RCT1 locus to M. truncatula chromosome 4, based on an F2 mapping population derived from the cross between the resistant genotype Jemalong A17 and the susceptible genotype F83005.5 (16). Fine mapping using 466 susceptible individuals (rct1/rct1) selected from the F2 population identified an EST (Expressed Sequence Tag)-based CAPS (Cleaved Amplified Polymorphic Sequence) marker, AW257289, that co-segregated with the RCT1 locus (Fig. 1). AW257289 anchors one end of the M. truncatula BAC (Bacterial Artificial Chromosome) clone H2–152N14, which is located on the physical map of M. truncatula within the ≈700 kb contig 1357 (http://www.medicago.org).
Fig. 1.
Map-based cloning of RCT1. The position of RCT1 was delimited to a genomic region between markers CAP29 and 71O16R. Numbers indicate the number of recombination breakpoints separating the marker from RCT1. Candidate genes of RCT1 are indicated. Arrows point to the transcriptional direction of each candidate gene. TNL = TIR-NBS-LRR; NL = NBS-LRR lacking a TIR domain; TN = TIR-NBS lacking a LRR domain. Map is drawn to scale.
To more precisely delimit the RCT1 locus within a physical interval, we used DNA sequence information from contig 1357 to develop new CAPS markers that flank AW257289 (Fig. 1). Through this process, we identified a total of three flanking recombination events: one between AW257289 and CAP29, and two between AW257289 and H2–71O16R. No recombination events were detected between AW257289 and markers H2–71O16L (CG959746), H2–61P8R (CG928897), and H2–144L3R (CR501753). We therefore determined that the RCT1 locus resides within an ≈200 kb window between 71O16R and CAP29. Sequencing and annotation of the BACs H2–144L3 (AC203223) and H2–152N14 (AC203224) identified five tandemly arrayed TIR-NBS-LRR (TNL) type R gene homologs (16). Three of the five NBS-LRR genes contain complete ORFs and share ≈80% identity at amino acid level, whereas the other two R gene homologs are truncated genes lacking either a TIR or an LRR domain. The three TNL genes, hereafter referred to as TNL-1, TNL-2, and TNL-3, respectively, were considered as candidate genes of RCT1.
RCT1 Locus Co-Segregates with Resistance to C. trifolii Races 2 and 4 in M. truncatula.
M. truncatula genotype Jemalong A17 was resistant to all three known races of C. trifolii, whereas F83005.5 was susceptible to the same three races. Parallel to mapping and cloning of RCT1, we also phenotyped two independent A17 X F83005.5 F2 mapping populations for resistance to C. trifolii races 2 and 4. Segregation data suggested that resistance to C. trifolii race 2 is possibly controlled by two independent dominant genes, as only 76 susceptible individuals were identified from a total of 1,166 F2 plants, which fits the 15:1 (resistant-to-susceptible) ratio (χ2 = 0.10, df = 1, P = 0.75). Genotyping of these 76 susceptible plants revealed complete linkage between the resistance phenotype to C. trifolii race 2 and marker AW257289; that is, all susceptible plants have the allele coming from the susceptible parent. Similar analyses were performed for resistance to C. trifolii race 4 in an F2 mapping population consisting of 262 F2 individuals. Of the 262 F2 individuals, the ratio of resistant-to-susceptible (206:56) statistically fits 3:1 (χ2 = 1.65, df = 1, P = 0.20), suggesting that resistance to C. trifolii race 4 is controlled by a single dominant gene. Strikingly, resistance to C. trifolii race 4 also co-segregated with the RCT1 locus based on mapping with the AW257289 marker. Taken together, our data suggest that resistance to the three C. trifolii races is controlled by either tightly linked genes or a single RCT1 gene in M. truncatula.
RCT1 Confers Broad-Spectrum Resistance to Anthracnose Disease When Transferred to Susceptible Alfalfa Clones.
To validate candidate genes from the RCT1 locus, genomic constructs (i.e., introns included) of TNL-1, TNL-2 and TNL-3 were cloned under control of their native promoters. The susceptible genotype F83005.5 was recalcitrant to transformation and regeneration, and thus we selected two independent clones from the alfalfa cultivar Regen SY (18) as a study system. The two selected clones, designated as Regen SY-6 and Regen SY-11, were susceptible to all three races of C. trifolii, a feature that enabled us to test whether RCT1 confers broad-spectrum resistance, as suggested by linkage mapping in M. truncatula.
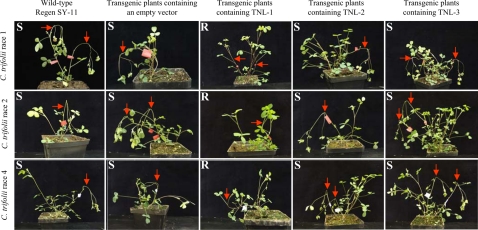
All transgenic alfalfa plants developed from the three candidate gene constructs were first inoculated with C. trifolii race 1. Independent transgenic plants containing TNL-2 (n = 55), TNL-3 (n = 15) or the empty vector pCAMBIA2300 (n = 26), as well as untransformed clones of the Regen SY-6 and Regen SY-11 (n = 10), were all susceptible to C. trifolii race 1 (Fig. 2). Three to four days post inoculation (dpi), the inoculated stems of these susceptible plants possessed a large lesion at the inoculation site and by 7 dpi these inoculated stems collapsed and died. In contrast, independent transformants containing the TNL-1 transgene (n = 42) were completely resistant to the pathogen. Thus, we conclude that TNL-1 is the RCT1 gene.
Fig. 2.
Complementation test of the RCT1 candidate genes. Transgenic plants containing individual candidate genes and the empty vector (pCAMBIA2300) as well as wild-type plants were inoculated with the races 1, 2, and 4 of C. trifolii. Only TNL-1 transgenic plants were resistance to C. trifolii. Arrows indicate inoculated stems. S = susceptible; R = resistance.
For purposes of evaluating resistance to C. trifolii races 2 and 4, vegetative clones were propagated from all transgenic lines and rated for disease phenotypes following pathogen inoculation. Strikingly, all transgenic plants containing the TNL-1 transgene were resistant to races 2 and 4, whereas all transgenic plants containing either TNL-2 or TNL-3 transgenes, as well as control vector only and non-transgenic plants, were susceptible. These data, along with the observation that resistance to C. trifolii races 1, 2 and 4 co-segregates with the RCT1 locus in M. truncatula, are consistent with a role for RCT1 in broad-spectrum resistance to C. trifolii.
RCT1 Is Constitutively Expressed and Alternatively Spliced.
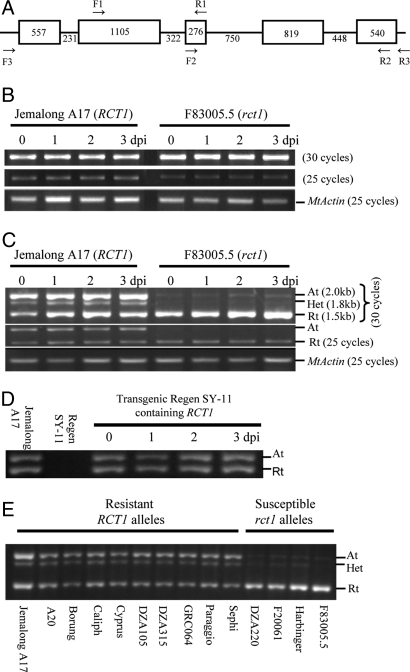
We deduced the structure of the RCT1 transcription unit based on a combination of ab initio predictions using FGENSH (19) and alignment of genomic and cDNA sequences. These analyses reveal a gene composed of five exons (Fig. 3A), with inferred intron positions typical of many TIR-NBS-LRR type R gene homologs described in Arabidopsis (20) and M. truncatula (5).
Fig. 3.
Expression analysis of RCT1 in M. truncatula and transgenic alfalfa by RT-PCR. (A) Gene structure of RCT1. The exons and introns are indicated by boxes and lines, respectively. Numbers indicate length of individual exons and introns. Arrows indicate the position of the primers used for RT-PCR analysis. (B) Constitutive expression of the resistant (RCT1) and susceptible (rct1) alleles in Jemalong A17 and F83005.5, respectively. Primers used were F1 and R1 that span the intron 2. (C) Alternative splicing of intron 4 of the RCT1 alleles in Jemalong A17 and F83005.5. The M. truncatula Actin gene was used as a control. Primers used were F2 and R2 that span the intron 3 and intron 4. At = alternative transcript that retained intron 4 (≈2.0 kb); Rt = regular transcript with intron 4 spliced out (≈1.5 kb); Het = heteroduplex (≈1.8 kb) resulting from RT-PCR of alternatively spliced mRNAs of RCT1. (D) Expression and alternative splicing of the transgene RCT1 in alfalfa. The primers used were F3 and R3 from the 5′-and 3′-UTR regions, respectively. This primer pair only amplified the transgene RCT1 but not homologs of alfalfa. (E) Expression analysis of additional resistant and susceptible alleles in M. truncatula. Primers used were F2 and R2, the same as in (C).
Semiquantitative reverse transcriptase (RT)-PCR using the RCT1-specific primers (F1 and R1 as indicated in Fig. 3A) was performed to analyze the expression profile of RCT1 in leaf tissue of susceptible and resistant genotypes, following inoculation with C. trifolii race 1. As shown in Fig. 3B, RCT1 was constitutively expressed in the resistant parent Jemalong A17, without apparent influence by fungal infection. The expression of RCT1 in healthy, non-infected tissue was further supported by analysis of the M. truncatula gene index (MtGI) database (http://compbio.dfci.harvard.edu), in which all ESTs with identity to RCT1 (i.e., TC96909, TC97262, and BF643292) originate from non-infected tissues.
Alignment of the RCT1 genomic sequence with EST assemblies present in the MtGI database revealed that two partial EST contigs (TC97262 and TC96909) can be assembled into a single predicted transcript. Interestingly, this deduced transcript contains the entire fourth intron of 448 bp. Retention of intron 4 is not a consequence of DNA contamination, because certain sequences within TC96909 and TC97262 derive from paired-end reads of single cDNA clones in which the second and third introns are spliced, but intron 4 is retained. RT-PCR using exonic primers spanning the third and fourth introns (primers F2 and R2 as indicated in Fig. 3A) confirmed the presence of two transcripts, which based on sequence analysis correspond to variant transcripts with or without intron 4 (Fig. 3C). The fully processed (intron 4 spliced out) and alternative (intron 4 retained) transcripts were present at comparable levels in the RNA profile of Jemalong A17, based on the semiquantitative RT-PCR analysis (Fig. 3C), and splicing of intron 4 was not obviously regulated by pathogen infection. A similar expression pattern was observed for the RCT1 transgene in transgenic alfalfa plants (Fig. 3D). A weak band of ≈1.8 kb was detected only at high cycle numbers (Fig. 3C), and sequence analysis indicated that it was an artifact of heteroduplex formation resulting from RT-PCR of alternatively spliced mRNAs of RCT1, consistent with observations of Eckhart et al. (21).
We obtained cDNA sequences from the 5′- and 3′-untranslated regions (UTR) of RCT1 by means of 5′ and 3′-rapid amplification of cDNA ends (RACE) experiments. Based on comparison to genomic sequence, the 5′ UTR is composed of a single 188-bp exon [supporting information (SI) Fig. S1]. By contrast, the 3′-UTR was represented by at least 3 transcript variants of 721, 734, and 801 bp (Fig. S2). Alignment of the 721-bp fragment with genomic sequence revealed 3 additional introns of 203, 95, and 80 bp, respectively. For example, the 801-bp fragment results from retention of the 80-bp intron. These results document multiple transcript variants present in the RCT1 transcript profile, with added complexity possible if alternative splicing events in the coding and non-coding regions occur independently.
Structure of Inferred RCT1 Proteins.
The fully processed RCT1 transcript is predicted to encode a protein of 1098 aa with a molecular weight of ≈125 kDa, consisting of an N-terminal TIR domain, a centrally located NBS domain with typical conserved motifs (22), and seven degenerate LRRs C-terminal to the NBS domain (Fig. 4). The extreme C terminus of RCT1 is highly conserved with members of TIR-NBS-LRR genes in M. truncatula but less conserved between species. The alternatively spliced transcript results in a shift in the reading frame and is predicted to encode a truncated protein of 936 aa with a molecular weight of ≈106 kDa. The first 920 aa of the truncated protein are identical to those of the full-length protein, including the entire TIR, NBS, and LRR domains, but lacking the C-terminal domain of the full-length RCT1 protein (Fig. 4).
Fig. 4.
Structure of the RCT1 protein(s). The conserved motifs within the TIR and NBS domains are underlined (20). The 7 predicted LRRs are highlighted in red color. The alternatively spliced transcript is predicted to encode a truncated protein lacking 178 aa in the N-terminal domain (green).
Expression- and Sequence-Level Polymorphisms between Resistant and Susceptible Alleles.
To explore the molecular nature of resistance and susceptible alleles, we characterized the expression and carried out sequence analysis of the rct1 allele from the susceptible genotype F83005.5. RT-PCR using RCT1-specific primers revealed that the fully spliced rct1 transcript was constitutively transcribed in the susceptible parent F83005.5 (Fig. 3B). By contrast, expression of the alternatively spliced transcript that retains intron 4 was undetectable at 25 cycles of RT-PCR. The correlation between an absence of alternative splicing and disease susceptibility was further examined by sequencing of RCT1 alleles from 12 additional genotypes of M. truncatula (9 resistant and 3 susceptible). As shown in Fig. 3E, the alternative transcript isoform was common to all resistant genotypes, but undetectable or very low in susceptible genotypes. Thus, alternative splicing of RCT1 is correlated with disease resistance to C. trifolii.
cDNA sequencing of the rct1 allele from F83005.5 reveals a total of 27 single nucleotide polymorphisms (SNPs) relative to the 3294-bp coding sequence of the RCT1 allele from Jemalong A17. Included among these polymorphic sites is a 2-bp deletion within the first exon (Fig. S3), which shifts the ORF and results in an immediate stop codon. Translation of rct1 from the next available in-frame start codon, would yield an NBS-LRR protein lacking the first 115 aa of the TIR domain (data not shown). Sequence polymorphisms were also detected in the 5′- and 3′-UTRs of rct1 (Fig. S1 and S4). The 5′-UTR of rct1 differs from that of RCT1 based on the presence of a single SNP and a 48-bp deletion of genomic sequence (Fig. S1). The 3′-UTR regions of rct1 and RCT1 are identical throughout the previously characterized intron-containing region, with alternative splicing of the 80-bp intron also observed in the rct1 allele (Fig. S5). However, the extreme 3′termini of RCT1 and rct1 cDNAs are not shared. In particular, a 119-bp fragment is present in rct1 but absent in the RCT1 genomic region in Jemalong A17. Analysis of the genomic sequence of F83005.5 around the 3′-UTR revealed an insertion of an ≈10-kb DNA fragment that contains the novel transcript terminus of rct1.
Discussion
The model legume M. truncatula is native to the Mediterranean basin and has long been cultivated as winter forage in Australia. The past decade has seen the development of abundant genetic and genomic tools for this model species, which has greatly facilitated our understanding of legume genomics and biology (23). The value of this model system has been enhanced by its close relationship with crop legumes, which is reflected in similar genome structures and conserved phenotypes such as legume-rhizobial symbiosis (23). Of crop legumes, alfalfa has become an immediate beneficiary from the study of the M. truncatula genomics, not only because alfalfa is a close relative of M. truncatula, but also because alfalfa itself is not amenable to genetic analysis. In addition to a focus on symbiotic plant-microbe interactions, significant efforts have taken advantage of M. truncatula as a model system to characterize legume-pathogen interactions (24). Importantly, most alfalfa pathogens also are pathogens of M. truncatula, leading to two key predictions: (1) that M. truncatula can serve as a tool to clone disease resistance genes for common pathogens of alfalfa, and (2) that functional disease resistance will be maintained when genes are moved across species boundaries by transgenic approaches.
Here we validate these predictions by isolating and characterizing the M. truncatula R gene RCT1. Genetic linkage analysis in M. truncatula and transgenic tests performed in alfalfa indicated that RCT1 confers broad-spectrum resistance to the three known races of C. trifolii. Broad-spectrum disease resistance conferred by NBS-LRR type R genes has been reported from other plant hosts. For example, the RB and RPI genes from wild potato species confer broad-spectrum resistance to nearly all known races of the late blight pathogen Phytophthora infestans in cultivated potato (25–27). In alfalfa, resistance to the three races of C. trifolii was reported to be controlled by two independent dominant genes, namely An1 and An2 (7, 9). An1 confers resistance to race 1 and likely race 4, whereas An2 confers resistance to races 1 and 2. Thus, only plants carrying genes An1 and An2 are resistant to all three races (7). By contrast, we demonstrate that M. truncatula RCT1 confers broad-spectrum anthracnose resistance in cultivated alfalfa. These results highlight a fundamental difference between these two species and demonstrate the potential of using M. truncatula genes for genetic improvement of alfalfa.
The successful inter-species transfer of R genes has been reported in other plant species (28–32). While there are examples that R genes are functional when transferred across family boundaries, R genes generally exhibit restricted taxonomic functionality (29), conferring a resistance response only in closely related species within a family. Given the close phylogenetic relationship between M. truncatula and alfalfa, functional R gene transfer in Medicago should not be a challenge.
Based on highly similar NBS-LRR sequences between M. truncatula and alfalfa (3, 4), one might predict that many disease resistance genes identified in M. truncatula will be conserved and located in syntenic regions of M. sativa. In the case of anthracnose, Medicago (Medicago spp.) and clovers (Trifolium spp.) share the same races of C. trifolii as pathogens, suggesting that anthracnose resistance may have originated before speciation within the Trifolieae tribe. Under such a scenario, with pressure from a common pathogen gene pool, RCT1 might represent a slow-evolving R gene (33). This prediction is evidenced by the isolation of a closest homolog of RCT1 (EU812207) from an alfalfa clone (the cultivar “ARC”) that is resistant to C. trifolii races 1 and 4 but susceptible to race 2. RCT1 is more similar to the alfalfa homolog (sharing ≈87% global identity at amino acid level) than to the two tandemly duplicated paralogs in M. truncatula (e.g., TNL-2 and TNL-3). It is interesting, therefore, that the genetic basis of resistance to C. trifolii differs between M. truncatula and M. sativa. In particular, resistance to races 1, 2 and 4 of C. trifolii is determined by two unlinked genes in tetraploid alfalfa, whereas only a single gene confers resistance to all three races in diploid M. truncatula. If broad spectrum resistance of RTC1 is ancestral to Medicago spp, then RTC1 function may have been partitioned between homeologous genes during the evolution of the tetraploid genome. Alternatively, RCT1 may have recently acquired novel specificities that are resistant to all three races. Further work is needed to address the evolutionary relationship between RCT1 in M. truncatula and the An1 and An2 genes in cultivated alfalfa, and the possible impact of polyploidy.
RCT1 is a member of the TIR-NBS-LRR family of R genes and falls within an extensive cluster of R gene loci on the top arm of M. truncatula chromosome 4 (5, 16). We demonstrated that RCT1 was constitutively expressed and alternatively spliced. Alternative splicing has been frequently detected for TIR-NBS-LRR type R genes, such as the tobacco N and the Arabidopsis RPS4 genes (34–36). The alternative transcripts of R genes generally possess premature termination codons and thus encode putative truncated proteins lacking the LRR and/or C-terminal domains (37). Interestingly, alternative transcripts of the tobacco N and the Arabidopsis RPS4 genes are both required for complete disease resistance (34, 35). Furthermore, the expression of alternatively spliced transcripts of the N and RPS4 genes was both up-regulated by pathogen infection (34, 38); such changes in expression profiles of R genes were correlated with resistance responses (34). In the case of RCT1, which shares similar gene structure with those of the N and RPS4 genes, alternative splicing was detected at both coding and 3′-UTR regions. Thus, there are likely multiple transcript variants present in the RCT1 expression profiles. In contrast to the tobacco N and the Arabidopsis RPS4 genes for which alternative splicing involves intron 2 and/or intron 3, alternative splicing of RCT1 in the coding region attributes to the retention of intron 4. The alternatively spliced transcript is predicted to encode a truncated protein consisting of the entire portion of the TIR, NBS, and LRR domains but lacks the C-terminal domain of the full-length RCT1 protein. It is unknown whether the alternative splicing events in the coding and non-coding regions are correlated. It is also unclear whether the alternatively spliced transcripts are required for the functionality of RCT1. Nevertheless, we detected expression-level polymorphisms for the alternatively spliced transcript involving intron 4 between the resistant and susceptible alleles. Moreover, the alternative splicing of RCT1 appears to be conserved between M. truncatula and alfalfa (data not shown). These observations suggest that alternative splicing of RCT1 may play a role in RCT1-mediated immunity in M. truncatula.
Sequence comparison between the coding regions of resistant (Jemalong A17) and susceptible (F83005.5) alleles identified 27 single nucleotide polymorphisms (SNPs), including a 2-bp deletion in the first exon. The 2-bp deletion changes the ORF and leads to an immediate stop codon. Thus, this deletion presumably abolishes the RCT1 function, resulting in the susceptible allele in F83005.5. However, this deletion appears to be unique for the F83005.5 allele and does not represent a conserved mechanism to generate susceptible alleles in M. truncatula, because sequencing additional susceptible alleles at this site did not detect such a deletion (data not shown). We also observed more sequence polymorphisms in the 5′- and 3′-UTRs; for example, the 5′-UTR of the rct1 allele in F83005.5 contains a 48-bp fragment deletion resulting from the deletion of genomic sequence, and the 3′-UTR regions differ significantly near the polyA site. Taken together, our data suggest that alternative splicing- and/or sequence-level polymorphisms may explain the molecular mechanisms underlying the evolution of resistant and susceptible alleles of RCT1.
Colletotrichum spp. are one of the most widespread and important disease-causing fungi of plants worldwide. The genus contains >35 species which cause anthracnose or blight on a wide range of temperate and tropical plants, including grain and pasture legumes, cereals, and fruits (39). During colonization of plant hosts, many species of Colletotrichum, including C. trifolii, use a hemibiotrophic infection strategy, in which the pathogen initially develops inside living host cells before switching to a destructive necrotrophic mode of infection (40). To date, cloning of resistance genes against the genus Colletotrichum has not been reported in any plant hosts. Thus, our work presented here will contribute significantly to our understanding of molecular mechanisms underlying host resistance against the hemibiotrophic fungal pathogens in the genus Colletotrichum.
Materials and Methods
Plant Materials and Disease Resistance Assay.
The F2 mapping populations were derived from the cross between M. truncatula genotypes Jemalong A17 (resistant) and F83005.5 (susceptible). Seedlings were grown in growth chambers programmed for 16h light at 23°C and 8h dark at 20°C. C. trifolii race 1 (isolate 2sp2), race 2 (isolate H4–2) and race 4 (isolate OH-WA-520) were used for inoculation as described by Yang et al. (16).
DNA Sequencing and Sequence Analysis.
Sequencing of BACs H2–144L3 (AC203223) and H2–152N14 (AC203224) were carried out at the Advanced Center for Genome Technology, Department of Chemistry and Biochemistry, University of Oklahoma. Gene prediction was performed using the FGENESH program (19). Domains were predicted using Pfam 21.0 (41). Sequence alignments were performed using the ClustalX (42).
Complementation Test.
Genomic DNA constructs that contained the individual candidate genes under the control of their native promoters were used for the transformation experiments. The genomic DNA of BAC H2–144L03 was digested with SacI and KpnI to obtain a 12.9-kb genomic fragment that contained the ≈5.0-kb TNL-1 coding region plus ≈3.6 kb upstream of the start codon and ≈4.7 kb downstream of the stop codon. The same BAC also was digested with StuI and BglII to obtain a 10.3-kb genomic fragment that covered the TNL-2 coding region (≈5.0 kb) plus ≈3.0 kb and ≈2.2 kb up- and down-stream sequence, respectively. The TNL-3 genomic fragment was obtained by digestion of the DNA of BAC H2–152N14 with SpeI and SgrAI. This digestion produced ≈10.0-kb fragment that contained TNL-3 coding region plus ≈3-kb promoter region and ≈300-bp 3′-UTR. The genomic fragments were cloned into the transformation vector pCAMBIA2300 and transformed into Agrobacterium tumefaciens strain LBA4404. Transformation of alfalfa followed the protocol developed by Samac and Austin-Phillips (43).
Analysis of Gene Expression by RT-PCR.
For gene expression analysis, plants were inoculated with C. trifolii race 1 by spraying spore suspension (2 × 106/ml) to the seedlings and maintained in a growth chamber conditioned as described before. Leaves at 0, 1, 2 and 3 dpi were collected for RNA isolation. Total RNA was isolated by the Qiagen Plant RNeasy. Two micrograms of RNA was used to perform RT reactions using M-MLV reverse transcriptase (Invitrogen) in a 20-μl reaction mixture. Two microliters of the RT reaction was used as a template in a 20-μl PCR solution. The PCR primers were as follows: MtActin, 5′-GGAGAAGCTTGCATATGTTG-3′ and 5′-TTAGAAGCACTTCCTGTGGA-3′; RCT1, F1: 5′-AAATGGTTTGCTCCAGGTAG-3′, F2: 5′-CAAAAGCTGTTGAGGGACTG-3′, F3: 5′-CCATAGATCTCTTCCTTTCTTTTCC-3′, R1: 5′-TTTCCACACAAGTTTAGCATTG-3′, R2: 5′-ATTTCGACGACTGGTTCATC-3′, and R3: GCCACCAATGTAAGCATAAAATCTGCAA.
Rapid Amplification of cDNA Ends (RACE).
One microgram of RNA was used in a 5′ and 3′ RACE amplification, using the SMART RACE cDNA amplification kit (Clontech). 5′ RACE primers used were: 5′-CAACAAATCCAGCAAGGCCAGCCGCAAC-3′ (first-round), 5′-AGCCGCAACACGAAGCTCATTTCTCCAC-3′ (second-round), and 5′-TGCGCGAGTGTCTTCTCCTCGGAAACTC-3′ (third-round). 3′ RACE primers used were: 5′-TGCCCAAGGCTGTCTCAGGTTTCCCATA-3′ (first-round), 5′-AAGCCTTTGGCTGACATGTGGATCAGAA-3′ (second-round), and 5′-TGCTGCCATGAGGATCTCTCGCCAAG-3′ (third-round). The resulting PCR products were cloned into pGEM-T Easy Vector System (Promega) and independently amplified clones were sequenced.
Supplementary Material
Acknowledgments.
We thank Douglas Cook and Steve Cannon for critical reading of the manuscript; Martin Dickman (Texas A & M University, College Station), Nichole O'Neill (U.S. Department of Agriculture, Beltsville, MD), and Landon Rhodes (Ohio State University, Columbus) for providing C. trifolii races; and Dr. J. M. Prosperi (Diversité et Adaptations des Plantes Cultivées, Paris) for supplying seed of M. truncatula. This work was supported by United States Department of Agriculture-National Research Initiative Competitive Grants Program Grants 2005-35301-15697 and 2005-35300-15461 (to H.Z.) and National Science Foundation Grant 0604966 (to B.A.R.). This article is published with the approval of the Director of the Kentucky Agricultural Experiment Station.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. EU812206 and EU812207).
This article contains supporting information online at www.pnas.org/cgi/content/full/0802518105/DCSupplemental.
References
- 1.Nutter FW, et al. Quantifying alfalfa yield losses caused by foliar diseases in Iowa, Ohio, Wisconsin, and Vermont. Plant Dis. 2002;86:269–277. doi: 10.1094/PDIS.2002.86.3.269. [DOI] [PubMed] [Google Scholar]
- 2.Cook DR. Medicago truncatula—a model in the making! Curr Opin Plant Biol. 1999;2:301–304. doi: 10.1016/s1369-5266(99)80053-3. [DOI] [PubMed] [Google Scholar]
- 3.Choi HK, et al. A sequence-based genetic map of Medicago truncatula and comparison of marker colinearity with M sativa. Genetics. 2004;166:1463–1502. doi: 10.1534/genetics.166.3.1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu H, Cannon SB, Young ND, Cook DR. Phylogeny and genomic organization of the TIR and non-TIR NBS-LRR resistance gene family in Medicago truncatula. Mol Plant Microbe Interact. 2002;15:529–539. doi: 10.1094/MPMI.2002.15.6.529. [DOI] [PubMed] [Google Scholar]
- 5.Ameline-Torregrosa C, et al. Identification and characterization of nucleotide-binding site-leucine-rich repeat genes in the model plant Medicago truncatula. Plant Physiol. 2008;146:5–21. doi: 10.1104/pp.107.104588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ostazeski SA, Elgin JH, McMurtrey JE. Occurrence of anthracnose on formerly anthracnose-resistant “Arc” alfalfa. Plant Dis Rep. 1979;63:734–736. [Google Scholar]
- 7.Mackie JM, Musial JM, O'Neill, Irwin JAG. Pathogenic specialisation within. Colletotrichum trifolii in Australia, and lucerne cultivar reactions to all known Australian pathotypes. Aust J Agric Res. 2003;54:829–836. [Google Scholar]
- 8.Ariss JJ, Rhodes LH. A new Colletotrichum trifolii race identified in Ohio. Phytopathology. 2006;96:S6. doi: 10.1094/PDIS-91-10-1362B. [DOI] [PubMed] [Google Scholar]
- 9.Elgin JH, Jr., Ostazeski SA. Inheritance of resistance to race 1 and race 2 anthracnose in Arc-1 and Saranac AR alfalfa. Crop Sci. 1985;25:861–865. [Google Scholar]
- 10.Allen SJ, Barnes GL, Caddel JL. A new race of Colletotrichum trifolii on alfalfa in Oklahoma. Plant Dis. 1982;66:922–924. [Google Scholar]
- 11.O'Neill NR. Pathogenic variability and host resistance in the Colletotrichum trifolii/Medicago sativa pathosystem. Plant Dis. 1996;80:450–457. [Google Scholar]
- 12.O'Neill NR. Defense expression in protected tissues of Medicago sativa is enhanced during compatible interactions with Colletotrichum trifolii. Phytopathology. 1996;86:1045–1050. [Google Scholar]
- 13.Mould MJR, Robb J. The. Colletotrichum trifolii-Medicago sativa interface, in culture: A cytological analysis. Can J Bot. 1992;70:114–124. [Google Scholar]
- 14.O'Neill NR, Bauchan GR. Sources of resistance to anthracnose in the annual Medicago core collection. Plant Dis. 2000;84:261–267. doi: 10.1094/PDIS.2000.84.3.261. [DOI] [PubMed] [Google Scholar]
- 15.Torregrosa C, et al. Cytological, genetic, and molecular analysis to characterize compatible and incompatible interactions between. Medicago truncatula and Colletotrichum trifolii. Mol Plant Microbe Interact. 2004;17:909–920. doi: 10.1094/MPMI.2004.17.8.909. [DOI] [PubMed] [Google Scholar]
- 16.Yang S, et al. Genetic and physical localization of an anthracnose resistance gene in. Medicago truncatula Theor Appl Genet. 2007;116:45–52. doi: 10.1007/s00122-007-0645-7. [DOI] [PubMed] [Google Scholar]
- 17.Salles II, Blount JW, Dixon RA, Schubert K. Phytoalexin induction and β-1,3-glucanase activities in Colletotrichum trifolii infected leaves of alfalfa (Medicago sativa L) Physiol Mol Plant Pathol. 2002;61:89–101. [Google Scholar]
- 18.Bingham ET. Registration of alfalfa hybrid Regen SY germplasm for tissue culture and transformation research. Crop Sci. 1991;31:1098. [Google Scholar]
- 19.Solovyev V, Salamov A. The Gene-Finder computer tools for analysis of human and model organisms genome sequences. Proc Int Conf Intell Syst Mol Biol. 1997;5:294–302. [PubMed] [Google Scholar]
- 20.Meyers BC, Kozik A, Griego A, Kuang H, Michelmore RW. Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell. 2003;15:809–834. doi: 10.1105/tpc.009308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eckhart L, Ban J, Ballaun C, Weninger W, Tschachler E. Reverse transcription-polymerase chain reaction products of alternatively spliced mRNAs form DNA heteroduplexes and heteroduplex complexes. J Biol Chem. 1999;274:2613–2615. doi: 10.1074/jbc.274.5.2613. [DOI] [PubMed] [Google Scholar]
- 22.Meyers BC, et al. Plant disease resistance genes encode members of an ancient and diverse protein family within the nucleotide-binding superfamily. Plant J. 1999;20:317–332. doi: 10.1046/j.1365-313x.1999.t01-1-00606.x. [DOI] [PubMed] [Google Scholar]
- 23.Zhu H, Choi HK, Cook DR, Shoemaker RC. Bridging model and crop legumes through comparative genomics. Plant Physiol. 2005;137:1189–1196. doi: 10.1104/pp.104.058891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tivoli B, Baranger A, Sivasithamparam K, Barbetti MJ. Annual Medicago: From a model crop challenged by a spectrum of necrotrophic pathogens to a model plant to explore the nature of disease resistance. Ann Bot (Lond) 2006;98:1117–1128. doi: 10.1093/aob/mcl132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song J, et al. Gene. RB cloned from Solanum bulbocastanum confers broad spectrum resistance to potato late blight. Proc Natl Acad Sci USA. 2003;100:9128–9133. doi: 10.1073/pnas.1533501100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van der Vossen E, et al. An ancient R gene from the wild potato species Solanum bulbocastanum confers broad-spectrum resistance to Phytophthora infestans in cultivated potato and tomato. Plant J. 2003;36:867–882. doi: 10.1046/j.1365-313x.2003.01934.x. [DOI] [PubMed] [Google Scholar]
- 27.van der Vossen E, et al. The. Rpi-blb2 gene from Solanum bulbocastanum is an Mi-1 gene homolog conferring broad-spectrum late blight resistance in potato. Plant J. 2005;44:208–222. doi: 10.1111/j.1365-313X.2005.02527.x. [DOI] [PubMed] [Google Scholar]
- 28.Thilmony RL, Chen ZT, Bressan RA, Martin GB. Expression of the tomato Pto gene in tobacco enhances resistance to Pseudomonas syringae pv tabaci expressing AvrPto. Plant Cell. 1995;7:1529–1536. doi: 10.1105/tpc.7.10.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tai TH, et al. Expression of the. Bs2 pepper gene confers resistance to bacterial spot disease in tomato. Proc Natl Acad Sci USA. 1999;96:14153–14158. doi: 10.1073/pnas.96.24.14153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiao SY, Charoenwattana P, Holcombe L, Turner JG. The Arabidopsis genes RPW81 and RPW82 confer induced resistance to powdery mildew diseases in tobacco. Mol Plant Microbe Interact. 2003;16:289–294. doi: 10.1094/MPMI.2003.16.4.289. [DOI] [PubMed] [Google Scholar]
- 31.Zhao B, et al. A maize resistance gene functions against bacterial streak disease in rice. Proc Natl Acad Sci USA. 2005;102:15383–15388. doi: 10.1073/pnas.0503023102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seo YS, et al. A viral resistance gene from common bean functions across plant families and is up-regulated in a non-virus-specific manner. Proc Natl Acad Sci USA. 2006;103:11856–11861. doi: 10.1073/pnas.0604815103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuang H, Woo SS, Meyers BC, Nevo E, Michelmore RW. Multiple genetic processes result in heterogeneous rates of evolution within the major cluster disease resistance genes in lettuce. Plant Cell. 2004;16:2870–2894. doi: 10.1105/tpc.104.025502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dinesh-Kumar SP, Baker BJ. Alternatively spliced N resistance gene transcripts: Their possible role in tobacco mosaic virus resistance. Proc Natl Acad Sci USA. 2000;97:1908–1913. doi: 10.1073/pnas.020367497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang XC, Gassmann W. RPS4-mediated disease resistance requires the combined presence of RPS4 transcripts with full-length and truncated open reading frames. Plant Cell. 2003;15:2333–2342. doi: 10.1105/tpc.013474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tan X, et al. Global expression analysis of nucleotide binding site-leucine rich repeat-encoding and related genes in Arabidopsis. BMC Plant Biol. 2007;7:56. doi: 10.1186/1471-2229-7-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jordan T, Schornack S, Lahaye T. Alternative splicing of transcripts encoding Toll-like plant resistance proteins—what's the functional relevance to innate immunity? Trends Plants Sci. 2002;7:392–398. doi: 10.1016/s1360-1385(02)02311-7. [DOI] [PubMed] [Google Scholar]
- 38.Zhang XC, Gassmann W. Alternative splicing and mRNA levels of the disease resistance gene RPS4 are induced during defense responses. Plant Physiol. 2007;145:1577–1587. doi: 10.1104/pp.107.108720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bailey JA, Jeger MJ. Colletotrichum: Biology, Pathology and Control. Wallingford, UK: CAB International; 1992. [Google Scholar]
- 40.O'Connell RJ, et al. Hemibiotrophic infection of Pisum sativum by Colletotrichum truncatum. Plant Pathol. 1993;42:774–783. [Google Scholar]
- 41.Bateman A, et al. The Pfam protein families database. Nucleic Acids Res. 2004;32:D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Samac DA, Austin-Phillips S. Alfalfa (Medicago sativa L) Methods Mol Biol. 2006;343:301–311. doi: 10.1385/1-59745-130-4:301. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.