Abstract
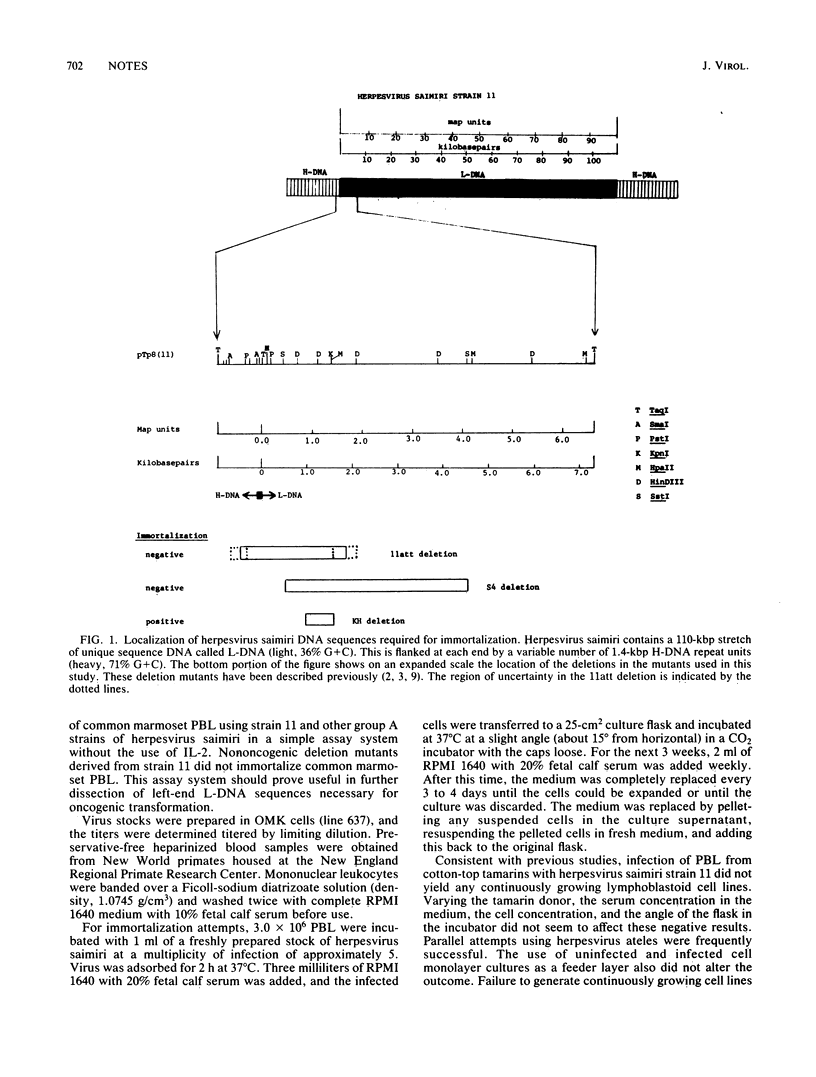
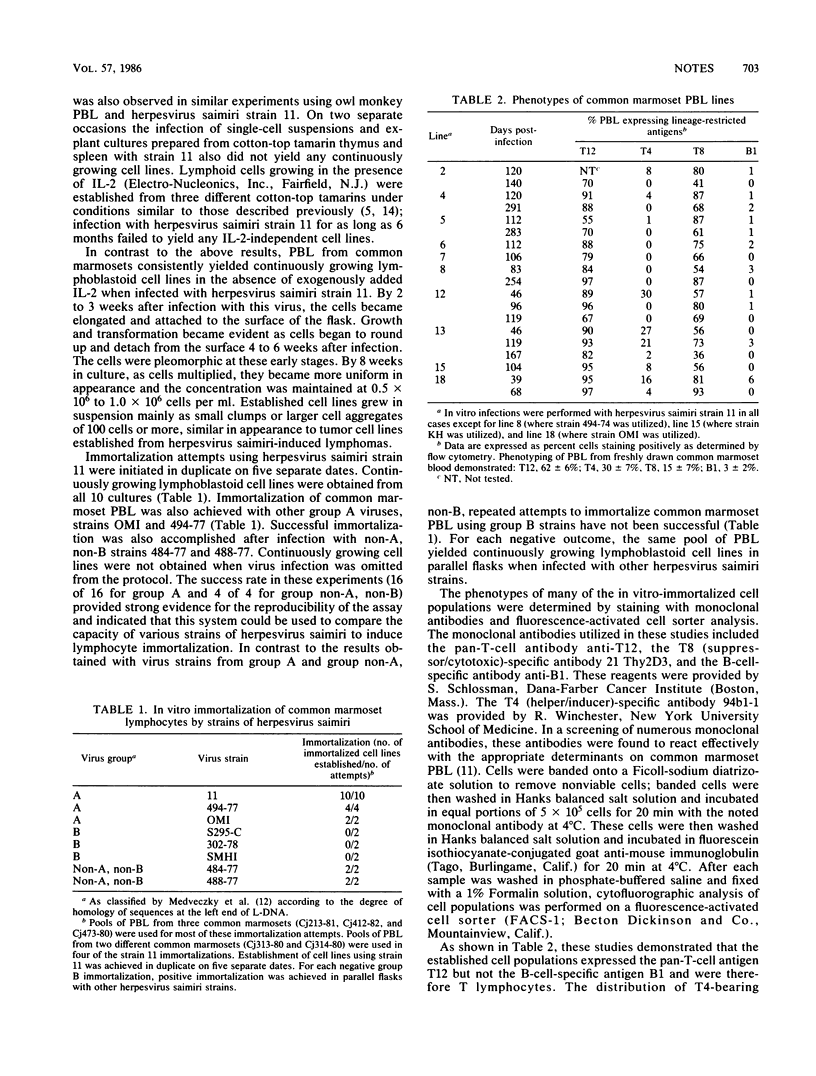
Herpesvirus saimiri L-DNA sequences between 0.0 and 4.0 map units (4.5 kilobase pairs) are required for oncogenicity; these sequences are not required for replication of the virus. To investigate the basis for the lack of oncogenicity of mutants with deletions in this region and to study the function of this region, we developed a reliable system for in vitro immortalization by herpesvirus saimiri. In contrast to peripheral blood lymphocytes from cotton-top tamarins (Saguinus oedipus) and owl monkeys (Aotus sp.), infection of peripheral blood lymphocytes from common marmosets (Callithrix jacchus) in vitro with herpesvirus saimiri consistently yielded continuously growing lymphoblastoid cell lines. Such cell lines were established using strains of herpesvirus saimiri from group A and group non-A, non-B; however, repeated attempts to immortalize common marmoset peripheral blood lymphocytes using strains from group B were not successful. Common marmoset cell lines immortalized by herpesvirus saimiri were T12+, T8+, T4-, and B1-, indicating that they were derived from suppressor/cytotoxic T lymphocytes. Cell lines could not be established using the nononcogenic mutants 11att and S4, both of which were derived from the group A strain 11 virus. Strain 11att has a spontaneous deletion and S4 has a constructed deletion in the 0.0 to 4.0 map unit region. Constructed strains which had these deleted sequences restored did immortalize common marmoset peripheral blood lymphocytes. Thus, the nononcogenic deletion mutants are defective for immortalization. This system should facilitate attempts to define the sequences responsible for immortalization and to determine their function.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ablashi D. V., Pearson G., Rabin H., Armstrong G., Easton J., Valerio M., Cicmanec J. Experimental infection of Callithrix Jacchus marmosets with Herpesvirus ateles, Herpesvirus saimiri, and Epstein Barr virus. Biomedicine. 1978 Feb;29(1):7–10. [PubMed] [Google Scholar]
- Desrosiers R. C., Bakker A., Kamine J., Falk L. A., Hunt R. D., King N. W. A region of the Herpesvirus saimiri genome required for oncogenicity. Science. 1985 Apr 12;228(4696):184–187. doi: 10.1126/science.2983431. [DOI] [PubMed] [Google Scholar]
- Desrosiers R. C., Burghoff R. L., Bakker A., Kamine J. Construction of replication-competent Herpesvirus saimiri deletion mutants. J Virol. 1984 Feb;49(2):343–348. doi: 10.1128/jvi.49.2.343-348.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk L., Johnson D., Deinhardt F. Transformation of marmoset lymphocytes in vitro with Herpesvirus ateles. Int J Cancer. 1978 May 15;21(5):652–657. doi: 10.1002/ijc.2910210517. [DOI] [PubMed] [Google Scholar]
- Fleckenstein B., Bornkamm G. W., Mulder C., Werner F. J., Daniel M. D., Falk L. A., Delius H. Herpesvirus ateles DNA and its homology with Herpesvirus saimiri nucleic acid. J Virol. 1978 Jan;25(1):361–373. doi: 10.1128/jvi.25.1.361-373.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamine J., Bakker A., Desrosiers R. C. Mapping of RNA transcribed from a region of the Herpesvirus saimiri genome required for oncogenicity. J Virol. 1984 Nov;52(2):532–540. doi: 10.1128/jvi.52.2.532-540.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koomey J. M., Mulder C., Burghoff R. L., Fleckenstein B., Desrosiers R. C. Deletion of DNA sequence in a nononcogenic variant of Herpesvirus saimiri. J Virol. 1984 May;50(2):662–665. doi: 10.1128/jvi.50.2.662-665.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laufs R., Fleckenstein B. Susceptibility to Herpesvirus saimiri and antibody development in old and new world monkeys. Med Microbiol Immunol. 1973 Mar 8;158(3):227–236. doi: 10.1007/BF02120558. [DOI] [PubMed] [Google Scholar]
- Letvin N. L., King N. W., Reinherz E. L., Hunt R. D., Lane H., Schlossman S. F. T lymphocyte surface antigens in primates. Eur J Immunol. 1983 Apr;13(4):345–347. doi: 10.1002/eji.1830130414. [DOI] [PubMed] [Google Scholar]
- Medveczky P., Szomolanyi E., Desrosiers R. C., Mulder C. Classification of herpesvirus saimiri into three groups based on extreme variation in a DNA region required for oncogenicity. J Virol. 1984 Dec;52(3):938–944. doi: 10.1128/jvi.52.3.938-944.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neubauer R. H., Briggs C. J., Noer K. B., Rabin H. Identification of normal and transformed lymphocyte subsets of nonhuman primates with monoclonal antibodies to human lymphocytes. J Immunol. 1983 Mar;130(3):1323–1329. [PubMed] [Google Scholar]
- Rabin H., Hopkins R. F., 3rd, Desrosiers R. C., Ortaldo J. R., Djeu J. Y., Neubauer R. H. Transformation of owl monkey T cells in vitro with Herpesvirus saimiri. Proc Natl Acad Sci U S A. 1984 Jul;81(14):4563–4567. doi: 10.1073/pnas.81.14.4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirm S., Müller I., Desrosiers R. C., Fleckenstein B. Herpesvirus saimiri DNA in a lymphoid cell line established by in vitro transformation. J Virol. 1984 Mar;49(3):938–946. doi: 10.1128/jvi.49.3.938-946.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright J., Falk L. A., Collins D., Deinhardt F. Mononuclear cell fraction carrying Herpesvirus saimiri in persistently infected squirrel monkeys. J Natl Cancer Inst. 1976 Oct;57(4):959–962. doi: 10.1093/jnci/57.4.959. [DOI] [PubMed] [Google Scholar]
- Wright J., Falk L. A., Wolfe L. G., Ogden J., Deinhardt F. Susceptibility of common marmosets (Callithrix jacchus) to oncogenic and attenuated strains of Herpesvirus saimiri. J Natl Cancer Inst. 1977 Nov;59(5):1475–1478. [PubMed] [Google Scholar]


