Abstract
Rtt109 is a protein acetyltransferase (PAT) that is responsible for the acetylation of lysine-56 of histone 3 (H3K56) in yeast. H3K56 acetylation has been implicated in the weakening of the interaction between the histone core and the surrounding DNA in the nucleosomal particle. Rtt109, in cooperation with various histone chaperones, promotes genomic stability and is required for resistance to DNA damaging agents. Here, we present the crystal structure of Rtt109 in complex with acetyl-CoA at a 2.0-Å resolution. Rtt109 consists of a core PAT domain, which binds the acetyl-CoA cofactor. A second domain, the activation domain, is tightly associated with the PAT domain. Autoacetylation of lysine-290 within the activation domain is required for stabilizing the interaction between the two domains and is essential for catalysis. Biochemical analysis demonstrates the requirement of a loop within the PAT domain for the binding of the histone chaperone Vps75, and mutational analysis identifies key residues for catalysis. We propose a model in which the autoacetylation of Rtt109 is crucial for the regulation of its catalytic activity.
Keywords: DNA repair, histone, structure
Eukaryotic genomic DNA is organized into a compact structure called chromatin. Chromatin formation is mainly established by a set of proteins termed histones. Histones are small globular proteins that are positively charged with flexible, N-terminal tails ≈40 residues in length. An octamer of histones, namely a H3/H4 tetramer that assembles with two H2A/H2B dimers, interacts with 147 bp of helical DNA to form the fundamental unit of chromatin, the nucleosomal particle (1). Histones undergo a variety of posttranslational modifications that are either attached or removed by enzyme families of counteracting activity (2, 3). The majority of the modifications are concentrated on the flexible N-terminal tails, although residues within the histone fold have also been found to be modified (4).
Recently, the functionally important acetylation of lysine-56 of histone H3 (Ac-H3K56), located within the folded histone core, has been described (5–9). Structural studies have revealed that H3K56 contributes to the formation of the stable quaternary structure of the nucleosome. The acetylation of H3K56 neutralizes the positive charge of lysine-56, disrupting the electrostatic interaction between the helical DNA of the nucleosomal complex, potentially resulting in a more flexible wrapping of DNA around the proteinaceous histone core (1). Acetylation of H3K56 is predominately seen on newly synthesized histones during the S phase of the meiotic or mitotic cell cycle (6, 9, 10). Yeast mutants that cannot acetylate H3K56 because of the expression of an unmodifiable histone H3 variant in which arginine replaces lysine-56 (H3-K56R) are sensitive to genotoxic compounds during the S phase of the cell cycle and display an accumulation of stalled replication forks (6, 9–11). Moreover, the acetylation of H3K56 has been associated with the repair of DNA lesions that occur during replication in the S phase (6).
A protein acetyltransferase, Rtt109, has been found to be responsible for H3K56 acetylation (11–13). Rtt109 was originally discovered in a genetic screen that selected for proteins, which increased the transposition rate of the Ty1 transposon (14). Subsequently, Rtt109 has been found to be required for tolerance of DNA damage during the S phase of the cell cycle. Moreover, Rtt109 deletion strains are hypersensitive to agents that generate replication stress, display an increase in gross chromosomal rearrangements, and exhibit a moderate hyperrecombination phenotype (11–13, 15).
Biochemical analysis of Rtt109 has revealed autoacetyltransferase activity as well as acetyltransferase activity toward H3K56. In vitro, the activity of Rtt109 is greatly enhanced in the presence of the histone chaperones Asf1 or Vps75. Rtt109 and Asf1 deletion strains, as well as strains expressing the unmodifiable H3K56R variant, are phenotypically indistinguishable, suggesting their involvement in the same cellular processes (11–13, 15–18).
To improve our understanding of the mechanism of protein acetylation by Rtt109, we identified the autoacetylation site and determined the crystal structure of Saccharomyces cerevisiae Rtt109 in its autoacetylated state. We present a mutational analysis that highlights not only the importance of several residues for in vivo activity but also the interaction with the histone chaperone Vps75, and we propose a mechanism in which Rtt109 is activated by autoacetylation.
Results
Rtt109 Is Autoacetylated on Lysine-290.
To investigate and clarify the domain organization of Rtt109, we performed primary structure and sequence conservation analysis, as well as secondary structure predictions for the 436-residue S. cerevisiae Rtt109 (referred to as Rtt109 in the following text). This analysis suggested that the entire coding region of Rtt109 is structured with the exception of the C-terminal ≈30 residues. Tryptic digestion of purified recombinant full-length Rtt109 expressed in Escherichia coli, followed by mass spectrometric analysis demonstrated that lysine-290 undergoes acetylation (data not shown).
Structure Determination.
Limited proteolysis experiments carried out with full-length Rtt109 revealed an internal proteolytically sensitive region flanked by two stable regions. The two stable regions comprised residues 1–127 and 171–436. Based on these results, we designed a series of expression constructs, in which the proteolytically sensitive region was sequentially shortened and the deleted region replaced by a flexible linker. The resulting proteins were then subjected to crystallization trials. Crystals of the ≈45-kDa Rtt109 variant Rtt109ΔL, containing residues 1–127 and 171–436 that were linked by a four-residue linker, appeared in the monoclinic space group C2 with one molecule in the asymmetric unit (Fig. 1A). The structure was solved by single-wavelength anomalous dispersion (SAD), using x-ray diffraction data from seleno-l-methionine-labeled crystals. The final model contains residues 1–403 and was refined to a 2.0-Å resolution with an Rwork and an Rfree of 15.6% and 18.6%, respectively. No electron density was observed for residues 404–436; these residues are presumed to be disordered and, therefore, have been omitted from the final model. For details of the data collection and refinement statistics, see supporting information (SI) Table S1.
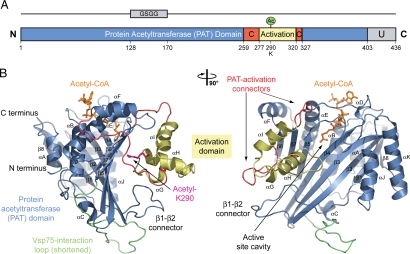
Fig. 1.
Structure of S. cerevisiae Rtt109ΔL. (A) Domain structure. Blue, protein acetyltransferase (PAT) domain; yellow, activation domain; red, PAT-activation domain connectors; gray, unstructured C-terminal region (U). The bar above the domain structure marks the crystallized fragment. (B Left) Structure of Rtt109ΔL in ribbon representation, colored as in A. (Right) A 90°-rotated view is shown.
Architectural Overview.
The Rtt109ΔL polypeptide chain folds into two distinct structural domains, a protein acetyltransferase domain and a smaller internal domain that we term the activation domain (Fig. 1B and Movie S1). The PAT domain provides a large tunnel for the acetyl-CoA cofactor, exposing the acetyl moiety at a small circular aperture in the front of the domain, whereas the adenosine ring at the other end of the cofactor molecule is exposed on the back of the PAT domain. The PAT domain adopts a structure that is related to the catalytic domain of p300/CBP, loosely resembling the catalytic domains of Gcn5 and Esa1 (19–21). The ≈45-residue activation domain adopts an α-helical fold composed of a three-helix bundle and is connected via two connector segments to the PAT domain. The PAT and activation domains closely pack against each other, positioning the acetylated residue, lysine-290 (Ac-K290), in the center of their interface. Although sequence analysis fails to predict a relationship between Rtt109 and other PATs, our structure clearly reveals an overall architectural similarity between Rtt109 and p300/CBP (21).
Protein Acetyltransferase Domain of Rtt109.
The core of the PAT domain comprises a central, almost flat, extensive β-sheet constructed of eight β-strands, forming a rectangular plate that is surrounded by eight α-helices (αA–αF and αJ–αK) and five extensive surface loops that are inserted between β1 and β2, αC and β5, β5 and αE, αF and αG, and αI and β7 (Fig. 1B). The PAT domain contains an insertion between αF and β7 composed of the three α-helices αG, αH, and αI. This insertion forms a small domain, the activation domain. The overall dimensions of the PAT domain are ≈60 Å × 40 Å × 40 Å.
A tunnel within the PAT domain tightly encapsulates the acetyl-CoA cofactor (Fig. 1B). This cofactor-binding tunnel is created on its base by a cavity within the PAT domain that is formed by the N-terminal portions of helices αB and αE as well as the C-terminal portions of the β-strands β4 and β5. In addition, the β5–αE connector forms a lid that covers the central portion of the acetyl-CoA. This binding site results in the tight association and precise positioning of the cofactor, in which both ends of the cofactor are exposed from the PAT domain. The acetyl head group of the cofactor is located at the base of an ≈7-Å-deep, water-filled, hydrophobic tunnel that ends in a small circular opening on the surface of the PAT domain. The circular opening and the hydrophobic tunnel provide only enough space for the side-chain atoms of the substrate lysine to be inserted, allowing for the ε-amino group to be positioned close to the acetyl moiety of the cofactor molecule. This finding is in line with the structural analysis of the p300/CBP PAT domain that has been crystallized in complex with the bisubstrate inhibitor lysine-CoA. In that structure, the lysine moiety of the inhibitor is accommodated by the hydrophobic tunnel, presumably mimicking the lysine side chain of native protein substrates (21).
Rtt109 Activation Domain.
The polypeptide chain of the Rtt109 PAT domain is interrupted between αF and β7 by the activation domain (Fig. 1B, Movie S1, and Movie S2). The activation domain comprises ≈45 residues that form three α-helices, αG, αH, and αI, which together fold into a small α-helical bundle. Furthermore, the activation domain is tethered via the 19-residue αF-αG and eight-residue αI-β7 connectors to the core of the PAT domain.
The PAT and activation domains tightly interact via two distinct surfaces. The first interface is formed between the 20-residue β1–β2 loop of the PAT domain that makes contact with αG and αH of the activation domain (Fig. 1B). The second interface is substantially larger and is formed between three β-strands of the core β-sheet of the PAT domain (β4, β5, and β7) and all three helices of the activation domain (αG, αH, and αI), burying ≈2,500 Å2 of surface area (Fig. 1B). The acetylated lysine-290 is located at the N-terminal end of helix αH of the activation domain and extends toward the front face of the PAT domain, stabilized by hydrogen-bond interactions with D288. Strikingly, this interface between the two domains is primarily generated by 11, highly conserved hydrophobic residues that form a hydrophobic cage around Ac-K290 (Fig. 2 and Movie S3). Hence, the neutralization of the positive charge of the terminal amino group of lysine-290 by its acetylation appears to be essential for the formation and the stability of the interface.
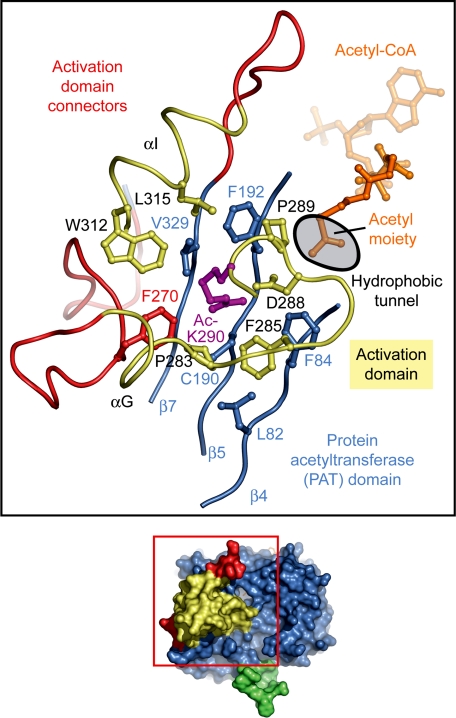
Fig. 2.
Interface between PAT and activation domains. Ribbon representation of the interface between the PAT and the activation domains colored according to Fig. 1A. The activation domain is tightly bound to the PAT domain. The acetylated lysine-290 of the activation domain is located in the center of the interface between the two domains and is inserted into a hydrophobic cavity of the PAT domain. The active-site cavity is located ≈10 Å away from the Ac-K290-binding site.
Active Site of Rtt109.
The front face of the Rtt109 surface contains a circular aperture that leads into a hydrophobic tunnel, exposing the acetyl moiety of the acetyl-CoA cofactor at its deepest confines (Fig. 3). This aperture is ≈6 Å wide and is located between the β-strands β4 and β5 and the surface loops β5–αE and αG–αH. The activation domain is positioned closely to the hydrophobic tunnel and is essential for its formation and stability. Furthermore, the bound activation domain together with helices αJ and αK of the PAT domain create a vertical groove that stretches over the entire surface of the enzyme. The opening of the hydrophobic tunnel is located within the groove. The predicted importance of the groove is highlighted by the striking evolutionary invariance of residues that dominate its surface properties (Fig. 3 and Fig. S1).
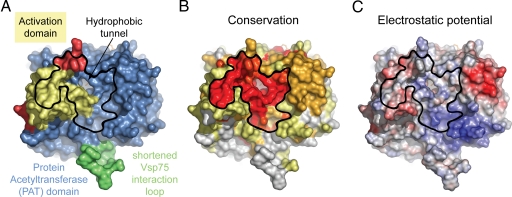
Fig. 3.
Surface properties of Rtt109ΔL. (A) Surface representation colored according to the participation of the various domains as in Fig. 1B. (B) Surface representation colored according to a multispecies sequence alignment (Fig. S1). The conservation at each position is mapped onto the surface and is shaded in a color gradient from light yellow (40% similarity) to dark red (100% identity). (C) Surface representation colored according to the electrostatic potential. The electrostatic potential is plotted onto the surface and colored in a gradient from red (−10 kBT/e) to blue (+10 kBT/e). The orientation of all surface representations is identical.
Comparison with Other Protein Acetyltransferases.
Over the last decade, several PAT structures have been determined (22). These PATs belong to a small number of distinct families that are principally divided into two classes, the nuclear and the cytoplasmic PATs. Several structures of nuclear PATs and their complexes with substrates and substrate analogs have been determined and have revealed a striking structural divergence. However, the close inspection of a structural superposition between p300/CBP, Gcn5, and Esa1 has uncovered a core region that appears to be conserved in all nuclear PATs (21). It has been proposed that the structural divergence between various PATs, in particular the extensive decoration of the presumed catalytic core with additional structural elements is crucial for the precise selection of the native protein substrates (21).
Our structural analysis of Rtt109 has revealed an unexpected overall architectural resemblance to p300/CBP, whereas Gcn5 and Esa1 are more distantly related (Fig. 4A). Both Rtt109 and p300/CBP are constructed of an extended, seven-stranded β-sheet core that is surrounded by a comparable set of additional structural elements (Fig. 4B). Moreover, the two structures exhibit strong similarity in the placement of the α-helices on the back face of the enzyme as well as in the design of the cofactor-binding tunnel. However, it is noteworthy to point out that although the binding modes of the pantetheine arms of the CoA molecules are similar in the two structures, the remainder of the CoA molecule exhibits conformational plasticity (Fig. 4B Right).
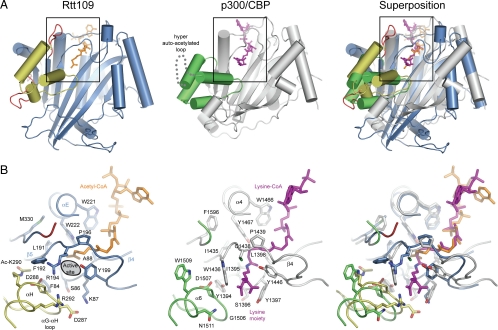
Fig. 4.
Structural comparison of Rtt109ΔL to p300/CBP. (A) Superposition of the structure of Rtt109ΔL (left, colored as in Fig. 1A) in complex with acetyl-CoA (orange) with p300/CBP (middle, PAT domain in gray and the activation domain in green) in complex with the bisubstrate inhibitor lysine-CoA (purple). The Inset illustrates the position of the active site and is expanded in B. (B Right) Superposition of the active sites of Rtt109 (Left) with p300/CBP (Center). Critical active site residues are shown in stick representation, and the Cα-traces are shown in coil representation, according to the coloring scheme in A. The orientation of all active sites is identical. The PDB ID code of the p300/CBP structure is 3BIY (21).
On the front face of the enzyme, the two structures exhibit a striking similarity with respect to the placement of a small α-helical bundle that we termed the activation domain of Rtt109. In Rtt109, the nature of the association between the activation and PAT domains strongly suggests that their interaction is regulated by the autoacetylation of lysine-290. In contrast, the placement of the activation domain equivalent in p300/CBP appears to be permanent because of the formation of an extensive hydrophobic interface. The permanent interaction is achieved by the replacement of the autoacetylation site K290 in Rtt109 by W1509 in p300/CBP (Fig. 4B). In both enzymes, the placement of the activation domain orients a short loop, the αG–αH connector, in close proximity to the hydrophobic tunnel (Fig. 4B). A superposition and the close inspection of the putative active sites of the two enzymes reveals the conservation of a number of residues in the vicinity of the hydrophobic pocket that are potentially important for catalysis (see below) (Fig. 4B).
A key difference between the two structures is the presence and absence of a number of surface loops. For example, p300/CBP contains an insertion in the activation domain not present in Rtt109 that has been found to be hyperacetylated and implicated in autoregulation of the enzyme (Fig. 4A Center). Rtt109, on the other hand, contains an additional surface loop that is required for the binding of histone chaperone Vps75 (see below).
It is important to note that the definition of the C-subdomain of p300/CBP is not identical to the definition of the activation domain in Rtt109. The C-subdomain in p300/CBP has been defined based on a limited proteolysis experiment, in which the molecule is cleaved within the hyperacetylated loop. Based on the presence and the involvement of this domain in the autoregulation of the two enzymes, we propose to rename this region in p300/CBP to “activation domain”. The architectural similarity between Rtt109 and p300/CBP and their proposed distinct activation mechanisms hints toward a diversity of autoregulatory mechanisms within this important family of PATs, similar to the diversity observed for protein kinases (23, 24).
Mutational Analysis.
To determine the importance of various residues within the two domains of Rtt109, we created 14 point mutants of Rtt109ΔL and assessed their phenotype in an in vivo reconstitution assay for resistance to DNA damage hypersensitivity (Fig. S2 and Fig. S3). The mutants can be divided into three categories: (1) mutants that interfere with the interaction between the activation and PAT domains, (2) mutants of the acetylated lysine-290, and (3) surface mutants in the vicinity of the putative active site. To rule out that the observed effects were due to misfolding, we purified all mutants to homogeneity in milligram quantities. All of the mutants we examined were indistinguishable from the full-length Rtt109 in their behavior on a gel-filtration column (data not shown).
The introduction of full-length, wild-type Rtt109 and the αC-β5 loop deletion mutant Rtt109ΔL into rtt109Δ cells greatly decreased their sensitivity to DNA damaging agents. Within the interface between the PAT and activation domains, we mutated five residues that are important for the formation of the hydrophobic cage surrounding Ac-K290. With the exception of C190S, all mutants in the interface (F84A, F285A, D288A, and W312A) were incapable of restoring DNA damage resistance to the rtt109Δ cells. We demonstrated that K290 undergoes autoacetylation. To determine whether autoacetylation is essential for catalytic activity, we generated four K290 mutants: K290A, K290R, K290E, and K290W. Strikingly, the mutation of the acetylation site K290 resulted in a complete loss of DNA damage resistance. On the Rtt109 surface in the vicinity of the active site, we mutated five residues. Whereas the D287A mutant resulted in an almost complete loss of resistance to DNA damage, only a very mild effect was detected in the E66A mutant, and no effect was seen in K87A, Q50A, and Q296A. Together, these experiments strongly suggest that the precise positioning of the activation domain is crucial for the activity of Rtt109. In particular, the acetylation of K290 cannot be substituted with the introduction of a bulky hydrophobic residue. Moreover, the precise positioning of D287 appears to be essential for survival during conditions in which its enzymatic activity is required. The lack of significant phenotypic effects for single mutants located in the putative substrate-binding groove of Rtt109 may be a result of only mild kinetic effects on the enzymatic activity that could be compensated for in our in vivo assay.
The histone chaperone Vps75 has been shown to interact with Rtt109. To identify the interaction site between Vps75 and Rtt109, we performed limited proteolysis experiments on Rtt109 in the absence and presence of Vps75. Unlike the proteolytic pattern that was observed for Rtt109 in isolation, the presence of Vps75 provided protection to several cleavage sites in the region between residues 128 and 170 located in the αC-β5 surface loop (data not shown). Size-exclusion chromatographic analysis of full-length Rtt109 and Rtt109ΔL in the presence and absence of Vps75 clearly demonstrates that this fragment is largely responsible for the Rtt109–Vps75 interaction (Fig. S2C). Similar experiments were carried out with the histone chaperone Asf1. However, neither full-length Rtt109 nor Rtt109ΔL in both the presence and absence of Vps75 exhibited detectable complex formation in our size-exclusion experiments (data not shown). This result may be due, however, to the strength of the interaction, which may be transient in nature and may require a more sensitive means of detection.
Discussion
Rtt109 is a PAT that is responsible for the acetylation of H3K56. We have demonstrated that Rtt109 is autoacetylated at lysine-290 and determined the crystal structure of Rtt109 in its autoacetylated state. Surprisingly, the structure reveals that Rtt109 has an overall architecture similar to p300/CBP, in which an almost flat PAT domain is connected to a smaller activation domain. The principal finding of our study is that Ac-K290 is located in the center of a hydrophobic cage that is formed by the surface of the PAT and activation domains, tightly associating the two domains. We conducted a mutational analysis that uncovered the Vps75-binding region and identified a number of crucial residues that are essential for Rtt109 activity in vivo.
Extensive studies have been carried out in search of a catalytic mechanism by which PAT domains acetylate lysine residues of native protein substrates (22). However, the precise reaction mechanism remains poorly understood. A structural comparison between the active sites of Rtt109 and p300/CBP identifies a number of conserved residues that potentially could be involved in catalysis. However, the mutation of these residues does not abolish the catalytic activity (21), suggesting that these residues primarily fulfill a structural role in the generation of the active site, or, alternatively, in the binding and recognition of the substrate. This finding supports the “hit and run” or “Theorell–Chance” reaction mechanism proposed for p300/CBP, in which the substrate lysine directly reacts with the acetyl-CoA cofactor (21).
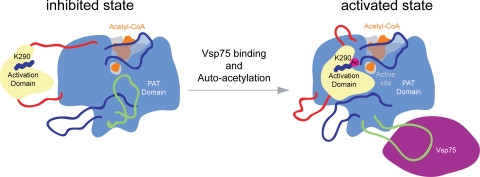
An interesting finding is that Rtt109 and p300/CBP contain an activation domain that is crucially involved in the creation and stabilization of the active site by providing a number of conserved residues. One key difference between the two structures is the nature of the interfaces that facilitate the interaction between the activation and PAT domains. Although the interface is exclusively hydrophobic in p300/CBP, it contains the Ac-K290 located in a hydrophobic cage in Rtt109. This suggests that the attachment of the activation domain is regulated by autoacetylation. Furthermore, we propose that Rtt109 exists in two states, an inactive state in which the activation domain is disengaged from the PAT domain and an active state in which the two domains engage each other, facilitated by autoacetylation (Fig. 5). We cannot rule out completely that the acetylated state of lysine-290 is a catalytic intermediate. However, the findings that Ac-K290 is deeply buried in a hydrophobic cage, is shielded from the substrate, and is not conserved in p300/CBP do not support its role as a catalytic residue.
Fig. 5.
Model for the activation mechanism of Rtt109. The activation of the S. cerevisiae protein acetyltransferase Rtt109 depends on the autoacetylation of lysine-290. We propose that Rtt109 exists in two states, an inactive, autoinhibited state and an activated state. In the inactive state, the activation domain is loosely tethered to the PAT domain. Upon autoacetylation, the PAT domain engages the acetylated lysine-290, resulting in the tight interaction between the activation and PAT domains. The catalytic activity of Rtt109 may be further enhanced by the sequestration of the Vps75 interaction loop, removing it from the active-site groove.
We demonstrated that Vps75 interacts with an ≈50-residue surface loop of Rtt109. To obtain crystals, we genetically shortened the loop, and, hence, no structural information for this loop was obtained. However, the proteolytic sensitivity and the location of this loop lead us to propose that this region of Rtt109 is involved in the regulation of the enzymatic activity. It is interesting to speculate that this loop is bound dynamically to the active site groove, preventing the efficient binding of the substrate. The interaction of Rtt109 with Vps75 may result in its removal from the active-site cleft and, therefore, may promote efficient substrate binding (Fig. 5). Although Vps75 appears not to be required for cellular survival (25), it nevertheless may be required for the efficient activation of Rtt109.
Short Methods
The details of molecular cloning, expression, purification, crystallization, x-ray diffraction data collection, structure determination, and yeast experiments are described in SI Text. In short, the Rtt109ΔL variant was expressed by using a modified pET28a vector (Novagen) that contained an N-terminal hexahistidine-tagged SUMO. Recombinant protein was purified by using several chromatographic techniques. Anomalous diffraction data of Rtt109ΔL crystals were collected at the X3A beamline at the National Synchrotron Light Source, Brookhaven National Laboratory, Upton, NY. The positions of the selenium atoms were determined by using SHELXD (26), and phases were determined in SHARP (27), followed by density modification and histogram matching in DM (28). A model was built with the program O (29) and refined with CNS (30). Data collection and refinement statistics are summarized in Table S1. The structure factors and atomic coordinates have been deposited to the Protein Data Bank with the PDB ID code 3CZ7.
Supplementary Material
Acknowledgments.
We thank A. Patke for discussions and comments on the manuscript, S. Etherton for help with editing the manuscript, M. Kampmann for advice on the yeast experiments, D. King for mass spectrometry analysis, T. Noriega for technical support, and B. Manjasetty and W. Shi for support during data collection. A.H. was supported by a grant from the Leukemia and Lymphoma Society.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3CZ7).
This article contains supporting information online at www.pnas.org/cgi/content/full/0805813105/DCSupplemental.
References
- 1.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 2.Stavropoulos P, Hoelz A. Lysine-specific demethylase 1 as a potential therapeutic target. Exp Opin Ther Targets. 2007;11:809–820. doi: 10.1517/14728222.11.6.809. [DOI] [PubMed] [Google Scholar]
- 3.Stavropoulos P, Blobel G, Hoelz A. Crystal structure and mechanism of human lysine-specific demethylase-1. Nat Struct Mol Biol. 2006;13:626–632. doi: 10.1038/nsmb1113. [DOI] [PubMed] [Google Scholar]
- 4.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 5.Xu F, Zhang K, Grunstein M. Acetylation in histone H3 globular domain regulates gene expression in yeast. Cell. 2005;121:375–385. doi: 10.1016/j.cell.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 6.Masumoto H, Hawke D, Kobayashi R, Verreault A. A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature. 2005;436:294–298. doi: 10.1038/nature03714. [DOI] [PubMed] [Google Scholar]
- 7.Ozdemir A, et al. Characterization of lysine 56 of histone H3 as an acetylation site in Saccharomyces cerevisiae. J Biol Chem. 2005;280:25949–25952. doi: 10.1074/jbc.C500181200. [DOI] [PubMed] [Google Scholar]
- 8.Hyland EM, et al. Insights into the role of histone H3 and histone H4 core modifiable residues in Saccharomyces cerevisiae. Mol Cell Biol. 2005;25:10060–10070. doi: 10.1128/MCB.25.22.10060-10070.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou H, Madden BJ, Muddiman DC, Zhang Z. Chromatin assembly factor 1 interacts with histone H3 methylated at lysine 79 in the processes of epigenetic silencing and DNA repair. Biochemistry. 2006;45:2852–2861. doi: 10.1021/bi0521083. [DOI] [PubMed] [Google Scholar]
- 10.Recht J, et al. Histone chaperone Asf1 is required for histone H3 lysine 56 acetylation, a modification associated with S phase in mitosis and meiosis. Proc Natl Acad Sci USA. 2006;103:6988–6993. doi: 10.1073/pnas.0601676103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Han J, et al. Rtt109 acetylates histone H3 lysine 56 and functions in DNA replication. Science. 2007;315:653–655. doi: 10.1126/science.1133234. [DOI] [PubMed] [Google Scholar]
- 12.Driscoll R, Hudson A, Jackson Yeast Rtt109 promotes genome stability by acetylating histone H3 on lysine 56. Science. 2007;315:649–652. doi: 10.1126/science.1135862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsubota T, et al. Histone H3–K56 acetylation is catalyzed by histone chaperone-dependent complexes. Mol Cell. 2007;25:703–712. doi: 10.1016/j.molcel.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scholes DT, Banerjee M, Bowen B, Curcio MJ. Multiple regulators of Ty1 transposition in Saccharomyces cerevisiae have conserved roles in genome maintenance. Genetics. 2001;159:1449–1465. doi: 10.1093/genetics/159.4.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schneider J, Bajwa P, Johnson FC, Bhaumik SR, Shilatifard A. Rtt109 is required for proper H3K56 acetylation: A chromatin mark associated with the elongating RNA polymerase II. J Biol Chem. 2006;281:37270–37274. doi: 10.1074/jbc.C600265200. [DOI] [PubMed] [Google Scholar]
- 16.Han J, Zhou H, Li Z, Xu RM, Zhang Z. Acetylation of lysine 56 of histone H3 catalyzed by RTT109 and regulated by ASF1 is required for replisome integrity. J Biol Chem. 2007;282:28587–28596. doi: 10.1074/jbc.M702496200. [DOI] [PubMed] [Google Scholar]
- 17.Han J, Zhou H, Li Z, Xu RM, Zhang Z. The Rtt109-Vps75 histone acetyltransferase complex acetylates non-nucleosomal histone H3. J Biol Chem. 2007;282:14158–14164. doi: 10.1074/jbc.M700611200. [DOI] [PubMed] [Google Scholar]
- 18.Xhemalce B, et al. Regulation of histone H3 lysine 56 acetylation in Schizosaccharomyces pombe. J Biol Chem. 2007;282:15040–15047. doi: 10.1074/jbc.M701197200. [DOI] [PubMed] [Google Scholar]
- 19.Trievel RC, et al. Crystal structure and mechanism of histone acetylation of the yeast GCN5 transcriptional coactivator. Proc Natl Acad Sci USA. 1999;96:8931–8936. doi: 10.1073/pnas.96.16.8931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yan Y, Barlev NA, Haley RH, Berger SL, Marmorstein R. Crystal structure of yeast Esa1 suggests a unified mechanism for catalysis and substrate binding by histone acetyltransferases. Mol Cell. 2000;6:1195–1205. doi: 10.1016/s1097-2765(00)00116-7. [DOI] [PubMed] [Google Scholar]
- 21.Liu X, et al. The structural basis of protein acetylation by the p300/CBP transcriptional coactivator. Nature. 2008;451:846–850. doi: 10.1038/nature06546. [DOI] [PubMed] [Google Scholar]
- 22.Hodawadekar SC, Marmorstein R. Chemistry of acetyl transfer by histone modifying enzymes: Structure, mechanism and implications for effector design. Oncogene. 2007;26:5528–5540. doi: 10.1038/sj.onc.1210619. [DOI] [PubMed] [Google Scholar]
- 23.Nolen B, Taylor S, Ghosh G. Regulation of protein kinases; controlling activity through activation segment conformation. Mol Cell. 2004;15:661–675. doi: 10.1016/j.molcel.2004.08.024. [DOI] [PubMed] [Google Scholar]
- 24.Pellicena P, Kuriyan J. Protein–protein interactions in the allosteric regulation of protein kinases. Curr Opin Struct Biol. 2006;16:702–709. doi: 10.1016/j.sbi.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 25.Selth L, Svejstrup JQ. Vps75, a new yeast member of the NAP histone chaperone family. J Biol Chem. 2007;282:12358–12362. doi: 10.1074/jbc.C700012200. [DOI] [PubMed] [Google Scholar]
- 26.Schneider TR, Sheldrick GM. Substructure solution with SHELXD. Acta Crystallogr D. 2002;58:1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- 27.La Fortelle ED, Bricogne G. Maximum-likelihood heavy-atom parameter refinement in the MIR and MAD methods. Methods Enzymol. 1997;276:476–494. doi: 10.1016/S0076-6879(97)76073-7. [DOI] [PubMed] [Google Scholar]
- 28.CCP4. Collaborative Computational Project, Number 4, The CCP4 Suite: Programs for Protein Crystallography. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 29.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 30.Brünger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.