Abstract
Human XPA is an important DNA damage recognition protein in nucleotide excision repair (NER). We previously observed that XPA binds to DNA lesion as a homodimer (1). Herein we report that XPA recognized undamaged DNA doublestrand/ single-strand (ds-ssDNA) junctions containing ssDNA branches with binding affinity (Kd = 49.1±5.1 nM) much higher than its ability to bind to DNA damage. The recognized DNA junction structures include Y-shape junction (with both 3′- and 5′- ssDNA branches), 3′-overhang junction (with a 3′-ssDNA branch), and 5′-overhang junction (with a 5′-ssDNA branch). Using gel filtration chromatography and gel mobility shift assays, we showed that the highly efficient binding appeared to be carried out by the XPA monomer and that the binding was largely independent of RPA. Furthermore, XPA efficiently bound to six-nucleotide mismatched DNA bubble substrates with or without DNA adducts including C8 guanine adducts of AF, AAF, and AP, and the T[6,4] T photo products. Using a set of defined DNA substrates with varying degrees of DNA bending, we also found that the XPC-HR23B complex recognized DNA bending, whereas neither XPA nor the XPA-RPA complex could bind to bent DNA. We propose that besides DNA damage recognition, XPA may also play a novel role in stabilizing, via its high affinity to ds-ssDNA junctions, the DNA strand opening surrounding the lesion for stable formation of pre-incision NER intermediates. Our results provide a plausible mechanistic interpretation for the indispensable requirement of XPA for both global genome and transcription-coupled repairs. Since ds-ssDNA junctions are common intermediates in many DNA metabolic pathways, the additional potential role of XPA in cellular processes is discussed.
INTRODUCTION
Nucleotide excision repair (NER) is an important DNA repair pathway that removes a broad variety of bulky or distorting DNA damages induced by chemicals and UV light (2-4). XPA (xeroderma Pigmentosum group A) is an indispensable factor for both global genome repair (GGR) and transcription-coupled repair (TCR), the two subpathways of NER (2-5). It is believed that XPA is involved in the DNA damage recognition in NER (1, 2, 4). XPA-DNA damage interaction is carried out in a cooperative manner by the XPA dimer (1, 6). Other DNA damage recognition proteins in NER include RPA (replication protein A) and the XPC-HR23B complex. The latter is believed to be the initial damage recognition factor, though it is required only for GGR (7). In TCR, where the XPC-HR23B is not required, the RNA polymerase complex plays a role for initial damage recognition, while the role of XPA remains unclear. XPA is believed to form a complex with RPA to stabilize or mediate the RPA-DNA interaction (8-10). However, recent studies indicate that XPA may be loaded as a separate factor to DNA lesions in cells (11, 12) and the presence of XPA or RPA has little effect on damaged DNA binding affinity of either protein (1, 13). Furthermore, XPA acts as a co-recognition factor with the XPC-HR23B complex in recognizing triplex-directed psoralen DNA interstrand crosslinks (14). A crucial role of XPA in NER was demonstrated by its involvement in both GGR and TCR, its presence in the final excision complex, and its being a limiting factor for NER and UV sensitivity in human cells (15).
Although the functions of XPA have been extensively studied, its exact role in NER remains elusive. Recent studies have showed that XPA efficiently binds to double-stranded three-way or four-way (Holliday junction-type) DNA structures (16), which appears to be correlated with repair efficiency (17). However, these structures are absent in NER intermediates, although the results have been suggested to support XPA recognition of the helical kinks induced by DNA damage (17). Besides its role in DNA damage recognition, XPA also has been suggested to be important for recruitment of other NER factors for nuclease assembly, which repairs the lesion (4, 18-21). The recruitment of Rad1-Rad10, the yeast homolog of XPF-ERCC1, by Rad14, the yeast homolog of XPA (22), has recently been demonstrated in vivo, even though the mechanism for this process remains unclear.
In the present study we report a potential novel function of XPA in NER. Using a homogeneous form of XPA purified from baculovirus-infected insect cells (1), we demonstrated that in addition to efficient interaction with damaged DNA substrates containing C8 guanine adducts of AF, AAF, or AP and the G[8,5-Me]T vicinal crosslink, XPA has an unusually high affinity for ds-ssDNA junctions with 3′- and/or 5′-ssDNA overhangs (including Y-structure junction and the junctions with either a 3′- or 5′-ssDNA branch). This DNA junction binding was carried out by the XPA monomer, whereas the DNA damage recognition is performed by the XPA dimer. Also noteworthy is the finding that XPA exhibited no significant difference in its affinity for different types of DNA adducts when contained in a six-nucleotide bubble structure. Furthermore, using a set of structure-specific DNA substrates with various degrees of bending angle, we showed that XPA had almost no affinity and specificity for bent DNA. By contrast, XPC-HR23B complex could bind efficiently and with high specificity to the bent DNA substrates. Our data suggest a novel major role for XPA in NER to stabilize the unwound DNA intermediate surrounding the lesion, which may also facilitate the recruitment of NRE factors for nuclease assembly.
MATEREIALS AND METHODS
Chemicals and Reagents
All chemicals, including Tris-HCl, Hepes, NaCl, ZnCl2, CaCl2, MgCl2, boric acid, EDTA, glycerol, ammonium persulfate, phenylmethylsulphonyfluoride (PMSF), N, N, N′, N′-tetramethylethylenediamine (TEMED), β-mercaptoethanol, and imidazole were purchased from Fisher Scientific Co. or Sigma Co. Ni-NTA beads were purchased from Qiagen. Chitin beads were purchased from New England Biolabs. Polyacrylamides and urea were obtained from Bio-Rad and National Diagnostics. Protease inhibitor cocktail was obtained from Roche Co. XPA antibody was obtained from Kamiya Biotech Inc. and anti-RPA 70, 32 and 14 antibodies were obtained from Santa Cruz Biotechnology Inc.
XPA and RPA protein purification
[His]6-XPA protein was purified from sf-21 insect cells infected with recombinant baculovirus pBac-XPA virus (graciously provided by J. J. Turchi, Wright State University School of Medicine) at a multiplicity of infection (moi) of 10 as described previously (1). RPA protein was expressed in BL21- (DE3)-RP competent cells (Stratagene) and induced with 0.7 mM IPTG at 25°C for 3 h. Recombinant RPA was purified using chitin affinity beads in a one-step procedure as described previously (6, 23). Briefly, chitin-bound RPA was eluted in 30 mM DTT cleavage buffer and RPA-positive fractions were pooled and concentrated. The eluted fractions containing RPA trimer were collected and dialyzed in RPA storage buffer containing 50% glycerol. XPA and RPA protein concentrations were determined using the Bio-Rad protein assay following the manufacture instructions.
XPC-HR23B protein complex purification
XPC-HR23B protein complex was purified as described previously (24). XPC/HR23B protein was purified from Sf21 insect cells infected with recombinant baculovirus expressing XPC and HR23B proteins (graciously provided by A. Sancar, University of North Carolina, Chapel Hill). Briefly, 500 ml Sf21 cells were infected with recombinant pBacXPC-HR23B baculovirus at a multiplicity of infection of about 10, and incubated at 27 °C for 48 h. Cells were harvested by low speed (2000 rpm) centrifugation, and cell free extracts were prepared (25, 26). Cell pellets were resuspended in 20 mL lysis buffer (10mM Tris•HCl pH 7.9, 1 mM EDTA, 5 mM dithiothreitol (DTT) and 0.5 mM PMSF, 1x Roche protease inhibitor cocktail), incubated on ice for 20 min, and homogenized using a Dounce homogenizer by eight strokes with a “B” pestle. To this suspension, 20 ml buffer (50 M Tris•HCl, pH 7.9, 10 mM MgCl2, 2 mM DTT, and 50% glycerol v/v) and sucrose were added, followed by gentle stirring until sucrose completely dissolved in the buffer (final concentration of sucrose is 12.5%). Subsequently, 6 ml of saturated (NH4)2SO4 was added drop by drop with gentle stirring over 20-min. The extracts were clarified by centrifugation for 3 h at 50,000 rpm with in a Beckman ultracentrifuge using a 50 Ti rotor at 4°C. The resulting supernatant was dialyzed against Buffer A (25 mM Tris-HCl, pH7.5, 1 mM EDTA, 1 mM DTT, 0.01% Triton X-100, 10% glycerol) containing 0.3 M KCl for 2 h. Dialyzed extracts were further centrifuged for 10 min at 12,000 g at 4°C and the supernatant was loaded onto a 12-ml cellulose phosphate column (Whatman®) equilibrated in the same buffer. The column was washed with 100 ml Buffer A (plus 0.3 M KCl) and eluted with 50 ml Buffer A (plus 1 M KCl). Eluted protein was pooled and diluted to 0.6 M KCl with Buffer A without KCl. Diluted protein was applied to a 5-ml ssDNA cellulose column (Sigma) equilibrated with buffer A (0.6 M KCl) and the column was washed with 100 ml buffer A (0.6 M KCl). Eluted protein was diluted with Buffer A to 0.3 M KCl, and applied to a 1 ml HiTrap™ DEAE FF column (Amersham Biosciences). Flow through fractions containing XPC/HR23B were collected and dialyzed against storage buffer and stored at −80°C in aliquots. Protein concentration was determined using the Bio-Rad protein assay. The purity of the XPC/HR23B complex was confirmed by SDS-PAGE (10%) and Western Blotting.
Substrate Construction
Substrates were constructed as described previously (27, 28). Briefly, 50-bp substrates were constructed by annealing equimolar amounts of either damaged or undamaged 10-mer or 11-mer with flanking 19-mer and 5′ 32P-radiolabled 20-mer in the presence of a complementary 44-mer (bottom strand), followed by ligation with 0.5 U T4-DNA ligase overnight at 16 °C. The ligation products were purified on a 12% denaturing gel, reannealed with a complementary 50-mer (bottom strand), which was again purified on an 8% native gel. Fifty-bp bubble substrates were constructed using the same protocol with the following modifications: the purified ligated 50-mer containing a DNA lesion (top strand) was reannealed with a partially complementary 50-mer containing indicated number of mismatched nucleotides. The 49-bp T[6,4]T and G[8,5-Me]T intrastrand crosslink substrates were constructed as reported (29). The substrates containing a G[nc]G intrastrand crosslink were synthesized and constructed as described previously (30). The G[nc]G lesion is the guanine-guanine binucleotides crosslinked with a two-, three-, or four-carbon tether (as shown below). The n in the G[nc]G stands for the number of carbons in the tether, while the c stands for carbon. 
The ds-ssDNA junction substrates with ssDNA overhangs were constructed by annealing 5′-terminally 32P-radiolabeled top strand with different bottom strands, followed by purification on an 8% native gel. All substrates were subjected to restriction enzyme analysis with HaeIII and/or RasI to verify proper alignment of the final products.
Electrophoretic Mobility Shift Assay (EMSA)
Binding of XPA, RPA, XPA-RPA, and XPC-HR23B to various DNA substrates was analyzed by gel mobility shift assay as described previously (1, 6). Typically, substrates (0.5-1 nM) were incubated with varying concentrations of protein at 30 °C in 20 μL of binding buffer (20 mM Hepes-KOH, pH 7.9, 75 mM KCl, 5 mM MgCl2, 1 mM DTT, 5% glycerol, 100 μg/mL acetylated BSA, Promega). Reactions were then placed on ice, 2 μL 80% (v/v) glycerol was added, and the mixture was immediately loaded onto a 3.5% native polyacrylamide gel and electrophoresed at 80V in 1x TBE buffer for 2 h at 4 °C. The gels were dried and exposed to phosphoimage screens overnight. Quantification of the radioactivity was carried out using a Fuji FLA-5000 scanner with the ImageGuage software.
Fluorescence Anisotropy Analysis
Anisotropy titrations were performed as described previously (23) to characterize the XPA binding to ds-ssDNA junction (5′-terminally labeled with a fluorescein on top strand) in the buffer containing 20 mM Hepes-KOH, pH 7.9, 75 mM KCl, 5 mM MgCl2, 2 mM DTT, 5% glycerol, 200 μg/mL human serum albumin (a gracious gift from Dr. David Johnson, East Tennessee State University). The data were analyzed using a one-site binding model and the non-linear least-squares method for the determination of the dissociation constants (1, 31).
DNase I and Permanganate Footprinting Assays
The DNase I footprinting assay was carried out as described previously (32). Prior to DNase I digestion, 3′-terminally-labeled Y-shape substrates (8 nM) were incubated in the presence and absence of 200 nM XPA (equal amount of XPA storage buffer was added for no XPA conditions) at 30 °C for 15 min in 10 μL of binding buffer (20 mM Hepes-KOH, pH 7.9, 75 mM KCl, 5 mM MgCl2, 1 mM EDTA, 1 mM DTT, 5% glycerol, and 100 μg/mL BSA (Promega). After incubation, CaCl2 was added to10 mM final concentration. Substrates were then digested with 0.7 ng of DNase I (Invitrogen) at room temperature for 1 min. The reactions were terminated by adding 10 μL of formamide DNA-denaturing buffer containing 40 mM EDTA, and the samples were immediately frozen in the dry ice-ethanol mixture. The samples were heated to 90 °C for 5 min, immediately chilled on ice, and subjected to electrophoresis in 12% polyacrylamide sequencing gel (8 M urea) under denaturing conditions.
The chemical footprinting assay was conducted according to procedures described previously (33). Briefly, labeled Y-shape substrates (8 nM) were incubated in 10 μL binding buffer with 200 nM XPA or without XPA as described in the previous section. Then 2.5 L of KMnO4 was added (0.2 mM final concentration) and the reactions were conducted at room temperature for 1 min. Reactions were terminated by adding equal volume of 2 M 2-mercapthoethanol containing 40 mM EDTA, immediately followed by chilling on ice. The DNA was precipitated with 70% cold ethanol and then incubated with 25 μL of 1 M piperidine at 90 °C for 25 min. Samples were spin-vacuumed and analyzed on a 12% sequencing gel under denaturing conditions.
Size Exclusion Chromatography/liquid Scintillation Assay
Gel filtration chromatography coupled with scintillation analysis was employed to analyze the stoichiometry of XPA binding to the ds-ssDNA junction substrate. Briefly, the substrate (1 nM) was incubated with varying concentrations of protein at 30 °C in 100 μL of binding buffer for 30 min. After incubation, the reactions were loaded onto a Superdex®-200 size exclusion column (Amersham Pharmacia Biosciences, Uppsala, Sweden), and eluted with XPA binding buffer (20 mM Hepes-KOH, pH 7.9, 75 mM KCl, 5 mM MgCl2, 1 mM DTT, 5% glycerol). 0.2-0.5mL fractions were collected using a Frac-900 auto collector (Amersham Pharmacia Biosciences, Uppsala, Sweden). 100 uL of each fraction was mixed with 5 mL ScintiSafe 30 scintillation cocktail (Fisher Scientific Inc.) and radioactivity was counted using a Beckman LS 3801 Liquid Scintillation System. Under the same conditions, 100 uL standard protein markers also were load onto Superdex® 200 size-column and eluted by the same buffer. The standard proteins markers included ribonuclease A (13.7 KDa), chymotrypsinogen A (25 KDa), ovalbumin (43 kDa), bovine albumin (67 kDa), aldolase (158 kDa), catalase (232 kDa), ferritin (440 kDa), and thyroglobulin (669 kDa). A linear relationship between the protein molecular weights (log MW) and retention volume (mL) was established by linear regression.
RESULTS
Protein Purification
Recombinant XPA and XPC-HR23B proteins overexpressed in baculovirus-infected insect cells were purified following the procedures as described in Materials and Methods. Purified XPA exhibited homogeneity as analyzed on 10% SDS-PAGE (1). RPA was purified from E. coli BL21(DE3)-RP cells transformed with the overexpressing plasmid pTYB-RPA containing RPA cDNA (6, 23). All these proteins have the purity greater than 95% (data not shown). The XPA and XPC/HR23B proteins were fully active to recognize damaged DNA substrates with various adducts (see below). Purified RPA also had full activity towards single-stranded DNA (data not shown).
Binding of XPA to DNA substrate containing a G[8,5-Me]T crosslink lesion
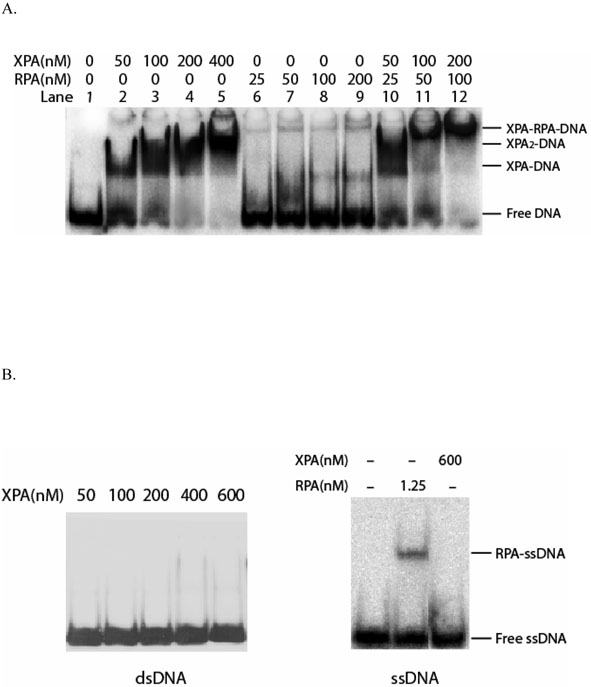
Electrophoretic mobility shift assay (EMSA) was used to examine the affinity and specificity of the XPA protein for damaged versus undamaged DNA. As shown in Figure 1, XPA had a high specific affinity for damaged DNA substrates as compared to undamaged DNA. Well-resolved XPA-DNA complexes were observed with the defined DNA substrate containing a γ-radiation induced G[8,5-Me]T guanine-thymine crosslink lesion, and the complex formation was dependent on the concentrations of XPA. Since XPA binds to damaged DNA in a cooperative manner (1), XPA2 dimer-DNA complexes formed at higher protein concentrations (Figure 1A, lanes 3-5) as determined previously (1). In contrast, RPA showed little affinity for the G[8,5-Me]T-containing substrate at protein concentrations ranging from 25-200 nM (Figure 1A, lanes 6-9), whereas RPA did interact with XPA-bound G[8,5-Me]T substrate to form a slower migrating band (Figure 1A, lanes 10-12). Evidently, the XPA protein efficiently recognized the G[8,5-Me]T lesion. By contrast, there was no detectable binding of XPA to undamaged DNA duplex (dsDNA) or single-stranded DNA (ssDNA) even at 600 nM concentration of the protein (Figure 1B).
Figure 1.
XPA binding to DNA duplex containing a G[8,5-Me]T crosslink lesion. Panel A: Homogeneous XPA protein purified from baculovirus-infected insect cells showed high specific affinity to 50-bp DNA substrate containing a γ-radiation-induced G[8,5-Me]T crosslink lesion in the presence and absence of RPA in gel mobility shift assays. RPA protein itself displayed no affinity for the DNA crosslink adduct (lanes 6-9), although the presence of RPA resulted in a super-shift of XPA-DNA bands corresponding to the XPA-RPA-DNA interaction (lanes 10-12). Panel B: XPA protein showed little or no affinity for undamaged DNA duplex or ssDNA. Gel mobility shift assays were performed on 3.5% native polyacrylamide gel electrophoresed at 4°C.
XPA binding to mismatched nucleotide bubble DNA substrates
EMSA analysis of XPA interaction with six-nucleotide mismatched DNA substrates revealed that XPA had a high affinity for DNA bubble structure with or without DNA damage (Figure 2A). For comparison, we examined the XPA interactions with DNA substrates containing an AF, AAF, AP, or T[6,4]T lesion in the six-nucleotide bubble structure (34). No significant difference in XPA binding was observed among these adducts. In addition, there was no difference in XPA binding between damaged or undamaged (ND) bubble substrates (Figure 2A). Binding of XPA to the bubble substrates was observed at protein concentrations as low as 2.5 nM (Figure 2A, lanes 2, 6, 10, 14, 18); such a high affinity of XPA-DNA (damaged or undamaged) interaction has not been previously reported. The effects of the DNA bubble size on the affinity of XPA also were investigated. Our results indicated that XPA recognized bubble structures with the size of four mismatched nucleotides or larger (Figure 2B, lanes 6-24). The protein appeared to have a higher affinity for the DNA substrate with an 8-base bubble or larger. XPA, however, did not efficiently recognize bubble structures smaller than four mismatched nucleotides (Figure 2B, Lanes 1-4). A possible explanation we considered for this observation is that XPA recognizes ds-ssDNA junctions.
Figure 2.
Binding of XPA protein to partially mismatched DNA bubble substrates with or without a lesion. Panel A: XPA bound to six base-mismatched DNA bubble substrates with or without AF, AAF, AP or T[6,4]T adduct at varying concentrations. ND-B6 stands for undamaged DNA substrate containing a six-base mismatched bubble. Panel B: XPA bound to DNA substrates with various sizes of bubble. The underline in the sequences indicates the mismatched bases for bubble formation.
XPA binds DNA substrates with a 3′- or 5′-overhang and stabilizes RPA-DNA interaction
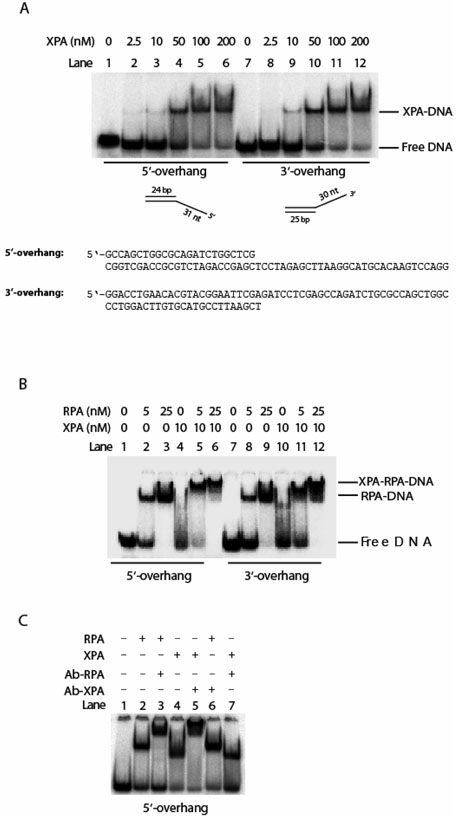
We next investigated the affinity of XPA for 3′-overhang and 5′-overhang DNA substrates, two types of ds-ssDNA junctions. Weak binding was observed for each substrate at XPA protein concentrations lower than 50 nM (Figure 3A, lanes 3 and 9). However, at 100 nM concentration quantitative binding of the substrates with XPA was noted (Figure 3A, lanes 5 and 11), and no detectable difference in binding affinity was observed between the two substrates. RPA also interacted with the substrates due to its high affinity for the ssDNA regions (Figure 3B, lanes 2, 3, 8, and 9). Addition of XPA at a concentration of 10 nM appeared to enhance the binding of RPA for both overhang substrates (Figure 3B, lanes 5, 6, 11, and 12). As XPA itself exhibited only weak binding affinity for each of the substrates at this concentration, the enhancement was likely due to the stabilization of the RPA-DNA complex through XPA-RPA interactions. However, the contribution from the combined but separate bindings of XPA to the junction and RPA to the ssDNA region also cannot be ruled out. To further confirm that the band shift was due to the XPA-DNA complex formation, monoclonal anti-XPA antibody (Kamiya) was added to the reaction after incubation of XPA with the 5′-overhang DNA substrate. As shown in Figure 3C (lanes 4 and 5), the protein-DNA complex band was supershifted by the antibody, indicating that the protein binding to the DNA substrate indeed was XPA. As controls, antibody against RPA32 subunit of RPA (Kamiya) also caused a supershift for RPA-DNA complex (lanes 2 and 3) and there was no cross-interaction of anti-XPA antibody with this complex (lane 6) and vice versa (lane 7).
Figure 3.
Interactions of XPA and/or RPA with 3′- and 5′-single strand overhang DNA substrates. Panel A: XPA bound to 3′-ssDNA overhang and 5′-ssDNA overhang DNA substrates at various concentrations. Panel B: XPA and RPA interacted with the 3′-overhang and 5′-overhang DNA substrates. Panel C: XPA and RPA interacted with 5′-overhang DNA substrate, and the complexes were supershifted with corresponding antibodies, respectively.
XPA interaction with ds-ssDNA junctions containing both 3′- and 5′-ssDNA overhangs (Y-shape structures)
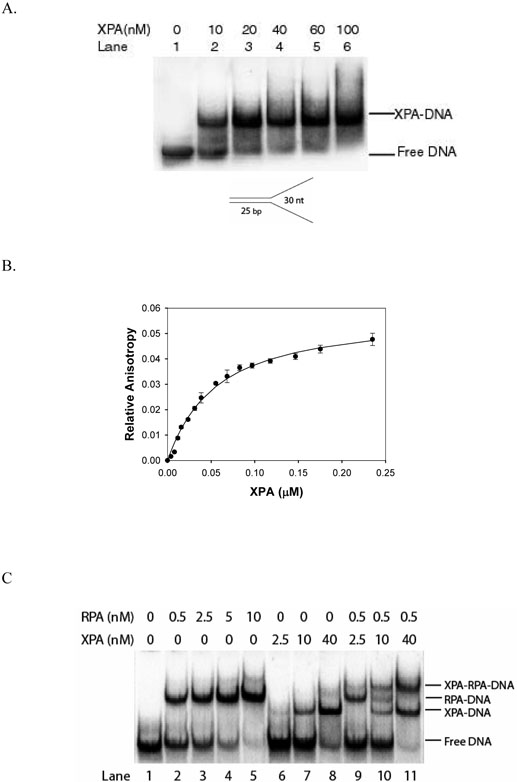
In order to examine the binding of XPA to the ds-ssDNA junction structure formed as intermediates in NER, we constructed a Y-shape ds-ssDNA junction substrate (Figure 4). EMSA analysis revealed that XPA efficiently bound to the Y-shape substrate (Figure 4A). Further determination by fluorescence anisotropy indicated that the binding had a Kd of 49.1±5.1 nM (Figure 4B). Evidently, this binding affinity of XPA for Y-shape DNA is much higher than for damaged DNA (without bubble) that has a Kd of hundreds of nM. Indeed, as shown in Figure 4C, although RPA had a higher affinity for the two ssDNA regions in the Y-shape substrate than XPA for the single junction (Figure 4C, lanes 2-5), XPA efficiently bound to the ds-ssDNA junction of the same substrate at a concentration as low as 10 nM (Figure 4C, lanes 10-11). It should be noted that the XPA had little affinity for dsDNA (Figure 1A) or ssDNA (data not shown). Interestingly, XPA appeared to slightly prefer binding to the ds-ssDNA junction of the Y-shape DNA molecules that were already bound by RPA (Figure 4C, lane 10), although there was a large excess of free DNA available for binding.
Figure 4.
The binding of XPA to Y-shape DNA substrate in the presence and absence of RPA. Panel A: XPA bound to the Y-shape DNA substrate in an XPA-concentration dependent manner. Panel B: Fluorescence anisotropy measurements of XPA binding to ds-ssDNA junction substrate. The binding isotherms were generated by titrating the substrate with increasing concentrations of XPA. Panel C: XPA and/or RPA bound to Yshape DNA. RPA bound to the single strand regions of the Y-shape substrate (lanes 2-5), while XPA bound to the ds-ssDNA junction of the substrate (lanes 6-8). When XPA and RPA both were present, formation of three complexes was observed corresponding to XPA binding to ds-ssDNA junction (XPA-DNA), RPA binding to the ssDNA regions of the substrate (RPA-DNA), and XPA and RPA concurrently binding to the junction and ssDNA regions of the same substrate molecules (XPA-RPA-DNA), respectively (lanes 10 and 11).
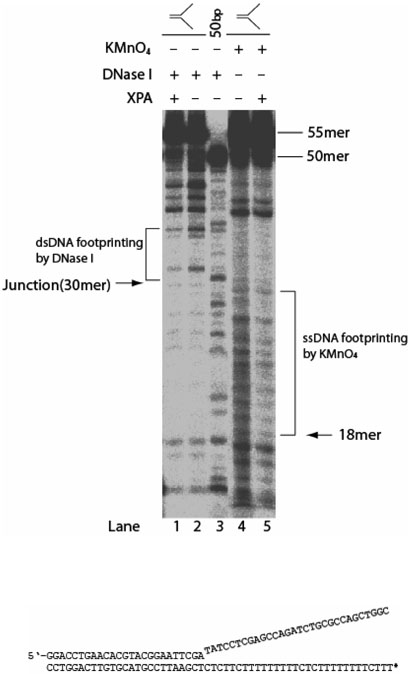
To confirm the direct contact of XPA with the ds-ssDNA junction with ssDNA overhangs and to define the size of the binding, we mapped the binding sites of XPA to the DNA substrate using both DNase I and permanganate (KMnO4) footprinting assays. The DNase I is best for dsDNA footprinting as it has a much higher nuclease activity towards dsDNA than ssNDA (33). In contrast, KMnO4 selectively attacks thymines in ssDNA. As shown in Figure 5, the Y-shape ds-ssDNA junction substrate contains a string of thymine in the ssDNA region of bottom strand. Thus, the substrate was radio-labeled with 32P at the 5′-end of the bottom strand for use in the footprinting assays. As shown in Figure 5, binding of XPA to the substrate resulted in a significant protection from DNase I of the dsDNA region of about 7 nucleotides 3′ (labeled bottom strand) to the junction (lanes 1 and 2), while about 12 nucleotides were shielded from KMnO4 oxidation in the ssDNA region 5′ to the junction (lanes 4 and 5). Lane 3 showed the DNase I digestion of a double-stranded 50-bp DNA for control. The footprinting results indicated that the XPA did bind to the ds-ssDNA junction.
Figure 5.
Footprinting of XPA binding to the ds-ssDNA junctions in the Y-shape substrate with 3′- and 5′-ssDNA overhangs. Lanes 1-3, dsDNA footprinting by DNase I. Lanes 4 and 5, ssDNA footprinting by KMnO4. Y-shape substrates with 32P-labeled 5′-end of the bottom strand (indicated by the asterisk) were incubated at 30 °C for 15 min in binding buffer with or without XPA. After the incubation, substrates were digested with 0.7 ng DNase I (lanes 1-3) or probed with 0.2 mM KMnO4 (lanes 5 and 6), followed by hot piperidine treatment. Both footprinting reactions were performed at room temperature for 1 min. The samples were analyzed on a 12% denaturing polyacrylamide gel.
Since XPA binds to damaged DNA as a protein homodimer as described previously (1), next we asked the question of whether XPA binds to ds-ssDNA junction in the same or different oligomeric forms. The above results of gel mobility shift assays in which the XPA-DNA junction complex migrated much faster than the XPA dimer-damaged DNA complex (the second band shift in Figure 1) (1) suggests that XPA may interact with ds-ssDNA junction as a monomer. This was further supported by a gel filtration assay combined with the liquid scintillation counting for determination of the stoichiometry of XPA-Y-shape DNA substrate binding. As shown in Figure 6, a retention volume of 13.2 mL was obtained for the XPA-Y-shape DNA complex when eluted from a Superdex®-200 column. In contrast, the XPA dimer-damaged DNA complex was eluted earlier at 12.2 mL (Figure 6) (1), while free DNA was eluted much later at a retention volume of about 22 mL (Figure 6). The corresponding apparent molecular weight of the complex was calculated to be ~70 kDa based on a standard plot of retention volumes versus the log of the molecular weights of standard protein markers (1). This suggested that the molecular ratio of protein to DNA is 1:1 (XPA MW: 36 kDa, Y-shape DNA MW: 33.7 kDa) (Figure 6). It should be noted, however, that this determination was based on the assumption that the protein-DNA complex primarily has a globular structure with the bound DNA as part of it, whereas the free DNA was eluted anomalously due to its linear structure. Nonetheless, the fact that XPA-Y-shape DNA complex has a larger retention volume than the complex of XPA dimer with damaged DNA, supports a monomeric form of XPA binding to ds-ssDNA junction. The same results were also obtained for XPA interaction with the 3′-overhang DNA (data not shown). As compared with the dimeric form of XPA needed for DNA damage recognition, these results support a new function for the protein. The XPA interaction with ds-ssDNA junctions was further supported by our in vivo evidence in which significant amount of immunofluorescent XPA nuclear foci formed in human diploid fibroblasts BJ cells with the PCNA knockdown by siRNA (submitted elsewhere). By contrast, no XPA nuclear foci formed in the same cells transfected with GFP siRNA. PCNA is an essential factor required for protein binding of ds-ssDNA junction intermediates at replication forks and many other DNA metabolic processes.
Figure 6.
Gel filtration-scintillation analysis for determination of the stoichiometry of XPA binding to Y-shape DNA. XPA of 40 and 1000 nM was incubated with radiolabeled Y-shape and AAF-50bp DNA substrates, respectively, and analyzed by gel filtration chromatography on Superdex® 200 column. The eluted fractions were then subjected to scintillation counting. Radioactivity profiles of the XPA–DNA complexes versus retention volume show that XPA-Y-shape DNA complex was eluted at 13.2 mL (▼), as compared to XPA-AAF DNA complex at 12.2 mL (●) (1). Free DNA was eluted at ~22 mL (◦). Determination of apparent molecular weights of the XPA-DNA complexes based on a linear relationship between the molecular weights of protein markers and their retention times suggests that XPA may bind to the Y-shape DNA substrate as a monomer.
XPC-HR23B, not XPA or XPA-RPA, recognizes DNA bending
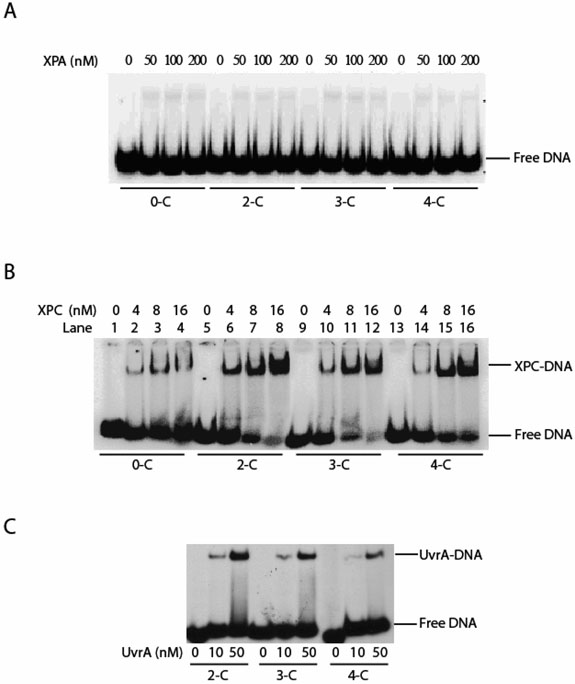
Although XPC or XPC-HR23B was previously reported to recognize DNA bending structures (35), the XPA protein also has been proposed as a recognition factor for DNA bending (16). In order to determine which protein/complex might be responsible for recognition of DNA bending induced by DNA damage, and the DNA structural determinants recognized by XPA, we constructed a set of DNA substrates (50-bp) containing different degree of DNA bending. These bendings were introduced through a simple two-, three-, or four-carbon tether crosslinking two adjacent guanines in the middle of the sequence (30) (Materials and Methods). The length of the tether in these substrates was indicated as 2-C, 3-C, and 4-C, etc. The bending angles of the DNA substrates were determined to be 30° for 2-C, 11.7° for 3-C, and 7.4° for 4-C (30). Thus, EMSA was employed to examine the binding of homogeneous XPA and XPC-HR23B to each of the bent DNA substrate. Neither XPA (Figure 7A) nor XPA-RPA (data not shown) showed any detectable affinity for these bent DNAs. By clear contrast, XPC-HR23B efficiently recognized DNA bending with the affinity in the following order: 2-C > 3-C > 4-C > 0-C (i.e., no tether) (Figure 7B), which correlated with the degrees of bending angle of these substrates. To further confirm the results, the same substrates were subjected to the binding with UvrA, the E. coli NER DNA damage recognition protein, which has been known to recognize DNA bending. As shown in Figure 7C, UvrA did bind to the substrates with the affinity increasing with larger bending angles. These results strongly suggest that XPC-HR23B complex, rather than XPA, is the factor that recognizes DNA bending, further supporting the XPA recognition of other types of DNA structures such as ds-ssDNA junctions.
Figure 7.
Recognition of DNA bending by XPA or XPC-HR23B complex. Pane A: XPA displayed no significant affinity for any of the substrates with DNA bending induced by a two carbon (2-C), three carbon (3-C), or four carbon (4-C) tether crosslinking two adjacent guanines. The 0-C stands for unmodified DNA with no bending (no carbon tether). Pane B: XPC-HR23B complex (labeled as XPC) had high affinities to DNA bending with the following order: 2-C > 3-C > 4-C > 0-C, which is consistent with the order of their bending-angles: 2-C > 3-C > 4-C > 0-C. Panel C: Binding of E. coli UvrA to the same DNA bending substrates (27-29).
DISCUSSION
DNA damage recognition in human NER utilizes cooperative binding and kinetic proofreading to provide a physiologically relevant mechanism by which DNA damage is efficiently discriminated and identified from a vast excess of undamaged DNA (4, 36). In comparison with XPC-HR23B, XPA appears to play a supporting role in DNA damage recognition. This appears to be inconsistent with the physiological significance of XPA in NER as XPA deficient cells are extremely sensitive to UV and also much more sensitive than the cells deficient in XPC, the major damage recognition protein in NER. Although the functions of XPA in NER have been extensively studied, the details of its role, particularly in TCR where XPC-HR23B is not required, remain unclear. All these imply that XPA may have other additional functions in NER. In the present study, we report that in addition to its involvement in DNA damage recognition, XPA demonstrated a novel biochemical activity in binding the biologically important DNA structures: the undamaged ds-ssDNA junctions with ssDNA branches, including the ssDNA-branched Y-structure junction and the junctions with either a 3′- or 5′-ssDNA overhang. While XPA was previously shown to be able to efficiently bind double-stranded three-way or four-way (Holliday junction-type) DNA structures, no affinity of XPA for the Y-shape ds-ssDNA junctions with ssDNA overhangs was observed (16, 17). The discrepancy is likely due to the use of different experimental conditions for the protein-DNA binding and the XPA proteins purified from different systems. In the present study, the human XPA was expressed in and purified to homogeneity from insect cells instead of E. coli. In addition, the three- and four-way dsDNA structures were used to mimic the helical kinks induced by DNA damage which were believed to be recognized by XPA, although no such structures formed as intermediates during NER (16, 17). By contrast, the undamaged ds-ssDNA junctions with ssDNA branches are the intermediate DNA structures formed during NER.
The binding of XPA to ds-ssDNA junction may mimic its interaction with the ds-ssDNA junctions formed during NER. Following initial DNA damage recognition, an opened DNA structure of ~25 nt around the lesion forms with TFIIH as a crucial intermediate in NER (37). In TCR, such DNA strand opening is present through the entire process of transcription. It is clear that ds-ssDNA junction is a major structural characteristic of the intermediate. Therefore, we wished to determine how the size of the DNA opening is defined and how an opened DNA structure stabilized in NER. We were also interested in elucidating how the nucleases are recruited to the right sites for incisions and the role of XPA in TCR where RNA polymerase complex is believed to carry out the initial DNA damage recognition. Our finding of XPA binding to ds-ssDNA junctions shed some light on these issues. The ability of XPA to recognize the ds-ssDNA junctions with such a high affinity (Kd = ~15 nM) suggests that the XPA-DNA junction binding may play a role in stabilizing the DNA opening following DNA unwinding at the lesion site by TFIIH. XPA may also stabilize the TFIIH-DNA interaction, and, thus, define the size of the strand opening. This is supported by the previous reports that the recruitment of TFIIH complex to the repair site was promoted by the presence of XPA (19, 21). On the other hand, XPA has been suggested to play a direct role in recruiting the ERCC1-XPF nuclease to the repair complex for 5′-incision of damaged DNA (18, 22) at the sites near the ds-ssDNA junction (37). It has been suggested that this recruitment was carried out by the XPA binding to the DNA lesion. The much higher affinity of XPA for the junctions relative to the DNA damage raises the question if DNA junction binding might be the major function of XPA in NER. This activity also provides an explanation for XPA's role in both GGR and TCR and the observation that XPA remains on the repair complex through the late stage of NER (12, 38).
Different from the dimeric form of XPA in DNA damage recognition, this XPA-ds-ssDNA junction binding activity appears to be exclusively delivered by XPA monomer, supporting that XPA may have at least two different DNA recognition activities: one is for damaged DNA while the other is for undamaged DNA. Interestingly, ds-ssDNA junctions are a common DNA intermediate structure formed in many DNA metabolic processes including DNA repair, replication and recombination, implying potential additional role of XPA in cellular processes. In support, a recent report indicated that ATR checkpoint signaling is compromised in UV-irradiated XPA deficient human cells during S-phase (39). Since activation of ATR checkpoint is replication dependent and a large number of ds-ssDNA junction intermediates form at replication forks, our finding provides some insights into the possible mechanism of the ATR signaling-XPA relationship.
It is well-established that XPA can form complexes with RPA. Since RPA is a ssDNA binding protein, the ssDNA regions next to a ds-ssDNA junction offers a biological platform for RPA to interact with XPA bound to the junction. Such protein-protein interaction may help stabilize, in a cooperative manner, the opened DNA structure of the repair complex and intermediates necessary for DNA damage incisions.
With respect to the role of XPA in DNA damage recognition as compared with that of XPC-HR23B complex, we also examined the interaction of these proteins with bent DNA, which is often induced by bulky DNA lesions. We synthesized a set of defined DNA bending substrates with defined bending angles. Our results indicated that XPA had little affinity towards DNA bending up to 30°. By contrast, XPC-HR23B complex efficiently recognized this DNA distortion. This is consistent with the role of XPC-HR23B in the initial DNA damage recognition in GGR. The results also are consistent with the fact that in TCR which requires XPA but not XPC-HR23B, no DNA bending could form at the lesion due to its location within a large DNA strand opening. Despite these, our results could not rule out the possibility of XPA binding to DNA bending with greater bending angles, such as those induced by cisplatin-GG intrastrand crosslink (50-60°) (40) and by (6-4) photoproducts (44°) (41). Nevertheless, these lesions concurrently introduce a large degree of DNA unwinding and twist, some of the major DNA helical distortions that are also induced by several DNA mono-adducts and recognized by DNA repair proteins. Taken together, our results indicate that XPC-HR23B complex has a much higher preference over XPA for recognition of DNA bending.
ACKNOWLEDGMENTS
We wish to thank Drs. Douglas P. Thewke and David Johnson for their technical help on the initial propagation of the recombinant viruses and optimizing protein-DNA interaction conditions.
This study was supported by NCI grant CA86927 (to Y.Z.) and NIEHS Grants ES09127 and ES013324 (to A.K.B.)
ABBREVIATIONS
- NER
Nucleotide excision repair
- XPA
Xeroderma Pigmentosum Complementation group A
- RPA
Replication protein A
- XPC
Xeroderma Pigmentosum Complementation group C
- IPTG
Isopropylthio-β-D-galactoside
- DTT
dithiothreitol
- EDTA
ethylenediaminetetraacetic acid
- AF
2-aminofluorene
- AAF
N-acetyl-2-aminofluorene
- AP
1-amionopyrene
- GT
G[8,5-ME]T tandem lesion
- T[6,4]T
TT crosslink 6, 4-photoproducts
- BD
Bending DNA substrates
- GGR
Global genome repair
- TCR
Transcription-coupled repair
- ds-ssDNA
double-strand/single-strand junctions of DNA
REFERENCES
- 1.Liu Y, Liu Y, Yang Z, Utzat C, Wang G, Basu AK, Zou Y. Cooperative interaction of human XPA stabilizes and enhances specific binding of XPA to DNA damage. Biochemistry. 2005;44:7361–7368. doi: 10.1021/bi047598y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wood RD. DNA damage recognition during nucleotide excision repair in mammalian cells. Biochimie. 1999;81:39–44. doi: 10.1016/s0300-9084(99)80036-4. [DOI] [PubMed] [Google Scholar]
- 3.Thoma BS, Vasquez KM. Critical DNA damage recognition functions of XPC-hHR23B and XPA-RPA in nucleotide excision repair. Mol Carcinog. 2003;38:1–13. doi: 10.1002/mc.10143. [DOI] [PubMed] [Google Scholar]
- 4.Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 5.Reardon JT, Sancar A. Nucleotide excision repair. Prog Nucleic Acid Res Mol Biol. 2005;79:183–235. doi: 10.1016/S0079-6603(04)79004-2. [DOI] [PubMed] [Google Scholar]
- 6.Yang ZG, Liu Y, Mao LY, Zhang JT, Zou Y. Dimerization of human XPA and formation of XPA2-RPA protein complex. Biochemistry. 2002;41:13012–13020. doi: 10.1021/bi026064z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sugasawa K, Ng JM, Masutani C, Iwai S, van der Spek PJ, Eker AP, Hanaoka F, Bootsma D, Hoeijmakers JH. Xeroderma pigmentosum group C protein complex is the initiator of global genome nucleotide excision repair. Mol Cell. 1998;2:223–232. doi: 10.1016/s1097-2765(00)80132-x. [DOI] [PubMed] [Google Scholar]
- 8.Wang M, Mahrenholz A, Lee SH. RPA stabilizes the XPAdamaged DNA complex through protein-protein interaction. Biochemistry. 2000;39:6433–6439. doi: 10.1021/bi000472q. [DOI] [PubMed] [Google Scholar]
- 9.Hermanson-Miller IL, Turchi JJ. Strand-specific binding of RPA and XPA to damaged duplex DNA. Biochemistry. 2002;41:2402–2408. doi: 10.1021/bi0112863. [DOI] [PubMed] [Google Scholar]
- 10.Patrick SM, Turchi JJ. Xeroderma pigmentosum complementation group A protein (XPA) modulates RPA-DNA interactions via enhanced complex stability and inhibition of strand separation activity. J Biol Chem. 2002;277:16096–16101. doi: 10.1074/jbc.M200816200. [DOI] [PubMed] [Google Scholar]
- 11.Rademakers S, Volker M, Hoogstraten D, Nigg AL, Mone MJ, Van Zeeland AA, Hoeijmakers JH, Houtsmuller AB, Vermeulen W. Xeroderma pigmentosum group A protein loads as a separate factor onto DNA lesions. Mol Cell Biol. 2003;23:5755–5767. doi: 10.1128/MCB.23.16.5755-5767.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Riedl T, Hanaoka F, Egly JM. The comings and goings of nucleotide excision repair factors on damaged DNA. Embo J. 2003;22:5293–5303. doi: 10.1093/emboj/cdg489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hey T, Lipps G, Krauss G. Binding of XPA and RPA to damaged DNA investigated by fluorescence anisotropy. Biochemistry. 2001;40:2901–2910. doi: 10.1021/bi002166i. [DOI] [PubMed] [Google Scholar]
- 14.Thoma BS, Wakasugi M, Christensen J, Reddy MC, Vasquez KM. Human XPC-hHR23B interacts with XPA-RPA in the recognition of triplex-directed psoralen DNA interstrand crosslinks. Nucleic Acids Res. 2005;33:2993–3001. doi: 10.1093/nar/gki610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koberle B, Roginskaya V, Wood RD. XPA protein as a limiting factor for nucleotide excision repair and UV sensitivity in human cells. DNA Repair (Amst) 2006. [DOI] [PubMed]
- 16.Missura M, Buterin T, Hindges R, Hubscher U, Kasparkova J, Brabec V, Naegeli H. Double-check probing of DNA bending and unwinding by XPA-RPA: an architectural function in DNA repair. Embo J. 2001;20:3554–3564. doi: 10.1093/emboj/20.13.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Camenisch U, Dip R, Schumacher SB, Schuler B, Naegeli H. Recognition of helical kinks by xeroderma pigmentosum group A protein triggers DNA excision repair. Nat Struct Mol Biol. 2006. [DOI] [PubMed]
- 18.Park CH, Sancar A. Formation of a ternary complex by human XPA, ERCC1, and ERCC4(XPF) excision repair proteins. Proc Natl Acad Sci U S A. 1994;91:5017–5021. doi: 10.1073/pnas.91.11.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park CH, Mu D, Reardon JT, Sancar A. The general transcription-repair factor TFIIH is recruited to the excision repair complex by the XPA protein independent of the TFIIE transcription factor. J Biol Chem. 1995;270:4896–4902. doi: 10.1074/jbc.270.9.4896. [DOI] [PubMed] [Google Scholar]
- 20.Guzder SN, Sung P, Prakash L, Prakash S. Nucleotide excision repair in yeast is mediated by sequential assembly of repair factors and not by a pre-assembled repairosome. J Biol Chem. 1996;271:8903–8910. doi: 10.1074/jbc.271.15.8903. [DOI] [PubMed] [Google Scholar]
- 21.Nocentini S, Coin F, Saijo M, Tanaka K, Egly JM. DNA damage recognition by XPA protein promotes efficient recruitment of transcription factor II H. J Biol Chem. 1997;272:22991–22994. doi: 10.1074/jbc.272.37.22991. [DOI] [PubMed] [Google Scholar]
- 22.Guzder SN, Sommers CH, Prakash L, Prakash S. Complex formation with damage recognition protein Rad14 is essential for Saccharomyces cerevisiae Rad1-Rad10 nuclease to perform its function in nucleotide excision repair in vivo. Mol Cell Biol. 2006;26:1135–1141. doi: 10.1128/MCB.26.3.1135-1141.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Y, Yang Z, Utzat CD, Liu Y, Geacintov NE, Basu AK, Zou Y. Interactions of human replication protein A with single-stranded DNA adducts. Biochem J. 2005;385:519–526. doi: 10.1042/BJ20041151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reardon JT, Mu D, Sancar A. Overproduction, purification, and characterization of the XPC subunit of the human DNA repair excision nuclease. J Biol Chem. 1996;271:19451–19456. doi: 10.1074/jbc.271.32.19451. [DOI] [PubMed] [Google Scholar]
- 25.Manley JL, Fire A, Cano A, Sharp PA, Gefter ML. DNAdependent transcription of adenovirus genes in a soluble whole-cell extract. Proc Natl Acad Sci U S A. 1980;77:3855–3859. doi: 10.1073/pnas.77.7.3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Manley JL, Fire A, Samuels M, Sharp PA. In vitro transcription: whole-cell extract. Methods Enzymol. 1983;101:568–582. doi: 10.1016/0076-6879(83)01038-1. [DOI] [PubMed] [Google Scholar]
- 27.Zou Y, Luo C, Geacintov NE. Hierarchy of DNA damage recognition in Escherichia coli nucleotide excision repair. Biochemistry. 2001;40:2923–2931. doi: 10.1021/bi001504c. [DOI] [PubMed] [Google Scholar]
- 28.Zou Y, Shell SM, Utzat CD, Luo C, Yang Z, Geacintov NE, Basu AK. Effects of DNA adduct structure and sequence context on strand opening of repair intermediates and incision by UvrABC nuclease. Biochemistry. 2003;42:12654–12661. doi: 10.1021/bi034446e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang Z, Colis LC, Basu AK, Zou Y. Recognition and incision of gamma-radiation-induced cross-linked guanine-thymine tandem lesion G[8,5- Me]T by UvrABC nuclease. Chem Res Toxicol. 2005;18:1339–1346. doi: 10.1021/tx050147+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kowalczyk A, Carmical JR, Zou Y, Van Houten B, Lloyd RS, Harris CM, Harris TM. Intrastrand DNA cross-links as tools for studying DNA replication and repair: two-, three-, and four-carbon tethers between the N(2) positions of adjacent guanines. Biochemistry. 2002;41:3109–3118. doi: 10.1021/bi010450j. [DOI] [PubMed] [Google Scholar]
- 31.Liu Y, Kvaratskhelia M, Hess S, Qu Y, Zou Y. Modulation of Replication Protein A Function by Its Hyperphosphorylation-induced Conformational Change Involving DNA Binding Domain B. J Biol Chem. 2005;280:32775–32783. doi: 10.1074/jbc.M505705200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zou Y, Liu TM, Geacintov NE, Van Houten B. Interaction of the UvrABC nuclease system with a DNA duplex containing a single stereoisomer of dG-(+)- or dG-(−)-anti-BPDE. Biochemistry. 1995;34:13582–13593. doi: 10.1021/bi00041a038. [DOI] [PubMed] [Google Scholar]
- 33.Zou Y, Van Houten B. Strand opening by the UvrA(2)B complex allows dynamic recognition of DNA damage. Embo J. 1999;18:4889–4901. doi: 10.1093/emboj/18.17.4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luo C, Krishnasamy R, Basu AK, Zou Y. Recognition and incision of site-specifically modified C8 guanine adducts formed by 2- aminofluorene, N-acetyl-2-aminofluorene and 1-nitropyrene by UvrABC nuclease. Nucleic Acids Res. 2000;28:3719–3724. doi: 10.1093/nar/28.19.3719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Janicijevic A, Sugasawa K, Shimizu Y, Hanaoka F, Wijgers N, Djurica M, Hoeijmakers JH, Wyman C. DNA bending by the human damage recognition complex XPC-HR23B. DNA Repair (Amst) 2003;2:325–336. doi: 10.1016/s1568-7864(02)00222-7. [DOI] [PubMed] [Google Scholar]
- 36.Reardon JT, Sancar A. Thermodynamic cooperativity and kinetic proofreading in DNA damage recognition and repair. Cell Cycle. 2004;3:141–144. [PubMed] [Google Scholar]
- 37.Evans E, Moggs JG, Hwang JR, Egly JM, Wood RD. Mechanism of open complex and dual incision formation by human nucleotide excision repair factors. Embo J. 1997;16:6559–6573. doi: 10.1093/emboj/16.21.6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tapias A, Auriol J, Forget D, Enzlin JH, Scharer OD, Coin F, Coulombe B, Egly JM. Ordered conformational changes in damaged DNA induced by nucleotide excision repair factors. J Biol Chem. 2004;279:19074–19083. doi: 10.1074/jbc.M312611200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bomgarden RD, Lupardus PJ, Soni DV, Yee MC, Ford JM, Cimprich KA. Opposing effects of the UV lesion repair protein XPA and UV bypass polymerase eta on ATR checkpoint signaling. Embo J. 2006;25:2605–2614. doi: 10.1038/sj.emboj.7601123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kozelka J, Petsko GA, G J, Q., Lippard SJ. High-salt and low-salt models for kinked adducts of cis-diamminedichloroplatinum(II) with oligonucleotide duplexes. Inorg. Chem. 1986;25:1075–1077. [Google Scholar]
- 41.Kim JK, Patel D, Choi BS. Contrasting structural impacts induced by cis-syn cyclobutane dimer and (6-4) adduct in DNA duplex decamers: implication in mutagenesis and repair activity. Photochem Photobiol. 1995;62:44–50. doi: 10.1111/j.1751-1097.1995.tb05236.x. [DOI] [PubMed] [Google Scholar]