Abstract
C4 plants have up to 10-fold higher apparent CO2 assimilation rates than the most productive C3 plants. This requires higher fluxes of metabolic intermediates across the chloroplast envelope membranes of C4 plants in comparison with those of C3 plants. In particular, the fluxes of metabolites involved in the biochemical inorganic carbon pump of C4 plants, such as malate, pyruvate, oxaloacetate, and phosphoenolpyruvate, must be considerably higher in C4 plants because they exceed the apparent rate of photosynthetic CO2 assimilation, whereas they represent relatively minor fluxes in C3 plants. While the enzymatic steps involved in the C4 biochemical inorganic carbon pump have been studied in much detail, little is known about the metabolite transporters in the envelope membranes of C4 chloroplasts. In this study, we used comparative proteomics of chloroplast envelope membranes from the C3 plant pea (Pisum sativum) and mesophyll cell chloroplast envelopes from the C4 plant maize (Zea mays) to analyze the adaptation of the mesophyll cell chloroplast envelope proteome to the requirements of C4 photosynthesis. We show that C3- and C4-type chloroplasts have qualitatively similar but quantitatively very different chloroplast envelope membrane proteomes. In particular, translocators involved in the transport of triosephosphate and phosphoenolpyruvate as well as two outer envelope porins are much more abundant in C4 plants. Several putative transport proteins have been identified that are highly abundant in C4 plants but relatively minor in C3 envelopes. These represent prime candidates for the transport of C4 photosynthetic intermediates, such as pyruvate, oxaloacetate, and malate.
C4 photosynthesis allows fast biomass accumulation with high nitrogen and water use efficiency (Leegood and Edwards, 1996; Sage, 2004) and is a desired trait to increase the productivity of crop plants (Matsuoka et al., 1998). To facilitate C4 photosynthesis in maize (Zea mays), a C4 plant of the NADP-malic enzyme type, the primary fixation and the reduction of carbon are spatially separated between two different cell types. Primary carbon fixation occurs in the mesophyll cells (Hatch, 1987). The mesophyll surrounds the bundle sheath cells, where CO2 is enriched around Rubisco and the reduction of carbon takes place. The chloroplasts of mesophyll and bundle sheath tissues are adapted to their respective roles (Slack et al., 1969; Edwards et al., 2001; Majeran et al., 2005). In addition to carbon fixation and reduction, several other pathways, such as nitrogen reduction and assimilation, are partitioned between mesophyll and bundle sheath chloroplasts (Renné et al., 2003; Majeran et al., 2005), and the adaptation of the soluble chloroplast proteome to C4 photosynthesis has been studied in considerable detail (Majeran et al., 2005). In maize, initial carbon assimilation in the mesophyll cell cytoplasm is accomplished by phosphoenolpyruvate carboxylase (PEPC), yielding oxaloacetate. Oxaloacetate is then imported into the chloroplasts, where it is reduced to malate, and subsequently exported to the cytosol again. After diffusion into bundle sheath cells, malate is decarboxylated in the chloroplasts, yielding CO2, NADPH, and pyruvate. While CO2 and NADPH enter the Calvin cycle in bundle sheath cells, pyruvate is returned to the mesophyll, where it is imported into the chloroplasts and converted to phosphoenolpyruvate (PEP) by phosphoenolpyruvate phosphate dikinase (PPDK), thus regenerating the primary CO2 acceptor, which is exported to the cytosol to enter a new round of CO2 assimilation (Fig. 1). In maize, carbon fixation is optimized beyond simply concentrating CO2 in the vicinity of Rubisco. The bundle sheath chloroplasts have limited PSII activity (Meierhoff and Westhoff, 1993) and produce less O2, which further reduces the oxygenation reaction of Rubisco. However, the absence of PSII activity prevents the operation of linear electron transport, limiting the production of reduction equivalents in the bundle sheath. Since CO2 assimilation in the Calvin cycle requires NADPH, this necessitates the shuttling of reduction equivalents between mesophyll and the bundle sheath by a 3-phosphoglycerate/triosephosphate shuttle (Fig. 1). Despite detailed knowledge about the soluble proteins involved in and necessary for C4 photosynthesis, the adaptation of integral and associated membrane proteins remains largely unknown. In this work, we focus on analyzing the quantitative and qualitative differences between chloroplast envelope membranes of C3 and C4 plants.
Figure 1.
Schematic representation of central carbon metabolism and associated transport processes in C3 chloroplasts and C4 PCA-type chloroplasts. In C3 chloroplasts, for three carbons fixed, at most one transport process is required; in C4 PCA-type chloroplasts, for three carbons fixed, at least 12 transport processes are required.
The plastids of green plants are separated from the cytosol by two membranes. Metabolite transport across the outer envelope is controlled by substrate-specific pore-forming proteins (Pohlmeyer et al., 1997, 1998; Bolter et al., 1999; Goetze et al., 2006). Solute transport across the inner envelope membrane is catalyzed by a large range of specific metabolite transporters (Weber, 2004; Weber et al., 2005; Weber and Fischer, 2007), some of which are capable of transporting metabolites against a concentration gradient. The spatial separation of initial carbon fixation and subsequent reduction in C4 plants requires a very high metabolite flow across the chloroplast envelope of both mesophyll and bundle sheath chloroplasts that exceeds the apparent rate of carbon assimilation (Laisk and Edwards, 2000). Pea (Pisum sativum) fixes about 17 μmol of carbon per square meter of leaf area per second (Grodzinski et al., 1998) and requires at most one transport process for three carbons fixed (i.e. the export of one molecule of triosephosphate from chloroplasts). Maize fixes about 27 μmol of carbon per square meter of leaf area per second (Grodzinski et al., 1998) and requires at least four transport processes (Fig. 1) for each carbon fixed. Consequently, the total metabolite transport rate across the chloroplast envelopes in C4 plants exceeds that in C3 plants by a factor of at least 18. High-velocity transport of all four metabolites involved in core C4 photosynthesis across the mesophyll chloroplast envelope has been demonstrated using isolated chloroplasts (Huber and Edwards, 1977a, 1997b; Hatch et al., 1984; Flügge et al., 1985; Aoki et al., 1992). While most transport proteins involved in core C4 photosynthesis in mesophyll chloroplasts have not yet been unequivocally identified at the molecular level, good candidates exist for PEP export, triosephosphate shuttling, and oxaloacetate/malate transport. The molecular nature of the pyruvate transporter, however, is unknown. Likewise, it is unknown whether the same or different transport proteins mediate pyruvate transport across the mesophyll and the bundle sheath chloroplast envelope. Adaptations of additional membrane proteins as a consequence of the spatial separation of photosynthesis, similar to what has been demonstrated for soluble proteins, are unknown. Since increasing the capacity for metabolite transport across the chloroplast envelope membrane is likely a key adaptation to C4 photosynthesis (Edwards et al., 2001), engineering efforts to introduce C4 photosynthesis in a C3 crop plant will likely critically depend on engineering not only the C4 pathway but also metabolite flux.
In this work, the protein complements of envelope membranes of C3 chloroplasts and C4 mesophyll chloroplasts are analyzed qualitatively and semiquantitatively. We hypothesized that analyzing chloroplasts with different modes of photosynthesis, such as the C3 and C4 types of carbon dioxide assimilation, will reveal the adaptations of the chloroplast envelope proteome to increased metabolite flow. Unfortunately, routine methods are not available to compare membrane proteins of different species quantitatively or even semiquantitatively. Membrane proteins are not amenable to two-dimensional gel electrophoresis, since extremely hydrophobic proteins, such as metabolite transporters, do not focus in the first dimension (Choe et al., 2005), and quantitative comparison relying on identical peptides, such as affinity tagging, are also not applicable, since there is considerable evolutionary distance between the C3 model pea and the C4 model maize. A direct quantification method, the total spectral count of proteins (the number of mass spectra that map to one protein), has been used to compare and even quantify proteins on a large scale (Liu et al., 2004; Lu et al., 2007; Majeran et al., 2008). This method was recently also applied to yeast membrane proteins, with results comparable to SILAC (Zybailov et al., 2005). We applied this strategy to compare the relative abundance of proteins in the chloroplast envelopes of C3 and C4 plants. We demonstrate that the massive metabolite fluxes across the chloroplast envelope required for maintaining the high photosynthetic rates of C4 plants are associated with significant increases in the relative abundance of several metabolite transporters, thus pinpointing apparent bottlenecks in metabolite flux across the chloroplast envelope membrane.
RESULTS
Envelope Proteome Coverage and Purity
The pea chloroplast envelope proteome was chosen to represent the envelope proteome of a C3 chloroplast. The proteins of the protein import complex found in our study were compared with those identified in earlier efforts (Froehlich et al., 2001, 2003; Ferro et al., 2003). We were able to identify all import complex components found by Froehlich et al. (2003) with the exception of Toc33, and we identified three additional import complex components that were not previously found. In comparison with Ferro et al. (2003), we also identified two additional components, while Toc33 was again missing. With regard to a major metabolite transport protein family, the phosphate translocators (Knappe et al., 2003a), we identified all members predicted to be present in the inner envelope of C3 chloroplasts (Eicks et al., 2002; Flügge et al., 2003). Based on these data and further analysis (data not shown), we conclude that the proteome of the pea chloroplast envelope has a similar qualitative composition as the proteomes from other C3 plants analyzed previously.
The maize chloroplast isolation protocol applied in this study was optimized for the isolation of maize mesophyll chloroplasts and thus C4 mesophyll chloroplast envelope membranes. Based on the virtual absence of Rubisco and the complete absence of malic enzyme, two markers for bundle sheath chloroplasts, and the relative abundance of mesophyll marker enzymes, such as PPDK and PEPC, the maize chloroplast envelope samples indeed represent a highly mesophyll-enriched preparation (Supplemental Table S1).
For each of the envelope proteome samples, the relative spectral abundance, likely resulting from extraplastidial sources such as mitochondria, the endomembrane system, cytosol, and nucleus, was determined. The level of contamination based on this measure was low; for the samples from maize, it was below 2.2%, and for the samples from pea, it was below 5.2%. In maize, no mitochondrial contamination was detected, and extraplastidial proteins were mostly residents of the cytosol and the endomembrane system. In pea, the main contaminant was mitochondrial proteins. The complete list of extraplastidial proteins can be extracted from Supplemental Tables S1 and S2. Relative abundance comparisons were performed with and without removing the contaminations from the samples, and the results were robust and therefore independent of the level and source of the contamination. We concluded that the samples are suitable for comparing a C3 with a C4 mesophyll chloroplast envelope.
The Envelope Proteomes of C3 and C4 Chloroplasts Are Qualitatively Similar
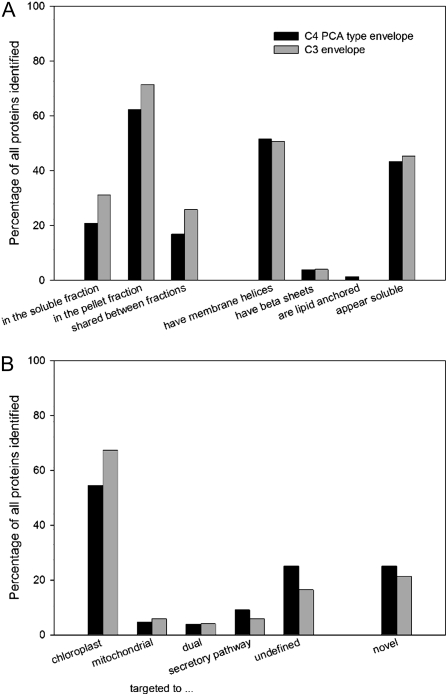
In the C4 mesophyll chloroplast envelope proteome, 231 nonredundant proteins were identified, and in the pea chloroplast envelope proteome, 322 nonredundant proteins were identified. Taken together, 420 unique proteins were identified, of which 368 (87.6%) were traditional chloroplast residents. In both samples, a similar percentage of proteins was soluble or insoluble in chloroform or methanol, respectively, with about one-third of all proteins being soluble in organic solvents to at least some degree (Fig. 2A). Likewise, a similar share of proteins could be detected in both fractions (Fig. 2A). The soluble fraction in organic solvents contained a number of proteins with high membrane helix content, but neither the hydrophobicity index nor the number of predicted membrane helices was strongly correlated with the solubility in organic solvent (data not shown).
Figure 2.
The envelope proteomes are similar when analyzed qualitatively. The percentage of proteins within one proteome is plotted. They are similar with regard to their physicochemical properties (solubility in organic solvents and the presence of membrane attachment structures; A) and with regard to their predicted targeting and percentage of novel proteins (B).
Little more than half of the proteins in both samples contain recognizable structures for membrane attachment (Fig. 2A). Most of these proteins have predicted α-helices that can span a membrane, some have demonstrated or predicted β-sheets, and very few are predicted to be anchored to the lipid bilayer by prenylation. The other half of the proteins have no obvious domains for membrane attachment or insertion.
The proteins in both envelope preparations are also very similar when their bioinformatically generated targeting predictions are compared. Most of the proteins identified in both the C4 mesophyll and the C3 envelope proteome samples possess a canonical target peptide for the protein import complex of chloroplasts (Fig. 2B; Emanuelsson et al., 1999, 2000; Schwacke et al., 2003). Less than 10% are predicted to be targeted to the mitochondria (Fig. 2B). For a surprisingly large group, no targeting signal can be identified within the N terminus of the protein sequence, and a number of proteins have strong bioinformatics support for targeting to the secretory pathway (Fig. 2B). These proteins include well-known residents of the chloroplast envelope, such as Toc64 and Toc159, as well as two outer envelope porins, OEP21 and OEP24.
Both envelope proteome samples yielded a comparable proportion of proteins not previously identified by proteomics, with 58 novel proteins from the C4 maize mesophyll envelope proteome sample and 69 novel proteins from the C3 pea envelope proteome sample.
Proteins with Similar Relative Abundance between Samples
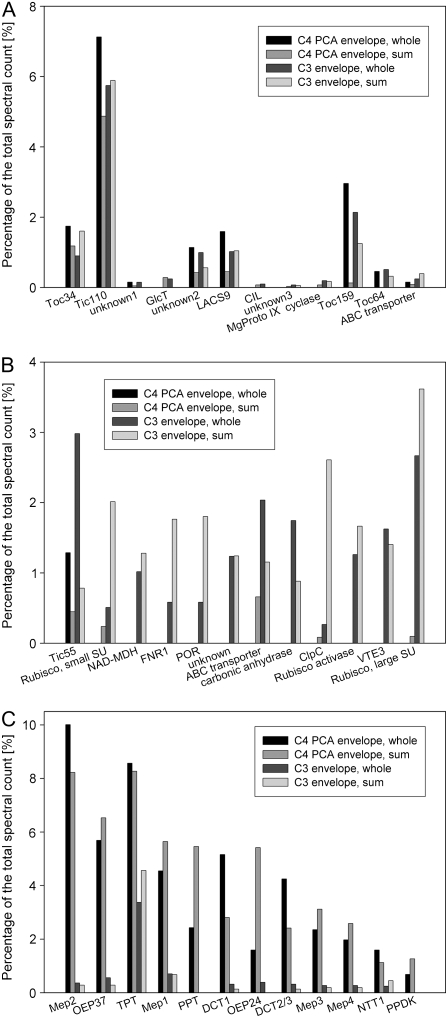
To visualize the compositional differences between C3 and C4 envelope membrane proteomes, the differences between the percentages of the total spectral count for each protein (the spectral count percentage in pea was subtracted from the spectral count percentage in maize) were plotted. Proteins that were identified in only a subset of the experiments were set to zero in the remaining experiments. Plotting the difference between the relative spectral abundance in C4 mesophyll and C3 chloroplast envelopes revealed that the majority of proteins do not differ by more than 0.5% in their relative spectral abundance (Fig. 3). This large group of proteins can be broken down into smaller groups of proteins; selected examples are shown in Figure 4A. In the first group, both relative spectral counts are high or intermediate, as is the case for proteins of the protein import complex components (Fig. 4A). As examples, part of the inner envelope pore, Tic110, and two outer envelope components, Toc34 and Toc64, are shown (Fig. 4A). A long-acyl-chain CoA synthase and a protein of unknown function also belong to this group of proteins, with high relative spectral abundance in all samples. A fourth import complex component, Toc159, inexplicably was reduced in one of two replicate experiments for C4 photosynthetic carbon assimilation (PCA)-type envelopes (Fig. 4A). Proteins that showed large variance between replicate experiments were considered unreliable and therefore were not considered further. The putative Glc transporter pGlcT, a putative ATP-dependent transporter, and an enzyme of chlorophyll biosynthesis are of intermediate relative abundance in both samples. There are also proteins that have a low absolute spectral count in one or both samples and therefore do not generate large differences, such as the transcription factor CIL or two proteins of unknown function (Fig. 4A; Supplemental Tables S1 and S2). A complete list of proteins with similar relative spectral abundance can be found in Supplemental Table S3.
Figure 3.
Quantitatively, the envelope proteomes differ in selected proteins. The relative abundance in C3-type envelopes was subtracted from the relative abundance in C4 PCA-type envelopes, and the difference was plotted for each protein. The dotted lines represent differences of ±0.5%.
Figure 4.
For the extremes and selected unchanged proteins from Figure 3, detailed results were plotted. A, Selected proteins that do not change significantly in relative abundance. B, The 12 proteins that are lowest in relative abundance in C4 PCA-type envelopes compared with C3-type envelopes. C, The 12 proteins that are highest in relative abundance in C4 PCA-type envelopes compared with C3-type envelopes.
Proteins with Different Relative Spectral Abundance in C4 Mesophyll Envelopes Compared with C3-Type Envelopes
Among the proteins with markedly decreased relative spectral abundance, only four of 12 contained membrane-spanning helices, whereas of the proteins with increased abundance, all but one were integral membrane proteins. Most of the proteins that were underrepresented in C4 mesophyll envelopes could not be detected at all in either of the replicate C4 experiments. The 12 proteins with the highest relative decreases are plotted in Figure 4B. There are four proteins involved in carbon fixation for the PCR cycle: Rubisco large and small subunits, the Rubisco activase, and a carbonic anhydrase, with Rubisco large subunit showing the highest relative decrease. In addition, there are three proteins associated with the protein import complex: Tic55, the ferredoxin:NADP reductase, and the import chaperone Hsp93/ClpC (Soll and Schleiff, 2004). There are also three enzymes: VTE3, a methyltransferase involved in vitamin E and plastoquinone biosynthesis (Cheng et al., 2003), protochlorophyllide reductase (Beale, 1999), and a NAD-dependent malate dehydrogenase. Finally, there are two proteins of unknown function, one of which is a putative ATP-dependent transporter.
The proteins that occupy a larger percentage of the spectral count in maize have high amplitudes of up to 9%, whereas proteins that occupy a larger percentage in the pea envelope have lower amplitudes of up to 3% (Fig. 3). Most of the following proteins that show major relative increases in maize belong to the classes of known and putative transport proteins, except for PPDK, the enzyme required for regenerating the CO2 acceptor PEP (Fig. 4C). The known transport proteins are two phosphate translocators, phosphoenolpyruvate phosphate translocator (PPT; Fischer et al., 1997) and triosephosphate phosphate translocator (TPT; Flügge and Heldt, 1984), two dicarboxylate translocators (DiTs), DCT1 and DCT2/3, and the ATP/ADP translocator NTT1 (Neuhaus et al., 1997). There are also two outer envelope proteins, OEP24 and OEP37 (Pohlmeyer et al., 1998; Goetze et al., 2006). Finally, we identified four proteins of unknown function in this group (mesophyll envelope proteins 1–4 [Mep1–Mep4]). Mep1 is predicted to have 12 membrane-spanning helices, and Mep2 is predicted to have a single membrane-spanning helix. Mep3 and Mep4 are paralogs, which map to the same Arabidopsis (Arabidopsis thaliana) ortholog, and both are predicted to have four membrane-spanning helices. Of the 12 proteins with the highest difference in spectral count compared with C3 envelopes, 10 have differential protein accumulation patterns between mesophyll and the bundle sheath (Majeran et al., 2008). Eight accumulate to higher levels in mesophyll membranes, and two accumulate to higher levels in bundle sheath membranes (Supplemental Table S4).
DISCUSSION
Pea was chosen to represent C3 plants because it has served as a model for C3 chloroplasts for a long time and high-purity chloroplast envelopes can be isolated with relative ease. Maize was chosen to represent C4 plants since most of the biochemical work on transport proteins has been published for maize chloroplasts compared with other C4 models (Huber and Edwards, 1977a, 1977b; Hatch et al., 1984; Ohnishi et al., 1990; Aoki et al., 1992). We established that the C3 envelope proteome from pea is comparable with earlier envelope proteomes prepared from the C3 plant Arabidopsis and confirmed the presence and absence, respectively, of marker proteins of C4 photosynthesis for C4 mesophyll envelope samples such as PPDK, PEPC, and Rubisco. The level of contamination was at most 5.2%. In the samples isolated from pea, the biggest contributors were mitochondrial outer envelope proteins such as porins. It is well known that, to foster metabolite exchange for photorespiration, chloroplasts of C3 plants are closely associated with mitochondria and peroxisomes (Schumann et al., 2007), thus explaining mitochondria being the major source of contaminating proteins. In the maize sample, mitochondrial contaminants were virtually absent. The contaminant with the highest relative spectral count was PEPC, a cytosolic enzyme that is required for initial CO2 fixation. Considering that the isolation protocols for pea and maize are almost identical, the marked difference in extraplastidial contaminants may result from the altered requirements in organelle association. Mesophyll chloroplasts do not photorespire, since Rubisco is virtually absent, and these chloroplasts therefore do not require a close association of chloroplasts, mitochondria, and peroxisomes, as is the case in C3 plants. Both envelope preparations also contain a number of proteins identified in previous thylakoid proteome studies (Supplemental Tables S1 and S2; Peltier et al., 2000, 2002, 2004). Currently, it remains unknown whether these proteins are trapped en route to the internal membrane system or whether they represent contaminations that are introduced during envelope isolation (Ferro et al., 2002; Froehlich et al., 2003). Relative abundance comparisons were performed with and without removing the contaminations from the samples, and the results were robust and therefore independent of the level and source of the contamination.
We analyzed whether detailed qualitative comparisons were possible. Solid judgments about the significance of the presence or absence of proteins require proteomics to be saturated to avoid false-negative calls. To determine whether the proteome identifications in either sample were saturated or whether a substantial number of proteins remained unidentified, the well-understood pathways of glycolipid biosynthesis were analyzed. They provide a number of housekeeping proteins that are expected to be identified in envelope proteomics studies if saturation was reached, such as two enzymes necessary for sulfolipid biosynthesis and two known enzymes and a three-partite transport protein involved in galactolipid biosynthesis (Benning et al., 2006). As in earlier efforts, the envelope proteomes in this study only identify a subset of proteins in each pathway, indicating that saturating coverage of the chloroplast envelope proteome remains to be achieved. It remains to be determined whether enzymes of glycolipid biosynthesis are difficult to detect with mass spectrometry or whether they are of too low absolute abundance. When additional replicates of the pea envelope proteome were tested, we also observed that enzymes with a low absolute spectral count disappeared and the enzyme catalyzing the next step appeared from replicate samples (data not shown). This may indicate that when proteins with a low number of spectra were analyzed, small variations during peptide separation, ionization, and detection may have determined whether or not they are present in any given sample. As a consequence, the envelope proteomes and not single proteins were the basis for the qualitative comparison.
We identified 231 and 322 nonredundant proteins in the C4 and C3 chloroplast envelopes, respectively. The higher number of proteins identified in the pea sample likely results from two reasons. (1) The total envelope sample from C4 mesophyll envelopes yielded a lower total spectral count, with fewer proteins identified (Supplemental Table S1), although the relative abundances for each protein remained similar (Fig. 4). Many proteins with a low absolute spectral count in the other experiments might have escaped detection. (2) The C4 mesophyll envelope sample contains some proteins with a very high relative spectral count compared with the C3 envelope sample, with up to 9% difference in relative abundance (Fig. 3). The peptides belonging to these proteins may have suppressed peptides of lesser abundance during ionization or detection in the mass spectrometer. The prefractionation by organic solvent extraction permitted the detection of additional proteins that could not be detected in a whole envelope preparation, as many of the proteins yielding high relative spectral counts fractionated into the organic solvent soluble fraction, thus removing the main source for ion suppression. Yet, total coverage did not reach the level obtained with C3 envelopes.
Analysis of the physicochemical properties revealed that the C4 mesophyll and the C3 envelope proteomes are remarkably similar. The fractionation pattern into soluble and insoluble in organic solvent was reproducible, as was the proportion of integral membrane proteins (Fig. 2A). Little more than half of the proteins in both samples contain recognizable structures for membrane attachment. In both envelope proteomes, this group of proteins included a number of proteins for which a close association with the membrane has been demonstrated, such as the membrane lipid-synthesizing and -modifying enzymes (Jarvis et al., 2000; Froehlich et al., 2001; Sanda et al., 2001; Yu et al., 2002). It cannot be excluded that the remaining seemingly soluble proteins also are closely associated with the chloroplast envelope, similar to what has been demonstrated for glycolytic enzymes at the mitochondrial membranes (Graham et al., 2007).
The proteins in both envelope preparations are also very similar when their bioinformatically generated targeting predictions are compared. About half of the proteins identified in both envelope proteome samples possess a chloroplast target peptide for the protein import complex (Fig. 2B; Emanuelsson et al., 1999, 2000; Schwacke et al., 2003), as was expected based on earlier results (Ferro et al., 2002; Froehlich et al., 2003). Some of the proteins that are predicted to possess a mitochondrial target peptide might be erroneously annotated as mitochondrial proteins by the prediction algorithm; a well-documented case in point is the most abundant metabolite transport protein on the C3 envelope, TPT, which is predicted to be targeted to the mitochondria. Alternatively, it may be due to contamination of the envelope preparation with true mitochondrial proteins. This, however, is unlikely, at least for the C4 mesophyll envelope sample, since no bona fide mitochondrial proteins could be identified in this preparation. For a surprisingly large group, no targeting signal can be identified within the N terminus of the protein sequence, and a number of proteins have strong bioinformatics support for targeting to the secretory pathway (Fig. 2B), including well-established plastid residents. Especially for proteins of the import complex, it has been established that not all of them require the canonical import machinery. Many of the proteins without classical chloroplast targeting peptides have been identified in multiple independent plastid proteomics studies (Supplemental Tables S1 and S2). The proteins identified in this and other studies might represent candidates for novel protein import pathways, as were recently reported for a carbonic anhydrase (Villarejo et al., 2005) and outer envelope proteins (Bae et al., 2008).
Both envelope proteome samples yielded a comparable proportion of proteins not previously identified in plastid proteome projects. Some of the novel identifications may be due to the instrumentation used in our study, since ultra-high-pressure HPLC coupled to Fourier-transform ion cyclotron resonance is capable of protein identification with very high resolution. Some proteins may have been identified because the sample was fractionated prior to proteome analysis, and some proteins, especially from the maize envelope sample, may have been identified because the chloroplast envelope is adapted to C4 photosynthesis and C4 chloroplast envelopes have not yet been analyzed by proteomics.
A Semiquantitative View of the Envelope Proteomes
For several reasons, a semiquantitative approach was needed to understand the differences between a C4 mesophyll and a C3-type chloroplast envelope. As pointed out earlier, qualitative analysis is hampered by unsaturated proteome identification; hence, some uncertainty is associated with the identification of proteins with low absolute spectral counts. The proteome sample from maize was compared with previous proteome samples, and more than 70% of the proteins identified in maize have been found previously in the plastid proteomes from other species (Fig. 2B), indicating that a large portion of the plastid envelope proteome is shared between different plastid species. Based on these results and on the adaptations of soluble proteins to C4 photosynthesis, we hypothesized that the differences between the C3 and C4 chloroplast envelopes are quantitative rather than qualitative. Unfortunately, no quantitative tools for comparing proteomes of different species are available. To overcome this limitation, we introduced percentage of the total spectral count as a measure for quantitative composition of the envelope proteome. This percentage is normalized to the total number of spectra identified within a single experiment, similar to the normalization procedures used for the interpretation of RNA hybridization experiments. This method enables comparisons between evolutionarily distant species. It is based on the assumption that orthologous proteins from different species have similar physicochemical properties and thus behave similarly throughout separation and identification when contained in similar samples, such as chloroplast envelopes. Although the percentage of total spectral counts is not an absolute measure of protein abundance, it is capable of capturing the relative contribution of a protein to the total, which enables comparison of nonrelated samples. The compositional differences between C3 and C4 envelope membrane proteomes were visualized by plotting the differences between the percentages of the total spectral count for each protein (the spectral count percentage in pea was subtracted from the spectral count percentage in maize). We chose to compare the difference in relative abundance over the fold change between the samples. Fold changes are likely a good measure if the proteins to be compared have high absolute spectral counts, which would allow a wide range of comparable values. In contrast, the envelope samples mainly consist of proteins of up to 10 absolute spectral counts each (Supplemental Tables S1 and S2), similar to results reported earlier (Bräutigam et al., 2008). A comparison based on fold changes would lead to many proteins of low absolute spectral count being erroneously identified as differentially expressed between C3 and C4 mesophyll envelopes and would fail to identify the protein with the second highest difference in relative spectral count, TPT (data not shown). We thus restricted analysis to the proteins with the highest relative changes in expression, which yielded comparable results in both experiments in each sample.
Marker Enzymes
A number of the proteins that were reduced or absent in the C4 sample are associated with functions that are expected to be absent from C4 mesophyll chloroplasts, such as the Rubisco large and small subunits, Rubisco activase, and carbonic anhydrase for photosynthetic carbon reduction (Fig. 4B). Since C4 mesophyll tissue has strongly reduced or absent Rubisco activity, the enzyme itself and its activase are also reduced. In mesophyll cells, a carbonic anhydrase, which quickly equilibrates CO2 and hydrogen carbonate, is needed in the cytosol for PEPC rather than in the chloroplast. The only soluble protein, which is massively increased in the C4 mesophyll envelope samples, is PPDK (Fig. 4C). The detection of this soluble enzyme, which occupies a large percentage of the spectral count within the C4 mesophyll envelope proteome sample, may result from its high abundance, due to its involvement in C4 photosynthesis and/or a close association with the membranes. It is likely absent from the pea sample because, in contrast to C4 plants, it represents a minor plastidic and cytosolic protein (Parsley and Hibberd, 2006) in C3 plants.
Proteins of the Protein Import Complex
At least two of the proteins that form the protein import complex seem to be housekeeping proteins, Tic110 and Toc75, which have a high relative spectral abundance in both samples. They form the pore in the inner and outer envelopes (Soll and Schleiff, 2004). The import receptor Toc159 was excluded from analysis because its relative abundance varied considerably between the biological replicates conducted on the C3 envelope. Two protein import complex components, Tic55 and ClpC/Hsp93, the import chaperone or protease subunit, were found among the proteins with a lower relative spectra abundance in C4 mesophyll envelopes. The remaining proteins, which are believed to be involved in the redox regulation of protein import, Tic32 and Tic62 (Kuchler et al., 2002), were identified in the C3 envelope sample and could not be identified from the C4 mesophyll envelope. Taken together with the results of import complex components, the two proteins involved in reduction equivalent synthesis and balancing, FNR (for ferredoxin:NADP + reductase) and a NAD-dependent malate dehydrogenase, may indicate a different mode of redox-dependent import in C3, as suggested by Kuchler et al. (2002), compared with C4 mesophyll envelopes. This difference may be explained by the spatial separation of reduction equivalent production between mesophyll and bundle sheath chloroplasts, which may result in a change of redox status regulation.
Transport Proteins
Phosphate Translocators
The transport protein TPT is one of the proteins with the highest relative spectral abundance in both envelope samples, but it is 2-fold more abundant in C4 mesophyll than in C3-type envelopes (Fig. 4C). TPT is the most abundant envelope transport protein in C3 chloroplast envelopes because it carries the major flux of carbon out of the C3 chloroplast during the day. In C4 mesophyll chloroplasts, the carbon fixation by Rubisco occurs in the bundle sheath plastids; therefore, carbon export cannot be the reason for the high relative abundance of TPT. However, since the bundle sheath chloroplasts are deficient in reduction equivalents due to limited PSII activity (Meierhoff and Westhoff, 1993), the reduction of 3-phosphoglycerate to triosephosphate occurs in mesophyll chloroplasts (Majeran et al., 2005). Since one exchange of the reduced for the oxidized form is necessary for each carbon fixed by Rubisco, the flux through the C4 TPT is at least 3-fold compared with the C3 TPT, in which only one exchange is required for three fixed carbon units for export (Fig. 1). Compared with the C3 TPT, the C4 TPT exchanges 3-phosphoglycerate rather than phosphate for triosephosphate (Fig. 1) and thus may be specifically adapted to its new role. Interestingly, this protein is reported to be more abundant in C4 mesophyll chloroplast membranes compared with bundle sheath chloroplast membranes (Majeran et al., 2008), although TPT also has to export carbon from bundle sheath chloroplasts. In C4 PCA-type chloroplasts, PEP has to be exported from the chloroplast with a rate slightly exceeding the rate of carbon fixation (Laisk and Edwards, 2000). In C3 chloroplasts, PEP transport is a minor flux and the PPT was initially identified in maize endosperm and characterized from cauliflower (Brassica oleracea) buds (Fischer et al., 1997). This maize PPT is highly expressed in roots and the female flower, but its expression level is not increased upon transition from etiolated tissue to green leaves (Supplemental Fig. S5). Orthologs of this PPT are expressed in leaf tissue of the C3 plant Arabidopsis (Knappe et al., 2003b; Voll et al., 2003), albeit at low levels. The maize mesophyll chloroplast envelope samples contain an isoform of PPT that is among the three most abundant proteins in this sample (Fig. 4C; Supplemental Table S1), while in pea, PPT belongs to the low-abundance group of proteins. In contrast to the PPT identified by Fischer et al. (1997), this PPT is expressed highly in leaves, is barely detectable in roots, and its expression level massively increases upon transition from etiolated to green leaves (Supplemental Fig. S5). The massive flux of PEP required for CO2 fixation is mediated by a specific PPT in the C4 leaf, and it can only be maintained by increasing the amount of PPT in the envelopes, compared with the envelope of the C3 species pea. This PPT protein is reported to be mesophyll specific (Majeran et al., 2008), in accordance with a role in C4 photosynthesis.
The demands for two of the four high-volume fluxes necessary for C4 photosynthesis (triosephosphate versus 3-phosphoglycerate and PEP versus inorganic phosphate) are thus accommodated by increased amounts of the respective transport proteins and hence increased Vmax. The Xyl-5-P translocator could only be identified in the C3 envelope sample, and a Glc-6-P translocator was not detected in either experiment.
Dicarboxylate Translocators
The envelope proteome of C4 mesophyll chloroplasts contains a higher percentage of proteins from the DiT family (Weber et al., 1995). Proteins of both the Glu/malate-exchanger type, DiT2 (Taniguchi et al., 2002; Renné et al., 2003), called DCT1 and DCT2/3 in maize (Taniguchi et al., 2004), and of the 2-oxoglutarate/malate-exchanger family, DiT1 (Weber et al., 1995), called OMT in maize (Taniguchi et al., 2004), are enriched. These transport proteins connect cytosolic and plastidic nitrogen metabolism through a two-translocator mechanism in C3 plants (Woo et al., 1987; Weber and Flügge, 2002; Renné et al., 2003), with DiT1 and DiT2 also playing a major role in photorespiration (Taniguchi et al., 2002; Renné et al., 2003; Schneidereit et al., 2006). Their function in C4 chloroplasts, such as those of maize mesophyll cells, is less well understood. There is controversial evidence with respect to their mRNA accumulation patterns in mesophyll cells (Renné et al., 2003; Taniguchi et al., 2004; Sawers et al., 2007). The protein accumulation pattern indicates higher expression of OMT and DCT1 in mesophyll chloroplast membranes and higher expression of DCT2/3 in bundle sheath chloroplast membranes (Majeran et al., 2008; Supplemental Table S4). The DiT family members have been proposed to play a role in central nitrogen metabolism (Renné et al., 2003) and, for OMT of the DiT1 family, to be the oxaloacetate/malate shuttle that is needed for core C4 photosynthesis (Fig. 1; Taniguchi et al., 2004). Currently, their in vivo function remains unclear in C4 plants, although their higher abundance in C4 compared with C3 may suggest that C4 photosynthesis causes higher fluxes of their cargo metabolites. Especially the role of the additional DiT2 family member DCT2/3 present in the bundle sheath of maize has not been elucidated, as the connection of nitrogen metabolism only requires two translocators and the function as an oxaloacetate/malate shuttle has only been proposed for OMT of the DiT1 family. In both samples, all members of the respective DiT families were identified.
Outer Envelope Porins
The higher metabolite flux across the inner envelope of C4 chloroplast necessitates a comparably high flow through the outer envelope. Two outer envelope porins, OEP24 and OEP37, occupy a larger percentage of the spectral count in C4 mesophyll envelopes. In vitro, OEP24 transports triosephosphates and dicarboxylates and thus is perfectly suited to accommodate the metabolite fluxes needed for core C4 photosynthesis (Pohlmeyer et al., 1998). OEP37 has been shown to transport inorganic cations in vitro but in vivo substrates have not yet been established, since the corresponding knockout mutant in the C3 plant Arabidopsis does not display an apparent phenotype (Goetze et al., 2006). In contrast, the regulated outer envelope porin, OEP21 (Bolter et al., 1999), is reduced in relative abundance (Supplemental Table S3), although not among the top 12 reduced proteins. It may be reduced because the regulation, which allows fine-tuning of the metabolite flow across the C3 envelope by the supply of ATP, 3-phosphoglycerate, and triosephosphate, may hinder metabolite exchange under C4 conditions. OEP16, an outer envelope protein that also forms a channel through the membrane, likely transports amino acids (Pohlmeyer et al., 1997). In contrast to the other outer envelope porins identified, this protein does not differ in relative spectral abundance between C3 and C4 PCA-type chloroplasts. The adaptations of the outer envelope proteins apparently reflect the changes in metabolite flux, indicating that the flux across the outer envelope might be limited and regulated by its proteins.
Other Transport Proteins
Although metabolite transport proteins appeared to be generally increased in C4 mesophyll chloroplast envelope over C3-type envelopes, two proteins with unknown function, a putative ATP-binding cassette-type transport protein and a protein of unknown function predicted to be anchored to the membrane, were absent from the C4 sample. In addition to the phosphate and dicarboxylate translocators, the C4 mesophyll envelopes contain more ATP/ADP translocator protein compared with C3 envelopes from pea. Mesophyll chloroplasts have a high demand for ATP, since the regeneration of the primary CO2 acceptor, PEP, from pyruvate requires two ATPs for each reaction. Since mesophyll chloroplasts are the source of reduction equivalents for both mesophyll and bundle sheath chloroplasts, cyclic electron transport may be limited in favor of linear electron transport, thus reducing the availability of ATP in the chloroplast stroma. This limitation could be overcome by importing ATP from other sources into PCA-type chloroplasts. Within the group of proteins that are more abundant in maize mesophyll compared with pea C3 chloroplast envelopes are also five proteins of unknown function. Of these proteins, one has one, three have four, and one has 12 predicted transmembrane helices. This study, in combination with results from other proteomics studies (Majeran et al., 2008), allows us to posit hypotheses about proteins catalyzing additional C4 metabolite fluxes. Mep1, a protein with 12 predicted transmembrane helices, is enriched in C4 mesophyll compared with C3 envelopes, and its protein accumulates evenly between mesophyll and bundle sheath. Moreover, its mRNA accumulates mainly in green leaves (Supplemental Fig. S5). This pattern of expression fits the pyruvate transport protein, which carries a higher load in C4 plants compared with C3 plants and is needed in both mesophyll and bundle sheath chloroplasts. Mep3 and Mep4, a pair of closely related proteins with four predicted transmembrane helices, of which one accumulated predominantly in the bundle sheath and the other in mesophyll tissue, are also candidates for the pyruvate transporter, since they are both elevated in C4 compared with C3. In contrast, Mep2 accumulates mainly in the mesophyll and hence is a candidate for an oxaloacetate/malate shuttle, in case members of the DiT family do not perform this function. The protein sequences and predicted structures of all candidate proteins are unrelated to any characterized protein.
Apart from being strong candidates for catalyzing metabolite fluxes across the maize mesophyll chloroplast envelope, which are increased to transfer core C4 photosynthesis metabolites, proteins of unknown function may carry fluxes that are increased as a by-product of the C4 syndrome. For example, sulfur metabolism seems to be differentially localized in C4 chloroplasts between mesophyll and bundle sheath (Majeran et al., 2005) and therefore may require abundant transfer proteins. A comparison of bundle sheath with C3 chloroplast envelope membranes may be necessary to identify a candidate for the malate importer of bundle sheath chloroplasts.
CONCLUSION
The comparison of the C4 mesophyll and C3 chloroplast envelope proteomes has revealed differences beyond the expected changes in metabolite transport proteins needed to support core C4 photosynthesis, including major changes in the outer envelope. The molecular nature of the phosphate translocators involved in C4 photosynthesis was established, and a number of candidate proteins for the additional fluxes were identified. Similar to what is observed during the transition from C3 to Crassulacean acid metabolism in Mesembryanthemum crystallinum (Häusler et al., 2000), the abundance of chloroplast envelope membrane transporters is adjusted to meet the high metabolic flux rates demanded by C4 photosynthesis. To date, metabolite transport proteins have not been included in efforts to reengineer C4 photosynthesis. This analysis points to a greater role of the chloroplast outer and inner envelope membranes, at least in mesophyll tissue, for establishing the C4 carbon-concentrating mechanism than was previously assumed. Limitations in metabolite exchange across the chloroplast envelope may have hampered efforts to establish C4 photosynthesis in C3 crop plants.
MATERIALS AND METHODS
Preparation of Chloroplast Envelope Protein Samples
Chloroplast envelope membranes were isolated from pea (Pisum sativum variety Little Marvel) plants as described previously (Douce and Joyard, 1979; Keegstra and Yousif, 1986) and from maize (Zea mays) plants grown on field sites. Briefly, fully expanded maize leaves were harvested, stored on ice, and cut into small pieces using razor blades. The leaves were homogenized in a Waring blender, and the resulting slurry was filtered through several layers of Miracloth to remove the bundle sheath strands. Chloroplasts and chloroplast envelopes were isolated as described (Douce and Joyard, 1979; Keegstra and Yousif, 1986). Envelope membranes were diluted in 10 volumes of ice-cold 1:1 (v/v) chloroform:methanol and stored on ice for 20 min. Insoluble proteins were sedimented by centrifugation at 20,000g for 20 min (“pellet fraction”), and both the protein pellet and the soluble fraction were dried and washed with hexane to remove residual membrane lipids. Envelope membrane samples and fractionated samples were mixed with SDS-PAGE loading buffer, incubated for 20 min on a reaction tube shaker at 15°C, and subsequently separated by 12.5% SDS-PAGE.
Protein Identification
After staining with Coomassie Brilliant Blue, each gel lane was cut into 10 equally sized slices. Proteins contained in the gel slices were subjected to tryptic cleavage as described by Shevchenko et al. (1996). Peptides were extracted and loaded onto a Waters Symmetry C18 peptide trap (5 μm, 180 μm × 20 mm) at a flow rate of 4 μL min−1 in 2% acetonitrile/0.1% formic acid for 5 min, using a Waters nanoAcquity Sample Manager. Using a Waters nanoAcquity UPLC system, the peptides were separated on a Waters BEH C18 nanoAcquity column (1.7 μm, 100 μm × 100 mm) over 90 min and fed into a ThermoElectron LTQ Fourier-transform ion cyclotron resonance mass spectrometer with a flow rate of 300 nL min−1 (buffer A = 99.9% water/0.1% formic acid, buffer B = 99.9% acetonitrile/0.1% formic acid: gradient of 5% B to 40% B from 0 to 63 min, 40% B to 90% B from 63 to 71 min, and 5% B from 71 to 90 min). Survey scans were taken at a resolution of 50,000, and the top 10 ions were dissociated by automated low-energy collision. The BioWorks Browser version 3.2 was used to convert the resulting tandem mass spectrometry (MS/MS) spectra to a peak list.
All mass spectra libraries were compared with a sequence database from pea (Bräutigam et al., 2008) that was generated by massively parallel pyrosequencing of cDNAs (Weber et al., 2007) and a maize cDNA database (ftp://occams.dfci.harvard.edu/pub/bio/tgi/data/Zea_mays) using the Mascot search algorithm, version 2.2 (www.matrixscience.com). Carbamidomethyl Cys was set as a fixed peptide modification, and oxidation of Met was permitted. Up to two missed tryptic sites were allowed. The peptide tolerance was set to ±10 μL L−1, and the MS/MS tolerance was set to 0.8 kD.
Protein Annotation and Bioinformatics
Mascot results were analyzed using an implementation of the peptide and protein prophet algorithms (Keller et al., 2002; Scaffold), with parameters set to 99% confidence for protein identification, requiring at least two unique peptides for each protein, and 95% confidence for all peptides counted. Where Scaffold reported multiple proteins identified for the same peptides, each match was manually inspected and low-scoring matches were discarded. The results were then exported to Microsoft Excel for further analysis. Cross-identifications in previous chloroplast proteomics projects were determined using BLASTX against the plprot database (Kleffmann et al., 2006), and the corresponding Arabidopsis (Arabidopsis thaliana) proteins were identified by BLASTX (Altschul et al., 1997) in The Arabidopsis Information Resource. Functional annotation and classification presented here were deduced from information in The Arabidopsis Information Resource (Swarbreck et al., 2008), ARAMEMNON (Schwacke et al., 2003), membranetransport.org (Ren et al., 2007), and manual curation of the pertinent literature. The predicted location and number of transmembrane helices were retrieved from ARAMEMNON. Tentative consensus sequences that identified the same Arabidopsis protein were aligned. If the sequences were identical, one of the identifications was discarded. If the sequences overlapped only partially, the annotations were unified and the number of peptides summed to generate a list of nonredundant identifications (Supplemental Tables S1 and S2). Before semiquantitative analysis, the spectral counts in each fraction were corrected for loading. The original data can be downloaded from PRIDE (http://www.ebi.ac.uk/pride/; Martens et al., 2005).
Semiquantitative Analysis of Protein Abundance
The semiquantitative analysis of protein abundance was based on the spectral count (i.e. the number of mass spectra mapping to a given protein in a single experiment). In the first experiment for each envelope preparation, all proteins in the sample were separated by SDS-PAGE and identified by liquid chromatography-electrospray ionization-MS/MS without prior fractionation (“whole envelopes”). In a second experiment, the proteins were first fractionated into a chloroform/methanol-soluble and an insoluble fraction. Proteins from both fractions were then separated by SDS-PAGE and subsequently identified by liquid chromatography-electrospray ionization-MS/MS. The spectral counts for each protein in both fractions were summed to yield the “sum” fraction. For all four experiments, the spectral count for each protein was normalized to the total number of spectra within the experiment (“percentage of the total spectral count”; Supplemental Tables S1 and S2). The robustness of the semiquantitative analysis was tested by introducing a number of disturbances into the experiment: omitting all proteins with a spectral count lower than 10 spectra identified, and including and excluding putative extraplastidial contaminations. The results were robust.
Accession Numbers
All proteomics data reported here have been submitted to the PRIDE data repository (http://www.ebi.ac.uk/pride/; Martens et al., 2005) and can be downloaded from PRIDE using the following PRIDE experiment accession numbers: maize C4 PCA-type chloroplast envelopes, pellet fraction (accession no. 3370); maize C4 PCA-type chloroplast envelopes, soluble fraction (accession no. 3371); maize C4 PCA-type chloroplast envelopes, whole (accession no. 3372); pea C3-type chloroplast envelopes, pellet fraction (accession no. 3376); pea C3-type chloroplast envelopes, soluble fraction (accession no. 3377); and pea C3-type chloroplast envelopes, whole (accession no. 3378).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. mRNA accumulation patterns of chloroplast envelope membrane transporters in different tissues of maize.
Supplemental Table S1. Proteins identified in C4 PCA-type maize chloroplast envelope membranes. Listed are proteins identified, number of spectra mapping to each maize accession number, annotation, classification, number of membrane-spanning domains, targeting prediction, and previous identifications in other proteomics studies.
Supplemental Table S2. Proteins identified in C3-type pea chloroplast envelope membranes. Listed are proteins identified, number of spectra mapping to each maize accession number, annotation, classification, number of membrane-spanning domains, targeting prediction, and previous identifications in other proteomics studies.
Supplemental Table S3. Percentage of total spectral counts for each protein identified in C4 PCA-type and C3-type chloroplasts of maize and pea, respectively.
Supplemental Table S4. Differential accumulation of maize chloroplast envelope membrane transporters in bundle sheath and mesophyll cells.
Supplementary Material
Acknowledgments
We are grateful to the Michigan State University Proteomics Core Facility for help with the proteomic identification of chloroplast envelope membrane proteins. We thank the Bioinformatics Core of the Michigan State University Research Technology Support Facility for assistance with database generation, sequence annotation, and data mining.
This work was supported by the National Science Foundation (grant no. IOB–0548610 to A.P.M.W. and S.H.-B.), the Deutsche Studienstiftung and the Barnett-Rosenberg-Foundation (to A.B.), and the Deutsche Forschungsgemeinschaft (grant no. WE 2231/4–1 to A.P.M.W.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Andreas P.M. Weber (andreas.weber@uni-duesseldorf.de).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki N, Ohnishi J, Kanai R (1992) 2 different mechanisms for transport of pyruvate into mesophyll chloroplasts of C4 plants: a comparative study. Plant Cell Physiol 33 805–809 [Google Scholar]
- Bae W, Lee YJ, Kim DH, Lee J, Kim S, Sohn EJ, Hwang I (2008) AKR2A-mediated import of chloroplast outer membrane proteins is essential for chloroplast biogenesis. Nat Cell Biol 10 220–227 [DOI] [PubMed] [Google Scholar]
- Beale SI (1999) Enzymes of chlorophyll biosynthesis. Photosynth Res 60 43–73 [Google Scholar]
- Benning C, Xu CC, Awai K (2006) Non-vesicular and vesicular lipid trafficking involving plastids. Curr Opin Plant Biol 9 241–247 [DOI] [PubMed] [Google Scholar]
- Bolter B, Soll J, Hill K, Hemmler R, Wagner R (1999) A rectifying ATP-regulated solute channel in the chloroplastic outer envelope from pea. EMBO J 18 5505–5516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bräutigam A, Shresta RP, Whitten D, Wilkerson CG, Carr KM, Froehlich JE, Weber APM (2008) Comparison of the use of a species-specific database generated by pyrosequencing with databases from related species for proteome analysis of pea chloroplast envelopes. J Biotechnol (in press) [DOI] [PubMed]
- Cheng ZG, Sattler S, Maeda H, Sakuragi Y, Bryant DA, DellaPenna D (2003) Highly divergent methyltransferases catalyze a conserved reaction in tocopherol and plastoquinone synthesis in cyanobacteria and photosynthetic eukaryotes. Plant Cell 15 2343–2356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe LH, Aggarwal K, Franck Z, Lee KH (2005) A comparison of the consistency of proteome quantitation using two-dimensional electrophoresis and shotgun isobaric tagging in Escherichia coli cells. Electrophoresis 26 2437–2449 [DOI] [PubMed] [Google Scholar]
- Douce R, Joyard J (1979) Isolation and properties of the envelope of spinach chloroplasts. In E Reid, ed, Plant Organelles. Ellis Horwood Publishers, Chichester, West Sussex, UK, pp 47–59
- Edwards GE, Furbank RT, Hatch MD, Osmond CB (2001) What does it take to be C4? Lessons from the evolution of C4 photosynthesis. Plant Physiol 125 46–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eicks M, Maurino V, Knappe S, Flügge UI, Fischer K (2002) The plastidic pentose phosphate translocator represents a link between the cytosolic and the plastidic pentose phosphate pathways in plants. Plant Physiol 128 512–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300 1004–1016 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, von Heijne G (1999) ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci 8 978–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferro M, Salvi D, Brugiere S, Miras S, Kowalski S, Louwagie M, Garin J, Joyard J, Rolland N (2003) Proteomics of the chloroplast envelope membranes from Arabidopsis thaliana. Mol Cell Proteomics 2 325–345 [DOI] [PubMed] [Google Scholar]
- Ferro M, Salvi D, Riviere-Rolland H, Vermat T, Seigneurin-Berny D, Grunwald D, Garin J, Joyard J, Rolland N (2002) Integral membrane proteins of the chloroplast envelope: identification and subcellular localization of new transporters. Proc Natl Acad Sci USA 99 11487–11492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer K, Kammerer B, Gutensohn M, Arbinger B, Weber A, Häusler RE, Flügge UI (1997) A new class of plastidic phosphate translocators: a putative link between primary and secondary metabolism by the phosphoenolpyruvate/phosphate antiporter. Plant Cell 9 453–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flügge UI, Häusler RE, Ludewig F, Fischer K (2003) Functional genomics of phosphate antiport systems of plastids. Physiol Plant 118 475–482 [Google Scholar]
- Flügge UI, Heldt HW (1984) The phosphate-triose phosphate-phosphoglycerate translocator of the chloroplast. Trends Biochem Sci 9 530–533 [Google Scholar]
- Flügge UI, Stitt M, Heldt HW (1985) Light-driven uptake of pyruvate into mesophyll chloroplasts from maize. FEBS Lett 183 335–339 [Google Scholar]
- Froehlich JE, Benning C, Dörmann P (2001) The digalactosyldiacylglycerol (DGDG) synthase DGD1 is inserted into the outer envelope membrane of chloroplasts in a manner independent of the general import pathway and does not depend on direct interaction with monogalactosyldiacylglycerol synthase for DGDG biosynthesis. J Biol Chem 276 31806–31812 [DOI] [PubMed] [Google Scholar]
- Froehlich JE, Wilkerson CG, Ray WK, McAndrew RS, Osteryoung KW, Gage DA, Phinney BS (2003) Proteomic study of the Arabidopsis thaliana chloroplastic envelope membrane utilizing alternatives to traditional two-dimensional electrophoresis. J Proteome Res 2 413–425 [DOI] [PubMed] [Google Scholar]
- Goetze TA, Philippar K, Ilkavets I, Soll J, Wagner R (2006) OEP37 is a new member of the chloroplast outer membrane ion channels. J Biol Chem 281 17989–17998 [DOI] [PubMed] [Google Scholar]
- Graham JWA, Williams TCR, Morgan M, Fernie AR, Ratcliffe RG, Sweetlove LJ (2007) Glycolytic enzymes associate dynamically with mitochondria in response to respiratory demand and support substrate channeling. Plant Cell 19 3723–3738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grodzinski B, Jiao JR, Leonardos ED (1998) Estimating photosynthesis and concurrent export rates in C-3 and C-4 species at ambient and elevated CO2. Plant Physiol 117 207–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch MD (1987) C-4 photosynthesis: a unique blend of modified biochemistry, anatomy and ultrastructure. Biochim Biophys Acta 895 81–106 [Google Scholar]
- Hatch MD, Droscher L, Flügge UI, Heldt HW (1984) A specific translocator for oxaloacetate transport in chloroplasts. FEBS Lett 178 15–19 [Google Scholar]
- Häusler RE, Baur B, Scharte J, Teichmann T, Eicks M, Fischer KL, Flügge UI, Schubert S, Weber A, Fischer K (2000) Plastidic metabolite transporters and their physiological functions in the inducible Crassulacean acid metabolism plant Mesembryanthemum crystallinum. Plant J 24 285–296 [DOI] [PubMed] [Google Scholar]
- Huber SC, Edwards GE (1977. a) Transport in C4 mesophyll chloroplasts: characterization of pyruvate carrier. Biochim Biophys Acta 462 583–602 [DOI] [PubMed] [Google Scholar]
- Huber SC, Edwards GE (1977. b) Transport in C4 mesophyll chloroplasts: evidence for an exchange of inorganic-phosphate and phosphoenolpyruvate. Biochim Biophys Acta 462 603–612 [DOI] [PubMed] [Google Scholar]
- Jarvis P, Dörmann P, Peto CA, Lutes J, Benning C, Chory J (2000) Galactolipid deficiency and abnormal chloroplast development in the Arabidopsis MGD synthase 1 mutant. Proc Natl Acad Sci USA 97 8175–8179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keegstra K, Yousif AE (1986) Isolation and characterization of chloroplast envelope membranes. Methods Enzymol 118 316–325 [Google Scholar]
- Keller A, Nesvizhskii AI, Kolker E, Aebersold R (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem 74 5383–5392 [DOI] [PubMed] [Google Scholar]
- Kleffmann T, Hirsch-Hoffmann M, Gruissem W, Baginsky S (2006) plprot: a comprehensive proteome database for different plastid types. Plant Cell Physiol 47 432–436 [DOI] [PubMed] [Google Scholar]
- Knappe S, Flügge UI, Fischer K (2003. a) Analysis of the plastidic phosphate translocator gene family in Arabidopsis and identification of new phosphate translocator-homologous transporters, classified by their putative substrate-binding site. Plant Physiol 131 1178–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knappe S, Löttgert T, Schneider A, Voll L, Flügge UI, Fischer K (2003. b) Characterization of two functional phosphoenolpyruvate/phosphate translocator (PPT) genes in Arabidopsis: AtPPT1 may be involved in the provision of signals for correct mesophyll development. Plant J 36 411–420 [DOI] [PubMed] [Google Scholar]
- Kuchler M, Decker S, Hormann F, Soll J, Heins L (2002) Protein import into chloroplasts involves redox-regulated proteins. EMBO 21 6136–6145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laisk A, Edwards GE (2000) A mathematical model of C-4 photosynthesis: the mechanism of concentrating CO2 in NADP-malic enzyme type species. Photosynth Res 66 199–224 [DOI] [PubMed] [Google Scholar]
- Leegood RC, Edwards GE (1996) Photosynthesis and the Environment, Vol 5. Kluwer Academic Publishers, Dordrecht, The Netherlands
- Liu H, Sadygov RG, Yates JR (2004) A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem 76 4193–4201 [DOI] [PubMed] [Google Scholar]
- Lu P, Vogel C, Wang R, Yao X, Marcotte EM (2007) Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation. Nat Biotechnol 25 117–124 [DOI] [PubMed] [Google Scholar]
- Majeran W, Cai Y, Sun Q, van Wijk KJ (2005) Functional differentiation of bundle sheath and mesophyll maize chloroplasts determined by comparative proteomics. Plant Cell 17 3111–3140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeran W, Zybailov B, Ytterberg AJ, Dunsmore J, Sun Q, van Wijk KJ (2008) Consequences of C4 differentiation for chloroplast membrane proteomes in maize mesophyll and bundle sheath cells. Mol Cell Proteomics (in press) [DOI] [PMC free article] [PubMed]
- Martens L, Hermjakob H, Jones P, Adamski M, Taylor C, States D, Gevaert K, Vandekerckhove J, Apweiler R (2005) PRIDE: The Proteomics Identifications Database. Proteomics 5 3537–3545 [DOI] [PubMed] [Google Scholar]
- Matsuoka M, Nomura M, Agarie S, Miyao-Tokutomi M, Ku MSB (1998) Evolution of C4 photosynthetic genes and overexpression of maize C4 genes in rice. J Plant Res 111 333–337 [Google Scholar]
- Meierhoff K, Westhoff P (1993) Differential biogenesis of photosystem II in mesophyll and bundle-sheath cells of monocotyledonous NADP-malic enzyme-type C-4 plants: the nonstoichiometric abundance of the subunits of photosystem II in the bundle-sheath chloroplasts and the translational activity of the plastome-encoded genes. Planta 191 23–33 [Google Scholar]
- Neuhaus HE, Thom E, Möhlmann T, Steup M, Kampfenkel K (1997) Characterization of a novel eukaryotic ATP/ADP translocator located in the plastid envelope of Arabidopsis thaliana L. Plant J 11 73–82 [DOI] [PubMed] [Google Scholar]
- Ohnishi J, Flugge UI, Heldt HW, Kanai R (1990) Involvement of Na+ in active uptake of pyruvate in mesophyll chloroplasts of some C4 plants: Na+/pyruvate cotransport. Plant Physiol 94 950–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsley K, Hibberd JM (2006) The Arabidopsis PPDK gene is transcribed from two promoters to produce differentially expressed transcripts responsible for cytosolic and plastidic proteins. Plant Mol Biol 62 339–349 [DOI] [PubMed] [Google Scholar]
- Peltier JB, Emanuelsson O, Kalume DE, Ytterberg J, Friso G, Rudella A, Liberles DA, Soderberg L, Roepstorff P, von Heijne G, et al (2002) Central functions of the lumenal and peripheral thylakoid proteome of Arabidopsis determined by experimentation and genome-wide prediction. Plant Cell 14 211–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltier JB, Friso G, Kalume DE, Roepstorff P, Nilsson F, Adamska I, van Wijk KJ (2000) Proteomics of the chloroplast: systematic identification and targeting analysis of lumenal and peripheral thylakoid proteins. Plant Cell 12 319–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltier JB, Ytterberg AJ, Sun Q, van Wijk KJ (2004) New functions of the thylakoid membrane proteome of Arabidopsis thaliana revealed by a simple, fast, and versatile fractionation strategy. J Biol Chem 279 49367–49383 [DOI] [PubMed] [Google Scholar]
- Pohlmeyer K, Soll J, Grimm R, Hill K, Wagner R (1998) A high-conductance solute channel in the chloroplastic outer envelope from pea. Plant Cell 10 1207–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohlmeyer K, Soll J, Steinkamp T, Hinnah S, Wagner R (1997) Isolation and characterization of an amino acid-selective channel protein present in the chloroplastic outer envelope membrane. Proc Natl Acad Sci USA 94 9504–9509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren QH, Chen KX, Paulsen IT (2007) TransportDB: a comprehensive database resource for cytoplasmic membrane transport systems and outer membrane channels. Nucleic Acids Res 35 D274–D279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renné P, Dressen U, Hebbeker U, Hille D, Flügge UI, Westhoff P, Weber APM (2003) The Arabidopsis mutant dct is deficient in the plastidic glutamate/malate translocator DiT2. Plant J 35 316–331 [DOI] [PubMed] [Google Scholar]
- Sage RF (2004) The evolution of C-4 photosynthesis. New Phytol 161 341–370 [DOI] [PubMed] [Google Scholar]
- Sanda S, Leustek T, Theisen MJ, Garavito RM, Benning C (2001) Recombinant Arabidopsis SQD1 converts UDP-glucose and sulfite to the sulfolipid head group precursor UDP-sulfoquinovose in vitro. J Biol Chem 276 3941–3946 [DOI] [PubMed] [Google Scholar]
- Sawers RJH, Liu P, Anufrikova K, Hwang JTG, Brutnell TP (2007) A multi-treatment experimental system to examine photosynthetic differentiation in the maize leaf. BMC Genomics 8 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneidereit J, Häusler RE, Fiene G, Kaiser WM, Weber APM (2006) Antisense repression reveals a crucial role of the plastidic 2-oxoglutarate/malate translocator DiT1 at the interface between carbon and nitrogen metabolism. Plant J 45 206–224 [DOI] [PubMed] [Google Scholar]
- Schumann U, Prestele J, O'Geen H, Brueggeman R, Wanner G, Gietl C (2007) Requirement of the C3HC4 zinc RING finger of the Arabidopsis PEX10 for photorespiration and leaf peroxisome contact with chloroplasts. Proc Natl Acad Sci USA 104 1069–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwacke R, Schneider A, van der Graaff E, Fischer K, Catoni E, Desimone M, Frommer WB, Flügge UI, Kunze R (2003) ARAMEMNON, a novel database for Arabidopsis integral membrane proteins. Plant Physiol 131 16–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Mann M (1996) Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal Chem 68 850–858 [DOI] [PubMed] [Google Scholar]
- Slack CR, Hatch MD, Goodchild J (1969) Distribution of enzymes in mesophyll and parenchyma-sheath chloroplasts of maize leaves in relation to C4-dicarboxylic acid pathway of photosynthesis. Biochem J 114 489–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soll J, Schleiff E (2004) Protein import into chloroplasts. Nat Rev Mol Cell Biol 5 198–208 [DOI] [PubMed] [Google Scholar]
- Swarbreck D, Wilks C, Lamesch P, Berardini TZ, Garcia-Hernandez M, Foerster H, Li D, Meyer T, Muller R, Ploetz L, et al (2008) The Arabidopsis Information Resource (TAIR): gene structure and function annotation. Nucleic Acids Res 36 D1009–D1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi M, Taniguchi Y, Kawasaki M, Takeda S, Kato T, Sato S, Tahata S, Miyake H, Sugiyama T (2002) Identifying and characterizing plastidic 2-oxoglutarate/malate and dicarboxylate transporters in Arabidopsis thaliana. Plant Cell Physiol 43 706–717 [DOI] [PubMed] [Google Scholar]
- Taniguchi Y, Nagasaki J, Kawasaki M, Miyake H, Sugiyama T, Taniguchi M (2004) Differentiation of dicarboxylate transporters in mesophyll and bundle sheath chloroplasts of maize. Plant Cell Physiol 45 187–200 [DOI] [PubMed] [Google Scholar]
- Villarejo A, Buren S, Larsson S, Dejardin A, Monne M, Rudhe C, Karlsson J, Jansson S, Lerouge P, Rolland N, et al (2005) Evidence for a protein transported through the secretory pathway en route to the higher plant chloroplast. Nat Cell Biol 7 1224–1231 [DOI] [PubMed] [Google Scholar]
- Voll LM, Häusler RE, Hecker R, Weber APM, Weissenböck G, Fiene G, Waffenschmidt S, Flügge UI (2003) The phenotype of the Arabidopsis cue1 mutant is not simply caused by a general restriction of the shikimate pathway. Plant J 36 301–317 [DOI] [PubMed] [Google Scholar]
- Weber A, Flügge UI (2002) Interaction of cytosolic and plastidic nitrogen metabolism in plants. J Exp Bot 53 865–874 [DOI] [PubMed] [Google Scholar]
- Weber A, Menzlaff E, Arbinger B, Gutensohn M, Eckerskorn C, Flügge UI (1995) The 2-oxoglutarate/malate translocator of chloroplast envelope membranes: molecular cloning of a transporter containing a 12-helix motif and expression of the functional protein in yeast cells. Biochemistry 34 2621–2627 [DOI] [PubMed] [Google Scholar]
- Weber APM (2004) Solute transporters as connecting elements between cytosol and plastid stroma. Curr Opin Plant Biol 7 247–253 [DOI] [PubMed] [Google Scholar]
- Weber APM, Fischer K (2007) Making the connections: the crucial role of metabolite transporters at the interface between chloroplast and cytosol. FEBS Lett 581 2215–2222 [DOI] [PubMed] [Google Scholar]
- Weber APM, Schwacke R, Flügge UI (2005) Solute transporters of the plastid envelope membrane. Annu Rev Plant Biol 56 133–164 [DOI] [PubMed] [Google Scholar]
- Weber APM, Weber KL, Carr K, Wilkerson C, Ohlrogge JB (2007) Sampling the Arabidopsis transcriptome with massively parallel pyrosequencing. Plant Physiol 144 32–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo KC, Flügge UI, Heldt HW (1987) A 2-translocator model for the transport of 2-oxoglutarate and glutamate in chloroplasts during ammonia assimilation in the light. Plant Physiol 84 624–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B, Xu CC, Benning C (2002) Arabidopsis disrupted in SQD2 encoding sulfolipid synthase is impaired in phosphate-limited growth. Proc Natl Acad Sci USA 99 5732–5737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zybailov B, Coleman MK, Florens L, Washburn MP (2005) Correlation of relative abundance ratios derived from peptide ion chromatograms and spectrum counting for quantitative proteomic analysis using stable isotope labeling. Anal Chem 77 6218–6224 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.