Abstract
Plants elaborate a vast array of enzymes that synthesize defensive secondary metabolites in response to pathogen attack. Here, we isolated the pathogen-responsive CaMNR1 [menthone: (+)-(3S)-neomenthol reductase] gene, a member of the short-chain dehydrogenase/reductase (SDR) superfamily, from pepper (Capsicum annuum) plants. Gas chromatography-mass spectrometry analysis revealed that purified CaMNR1 and its ortholog AtSDR1 from Arabidopsis (Arabidopsis thaliana) catalyze a menthone reduction with reduced nicotinamide adenine dinucleotide phosphate as a cofactor to produce neomenthol with antimicrobial activity. CaMNR1 and AtSDR1 also possess a significant catalytic activity for neomenthol oxidation. We examined the cellular function of the CaMNR1 gene by virus-induced gene silencing and ectopic overexpression in pepper and Arabidopsis plants, respectively. CaMNR1-silenced pepper plants were significantly more susceptible to Xanthomonas campestris pv vesicatoria and Colletotrichum coccodes infection and expressed lower levels of salicylic acid-responsive CaBPR1 and CaPR10 and jasmonic acid-responsive CaDEF1. CaMNR1-overexpressing Arabidopsis plants exhibited enhanced resistance to the hemibiotrophic pathogen Pseudomonas syringae pv tomato DC3000 and the biotrophic pathogen Hyaloperonospora parasitica isolate Noco2, accompanied by the induction of AtPR1 and AtPDF1.2. In contrast, mutation in the CaMNR1 ortholog AtSDR1 significantly enhanced susceptibility to both pathogens. Together, these results indicate that the novel menthone reductase gene CaMNR1 and its ortholog AtSDR1 positively regulate plant defenses against a broad spectrum of pathogens.
Plants constitutively synthesize a wide variety of secondary metabolites to aid fitness by preventing pathogen invasion and insect herbivory as well as by attracting pollinators and natural enemies of herbivores (Wink, 1998; Osbourn et al., 2003; Kliebenstein, 2004). Moreover, plants also induce defensive secondary metabolites in response to pathogen attack and stress (Litvak and Monson, 1998; Arimura et al., 2000; Biere et al., 2004), including compounds such as terpenoids, phytoalexins, and glucosinolates. It has been reported that the plant kingdom produces approximately 50,000 secondary metabolites of known structure, including 30,000 terpenoids, 12,000 alkaloids, 2,500 phenylpropanoids, and 2,500 other compounds (De Luca and St Pierre, 2000). In Arabidopsis (Arabidopsis thaliana), approximately 25% of the total 25,498 protein-encoding genes are predicted to be involved in secondary metabolism (Arabidopsis Genome Initiative, 2000). These findings suggest the significance of secondary metabolite production in plant defense responses to biotic and abiotic stresses. However, relatively little is known regarding the functions of the genes that control the biosynthesis of secondary metabolites.
Among these metabolites, terpenoids comprise one of the largest and most diverse groups, which include menthol, abscisic acid, chlorophyll, gibberellin, β-carotene, and rubber (Davis and Croteau, 2000). Terpenoids are frequently described as natural products active against a variety of herbivores (Litvak and Monson, 1998) and pathogens, including bacteria (Cichewicz and Thorpe, 1996; Trombetta et al., 2005; Schelz et al., 2006), fungi (Kubo et al., 1993; Rana et al., 1997), viruses (Harrigan et al., 1993; Sun et al., 1996), and protozoa (Ghoshal et al., 1996). For instance, conifer oleoresin, a complex mixture of terpenoids secreted in response to attack by insect predators, is toxic to insects and their symbiotic fungal pathogens (Philips and Croteau, 1999). The specific mechanisms of the antimicrobial action of terpenoids are not fully understood. However, the toxic effects of terpenoids on membrane structure and function have been studied (Mendoza et al., 1997; Trombetta et al., 2005; Pérez-Fons et al., 2006). The lipophilic property of terpenoids allows them to partition from the aqueous phase into membrane structures, resulting in membrane expansion, increased membrane fluidity and permeability, disturbance of membrane-embedded proteins, inhibition of respiration, and alteration of ion transport processes.
The significance of terpenoids in disease resistance has been reported for a number of plant-pathogen interactions. For instance, sesquiterpenoid phytoalexins, including 2,7-dihydroxycadalene, 2-hydroxy-7-methoxycadalene, lacinilene C, and lacinilene C 7-methyl ether, significantly accumulate in the leaves of resistant cotton (Gossypium hirsutum) lines but not in susceptible varieties after infection by Xanthomonas campestris pv malvacearum (Essenberg et al., 1982, 1990; Pierce et al., 1996). These compounds diffuse from cells exhibiting the hypersensitive response (HR) to arrest bacterial growth in resistant plants. Moreover, mutational analysis of a fungal pathogen of oat (Avena sativa) roots, Gaeumannomyces graminis, revealed that the triterpene saponin determines the host range of these pathogens (Bowyer et al., 1995). Mutation of G. graminis avenacinase, a saponin-detoxifying enzyme, leads to a loss of pathogenicity on oat, which produces saponins. However, the mutant fungal pathogen retains full pathogenicity on wheat (Triticum aestivum), which does not produce saponins. These findings underscore the significant roles of terpenoids in plant defense responses.
The biosynthetic pathway of terpenoids is well established in peppermint (Mentha piperita) by in vivo and cell-free studies (Davis and Croteau, 2000; Gershenzon et al., 2000; Mahmoud and Croteau, 2001; Ringer et al., 2005; Davis et al., 2005). Terpenoids (monoterpenes [C10], sesquiterpenes [C15], diterpenes [C20], triterpenes [C30], tetraterpenes [C40], and polyterpenes [>C40]) are synthesized by the condensation of isopentenyl diphosphate and its allylic isomer, dimethylallyl diphosphate, by prenyltransferase following conversion by terpenoid synthase and various enzymes, such as short-chain dehydrogenase/reductases (SDRs).
The primary pathway of monoterpene biosynthesis is well characterized, and a variety of enzymes is required in these reactions (Croteau et al., 2005; Davis et al., 2005; Ringer et al., 2005). The final product, (−)-menthol, which is synthesized as the most familiar monoterpene in this pathway, is used in a wide range of confectionery goods, pharmaceuticals, oral health-care products, cigarettes, and toiletries (http://www.leffingwell.com/menthol1/menthol1.htm). The conversion from (−)-menthone to (−)-menthol is catalyzed by stereospecific dehydrogenases, such as menthone reductases [MMR, (−)-menthone:(−)-(3R)-menthol reductase; MNR, (−)-menthone:(−)-(3S)-neomenthol reductase], in a NADPH-dependent manner (Davis et al., 2005). Sequence analysis of the cDNAs encoding MNR and MMR indicates that menthone reductases belong to a member of the SDR superfamily (Kallberg et al., 2002). SDRs are enzymes of great functional diversity. They typically share only 15% to 30% sequence identity, but specific sequence motifs are prominent, indicating common folding patterns (Kallberg et al., 2002). SDRs have approximately 250-residue subunits catalyzing NAD(P)(H)-dependent oxidation/reduction reactions. Most neomenthol is known to be glycosylated and transported to the rhizome in peppermint (Kjonaas et al., 1982). However, its biological function has not yet been determined in peppermint and other plant species.
Here, we isolated and functionally characterized a novel gene, menthone: (+)-(3S)-neomenthol reductase (CaMNR1), from pepper (Capsicum annuum) leaves infected by Xanthomonas campestris pv vesicatoria (Xcv) and its ortholog SDR gene from Arabidopsis, AtSDR1. The pepper CaMNR1 and its ortholog Arabidopsis AtSDR1 genes were overexpressed in Escherichia coli and purified to homogeneity. Enzyme activity of purified CaMNR1 and AtSDR1 was confirmed by gas chromatography-mass spectrometry (GC-MS) analysis of reaction products. The catalytic reactions of CaMNR1 and AtSDR1 yielded predominantly neomenthol as a reaction product at neutral pH. In this study, we used virus-induced gene silencing (VIGS) in pepper (Baulcombe, 1999) and ectopic expression in Arabidopsis (Clough and Bent, 1998) as efficient reverse genetics approaches to define the functions of the CaMNR1 gene in plant defense. CaMNR1-silenced pepper plants were highly susceptible to infection by Xcv and Colletotrichum coccodes, accompanied by significantly lowered expression levels of the salicylic acid (SA)-responsive CaBPR1 (basic PR1) and CaPR10 and jasmonic acid (JA)-responsive CaDEF1 (defensin) genes. Transgenic Arabidopsis plants that constitutively overexpressed the CaMNR1 gene also exhibited enhanced basal resistance to Pseudomonas syringae pv tomato (Pst) DC3000 and Hyaloperonospora parasitica. In contrast, a T-DNA insertion in AtSDR1, a putative ortholog of CaMNR1 in Arabidopsis, enhanced susceptibility to these pathogens.
RESULTS
CaMNR1 cDNA Encodes the MNR Protein
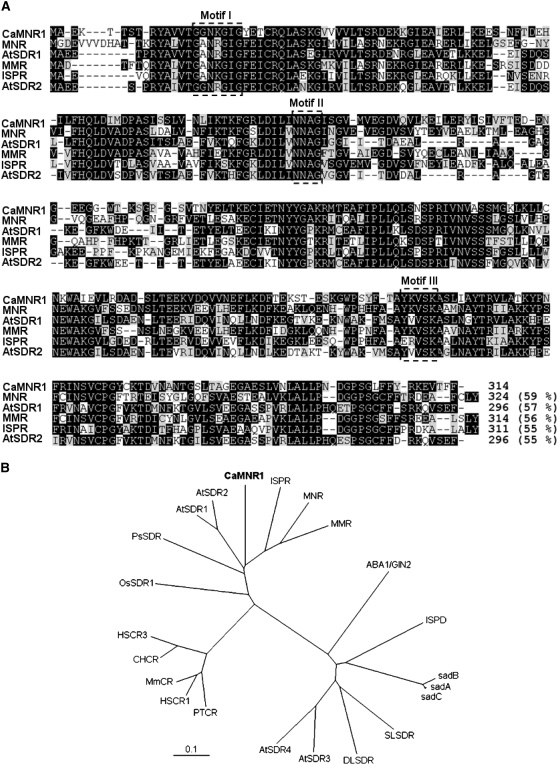
The CaMNR1 cDNA was isolated from a cDNA library made from pepper leaves infected with the Xcv avirulent strain Bv5-4a using a macro cDNA array method (Jung and Hwang, 2000; Chung et al., 2007). The CaMNR1 cDNA consists of 1,136 bp, including a poly(A) tail, and it encodes a protein of 314 amino acids with a predicted molecular mass of 34.7 kD and a pI of 5.39 (Supplemental Fig. S1). A database search (http://www.ncbi.nlm.nih.gov/blast/) using the translated CaMNR1 amino acid sequence as a query showed 59% identity to the MNR protein (accession no. AAQ55959) from peppermint (Davis et al., 2005). CaMNR1 shares moderate amino acid identity (55%–57%) with Arabidopsis and peppermint SDRs (Fig. 1A). Computational analysis of the CaMNR1 peptide sequence using PROSITE (www.expasy.org/prosite) revealed that CaMNR1 possesses conserved short-chain dehydrogenase domains, including a coenzyme-binding domain (motif I: GxxxGxG), a structural domain of undefined function (motif II: [N/C]NAG), and an active site element (motif III: YxxxK; Kallberg et al., 2002; Davis et al., 2005; Ringer et al., 2005). A phylogenetic tree of CaMNR1 and its closest relatives illustrates its proximity to peppermint SDRs, including MNR, MMR, and isopiperitenone reductase, and to uncharacterized SDRs from Arabidopsis (Fig. 1B).
Figure 1.
Amino acid sequence alignment and phylogenic analysis of CaMNR1. A, Comparison of the deduced amino acid sequences of the pepper CaMNR1 cDNA (CaMNR1) with menthone: (+)-(3S)-neomenthol reductase protein (MNR), (−)-menthone: (−)-(3R)-menthol reductase (MMR), isopiperitenone reductase (ISPR), and Arabidopsis SDRs (AtSDR1 and AtSDR2). The conserved coenzyme-binding domain (motif I: GXXXGXG), the structural domain of undefined function (motif II: [N/C]NAG), and the active site element (motif III: YXXXK) are shown above the alignment. Black boxes indicate identical amino acid residues. Dashes indicate spacing in the amino acid sequences for proper alignment. B, Alignment of full-length pepper CaMNR1 with MNR (accession no. AAQ55959), MMR (accession no. AAQ55960), ISPR (accession no. AAQ75422), (−)-trans-iso-piperitenol dehydrogenase (ISPD; accession no. AAU20370), Arabidopsis SDRs (AtSDR1, accession no. At3g61220; AtSDR2, accession no. At2g24190; AtSDR3, accession no. At2g47140; AtSDR4, accession no. At2g47130; and ABA1/GIN2, accession no. At1g52340), Papaver somniferum SDR (PsSDR; accession no. ABC47654), Oryza sativa SDR (OsSDR1; accession no. NP_001053402), Pisum sativum SDRs (sadA, accession no. AAF04193; sadB, accession no. AAF04194; and sadC, accession no. AAF04253), Solanum lycopersicum SDR (SLSDR; accession no. CAB91875), Digitalis lanata 3-β-hydroxysteroid-hydrogenase (DLSDR; accession no. CAC93667), human carbonyl reductases (HSCR1, accession no. J04056; and HSCR3, accession no. NP_001227), Mus musculus carbonyl reductase (MmCR; accession no. NP_031646), pig (Sus scrofa) carbonyl reductase (PTCR; accession no. M80709), and hamster (Cricetulus griseus) carbonyl reductase (CHCR; accession no. AB020238). The scale value of 0.1 represents 0.1 amino acid substitutions per site.
Overexpression and Purification of CaMNR1 in E. coli
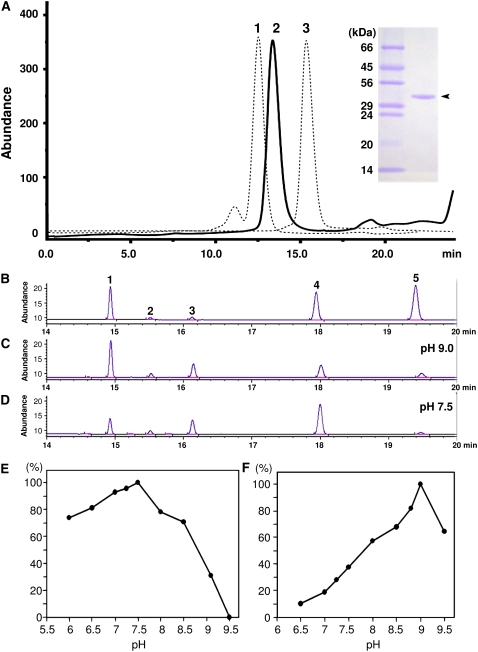
The CaMNR1 protein was overexpressed in E. coli and purified to homogeneity in several chromatographic steps (Fig. 2A). Neomenthol was the predominant reaction product generated by CaMNR1, while menthol was produced extremely slowly and at very low yield. CaMNR1 shares slightly higher sequence identity with peppermint MNR than with MMR. Therefore, we designated CaMNR1 as a menthone neomenthol reductase. The final yield of recombinant CaMNR1 was approximately 10 mg of over 99% purity from a 3-L bacterial culture (Fig. 2A). Purified CaMNR1 is a monomeric enzyme in solution with a molecular mass of approximately 34 kD, as indicated by gel filtration analysis (Fig. 2A). As described previously, CaMNR1 belongs to the SDR superfamily, members of which adopt a unique α/β structure known as the Rossmann fold (Oppermann et al., 2003). Their coenzyme specificity can be predicted very accurately by a hidden Markov model-based method (Kallberg and Persson, 2006). When the CaMNR1 sequence was submitted to a prediction server (http://www.ifm.liu.se/bioinfo/), a NADP-binding domain (residues 4–46) was identified, which indicates the same coenzyme specificity as that of biochemically characterized MMR and MNR (Davis et al., 2005).
Figure 2.
Purification and enzyme activity of CaMNR1. A, Gel filtration profile of recombinant CaMNR1 by Superose 12 column chromatography reveals that purified CaMNR1 (34 kD; peak 2) is a monomeric enzyme in solution. Standard molecular mass markers are bovine serum albumin (66 kD; peak 1) and chymotrypsinogen (25 kD; peak 3). The inset represents the final purity of recombinant CaMNR1 (arrowhead) and molecular mass markers. B, Gas chromatographic analyses of monoterpene standards: peak 1, menthone; peak 2, isomenthol; peak 3, camphor; peak 4, neomenthol; and peak 5, menthol. C and D, Chromatograms of enzymatic reaction products assayed at pH 9.0 (C) and pH 7.5 (D). The y axis is a detector response unit corresponding to the generated reaction products. E, Effects of pH on enzymatic activity using menthone as a substrate. F, Effects of pH on enzymatic activity using neomenthol as a substrate. Product values (y axis) are relative percentages when the maximal amount at optimal pH is taken as 100%. [See online article for color version of this figure.]
Enzyme Activity of CaMNR1
Monoterpene products were quantified and identified by their GC retention times in comparison with standards including (+)-camphor (Fig. 2B). CaMNR1 converted (−)-menthone to 93% (+)-(3S)-neomenthol and 7% (−)-(3R)-menthol at pH 7.5 and to 72% (+)-(3S)-neomenthol and 28% (−)-(3R)-menthol at pH 9.0 with NADPH as a cofactor. Although the amount of menthol generated was higher at pH 9.0 than at pH 7.5, neomenthol was the predominant reaction product (Fig. 2, C and D).
The oxidation of menthol isomers in the presence of NADP+ was also evaluated. Only neomenthol was converted into menthone in the presence of the NADP+ cofactor. The oxidation reaction displayed maximal activity at alkaline pH (9.0), whereas the reduction reaction was maximal at neutral pH 7.5 (Fig. 2, E and F). All kinetic parameters of CaMNR1 for monoterpene substrates and cofactors are summarized in Table I. Interestingly, the Km values of CaMNR1 for the forward reaction (reduction) were lower and its turnover rates were much higher than those for the reverse reaction (oxidation); however, the catalytic efficiencies of CaMNR1 for both reactions were similar.
Table I.
Kinetic parameters of CaMNR1
| Parameter | Forward Reaction
|
Reverse Reaction
|
||
|---|---|---|---|---|
| Menthone | NADPH | Neomenthol | NADP+ | |
| Km (μm) | 35.1 ± 4.3 | 21.7 ± 3.4 | 113.2 ± 14.9 | 76.7 ± 1.2 |
| kcat (s−1) | 0.039 ± 0.003 | 0.053 ± 0.01 | 0.13 ± 0.01 | 0.13 ± 0.001 |
| kcat/Km (μm−1 s−1) | 1.01 ± 0.05 × 10−3 | 2.44 ± 0.28 × 10−3 | 1.11 ± 0.04 × 10−3 | 1.74 ± 0.02 × 10−3 |
In Vitro Antimicrobial Activities of Menthone, Neomenthol, and Menthol
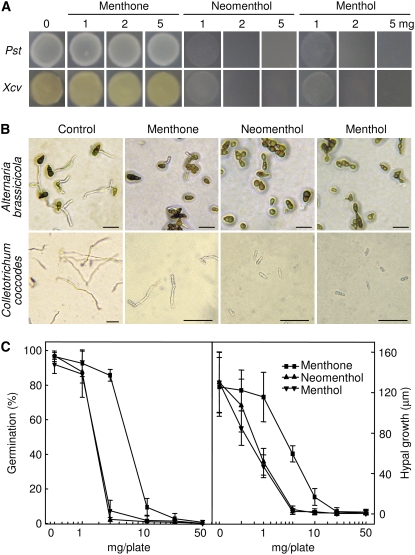
In vitro analysis of enzyme activity revealed that CaMNR1 converted (−)-menthone to (+)-(3S)-neomenthol and (−)-(3R)-menthol (Fig. 2). As described previously, most terpenoids are known to be active against a wide variety of microorganisms, including gram-positive and gram-negative bacteria and fungi (Cowan, 1999; Trombetta et al., 2005; Arfa et al., 2006). We analyzed the antimicrobial activities of (−)-menthone and its metabolites (+)-(3S)-neomenthol and (−)-(3R)-menthol using the microatmosphere method (Arfa et al., 2006). Interestingly, neomenthol and menthol strongly inhibited the growth of the phytopathogenic bacteria Pst DC3000 and Xcv at a concentration of 1 mg per plate (Fig. 3A). However, menthone, the precursor form of neomenthol and menthol, was not effective at inhibiting Pst and Xcv growth at a concentration of 5 mg per plate. Similarly, neomenthol and menthol completely inhibited spore germination and hyphal growth of the phytopathogenic fungi Alternaria brassicicola and C. coccodes at a concentration of 5 mg per plate; however, menthone was inhibitory against A. brassicicola and C. coccodes at higher concentrations (>10 mg per plate; Fig. 3, B and C).
Figure 3.
Antimicrobial activities of menthone, neomenthol, and menthol. A, Inhibition of growth of Pst DC3000 and Xcv at 48 h after treatment with various concentrations of menthone, neomenthol, and menthol. B, Micrographs of the inhibitory effects of menthone, neomenthol, and menthol on spore germination of A. brassicicola and C. coccodes at 24 h after treatment with 5 mg per plate (9 cm in diameter) of these compounds. Bars = 50 μm. C, Inhibitory effects of menthone, neomenthol, and menthol on spore germination and hyphal growth of C. coccodes at 12 and 24 h after treatment with various concentrations of these compounds. Data are means ± sd of three independent experiments.
CaMNR1 Is Expressed in an Organ-Specific Manner and Induced in Leaves by Bacterial Infection and Abiotic Elicitor Treatments
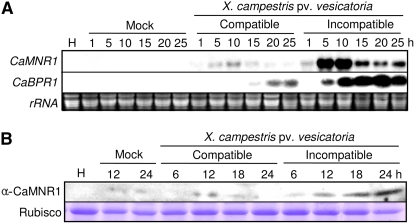
Expression of the CaMNR1 gene was examined in different pepper organs by RNA gel-blot analysis (Supplemental Fig. S2A). The CaMNR1 transcripts were detected in flowers and red fruit tissues of healthy pepper plants. However, transcripts were not present in healthy leaves, stems, roots, or green fruits. RNA gel-blot analysis of the CaMNR1 gene was performed to determine whether it is induced in pepper leaves during compatible and incompatible interactions with Xcv (Fig. 4A). The CaMNR1 gene was strongly induced in the leaves inoculated with the avirulent strain Bv5-4a (incompatible interaction). However, CaMNR1 transcripts were only weakly detected in leaves inoculated with the virulent strain Ds1 (compatible interaction). Protein expression is often indicative of gene function. We next determined CaMNR1 protein levels by protein gel-blot analysis to investigate the possible role of CaMNR1 in enhancing plant resistance to Xcv infection. As shown in Figure 4B, infection with the avirulent Xcv strain strongly induced the accumulation of CaMNR1 in pepper leaves, compared with that in leaves infected with the virulent Xcv strain. These data indicate that the CaMNR1 gene plays a role in regulating the resistance of pepper plants at the mRNA and protein levels.
Figure 4.
Pathogen-induced expression of CaMNR1 in pepper leaves. A, RNA gel-blot analysis of the expression of the CaMNR1 and CaBPR1 genes in pepper leaves at various time points after inoculation with the Xcv virulent strain Ds1 (compatible) and the avirulent strain Bv5-4a (incompatible) at the six-leaf stage. B, Protein gel-blot analysis of the expression of CaMNR1 in pepper leaves at various time points after Xcv inoculation. Equal loading of proteins was confirmed by comparing the band intensities of the Rubisco protein stained with Coomassie blue. H, Uninoculated healthy leaves; Mock, 10 mm MgCl2. [See online article for color version of this figure.]
To determine whether CaMNR1 expression is triggered by abiotic elicitors, pepper plants at the six-leaf stage were treated with SA, ethylene, methyl jasmonate (MeJA), and abscisic acid (ABA; Supplemental Fig. S2B). The CaMNR1 gene was strongly induced by SA, ethylene, and ABA treatment. In pepper leaves treated with ethylene and ABA, CaMNR1 transcripts began to significantly increase at 1 h after treatment, and they were markedly induced at 15 h after SA treatment. By contrast, MeJA slightly induced CaMNR1 gene expression at 5 h after treatment. To determine whether CaMNR1 expression is influenced by environmental stress, pepper plants were exposed to hydrogen peroxide (H2O2), drought, cold, and mechanical wounding stresses (Supplemental Fig. S2C). Under drought conditions, the CaMNR1 transcript was rapidly induced within 1 h and thereafter gradually increased to 25 h. However, H2O2, cold, and mechanical wounding treatment did not cause CaMNR1 transcripts to appear in treated pepper leaves.
Metabolite Profiles in Pepper Leaves
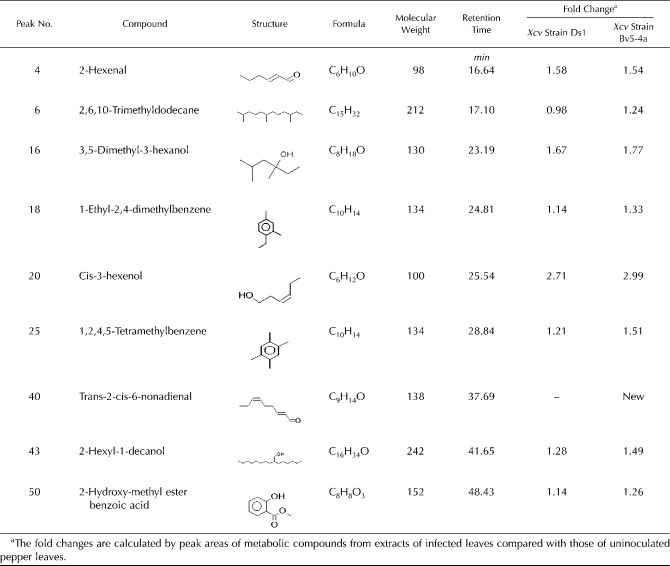
We tested the in vitro enzyme activities of CaMNR1 (Fig. 2) and the antimicrobial activities of menthone, neomenthol, and menthol (Fig. 3). The findings led us to check whether these monoterpenes are present in pepper leaves. We first used GC-MS analysis to evaluate the quantity of metabolic components induced by Xcv infection. In pepper plants, some pathogen-induced metabolic compounds were increased or newly induced at 18 h after inoculation with virulent and avirulent Xcv (Table II; Supplemental Figs. S3 and S4). The lipid-derived volatile compounds 2-hexenal, 3,5-dimethyl-3-hexanol, and cis-3-hexenol distinctly accumulated in pepper leaves infected with virulent and avirulent Xcv (Table II). The long-chain hydrocarbons 2,6,10-trimethyldodecane (farnesane) and 2-hexyl-1-decanol also accumulated in pepper leaves inoculated with Xcv. In particular, trans-2-cis-6-nonadienal, a product derived from a 9-hydroperoxide of linolenic acids (Matthew and Galliard, 1978), was de novo synthesized and induced mainly by infection with avirulent Xcv that triggered HR. Previously, HR-specific accumulation of trans-2-cis-6-nonadienal was reported in Phaseolus vulgaris leaves (Croft et al., 1993). Furthermore, some metabolic compounds with a benzene ring, including 1-ethly-2,4-dimethylbenzene, 1,2,4,5-tetramethyl benzene, and 2-hydroxy-methylester benzoic acid (methyl salicylate; MeSA), were increased during Xcv infection. Recently, MeSA was reported to be a key mobile signal in systemic acquired resistance in plants (Park et al., 2007). However, we failed to detect menthone, neomenthol, menthol, or their derivatives among the metabolic components of extracts of healthy or Xcv-infected pepper leaves (Supplemental Figs. S3 and S4).
Table II.
Secondary metabolites accumulated in pepper leaves infected with virulent strain Ds1 and avirulent strain Bv5-4a of Xcv
VIGS of the CaMNR1 Gene Alters Pepper Defense-Related Gene Expression
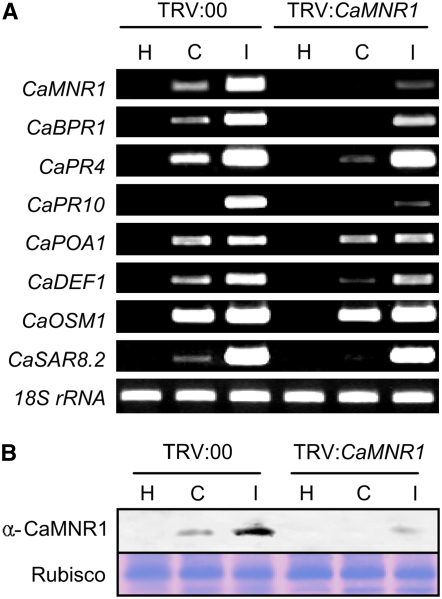
We revealed that the CaMNR1 gene was strongly induced in pepper leaves during the incompatible interaction of pepper plants with the Xcv avirulent strain Bv5-4a (Fig. 4). To examine the effect of loss of function of CaMNR1, we silenced the gene in pepper plants using the tobacco rattle virus (TRV)-mediated VIGS technique and the full-length open reading frame (Liu et al., 2002; Chung et al., 2004; Choi et al., 2007). CaMNR1 transcript levels were analyzed in unsilenced (TRV:00) and silenced (TRV:CaMNR1) leaves by reverse transcription (RT)-PCR (Fig. 5A). For the empty vector control, unsilenced leaves, the CaMNR1 gene was strongly induced at 18 h after inoculation with virulent and avirulent strains of Xcv (5 × 106 colony forming units [cfu] mL−1), but expression was compromised in CaMNR1-silenced pepper leaves. This suggests that silencing was effective for CaMNR1. We further tested whether the expression of several pepper pathogenesis-related (PR) genes was affected in CaMNR1-silenced leaves. Transcript levels of pepper CaBPR1, CaPR4, CaPR10, CaDEF1, and CaSAR8.2, but not of CaPOA1 (ascorbate peroxidase) and CaOSM1 (osmotin), were significantly lowered in CaMNR1-silenced leaves infected with Xcv, especially during the compatible interaction. Healthy pepper leaves did not produce any transcripts of the PR genes examined. In order to determine the relative abundance of the CaMNR1 protein, we next performed protein gel-blot analysis using rabbit polyclonal antibodies against CaMNR1 (Fig. 5B). Accumulation of the CaMNR1 protein was completely compromised in silenced leaves at 18 h after inoculation with virulent Xcv (compatible interaction); however, the CaMNR1 protein remained at a detectably visible level in the avirulent Xcv-infected leaves (incompatible interaction). These analyses indicated that TRV-mediated VIGS of CaMNR1 compromised the accumulation of CaMNR1 transcripts and protein and that this was accompanied by lower expression of some defense-related genes during the defense response of pepper plants.
Figure 5.
Expression of defense-related genes in CaMNR1-silenced pepper leaves. A, RT-PCR analyses of the expression of the CaMNR1, CaBPR1, CaPR4, CaPR10, CaPOA1, CaDEF1, CaOSM1, and CaSAR8.2 genes in unsilenced (TRV:00) and CaMNR1-silenced (TRV:CaMNR1) pepper plants at 18 h after inoculation with Xcv (108 cfu mL−1). 18S rRNA was the control. The experiment was replicated three times with similar results. H, Uninoculated healthy leaves; C, compatible interaction between the Xcv virulent strain Ds1 and pepper leaves; I, incompatible interaction between the Xcv avirulent strain Bv5-4a and pepper leaves. B, Protein gel-blot analysis of the expression of CaMNR1 in unsilenced (TRV:00) and CaMNR1-silenced (TRV:CaMNR1) pepper leaves at 18 h after inoculation with the Xcv virulent strain Ds1 and the avirulent strain Bv5-4a. Equal loading of proteins was confirmed by comparing the band intensities of the Rubisco protein stained with Coomassie blue. [See online article for color version of this figure.]
VIGS of CaMNR1 Enhances the Susceptibility of Pepper against Xcv and C. coccodes
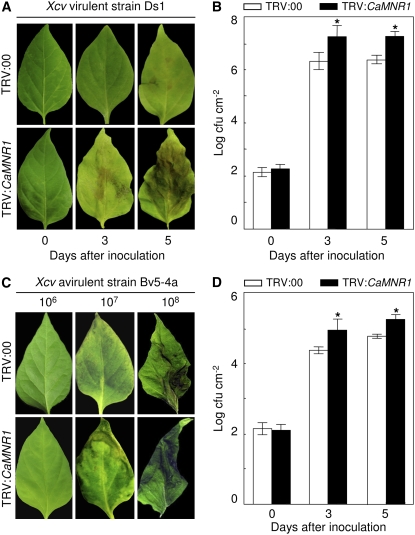
Silencing resulted in a susceptible response of pepper to infection by the Xcv virulent strain Ds1 and the avirulent strain Bv5-4a (Fig. 6). CaMNR1-silenced pepper leaves (TRV:CaMNR1) exhibit chlorosis at 3 d after virulent Xcv inoculation, whereas the empty vector, control leaves (TRV:00) became chlorotic at 5 d after inoculation (Fig. 6A). More severe chlorosis phenotypes appeared in CaMNR1-silenced leaves than in unsilenced leaves at 5 d after inoculation. However, we did not observe any apparent phenotypic changes in CaMNR1-silenced pepper leaves infected with the avirulent strain Bv5-4a compared with unsilenced leaves (Fig. 6C). Importantly, infection by virulent and avirulent strains of Xcv resulted in high levels of bacterial growth in CaMNR1-silenced plants compared with unsilenced plants at 3 and 5 d after inoculation (Fig. 6, B and D).
Figure 6.
Enhanced susceptibility of CaMNR1-silenced pepper plants to Xcv infection. A, Disease symptoms at 0, 3, and 5 d after inoculation with virulent Xcv (5 × 106 cfu mL−1). B, Bacterial growth in leaves of unsilenced (TRV:00) and CaMNR1-silenced (TRV:CaMNR1) pepper plants at 0, 3, and 5 d after inoculation (5 × 104 cfu mL−1). Data are means ± sd from three independent experiments. Asterisks indicate significant differences as determined by Student's t test (P < 0.05). C, Disease symptoms at 1 d after inoculation with avirulent Xcv suspensions of different concentrations (106, 107, and 108 cfu mL−1). D, Bacterial growth in leaves of unsilenced (TRV:00) and CaMNR1-silenced (TRV:CaMNR1) pepper plants at 0, 1, 3, and 5 d after inoculation (5 × 104 cfu mL−1). Data are means ± sd from three independent experiments. Asterisks indicate significant differences as determined by Student's t test (P < 0.05).
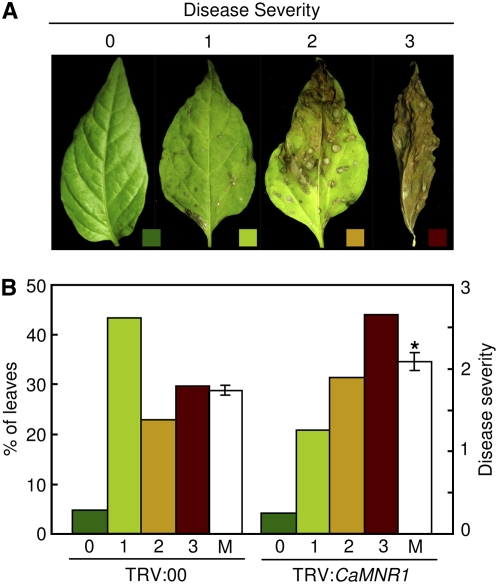
We also analyzed the response of CaMNR1-silenced pepper to C. coccodes, the causal agent of pepper anthracnose. CaMNR1-silenced pepper leaves exhibited enhanced susceptibility to C. coccodes infection (Fig. 7). Disease severity was rated on the basis of lesion numbers and areas of brown or dark brown spots at 5 d after inoculation (Fig. 7A). Approximately 45% of CaMNR1-silenced leaves showed severe disease symptoms (enlarged dark brown lesions with severe chlorosis), compared with 25% severely diseased, unsilenced leaves. These data indicated that CaMNR1 expression is required for basal resistance of pepper plants against the bacterial pathogen Xcv and the fungal pathogen C. coccodes.
Figure 7.
Enhanced susceptibility of CaMNR1-silenced (TRV:CaMNR1) pepper plants to C. coccodes infection. A, Disease severities were rated on a scale of 0 to 3 (0, no symptoms; 1, weakly visible symptoms; 2, dark brown lesions with mild chlorosis; 3, enlarged dark brown lesions with severe chlorosis). B, Percentages of pepper leaves exhibiting the indicated disease severities at 5 d after inoculation with C. coccodes. Disease severity was rated on more than 50 leaves. The asterisk indicates a significant difference between means as determined by Student's t test (P < 0.05). M, Means of disease severities.
Overexpression of CaMNR1 in Arabidopsis Confers Enhanced Expression of AtPR1 and AtPDF1.2
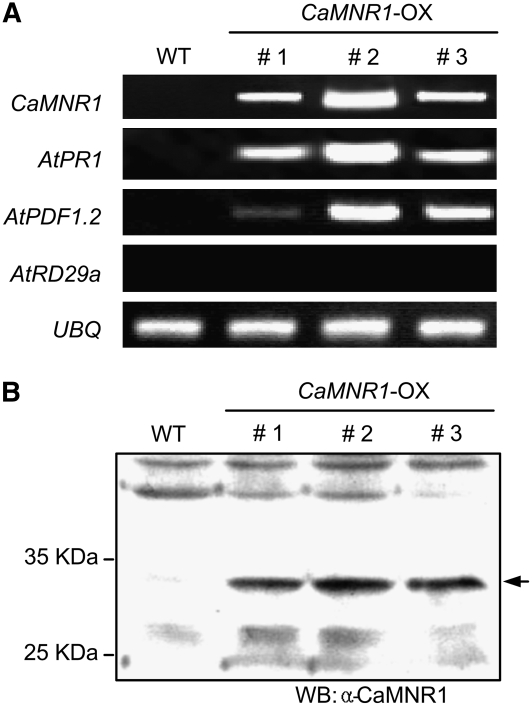
To investigate the effect of a gain of function of CaMNR1, transgenic Arabidopsis plants expressing CaMNR1 were generated. The CaMNR1 open reading frame was integrated between the cauliflower mosaic virus 35S promoter and the nos terminator region in the binary vector pBIN35S (Choi et al., 2007). Arabidopsis ecotype Columbia (Col-0) plants were transformed with the 35S:CaMNR1 construct by the floral dipping method (Clough and Bent, 1998). We did not observe any visible phenotype difference between wild-type and CaMNR1-overexpression (OX) transgenic lines. Three Arabidopsis transgenic T2 lines were tested to determine whether overexpression of CaMNR1 induces the expression of defense-related genes. The pathogenesis-related genes AtPR1 and AtPDF1.2 were strongly induced in the tested CaMNR1-OX transgenic lines (Fig. 8A). The transcript levels of the SA-responsive AtPR1 gene were very high in CaMNR1-OX transgenic plants. However, the JA-responsive AtPDF1.2 gene was induced at relatively low levels compared with AtPR1. In contrast, transcripts of the ABA-responsive gene AtRD29a were not detected in CaMNR1-OX transgenic plants. Wild-type plants produced no detectable transcripts for any of the genes examined. We also examined the expression of the CaMNR1 gene at the protein level in transgenic Arabidopsis plants using rabbit polyclonal antibodies against CaMNR1 (Fig. 8B). In CaMNR1-OX transgenic leaves, CaMNR1 protein was strongly expressed. However, we could not detect any CaMNR1 protein in the wild-type leaves. This led us to conclude that ectopic expression of CaMNR1 in a heterologous system triggers the expression of PR genes (AtPR1 and AtPDF1.2) but not of the ABA-responsive gene AtRD29a.
Figure 8.
Generation of CaMNR1-OX Arabidopsis transgenic plants. A, RT-PCR analyses of expression of the CaMNR1, AtPR1, AtPDF1.2, and AtRD29a genes in leaves of wild-type Arabidopsis (ecotype Col-0; WT) and CaMNR1-OX (lines 1, 2, and 3) plants. B, Protein gel-blot analysis of CaMNR1 expression in leaves of wild-type and CaMNR1-OX plants. The arrow indicates the CaMNR1 band detected by the anti-CaMNR1 polyclonal antibody.
Overexpression of CaMNR1 in Arabidopsis Enhances Basal Resistance against Pst and H. parasitica
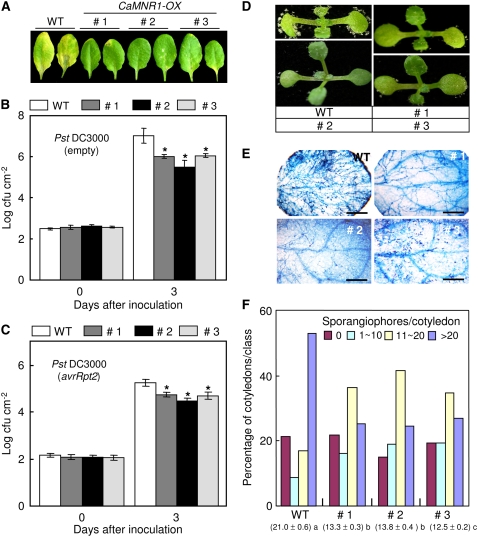
RT-PCR and protein gel-blot analysis showed that CaMNR1-OX transgenic lines constitutively expressed CaMNR1 and defense-related genes, which suggests a possible role for CaMNR1 in defense signaling activation in Arabidopsis (Fig. 8). To determine if the overexpression of CaMNR1 in Arabidopsis confers enhanced resistance, we examined bacterial growth using the virulent (empty vector) or a near-isogenic avirulent (avrRpt2) strain of Pst DC3000. Seven days after inoculation, severe chlorosis symptoms developed on the leaves of wild-type plants infected with virulent Pst DC3000. However, CaMNR1 transgenic lines exhibited only slight chlorotic symptoms compared with wild-type plants (Fig. 9A). Consistent with the increased expression of defense-related genes and no visible disease symptoms, CaMNR1 transgenic lines exhibited significantly lower bacterial growth at 3 d after inoculation than did wild-type plants (Fig. 9B). CaMNR1 transgenic lines also exhibited increased resistance to the avirulent strain Pst DC3000 carrying avrRpt2 (Fig. 9C). The avirulent strain Pst DC3000 (avrRpt2) grew approximately 10-fold better in wild-type plants compared with CaMNR1 transgenic lines.
Figure 9.
Enhanced basal disease resistance of CaMNR1-OX transgenic Arabidopsis. A, Disease symptoms on the leaves of wild-type and transgenic Arabidopsis plants at 5 d after spray inoculation with Pst DC3000 (108 cfu mL−1). B and C, Bacterial growth in the leaves of wild-type and transgenic plants at 0 and 3 d after inoculation with a virulent (empty) strain (B) and an avirulent (avrRpt2) strain (C) of Pst DC3000 (5 × 104 cfu mL−1). Data are means ± sd from three independent experiments. Asterisks indicate significant differences as determined by Student's t test (P < 0.05). D, Disease symptoms on the cotyledons of wild-type and transgenic Arabidopsis plants at 7 d after spray inoculation with H. parasitica isolate Noco2 (5 × 104 spores mL−1). E, Trypan blue staining of cotyledons of wild-type and transgenic Arabidopsis plants at 7 d after inoculation with H. parasitica isolate Noco2. Bars = 200 μm. F, Numbers of sporangiophores per cotyledon of wild-type and transgenic plants at 7 d after inoculation with H. parasitica. Cotyledons were classified as follows: no sporulation (no sporangiophores per cotyledon), light sporulation (approximately 1–10 sporangiophores per cotyledon), medium sporulation (approximately 11–20 sporangiophores per cotyledon), or heavy sporulation (more than 20 sporangiophores per cotyledon). Sporangiophores were counted on more than 50 cotyledons per genotype. Average numbers of sporangiophores produced on the cotyledons of wild-type and transgenic lines are shown at bottom for each of the lines tested. Statistically significant differences between means were determined employing the lsd (P = 0.05).
To determine whether the overexpression of CaMNR1 also enhances disease resistance to a biotrophic oomycete pathogen, we inoculated H. parasitica virulent isolate Noco2 onto cotyledons of 7-d-old Col-0 plants. Freshly harvested conidiospores (5 × 104 mL−1) of downy mildew were sprayed onto cotyledons of 7-d-old wild-type (ecotype Col-0) and CaMNR1-OX transgenic plants. Seven days after inoculation, sporangiophores were counted on over 50 cotyledons with a stereomicroscope. Wild-type plants exhibited a heavy infestation of sporangiophores on cotyledon surfaces (Fig. 9D). However, the number of sporangiophores was significantly reduced in CaMNR1 transgenic lines (Fig. 9F). Approximately 53% of wild-type cotyledons exhibited heavy sporulation (>20 sporangiophores per cotyledon). However, only 24% to 26% of CaMNR1 transgenic cotyledons exhibited heavy sporulation. The average number of sporangiophores was significantly lower in CaMNR1 transgenic lines (line 1, 13.36; line 2, 13.88; line 3, 12.59) than in wild-type plants (21.05). Trypan blue staining of infected leaves revealed that CaMNR1-OX transgenic lines exhibited not only reduced levels of sporulation but also retarded hyphal growth in cotyledons compared with wild-type plants (Fig. 9E). Thus, we conclude that overexpression of CaMNR1 confers enhanced basal resistance against at least two taxonomically unrelated hemibiotrophic bacterial and biotrophic oomycete pathogens in Arabidopsis plants.
Altered Responses of sdr1 Mutants to Pst and H. parasitica
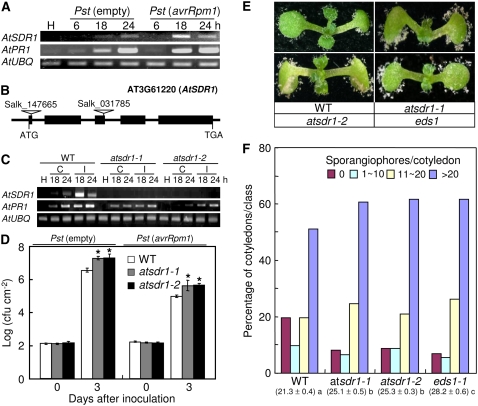
Alignment and phylogenetic analysis of the deduced amino acid sequence of CaMNR1 revealed its proximity to two uncharacterized SDR genes from Arabidopsis (Fig. 1). To determine the function of AtSDR1 and AtSDR2 during the defense response, we analyzed the expression of AtSDR1 and AtSDR2 in healthy and Pst DC3000-infected Arabidopsis leaves by RT-PCR. As shown in Figure 10A, we could not detect any AtSDR1 and AtSDR2 transcripts in noninoculated healthy leaves. Interestingly, infection by Pst DC3000 distinctly induced the expression of AtSDR1, but not AtSDR2. Expression of AtSDR1 was stronger in avirulent Pst DC3000 (avrRpm1)-infected plants than in virulent Pst DC3000 (empty)-infected plants. Expression patterns of AtSDR1, but not of AtSDR2, after infection with bacterial pathogens were similar to those of CaMNR1 (Figs. 4A and 10A). Furthermore, CaMNR1 shares higher sequence identity with AtSDR1 than with AtSDR2 (Fig. 1B). To test whether AtSDR1 has MNR activity, it was overexpressed in E. coli and purified to homogeneity following several chromatographic steps (Supplemental Fig. S6A). Purified AtSDR1 was a monomeric enzyme in solution, with a molecular mass of approximately 32 kD. Monoterpene products were quantified and identified by their GC retention times (Supplemental Fig. S6B). The optimal pH for AtSDR1 enzyme activity of menthone reduction to neomenthol was slightly higher than that of CaMNR1 (Fig. 2; Supplemental Fig. S6E). AtSDR1 converted (−)-menthone to 92% (+)-(3S)-neomenthol and 8% (−)-(3R)-menthol at pH 8.0 with NADPH as a cofactor (Supplemental Fig. S6, C and E). Neomenthol was the predominant reaction product catalyzed by AtSDR1, like CaMNR1, because the amount of menthol generated was less than 10% at all pH ranges. The oxidation of menthol isomers in the presence of NADP+ was also evaluated (Supplemental Fig. S6, D and F). Similar to the results of enzyme activity of CaMNR1, neomenthol was only converted into menthone by AtSDR1 in the presence of the NADP+ cofactor. The oxidation reaction of AtSDR1 displayed maximal activity at pH 9.0 (Supplemental Fig. S6F). Thus, these findings enabled us to further analyze the Arabidopsis AtSDR1 gene as a putative ortholog of CaMNR1.
Figure 10.
Enhanced disease susceptibility of sdr1 Arabidopsis mutants. A, RT-PCR analyses of the expression of Arabidopsis AtSDR1 and AtPR1 in the leaves of wild-type Col-0 plants at 6, 18, and 24 h after inoculation with virulent (empty) and avirulent (avrRpm1) Pst DC30000. B, Schematic diagram showing AtSDR1 and its T-DNA insertion mutants. C, RT-PCR analyses of transcript levels of AtSDR1 and AtPR1 in leaf tissues of wild-type Col-0 and sdr1 mutant plants at 18 and 24 h after inoculation with virulent (compatible [C]) and avirulent (avrRpm1; incompatible [I]) Pst DC30000 (106 cfu mL−1). The AtSDR1 transcript is absent in sdr1 plants. D, Bacterial growth in the leaves of wild-type and sdr1 mutant plants at 0 and 3 d after inoculation with virulent (empty) and avirulent (avrRpm1) strains of Pst DC3000 (5 × 104 cfu mL−1). Asterisks indicate significant differences as determined by Student's t test (P < 0.05). E, Disease symptoms on the cotyledons of wild-type and sdr1 mutant plants at 7 d after inoculation with H. parasitica (5 × 104 spores mL−1). F, Numbers of sporangiophores per cotyledon of wild-type and sdr1 mutant plants at 7 d after inoculation with H. parasitica. Diseased cotyledons were classified as follows: no sporulation (no sporangiophores per cotyledon), light sporulation (approximately 1–10 sporangiopores per cotyledon), medium sporulation (approximately 11–20 sporangiophores per cotyledon), or heavy sporulation (more than 20 sporangiophores per cotyledon). Sporangiophores were counted on more than 50 cotyledons per genotype. Average numbers of sporangiophores on the cotyledons of wild-type and mutant lines are shown at bottom for each of the lines tested. Statistically significant differences between means were determined employing the lsd (P = 0.05).
The sdr1-1 and sdr1-2 Arabidopsis mutants obtained from the Salk T-DNA populations (http://www.arabidopsis.org) have a T-DNA insertion within their first and third exons, respectively (Fig. 10B). In wild-type plants, AtSDR1 transcripts of expected sizes accumulated to a significant level at 18 and 24 h after inoculation with avirulent Pst DC3000. Therefore, we tested whether the sdr1 mutant exhibits altered responses to pathogens. In the sdr1 insertion homozygous mutants selected from the Salk T-DNA bulk lines, no transcripts of the expected size were detected at 18 and 24 h after avirulent Pst DC3000 infection (Fig. 10C). Furthermore, AtPR1 expression was distinctly reduced in the sdr1 mutant plants during infection with virulent and avirulent Pst DC3000 compared with wild-type plants. No obvious abnormal phenotypes in the sdr1-1 and sdr1-2 mutant plants were uncovered during plant growth or development (data not shown). To determine the role of AtSDR1 in plant defense, we first evaluated responses of the sdr1-1 and sdr1-2 mutant plants to virulent and avirulent strains of Pst DC3000. As shown in Figure 10D, the homozygous sdr1-1 and sdr1-2 mutants exhibited 5- to 10-fold more bacterial growth compared with wild-type plants at 3 d after inoculation with virulent and avirulent Pst DC3000. Significantly enhanced bacterial growth and reduced expression of AtPR1 in sdr1 mutant leaves indicated that AtSDR1 is required for basal defense or R gene-mediated resistance to the hemibiotrophic bacterial pathogen. We also examined the response of the sdr1 mutants to the biotrophic oomycete pathogen H. parasitica isolate Noco2 (Fig. 10, E and F). Interestingly, sdr1 mutant plants exhibited enhanced susceptibility to infection with H. parasitica isolate Noco2. Approximately 60% of the cotyledons of sdr1 mutants exhibited heavy sporulation (>20 sporangiophores per cotyledon), while 50% of the cotyledons of wild-type plants were heavily sporulated. The average number of sporangiophores in sdr1 mutant plants (sdr1-1, 25.1; sdr1-2, 25.3) was higher than in wild-type plants (21.3), but lower than in eds1 mutant plants (Landsberg erecta background, 28.2). These data indicate that AtSDR1 plays a crucial role in the defense response of Arabidopsis to the hemibiotrophic bacterial pathogen P. syringae and the biotrophic oomycete pathogen H. parasitica.
DISCUSSION
In this study, we identified a pepper CaMNR1 gene using an array-based differential screening method (Jung and Hwang, 2000) and its putative ortholog AtSDR1 in Arabidopsis. The amino acid sequences of CaMNR1 and AtSDR1 share 59% and 54% sequence identity, respectively, with peppermint MNR (Davis et al., 2005). The CaMNR1 and AtSDR1 proteins belong to the SDR family, based on length (300 ± 50 amino acids) and on conserved sequence motifs, including an N-terminal cofactor-binding domain (TGxxxGxG), a downstream structural domain (GxxDxxxNNAG) for stabilizing a central β-sheet, and a catalytic domain (YxxxK; Persson et al., 2003; Davis et al., 2005). SDRs are enzymes of great functional diversity with a wide substrate spectrum, ranging from steroids, alcohols, sugars, and aromatic compounds to xenobiotics (Kallberg et al., 2002; Persson et al., 2003).
CaMNR1 and AtSDR1 exhibited an enzymatic activity for menthone reduction, as predicted by significant sequence identities with peppermint MNR and MMR. Indeed, CaMNR1 shares 59% and 56% sequence identity with peppermint MNR and MMR, respectively. CaMNR1 and AtSDR1 possess both enzyme activities, but neomenthol generation is predominant. Compared with peppermint MNR, CaMNR1 showed not only greater substrate specificity but also greater catalytic power, indicating a much higher enzyme turnover number (Davis et al., 2005). The optimal pH of CaMNR1 and AtSDR1 was also different from that of peppermint MNR. CaMNR1 and AtSDR1 displayed maximal activities at neutral pH 7.5 and 8.0, respectively, whereas the activity of peppermint MNR enzyme was maximal at alkaline pH 9.3 (Davis et al., 2005). However, a direct comparison should not be overestimated, due to different assay conditions, including pH and the concentration of each component. The capability of CaMNR1 and AtSDR1 to oxidize monoterpenols was also tested in the presence of NADP+. Only oxidation of neomenthol to menthone was significantly detected by GC analysis. In contrast to peppermint MNR, the Km value of CaMNR1 for NADP+ was higher than that for NADPH. The cofactor binding affinity of CaMNR1 was significantly lower than that of peppermint menthone reductases. By contrast, the menthone-binding affinity of CaMNR1 was much higher than that of peppermint MNR but lower than that of peppermint MMR (Davis et al., 2005). Interestingly, CaMNR1 possessed significantly higher catalytic activity for neomenthol oxidation compared with peppermint menthone reductases (Davis et al., 2005). A very low level of neomenthol was detected at nearly all developmental stages, whereas a very high level of menthone was observed in peppermint oil. The very low activity of neomenthol oxidation by peppermint MNR is consistent with the in vivo level of each compound in peppermint (Davis et al., 2005). However, further studies on monoterpene metabolism in pepper and Arabidopsis plants are required to gain new insights into the enzymatic activity of CaMNR1 and AtSDR1 for neomenthol oxidation.
The enzymatic activities of CaMNR1 described in this study and the well-known antimicrobial activities of monoterpenes (Cowan, 1999; Dixon, 2001; Arfa et al., 2006) led us to test the antimicrobial activities of menthone, neomenthol, and menthol. Neomenthol and menthol strongly inhibited phytopathogenic bacterial and fungal growth compared with their precursor form, menthone. The differential antimicrobial activities of monoterpene compounds may be related to their different hydrophobicities, which can be expressed as their logP values (partitioning behavior of the compound in octanol/water; Lanciotti et al., 2003; Arfa et al., 2006). The estimated octanol/water partition coefficient (logP) was 2.87 for menthone and 3.38 for both neomenthol and menthol (http://www.syrres.com/esc/est_kowdemo.htm). Hydrophobic compounds with logP values higher than 3 exhibit efficient antimicrobial activities due to their high affinity for cell membranes (Arfa et al., 2006). The insertion of these compounds between cell membranes induces changes in membrane physicochemical properties, including lipid ordering and bilayer stability (Sikkema et al., 1995; Weber and de Bont, 1996). Interestingly, neomenthol and menthol, both of which have a logP of 3.38, were more effective at inhibiting phytopathogenic fungal and bacterial growth than menthone, which has a logP of 2.87. These findings support the idea that the production of neomenthol or menthol from menthone by pathogen-responsive CaMNR1 may enhance the immunity of pepper plants against pathogen infections.
In attempts to determine the metabolite profile of the terpenoid pathway in pepper leaves, we used GC-MS analyses to detect certain metabolic compounds that are de novo synthesized and accumulated by virulent and avirulent Xcv infections. Lipid-derived volatiles, including 2-hexenal and cis-3-hexenol identified in this study, were reported not only to increase in P. vulgaris leaves inoculated with Pseudomonas syringae pv phaseolicola but also to exhibit in vitro antifungal and antiprotozoal activity (Croft et al., 1993). Interestingly, MeSA was detected and its levels increased in extracts of pepper leaves inoculated with virulent and avirulent Xcv. Recently, Park et al. (2007) reported that MeSA is a critical mobile signal for systemic acquired resistance in tobacco (Nicotiana tabacum) plants and that silencing of salicylic acid methyl transferase, which converts SA to MeSA, compromised systemic acquired resistance. These findings support the notion that MeSA may play an important role in the defense response of pepper plants. De novo synthesis of trans-2-cis-6-nonadienal in pepper plants during an incompatible interaction with an avirulent Xcv strain is also consistent with HR-specific accumulation of this compound in P. vulgaris leaves (Croft et al., 1993). Trans-2-cis-6-nonadienal is known to be synthesized from the 9-hydroperoxide of linolenic acid by hydroperoxide lyase activity (Matthew and Galliard, 1978). The absence of trans-2-cis-6-nonadienal in uninfected healthy or virulent bacterial pathogen-infected P. vulgaris (Croft et al., 1993) and pepper leaves in this study suggests that a highly specific and common mode of lipid peroxidation occurs during HR in plants. Despite our various efforts to detect monoterpenes in pepper, we were unable to detect menthone, neomenthol, or menthol in the extracts of pepper leaves infected by Xcv. Only one terpenoid pathway-related metabolic compound, 2,6,10-trimethyldodecane (farnesane), the parent compound of approximately 10,000 sesquiterpenes (Breitmaier, 2007), was induced in Xcv-infected pepper leaves. This may be because these monoterpenes are present at extremely low levels or are absent in pepper plants. Plants elaborate a diverse array of secondary metabolites, many of which have evolved to confer resistance against pathogen attack (Grayer and Harborne, 1994; Pichersky and Gang, 2000; Dixon, 2001; Kliebenstein, 2004). However, little is known about the terpenoid pathways of members of the Solanaceae family, including pepper plants. The extremely low levels or the absence of these monoterpenes is probably why it has been difficult to analyze monoterpene pathways in pepper plants. Further detection of menthone metabolites in pepper plants will be required to gain insights into the in vivo function of CaMNR1 in defense responses to pathogen invasion.
To determine the expression patterns of CaMNR1 in pepper plants, we performed RNA and protein gel-blot analyses. CaMNR1 transcripts and proteins were strongly induced in pepper leaves inoculated with an avirulent strain of Xcv but not with a virulent strain. Avirulent strains of Xcv trigger resistance responses, including the oxidative burst, the HR, and PR gene induction in pepper plants (Lee and Hwang, 2005). Therefore, the increased expression of CaMNR1 suggests that it has a cellular function in the resistance response of pepper plants infected by avirulent strains of Xcv. To examine the effect of a loss of function of CaMNR1 in the defense response, we used the TRV-based VIGS system in pepper plants (Liu et al., 2002; Chung et al., 2004). Plants have evolved a two-branched defense system: pathogen-associated molecular patterns-triggered immunity (PTI) and effector-triggered immunity (ETI; Jones and Dangl, 2006). Recognition of pathogen-associated molecular patterns by plant receptor-like kinases triggers PTI responses, including the oxidative burst, callose deposition, and defense gene induction (Felix et al., 1999; Zipfel et al., 2004). To overcome PTI, pathogens deliver effectors to plant cells. Successful recognition of effectors by R proteins activates ETI responses, including the HR. However, unsuccessful recognition of effectors may enhance disease susceptibility. Interestingly, CaMNR1 gene-silenced pepper plants exhibited significantly enhanced susceptibility to virulent and avirulent strains of Xcv. These findings suggest that CaMNR1 may act as a common signaling factor for both PTI and ETI. However, its signal intensities were stronger in ETI than in PTI.
RT-PCR and protein gel-blot analyses showed that silencing of the CaMNR1 gene significantly reduced CaMNR1 transcript and protein levels in pepper leaves inoculated with virulent and avirulent strains of Xcv. Furthermore, expression levels of defense-related genes, including CaBPR1, CaPR4, CaPR10, CaDEF1, and CaSAR8.2, were significantly lower in CaMNR1-silenced leaves compared with unsilenced leaves after infection with Xcv. Local and systemic induction of some defense-related genes has been reported for pepper plants (Lee and Hwang, 2005). Defense-related proteins such as CaSAR8.2 and defensin exhibit antimicrobial activity (Broekaert et al., 1995; Lee and Hwang, 2006). The lowered expression of defense-related genes in CaMNR1-silenced leaves indicates that CaMNR1 expression is important for the induction of downstream defense-related genes. Consistent with these results, CaMNR1-silenced pepper plants exhibited enhanced susceptibility against the hemibiotrophic bacterial pathogen Xcv and the necrotrophic fungal pathogen C. coccodes.
To address the biological function of CaMNR1, we attempted to generate transgenic pepper plants that constitutively expressed the CaMNR1 gene. Unfortunately, this approach was unsuccessful due to extremely low efficiencies of transformation and regeneration. Therefore, we established transgenic Arabidopsis plants that overexpressed CaMNR1 from the cauliflower mosaic virus 35S promoter. Interestingly, ectopic overexpression of CaMNR1 induced the constitutive expression of PR genes in uninfected Arabidopsis plants. The expression levels of Arabidopsis AtPR1 and AtPDF1.2 correlated with CaMNR1 expression levels in transgenic Arabidopsis. The induction of defense responses is known to be activated by various signal transduction pathways, which are regulated by signaling molecules, such as SA, JA, and ethylene (Glazebrook, 1999; Lee and Hwang, 2005). In Arabidopsis, induction of AtPR1 occurs via a SA-dependent pathway (Ukness et al., 1992), whereas the induction of AtPDF1.2 (defensin) depends on the JA-dependent pathway (Penninckx et al., 1998; Thomma et al., 1998). This suggests that CaMNR1 plays a critical role in the regulation of PR genes through both SA- and JA-dependent pathways. Moreover, ectopic overexpression of CaMNR1 significantly enhanced basal resistance against infection by Pseudomonas syringae and H. parasitica. In contrast, T-DNA insertion mutations in the AtSDR1 gene, a putative ortholog of CaMNR1 in Arabidopsis plants, significantly enhanced the susceptibility of Arabidopsis plants to these pathogens, followed by lowered expression of AtPR1. These findings suggest that CaMNR1 and AtSDR1 may play roles as signaling factors that may increase the basal resistance of plants. In mammalian cells, carbonyl reductase (CR), a well-characterized SDR family enzyme, shows homology with CaMNR1. Reactive carbonyl groups, which are frequently found in endogenous or xenobiotic compounds, can covalently modify DNA or amino acids and initiate degenerative diseases or cancers (Oppermann, 2007). CRs detoxify these reactive carbonyl groups. Moreover, CRs are also important for the metabolic conversion of carbonyl groups of endogenous hormones, such as glucocorticoids, androgens, estrogens, and eicosanoids. Therefore, we can propose some possible mechanisms for increased and decreased basal resistance in the CaMNR1 transgenic lines and sdr1 mutants, respectively. CaMNR1 or AtSDR1 may catalyze the conversion of inactive endogenous or xenobiotic compounds into active forms that can exhibit antimicrobial activities or activate defense responses in plants. Alternatively, CaMNR1 may also detoxify xenobiotic effector molecules derived from pathogens to reduce the virulence of pathogens.
Taking all of the available evidence together, this study provides clues for the elucidation of the cellular functions of pepper CaMNR1 and its putative ortholog Arabidopsis AtSDR1 in plant defense responses. VIGS of CaMNR1 in pepper and the T-DNA insertion mutation of AtSDR1 in Arabidopsis led to significantly enhanced susceptibility to both bacterial and fungal pathogens. In contrast, ectopic overexpression of CaMNR1 significantly enhanced disease resistance in Arabidopsis. Interestingly, the expression levels of CaMNR1 correlated with those of SA- and JA-responsive PR genes in both pepper and Arabidopsis plants, suggesting the functional involvement of CaMNR1 in a broad spectrum of defense responses through the regulation of downstream defense-related genes.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Pepper (Capsicum annuum ‘Nockwang’) and Arabidopsis (Arabidopsis thaliana ecotype Col-0) plants were used in this study. Pepper plants were grown in a plastic tray (55 × 35 × 15 cm) containing a steam-sterilized soil mix (peat moss, perlite, and vermiculite, 5:3:2, v/v/v) and loam soil (1:1, v/v) at 28°C with a daylength of 16 h at a light intensity of 70 μmol photons m−2 s−1. Pepper plants at the six-leaf stage were used for pathogen infection and abiotic elicitor and environmental stress treatments. Arabidopsis plants were grown at 24°C with a photosynthetic flux of 130 μmol photons m−2 s−1 for 8 h of light and 60% relative humidity in a controlled-environment chamber. Plants were raised in pots containing peat moss, perlite, and vermiculite (1:0.5:1, v/v/v). Prior to sowing, wild-type (Col-0) and CaMNR1-OX mutant seeds were sterilized with 2% (v/v) sodium hypochlorite, followed by imbibition at 4°C for 3 d to overcome dormancy.
Pathogens and Inoculation Procedures
Pepper plants were inoculated with virulent (Ds1) and avirulent (Bv5-4a) strains of Xanthomonas campestris pv vesicatoria (Xcv). Bacteria were cultured overnight in yeast-nutrient broth (5 g of yeast extract and 8 g of nutrient broth per L) at 28°C. Pepper plants at the six-leaf stage were inoculated by infiltrating bacterial suspensions into the abaxial side of fully expanded leaves using a syringe without a needle. Infected plants were incubated in a controlled moist chamber at 28°C with 100% relative humidity for 16 h, and infected leaves were harvested at various time points for bacterial growth and RNA and protein gel-blot analyses (Kim et al., 2007).
Virulent (empty vector) and avirulent (avrRpm1 and avrRpt2) strains of Pseudomonas syringae pv tomato (Pst) DC3000 were used for infection of Arabidopsis. Pst DC3000 was grown overnight in King's B medium containing 50 μg mL−1 rifampicin and 50 μg mL−1 kanamycin. To measure bacterial growth, leaves of 4-week-old wild-type and CaMNR1 transgenic T2 plants were infiltrated with 105 cfu mL−1 (optical density at 600 nm [OD600] = 0.001) Pst DC3000 in 10 mm MgCl2 using a syringe without a needle. The infected plants were incubated in a moist chamber at 28°C for 18 h, and infected leaves were harvested at various time points for bacterial growth and RNA gel-blot analyses. The oomycete pathogen Hyaloperonospora parasitica isolate Noco2 was propagated at weekly intervals on susceptible Col-0 plants (Reignault et al., 1996). One-week-old seedlings of wild-type (Col-0) and CaMNR1-OX transgenic plants were spray inoculated using freshly harvested conidiospores (5 × 104 spores mL−1). The infected plants were incubated at 17°C in a controlled-environment chamber. Sporangiophores were counted at 7 d after inoculation. Cotyledons were stained with trypan blue to visualize host cell death and oomycete structures.
Antimicrobial Activity of Monoterpenes
The antimicrobial activities of the monoterpenes (−)-menthone, (+)-(3S)-neomenthol, and (−)-(3R)-menthol (Sigma-Aldrich) were analyzed in the vapor phase by the microatmosphere method (Arfa et al., 2006). To prepare bacterial inocula for evaluation of antimicrobial activity of these monoterpenes, Pst DC3000 and Xcv were cultured in nutrient broth for 18 h at 28°C. Bacterial cells were collected by centrifugation and resuspended in 10 mm MgCl2 solution to 107 cfu mL−1. The bacterial suspensions were uniformly spread on nutrient agar medium. Alternaria brassicicola and Colletotrichum coccodes grown in potato dextrose agar medium were sporulated on oatmeal agar medium at 28°C. After harvesting spores in sterile tap water, the concentrations of the spore suspensions were adjusted to 105 spores mL−1. These spore suspensions were then uniformly spread on 1% water agar. Different amounts of the monoterpenes (0.1, 0.5, 1.5, 10, and 50 mg per plate [9 cm in diameter]) used to inhibit the growth of plant pathogens were dissolved in 1 mL of ethyl acetate, impregnated homogeneously on sterilized filter paper (63 cm2), and dried for 1 min to evaporate ethyl acetate. Filter papers were placed on the lids of petri plates containing 20 mL of agar medium whose surfaces had been uniformly covered with the tested bacterial and fungal pathogens. Controls were made with papers impregnated with 1 mL of ethyl acetate alone. The petri plates were incubated for 48 h at 28°C for bacterial growth or for 12 and 24 h at 28°C for spore germination and hyphal growth of fungi, respectively.
Isolation and Sequence Analysis of Pathogen-Induced CaMNR1 cDNAs
For the construction of a pathogen-induced cDNA library, the Xcv avirulent strain Bv5-4a was inoculated into pepper leaves. A pepper cDNA library was constructed using 5 μg of poly(A)+ mRNA extracted from inoculated pepper leaves (Kim and Hwang, 2000). To isolate pathogen-inducible cDNAs from the pepper cDNA library, macroarray-based differential screening was performed according to the method of Jung and Hwang (2000). The CaMNR1 cDNA sequence was analyzed using BLAST programs provided by the National Center for Biotechnology Information databases (Altschul et al., 1997). Amino acid sequences were aligned using the ClustalW program (http://www.ebi.ac.uk/clustalw). The phylogenetic tree was generated using ClustalW and displayed with Treeview 1.6.6.
Subcloning of CaMNR1 and AtSDR1
Site-directed mutagenesis was utilized to remove 34 amino acid residues between the thrombin cleavage and EcoRI restriction sites of the vector pET-32a (Novagen). Forward (5′-CTGTCGCCACGCGGTTCTGAATTCAAAGAAACCGCTGC-3′) and reverse (5′-GCAGCGGTTTCTTTGAATTCAGAACCGGCGTGGCACCAG-3′) primers containing EcoRI restriction sites (underlined) were used for QuikChange site-directed mutagenesis (Stratagene). The products were amplified by the addition of 10 units of LA Taq DNA polymerase (TaKaRa) with approximately 10 ng of template DNA, 2.5 mm deoxynucleoside triphosphates, 10 mm of each primer, and 2× GC buffer in 50 μL using a Thermal Cycler 2720 (Applied Biosystems). The mutated plasmids were digested with EcoRI and religated with T4 DNA ligase (TaKaRa). Resultant DNAs were transformed into Escherichia coli BL21(DE3).
The AtSDR1 gene was cloned from the Arabidopsis cDNA library by PCR amplification using the forward (5′-GGATCCATGGCAGAGGAAACTCCAAGATATG-3′) and reverse (5′-CTCGAGTCAGAATTCTGAAACTTGCTGCG-3′) primers, which contain the BamHI and XhoI restriction sites for cloning. The resultant PCR product was cloned into modified pGEX-4T vector, which has an N-terminal glutathione S-transferase (GST) followed by a tobacco etch virus protease cleavage site. The cloned vector was transformed into E. coli Rosetta(DE3) cells.
Overexpression and Purification of CaMNR1 and AtSDR1
E. coli containing the CaMNR1- or AtSDR1-expressing plasmid was incubated with vigorous shaking at 37°C until the OD600 reached approximately 0.7. CaMNR1 and AtSDR1 were induced by the addition of 1 mm isopropylthio-β-galactoside at 25°C for 12 h. The cells were pelleted by centrifugation for 30 min (6,000 rpm; Beckman JA-10 rotor), resuspended in 50 mm Tris-HCl buffer (pH 8.5; CaMNR1) or 1× PBS buffer (pH 7.4; AtSDR1), and then disrupted by sonication. The lysate was centrifuged for 1 h (15,000 rpm; Beckman JA-20 rotor), and the supernatant was collected. To purify His-tagged thioredoxin-CaMNR1 protein, the soluble fraction was loaded onto a His-Trap HP column (GE Healthcare) preequilibrated with 50 mm Tris-HCl, pH 8.5, and 100 mm NaCl, and the target enzyme was eluted with 150 mm imidazole. To purify GST-tagged AtSDR1 protein, the soluble fraction was loaded onto the column packed with GST-agarose beads preequilibrated with 1× PBS buffer, pH 7.4, and GST-tagged target protein was eluted with 50 mm Tris-HCl, pH 8.0, 10 mm NaCl, and 20 mm reduced glutathione.
Fractions containing the His-tagged thioredoxin-CaMNR1 or GST-tagged AtSDR1 fusion protein were incubated with 120 units of human α-thrombin or tobacco etch virus protease at 22°C for 12 h (1:1,000 or 1:50 molar ratio, respectively). The thioredoxin- and GST-free target proteins were further purified by gel filtration chromatography (Superose 12) followed by anion-exchange chromatography (Resource Q or Mono Q column, respectively). The concentrations of purified CaMNR1 and AtSDR1 were determined by the Bradford assay (Bradford, 1976).
Activity Assay of CaMNR1 and AtSDR1
Preparatory identification and quantification were done with the enzyme preparation (400 μg of CaMNR1 and AtSDR1) in 1 mL of assay buffer (50 mm HEPES, pH 7.5, with 100 mm NaCl and 5 mm β-mercaptoethanol) in the presence of 1 mm menthone and 500 μm NADPH. Following incubation at 31°C for 12 h, monoterpene products were extracted with 0.5 mL of pentane as described (Davis et al., 2005). Reaction products were analyzed by GC and GC-MS (Hewlett-Packard model 6890 series gas chromatograph) with a 25-m × 0.2-mm × 0.33-μm film of polyethylene glycol coating (HP-FFAP 19091F-102; Agilent Technologies) as described. For quantification of the reaction products, 100 μm (+)-camphor was added as an internal standard. The enzymatic activity depending on pH was measured by the same procedure described above, except with an assay buffer of a different pH range (pH 6–10, with intervals of 0.5 pH units). The enzymatic activity for the reverse (oxidation) reaction was also analyzed in the presence of 1 mm neomenthol as a substrate and 500 μm NADP+ as a cofactor. All experimental procedures were essentially the same as for the forward (reduction) reaction, except for a buffer for optimal pH (50 mm Bicine, pH 9.0).
Analysis of CaMNR1 Enzyme Kinetics
Enzyme kinetics were analyzed as described with minor modifications (Davis et al., 2005). Assay mixtures containing 1 mL of 50 mm HEPES, pH 7.5, with 100 mm NaCl and 5 mm β-mercaptoethanol for the forward reaction and 1 mL of 50 mm Bicine, pH 9.0, with 100 mm NaCl and 5 mm β-mercaptoethanol for the reverse reaction were combined with 500 μm cofactor (NADPH or NADP+) and various concentrations (2.5–500 μm) of a monoterpene substrate (menthone or neomenthol) and preincubated at 31°C before initiation of the reaction by enzyme addition (15 μg of CaMNR1). The reactions were stopped after 15 min by the addition of 0.5 mL of pentane containing camphor as an internal standard, followed by vigorous mixing and cooling on ice. The reaction products in the pentane extract were quantitated by GC using the conditions described above with peak area integration in the chromatograph. The kinetic parameters for the cofactor were determined spectrophotometrically (ΔA340) based on the disappearance or appearance of NADPH. The same assay mixtures were combined with 100 μm monoterpene substrate and various concentrations (2.5–500 μm) of the cofactor (NADPH or NADP+). Kinetic parameters were determined by analyzing Lineweaver-Burk plots from four independent experiments.
Extraction of Metabolic Compounds from Pepper Leaves
The liquid-liquid extraction method was used to extract metabolic compounds from pepper leaves. Five grams of uninoculated and Xcv-inoculated pepper leaves was sampled, ground to a fine powder in liquid nitrogen, and mixed with 10 mL of 70% ethanol. Fifty micrograms of camphor was added to each sample as an internal standard. The mixture was stored for 1 d at room temperature. Then, 5 mL of the supernatant of the leaf extract was mixed with 5 mL of distilled water and 1.2 mL of organic solvent A (ethyl acetate:hexane:methylene chloride, 5:1:1, v/v/v; Gherman et al., 2000). The mixture was agitated for 1 h, and 1 mL of the supernatant was dried by nitrogen gas. The residual pellet was dissolved in 100 μL of solvent A, and 5 μL of the extract was used for GC-MS analysis.
GC-MS Analysis
An Agilent 6890N gas chromatograph (Agilent Technologies) coupled with an Agilent 5975A mass spectrometer (Agilent Technologies) was used in this study. The compound mixture was separated on a DB Wax-fused silica capillary column (60 m length, 0.25 mm diameter, and 0.25 μm film thickness; J&W Science) with a temperature program of 75°C (kept for 8 min) to 200°C (kept for 5 min) at a rate of 2°C min−1. The injector temperature was 270°C, and the flow rate of the carrier gas, helium, was 1 mL min−1. The Agilent mass spectrometer had an electron energy of 70 eV, electron emission of 300 μA, and ion source temperature of 100°C. The resulting chromatograms were analyzed using the National Institute of Standards and Technology mass spectral library version 2.0 d.
Treatment with Abiotic Elicitors and Environmental Stresses
Leaves from pepper plants at the six-leaf stage were sprayed with 5 mm SA, 100 μm MeJA, or 10 mm H2O2. Pepper plants treated with MeJA were tightly sealed in a plastic bag. For ethylene treatment, whole pepper plants were removed from soil and then placed in a water-containing small glass chamber, followed by injection of ethylene gas (5 μL L−1). For drought stress treatment, whole pepper plants were removed from soil, washed to remove soil particles attached to roots, and then dried. Mechanical wounding stress was performed by injuring leaves with needles. Plants were placed at 4°C for low-temperature treatment. Leaves treated with various elicitors or subjected to environmental stresses were harvested at various time points and stored at −70°C until used for RNA isolation.
RNA Isolation and RNA Gel-Blot Analysis
Total RNA was extracted from pepper leaves, stems, roots, flowers, and fruits using the guanidine isothiocyanate method (Chomczynski and Sacchi, 1987). Total cellular RNA of CaMNR1-OX Arabidopsis T2 plants was also extracted from the aerial portions using TRIzol Reagent (Invitrogen) according to the manufacturer's instructions. For probe generation, the coding region of the CaMNR1 gene was amplified using gene-specific primers (forward, 5′-ATGGCAGAGAAAACCACCAGC A-3′; reverse, 5′-TCAAAAAAAAGTGACCTCCTTTCTGT-3′). The amplified PCR product was labeled with 32P using a random priming kit (Boehringer Mannheim). Agarose gel electrophoresis, RNA transfer, and hybridization with the CaMNR1 fragment were performed using standard procedures.
Protein Preparation and Protein Gel-Blot Analysis
The His-tagged CaMNR1 fusion proteins were subjected to SDS-PAGE and purified from the excised bands before being injected into rabbits to generate immune sera against CaMNR1 (LabFrontier). Specific binding of immune sera to CaMNR1 was confirmed, and the antibodies were then used for western-blot analysis at a 1:2,000 dilution.
Total protein extracts were prepared by grinding 200 mg of leaf tissue of pepper and Arabidopsis in 1 mL of grinding buffer (20 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm EDTA, 1% Triton X-100, 0.1% SDS, 5 mm dithiothreitol, and Complete Protease Inhibitor Cocktail [Roche]), followed by pelleting the insoluble debris by centrifugation at 20,000g for 15 min at 4°C. The concentrations of proteins in the supernatants were determined by the Bradford protein assay (Bradford, 1976). About 20 μg of total protein was separated on 10% SDS-PAGE gels and transferred to polyvinylidene difluoride membranes. Western blotting was done by the standard method. Equal loading of proteins was confirmed by comparing the band intensities of the proteins on a Coomassie blue-stained SDS-PAGE gel.
VIGS
The TRV-based VIGS system was used for gene silencing in pepper plants as described by Liu et al. (2002). The full-length CaMNR1 open reading frame was amplified by PCR using gene-specific primers (forward, 5′-ATGGCAGAGAAAACCACCAGCA-3′; reverse, 5′-TCAAAAAAAAGTGACCTCCTTTCTGT-3′). The pepper CaMNR1 gene was cloned into the vector pTRV2 to yield pTRV2:CaMNR1. Agrobacterium tumefaciens strain GV3101 carrying pTRV1 or pTRV2:CaMNR1 was coinfiltrated into the fully expended cotyledons of pepper plants (OD600 = 0.2). Plants were placed in a growth room at 25°C with a 16-h-light/8-h-dark photoperiod for growth and viral spread.
Identification and Isolation of CaMNR1-OX Transgenic Lines and sdr1 Mutants
Transgenic Arabidopsis plants expressing the CaMNR1 gene were generated using the floral dipping method (Clough and Bent, 1998). Three lines of putative transgenic Arabidopsis plants (T1) harboring 35S:CaMNR1 were selected by plating seeds on Murashige and Skoog medium (Duchefa) containing 50 mg L−1 kanamycin.
T-DNA insertion mutants (Salk_147665 and Salk_031785) of the AtSDR1 gene were obtained from the ABRC (Ohio State University). The kanamycin-resistant plants were selected on Murashige and Skoog agar medium containing 50 mg L−1 kanamycin. To identify the homozygous mutant lines, PCR was performed with genomic DNA of sdr1-1 and sdr1-2 mutants using the following gene-specific primers: sdr1-1 LP (5′-GTGAACCGATGGATTGAATTG-3′), sdr1-1 RP (5′-TTGGGTTTTCACAAACTCAGC-3′), sdr1-2 LP (5′-CAAGACTAAAACAACGGCGTC-3′), sdr1-2 RP (5′-CCCATGGAGGATGATACATTG-3′), and left-border-specific primer LBb1 (5′-GCGTGGACCGCTTGCTGCAACT-3′).
RT-PCR Analysis
Total RNA was extracted from pepper and Arabidopsis leaves as described above. RT reactions were performed with total RNA (2 μg) and oligo p(dT)15 primer (Roche) at 42°C using avian myeloblastosis virus reverse transcriptase (Roche) in a 20-μL reaction volume. Aliquots (1 μL) of RT reaction products were used for RT-PCR analysis with the following gene-specific primers: CaBPR1F (5′-CAGGATGCAACACTCTGGTGG-3′) and CaBPR1R (5′-ATCAAAGGCCGGTTGGTC-3′) for CaBPR1 (accession no. AF053343); CaPR4F (5′-GCGGTAGATGCTTGAGGGT-3′) and CaPR4R (5′-CAATCTCGACAATAGTATGAAATCA-3′) for CaPR4 (accession no. AF244122); CaPR10F (5′-TGTCGAAGGTGGTCCAATAAA-3′) and CaPR10R (5′-TAGACAGAAGGATTGGCGAGG-3′) for CaPR10 (accession no. AF244121); CaPOA1F (5′-ATCTGTACCAGCTTGCACGTGT-3′) and CaPOA1R (5′-CCCTCACTGTGGCCTTGG-3′) for CaPOA1 (accession no. AF442387); CaDEF1F (5′-CAAGGGAGTATGTGCTAGTGAGAC-3′) and CaDEF1R (5′-TGCACAGCACTATCATTGCATAC-3′) for CaDEF1 (accession no. AF442388); CaOSM1F (5′-ACATTTCAGTAATCGATGGATTCA-3′) and CaOSM1R (5′-TAGTCCAACTTTGGCAAGTAAAT-3′) for CaOSM1 (accession no. AY262059); CaSAR8.2F (5′-CAGGGAGATGAATTCTGAGGC-3′) and CaSAR8.2R (5′-CATATGAACCTCTATGGATTTCG-3′) for CaSAR8.2 (accession no. AF313766); AtPR1F (5′-ATGAATTTTACTGGCTTCTCG-3′) and AtPR1R (5′-TTAGTATGGCTTCTCGTTCACAT-3′) for AtPR1 (accession no. At2G14610); AtPDF1.2F (5′-ATGGCTAAGTTTGCTTCCATC-3′) and AtPDF1.2R (5′-TTAACATGGGACGTAAGTAA-3′) for AtPDF1.2 (accession no. At5G44420); AtRD29AF (5′-GGTAGTGAATCAGGAGCTGAGC-3′) and AtRD29AR (5′-TCCACCTCCGGAGATAGGTA-3′) for AtRD29A (accession no. D13044); and AtUBQF (5′-GTAGTGCTAAGAAGAGCAAGA-3′) and AtUBQR (5′-TCAAGCTTATTCTT-3′) for AtUBQ (accession no. At3g62250). RT-PCR conditions were 95°C for 10 min and 30 cycles of 95°C for 30 s, 52°C for 30 s, and 72°C for 1.5 min. Single bands for PCR products were confirmed on an agarose gel.
Sequence data of CaMNR1 from this article have been deposited in the GenBank data library under accession number EF576664.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Nucleotide sequence and deduced amino acid sequence of the CaMNR1 cDNA encoding the MNR protein.
Supplemental Figure S2. RNA gel-blot analysis of the expression of CaMNR1 in pepper plants.
Supplemental Figure S3. Comparison of GC-MS total ion chromatograms of pepper leaf extracts with those of standard mixtures.
Supplemental Figure S4. GC-MS total ion chromatograms of extracts from pepper leaves infected with virulent (Ds1) or avirulent (Bv5-4a) Xcv.
Supplemental Figure S5. Mass spectra of various metabolic compounds obtained from leaf extracts of pepper plants.
Supplemental Figure S6. Purification and enzyme activity of AtSDR1.
Supplementary Material
Acknowledgments
We thank Dr. S.P. Dinesh-Kumar (Yale University) for the vectors pTRV1 and pTRV2, Dr. U. Bonas (Martin-Luther-Universität) for Agrobacterium strain GV3101, and Dr. Jonathan D.G. Jones (John Innes Centre) for H. parasitica isolate Noco2 and eds1 Arabidopsis mutants. We also thank Dr. B.S. Kim (Korea University) for helpful discussions and comments on GC-MS data analysis.
This work was supported by grants from the Crop Functional Genomics Center of the 21st Century Frontier Research Program funded by the Ministry of Science and Technology, Korea (B.K.H.), the Center for Plant Molecular Genetics and Breeding Research, Seoul National University, Korea (B.K.H.), the BioGreen21 Program, Rural Development Administration, Korea (H.K.S. and B.K.H.), and the Plant Signaling Network Research Center, Korea Science and Engineering Foundation, Korea University, Korea (H.K.S.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Byung Kook Hwang (bkhwang@korea.ac.kr).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408 796–815 [DOI] [PubMed] [Google Scholar]
- Arfa AB, Combes S, Preziosi-Belloy L, Gontard N, Chalier P (2006) Antimicrobial activity of carvacrol related to its chemical structure. Lett Appl Microbiol 43 149–154 [DOI] [PubMed] [Google Scholar]
- Arimura G, Ozawa R, Shomoda T, Nishioka T, Boland W, Takabayashi J (2000) Herbivory-induced volatiles elicit defense genes in lima bean leaves. Nature 406 512–515 [DOI] [PubMed] [Google Scholar]
- Baulcombe DC (1999) Fast forward genetics based on virus-induced gene silencing. Curr Opin Plant Biol 2 109–113 [DOI] [PubMed] [Google Scholar]
- Biere A, Marak HB, Van Damme MM (2004) Plant chemical defense against herbivores and pathogens: generalized defense or trade-offs? Oecologia 140 430–441 [DOI] [PubMed] [Google Scholar]
- Bowyer P, Clarke BR, Lunness P, Daniels MJ, Osbourn AE (1995) Host range of a plant pathogenic fungus determined by a saponin detoxifying enzyme. Science 267 371–374 [DOI] [PubMed] [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72 248–254 [DOI] [PubMed] [Google Scholar]
- Breitmaier E (2007) Sesquiterpenes. In E. Breitmaier, ed, Terpenes. Wiley-VCH, Weinheim, Germany, pp 22–51
- Broekaert WF, Terras FRG, Cammue BPA, Osborn RW (1995) Plant defensins: novel antimicrobial peptides as components of the host defense system. Plant Physiol 108 1353–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HW, Kim YJ, Lee SC, Hong JK, Hwang BK (2007) Hydrogen peroxide generation by the pepper extracellular peroxidase CaPO2 activates local and systemic cell death and defense response to bacterial pathogens. Plant Physiol 145 890–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 162 156–159 [DOI] [PubMed] [Google Scholar]
- Chung E, Seong E, Kim YC, Chung EJ, Oh SK, Lee S, Park JM, Joung YH, Choi D (2004) A method of high frequency virus-induced gene silencing in chili pepper (Capsicum annuum L. cv. Bukang). Mol Cells 17 377–380 [PubMed] [Google Scholar]
- Chung ES, Oho SK, Park MJ, Choi D (2007) Expression and promoter analyses of pepper CaCDPK4 (Capsicum annuum calcium dependent protein kinase 4) during plant defense response to incompatible pathogen. Plant Pathol J 23 76–89 [Google Scholar]
- Cichewicz RH, Thorpe PA (1996) The antimicrobial properties of chile peppers (Capsicum species) and their uses in Mayan medicine. J Ethnopharmacol 52 61–70 [DOI] [PubMed] [Google Scholar]
- Clough SH, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Cowan MM (1999) Plant products as antimicrobial agents. Clin Microbiol Rev 12 564–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft KPC, Jutter F, Slusrenko AJ (1993) Volatile products of the lipoxygenase pathway evolved from Phaseolus vulgaris (L.) leaves inoculated with Pseudomonas syringae pv. phaseolicola. Plant Physiol 101 13–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croteau R, Davis EM, Ringer KL, Wildung MR (2005) Menthol biosynthesis and molecular genetics. Naturwissenschaften 92 562–577 [DOI] [PubMed] [Google Scholar]
- Davis EM, Croteau R (2000) Cyclization enzymes in the biosynthesis of monoterpenes, sesquiterpenes, and diterpenes. Top Curr Chem 209 53–95 [Google Scholar]
- Davis EM, Ringer KL, McConkey E, Croteau R (2005) Monoterpene metabolism: cloning, expression, and characterization of menthone reductase from peppermint. Plant Physiol 137 873–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Luca V, St Pierre B (2000) The cell and developmental biology of alkaloid biosynthesis. Trends Plant Sci 5 168–173 [DOI] [PubMed] [Google Scholar]
- Dixon RA (2001) Natural products and plant disease resistance. Nature 411 843–847 [DOI] [PubMed] [Google Scholar]
- Essenberg M, Doherty MA, Hamilton BK, Henning VT, Cover EC, McFaul SJ, Johnson MW (1982) Identification and effects on Xanthomonas campestris pv. malvacearum of two phytoalexins from leaves and cotyledons of resistant cotton. Phytopathology 72 1349–1356 [Google Scholar]
- Essenberg M, Grover PB Jr, Cover EC (1990) Accumulation of antibacterial sesquiterpenoids in bacterially inoculated Gossypium leaves and cotyledons. Phytochemistry 29 3107–3113 [Google Scholar]
- Felix G, Duran JD, Volko S, Boller T (1999) Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J 18 265–276 [DOI] [PubMed] [Google Scholar]
- Gershenzon J, McConkey ME, Croteau RB (2000) Regulation of monoterpene accumulation in leaves of peppermint. Plant Physiol 122 205–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gherman C, Culea M, Cozar O (2000) Comparative analysis of some active principles of herb plants by GC/MS. Talanta 53 253–262 [DOI] [PubMed] [Google Scholar]
- Ghoshal S, Prasad BN, Lakshmi V (1996) Antiamoebic activity of Piper longum fruits against Entamoeba histolytica in vitro and in vivo. J Ethnopharmacol 50 167–170 [DOI] [PubMed] [Google Scholar]
- Glazebrook J (1999) Genes controlling expression of defense responses in Arabidopsis. Curr Opin Plant Biol 2 280–286 [DOI] [PubMed] [Google Scholar]
- Grayer RJ, Harborne JB (1994) A survey of antifungal compounds from higher plants. Phytochemistry 37 19–42 [DOI] [PubMed] [Google Scholar]
- Harrigan GG, Ahmad A, Baj N, Glass TE, Gunatilaka AAL, Kingston DGI (1993) Bioactive and other sesquiterpenoids from Porella cordeana. J Nat Prod 56 921–925 [DOI] [PubMed] [Google Scholar]
- Jones JD, Dangl JL (2006) The plant immune system. Nature 444 323–329 [DOI] [PubMed] [Google Scholar]
- Jung HW, Hwang BK (2000) Isolation, partial sequencing, and expression of pathogenesis-related cDNA genes from pepper leaves infected by Xanthomonas campestris pv. vesicatoria. Mol Plant Microbe Interact 13 136–142 [DOI] [PubMed] [Google Scholar]
- Kallberg Y, Oppermann U, Jornvall H, Persson B (2002) Short-chain dehydrogenases/reductases (SDRs). Eur J Biochem 269 4409–4417 [DOI] [PubMed] [Google Scholar]
- Kallberg Y, Persson B (2006) Prediction of coenzyme specificity in dehydrogenases/reductases: a hidden Markov model-based method and its application on complete genomes. FEBS J 273 1177–1184 [DOI] [PubMed] [Google Scholar]
- Kim BS, Kim YC, Shin KS, Kim JH (2007) Near-isogenic lines for genes conferring hypersensitive resistance to bacterial spot in chili pepper. Plant Pathol J 23 155–160 [Google Scholar]
- Kim YJ, Hwang BK (2000) Pepper gene encoding a basic pathogensis-related 1 protein is pathogen and ethylene inducible. Physiol Plant 108 51–60 [Google Scholar]
- Kjonaas R, Martinkus-Taylor C, Croteau RB (1982) Metabolism of monoterpenes: conversion of l-menthone to l-menthol and d-neomenthol by stereospecific dehydrogenases from peppermint (Mentha piperita) leaves. Plant Physiol 69 1013–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein DJ (2004) Secondary metabolites and plant/environment interactions: a view through Arabidopsis thaliana tinged glasses. Plant Cell Environ 20 675–684 [Google Scholar]
- Kubo IH, Muroi H, Himejima M (1993) Combination effects of antifungal nagilactones against Candida albicans and two other fungi with phenylpropanoids. J Nat Prod 56 220–226 [DOI] [PubMed] [Google Scholar]
- Lanciotti R, Belleti N, Patrignani F, Gianotti A, Gardini F, Guerzoni ME (2003) Application of hexanal, (E)-2-hexanal, and hexyl acetate to improve the safety of fresh-sliced apples. J Agric Food Chem 51 2958–2963 [DOI] [PubMed] [Google Scholar]
- Lee SC, Hwang BK (2005) Induction of some defense-related genes and oxidative burst is required for the establishment of systemic acquired resistance in Capsicum annuum. Planta 221 790–800 [DOI] [PubMed] [Google Scholar]
- Lee SC, Hwang BK (2006) CASAR82A, a pathogen-induced pepper SAR8.2, exhibits an antifungal activity and its overexpression enhances disease resistance and stress tolerance. Plant Mol Biol 61 95–109 [DOI] [PubMed] [Google Scholar]
- Litvak ME, Monson RK (1998) Patterns of induced and constitutive monoterpene production in conifer needles in relation to insect herbivory. Oecologia 144 531–540 [DOI] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Dinesh-Kumar SP (2002) Virus-induced gene silencing in tomato. Plant J 31 777–786 [DOI] [PubMed] [Google Scholar]
- Mahmoud SS, Croteau RB (2001) Metabolic engineering of essential oil yield and composition in mint by altering expression of deoxyxylulose phosphate reductoisomerase and menthofuran synthase. Proc Natl Acad Sci USA 98 8915–8920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthew JA, Galliard T (1978) Enzymic formation of carbonyls from linoleic acid in leaves of Phaseolus vulgaris. Phytochemistry 17 1043–1044 [Google Scholar]
- Mendoza L, Wilkens M, Urzua A (1997) Antimicrobial study of the resinous exudates and of diterpenoids and flavonoids isolated from some Chilean Pseudognaphalium (Asteraceae). J Ethnopharmacol 58 85–88 [DOI] [PubMed] [Google Scholar]
- Oppermann U (2007) Carbonyl reductases: the complex relationships of mammalian carbonyl- and quinone-reducing enzymes and their role in physiology. Annu Rev Pharmacol Toxicol 47 293–322 [DOI] [PubMed] [Google Scholar]
- Oppermann U, Filling C, Hult M, Shafqat N, Wu X, Lindh M, Shafqat J, Nordling E, Kallberg Y, Persson B, et al (2003) Short-chain dehydrogenases/reductases (SDR): the 2002 update. Chem Biol Interact 143–144 247–253 [DOI] [PubMed] [Google Scholar]
- Osbourn AE, Qi X, Townsend B, Qin B (2003) Dissecting plant secondary metabolism-constitutive chemical defenses in cereals. New Phytol 159 101–108 [DOI] [PubMed] [Google Scholar]
- Park SW, Kaimoyo E, Kumar D, Mosher S, Klessig DF (2007) Methyl salicylate is a critical mobile signal for plant systemic acquired resistance. Science 318 113–116 [DOI] [PubMed] [Google Scholar]
- Penninckx IA, Thomma BP, Buchala A, Métraux JP, Broekaert WF (1998) Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis. Plant Cell 10 2103–2114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Fons L, Aranda FJ, Guillén J, Villalaín J, Micol V (2006) Rosemary (Rosmarinus officinalis) diterpenes affect lipid polymorphism and fluidity in phospholipid membranes. Arch Biochem Biophys 453 224–236 [DOI] [PubMed] [Google Scholar]
- Persson B, Kallberg Y, Oppermann U, Jörnvall H (2003) Coenzyme-based functional assignments of short-chain dehydrogenase/reductases (SDRs). Chem Biol Interact 143–144 271–278 [DOI] [PubMed] [Google Scholar]
- Philips MA, Croteau RB (1999) Resin-based defenses in conifers. Trends Plant Sci 4 184–190 [DOI] [PubMed] [Google Scholar]
- Pichersky E, Gang DR (2000) Genetics and biochemistry of secondary metabolites in plants: an evolutionary perspective. Trends Plant Sci 5 439–445 [DOI] [PubMed] [Google Scholar]
- Pierce ML, Cover EC, Richardson PE, Scholes VE, Essenberg M (1996) Adequacy of cellular phytoalexin concentrations in hypersensitivity responding cotton leaves. Physiol Mol Plant Pathol 48 305–324 [Google Scholar]
- Rana BK, Singh UP, Taneja V (1997) Antifungal activity and kinetics of inhibition by essential oil isolated from leaves of Aegle marmelos. J Ethnopharmacol 57 29–34 [DOI] [PubMed] [Google Scholar]
- Reignault P, Frost LN, Richardson H, Daniels MJ, Jones JD, Parker JE (1996) Four Arabidopsis RPP loci controlling resistance to the Noco2 isolate of Peronospora parasitica map to regions known to contain other RPP recognition specificities. Mol Plant Microbe Interact 9 464–473 [DOI] [PubMed] [Google Scholar]
- Ringer KL, Davis EM, Croteau RB (2005) Monoterpene metabolism. cloning, expression, and characterization of (−)-isopiperitenol/(−)-carveol dehydrogenase of peppermint and spearmint. Plant Physiol 137 863–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schelz Z, Molnar J, Hohmann J (2006) Antimicrobial and antiplasmid activities of essential oils. Fitoterapia 77 279–285 [DOI] [PubMed] [Google Scholar]
- Sikkema J, de Bont JA, Poolman B (1995) Mechanisms of membrane toxicity of hydrocarbons. Microbiol Rev 59 201–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun HD, Qiu SX, Lin LZ, Wang ZY, Lin ZW, Pengsuparp T, Pezzuto JM, Fong HH, Cordell GA, Fransworth NR (1996) Nigranoic acid, a triterpenoid from Schisandra sphaerandra that inhibits HIV-1 reverse transcriptase. J Nat Prod 59 525–527 [DOI] [PubMed] [Google Scholar]
- Thomma BPHJ, Eggermont K, Penninckx IAMA, Mauch-Mani B, Vogelsang R, Cammue BPA, Broekaert WF (1998) Separate jasmonate-dependent and salicylate-dependent defense-response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc Natl Acad Sci USA 95 15107–15111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trombetta D, Castelli F, Sarpietro MG, Venuti V, Cristani M, Daniele C, Saija A, Mazzanti G, Bisignano G (2005) Mechanisms of antibacterial action of three monoterpenes. Antimicrob Agents Chemother 49 2474–2478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ukness S, Mauch-Mani B, Moyer M, Potter S, Williams S, Dincher S, Chandler D, Slusarenko A, Ward E, Ryals J (1992) Acquired resistance in Arabidopsis. Plant Cell 4 645–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber FJ, de Bont JA (1996) Adaptation mechanisms of microorganisms to the toxic effects of organic solvents on membranes. Biochim Biophys Acta 1286 225–245 [DOI] [PubMed] [Google Scholar]
- Wink M (1998) Plant breeding: importance of plant secondary metabolites for protection against pathogens and herbivores. Theor Appl Genet 75 225–233 [Google Scholar]
- Zipfel C, Robatzek S, Navarro L, Oakeley EJ, Jones JD, Felix G, Boller T (2004) Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428 764–767 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.