Abstract
A variety of mechanisms have been proposed to account for the extension of life span in seeds (seed longevity). In this work, we used Arabidopsis (Arabidopsis thaliana) seeds as a model and carried out differential proteomics to investigate this trait, which is of both ecological and agricultural importance. In our system based on a controlled deterioration treatment (CDT), we compared seed samples treated for different periods of time up to 7 d. Germination tests showed a progressive decrease of germination vigor depending on the duration of CDT. Proteomic analyses revealed that this loss in seed vigor can be accounted for by protein changes in the dry seeds and by an inability of the low-vigor seeds to display a normal proteome during germination. Furthermore, CDT strongly increased the extent of protein oxidation (carbonylation), which might induce a loss of functional properties of seed proteins and enzymes and/or enhance their susceptibility toward proteolysis. These results revealed essential mechanisms for seed vigor, such as translational capacity, mobilization of seed storage reserves, and detoxification efficiency. Finally, this work shows that similar molecular events accompany artificial and natural seed aging.
Before aging ultimately and irreparably leads to seed death, punctual and progressive accumulation of alterations during storage are likely to affect the potential ability of seeds to germinate. This deterioration process can occur even under the “best” storage conditions. The life span of seeds is determined by their genetic and physiological storage potential and by any deteriorating events that occur prior to or during storage as well as by the interaction with environmental factors (Bewley and Black, 1994). Since seed storage is often accompanied by a progressive loss of germination vigor, storage conditions must be optimized for both the preservation of genetic resources and commercial applications. For orthodox or desiccation-tolerant seeds, low seed moisture content, low temperature, or cryopreservation seems to result in an increase in storage life span (Abdalla and Roberts, 1968; Walters, 2004; Walters et al., 2004). However, a recent investigation reported a large heterogeneity and inequality for longevity between seeds descended from different plant species (Walters et al., 2005). Genetic approaches in rice (Oryza sativa; Miura et al., 2002) and Arabidopsis (Arabidopsis thaliana; Bentsink et al., 2000; Clerkx et al., 2004b) showed that seed longevity is controlled by several genetic factors. For both plants, several quantitative trait loci were identified as affecting viability, and these were located on different chromosomes. This behavior suggests that seed longevity is a multigenic trait including various seed traits, including germination under various stresses or Suc and seed oligosaccharide contents (Clerkx et al., 2004b). Several molecular studies also support this notion of the complex genetic basis of seed longevity, as diverse mechanisms were documented to play a role. For example, Arabidopsis seed mutants affected in flavonoid (Debeaujon et al., 2000) or in tocopherol (Sattler et al., 2004) biosynthetic pathways display reduced longevity, a finding that agrees with data showing that protection against reactive oxygen species (ROS) production and attack are important features of Arabidopsis seed longevity (Clerkx et al., 2004a). Furthermore, Bentsink et al. (2006) recently showed that mutations within the DOG1 gene, which specifically controls seed dormancy in Arabidopsis, were associated with a seed longevity phenotype, indicating that the absence of dormancy might also be a factor that reduces seed longevity. Also, transgenic Arabidopsis seeds overaccumulating a heat stress transcription factor exhibit enhanced accumulation of heat shock proteins (HSPs) and improved resistance to aging (Prieto-Dapena et al., 2006). On the contrary, a high level of a membrane lipid-hydrolyzing phospholipase D (PLDα1) seems detrimental for seed quality (Devaiah et al., 2007). Finally, the fantastic multicentenarian longevity of sacred lotus (Nelumbo nucifera) seeds (Shen-Miller et al., 1995; Shen-Miller, 2002) has been correlated with the extractible activity of the protein l-isoaspartyl methyltransferase, an enzyme repairing abnormal l-isoaspartyl residues accumulated in proteins during aging, notably during oxidative stress (Ingrosso et al., 2002; Clarke, 2003; Xu et al., 2004).
We are interested in determining the molecular basis of seed longevity. For that purpose, the model plant Arabidopsis can be viewed as a reference species allowing a molecular dissection of this trait. Indeed, achievement of the Arabidopsis genome sequence (Arabidopsis Genome Initiative, 2000) markedly increased our knowledge and understanding of the large complexity of plant growth regulation and development (Somerville and Koornneef, 2002). Global approaches such as transcriptome profiling (Ogawa et al., 2003; Nakabayashi et al., 2005; for review, see Holdsworth et al., 2008) have proved useful for the characterization of potential biomarkers of seed quality and germinative capacity. However, the functional components of a biological system are proteins and metabolites. Thanks to the availability of genomic sequence information and based on the progress achieved in sensitive and rapid separation of proteins and in their high-throughput identification by electrophoresis and mass spectrometry, proteomic approaches have opened up new perspectives to analyze the complex functions of model plants and crop species (Cánovas et al., 2004; Park, 2004; Agrawal et al., 2005a, 2005b, 2005c; Rossignol et al., 2006; Jorrín et al., 2007). In this way, previous proteomic studies revealed the requirements of RNA and protein synthesis for Arabidopsis seed germination (Gallardo et al., 2001, 2002a, 2002b; Rajjou et al., 2004, 2007a). In particular, these studies revealed that proteins and mRNAs stored in the dry mature seeds are sufficient for germination sensu stricto (Rajjou et al., 2004).
Based on these previous findings, in this work we have used proteomics and a seed deterioration treatment known as controlled deterioration (CDT) that is presumed to mimic natural aging (Delouche and Baskin, 1973; Bentsink et al., 2000; Clerkx et al., 2004b) to unravel the mechanisms of seed vigor loss during storage. Sensitivity of seeds to CDT has been successfully used for the rapid evaluation and prediction of seed vigor and longevity (Powell, 1995; TeKrony, 1995; Lanteri et al., 1996; McDonald, 1999; Bentsink et al., 2000; Halmer, 2000; Clerkx et al., 2004a, 2004b; Sattler et al., 2004; Job et al., 2005; Prieto-Dapena et al., 2006). Accordingly, this treatment is widely used by seed companies as a vigor assay for numerous seed species and has been described for Arabidopsis seeds (Tesnier et al., 2002). Here, we compared Arabidopsis seed samples submitted to CDT for different times up to 7 d. A comparison of the dry seed proteome for each sample was carried out to reveal changes in the accumulation of specific proteins during the treatment. The proteome of 1-d-imbibed seeds was also characterized for all seed samples to analyze the behavior of the treated seeds during early steps of the germination process. Since CDT and prolonged seed storage are known to entail an oxidative stress (Goel et al., 2003; Bailly, 2004; Job et al., 2005; Kibinza et al., 2006), which can lead to the formation of oxidatively modified proteins (Terskikh et al., 2008), we also analyzed the oxidized proteome in the deteriorated seeds. Finally, we discuss our results in comparison with natural aging conditions.
RESULTS AND DISCUSSION
CDT Entails a Seed Vigor Loss
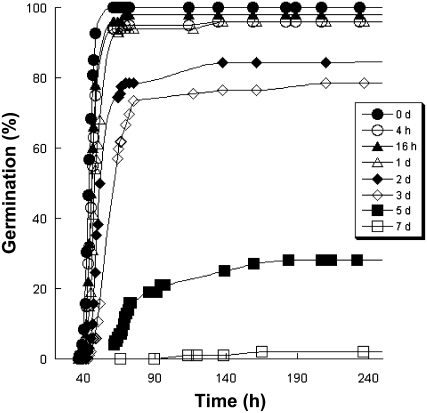
The CDT protocol described by Tesnier et al. (2002) was used to alter seed vigor of wild-type Arabidopsis ecotype Landsberg erecta (Ler) seeds for up to 7 d (see “Materials and Methods”). The germination ability of each seed sample was assessed by germination assays (Fig. 1). CDT led to a rapid decline of seed vigor, affecting the speed, homogeneity, and final extent of Arabidopsis seed germination (Fig. 1).
Figure 1.
Influence of CDT on Arabidopsis seed germination. Seeds were submitted to CDT for different periods (black circles, 0 d; white circles, 4 h; black triangles, 16 h; white triangles, 1 d; black diamonds, 2 d; white diamonds, 3 d; black squares, 5 d; white squares, 7 d) as described in “Materials and Methods.” The graph shows a representative experiment carried out three times in triplicate.
Rationale of the Proteomic Approach
To reveal molecular mechanisms associated with the loss of seed vigor induced by CDT, a differential proteomic approach was used under two different protocols. In the first, we hypothesized that CDT can directly affect the proteome of the seeds, and hence their vigor. In the second, we analyzed whether the loss in seed vigor imposed by CDT resulted from incapacity of the deteriorated seeds to appropriately set up the protein changes normally accompanying early germination (Gallardo et al., 2001; Rajjou et al., 2004).
Total soluble protein extracts from all seed samples (control and deteriorated seeds) were separated by two-dimensional (2D) PAGE. Following silver nitrate staining, protein patterns were determined by image analysis, and protein spots were quantified by Image Master 2D Elite software. Because of the very high reproducibility of 2D protein patterns compared with our previous work (Gallardo et al., 2001, 2002a; Rajjou et al., 2004, 2006a; Job et al., 2005), a number of proteins of interest could be readily identified using previously established reference maps for the Arabidopsis seed proteome (http://www.seed-proteome.com). Besides, this study allowed the identification of 87 novel proteins that accumulate in Arabidopsis seeds (Supplemental Table S2).
The Proteome Is Markedly Affected in Seeds Submitted to CDT
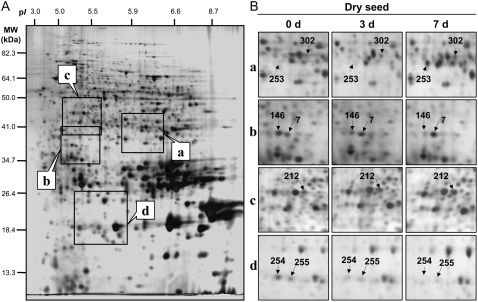
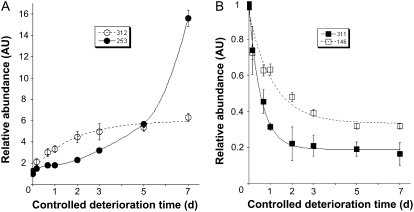
A typical 2D gel corresponding to the control nondeteriorated seeds is shown in Figure 2A. Image analysis of 2D protein patterns from control and deteriorated seed samples revealed 12 protein spots that were more abundant in the deteriorated seeds, corresponding to one chloroplastic and 10 nuclear genes. In parallel, six protein spots showed a lower abundance in the deteriorated seeds, corresponding to five genes (Fig. 2; Tables I and II). The time needed to reach 50% of the change in spot accumulation (T1/2) during the progress of CDT is a good index of the sensitivity of each of these protein spots toward deterioration. This kinetic analysis disclosed that proteins whose accumulation was altered by CDT displayed a wide range of sensitivity toward this treatment (Tables I and II), with T1/2 values ranging from 0.4 to more than 7 d. This finding is illustrated in Figure 3 for protein spots whose relative volume increased (spots 253 and 312) or decreased (spots 146 and 311) during the progress of CDT.
Figure 2.
Influence of CDT on the proteome of Arabidopsis seeds. An equal amount (150 μg) of total soluble protein extracts was loaded on each gel. A, Silver-stained 2D gel of total soluble proteins from nondeteriorated seeds (0 d, control seeds). The indicated portions of the gel (a, b, c, and d) are reproduced in B. B, Enlarged windows (a–d) of 2D gels as shown in A for nondeteriorated seeds (left), 3-d deteriorated seeds (middle), and 7-d deteriorated seeds (right). The seven labeled protein spots (spots 7, 146, 212, 253, 254, 255, and 302) were identified by mass spectrometry and by comparison with Arabidopsis seed protein reference maps (Gallardo et al., 2001, 2002a; Rajjou et al., 2004, 2006a; Job et al., 2005; http://www.seed-proteome.com; Tables I and II; Supplemental Table S2). Protein spot quantitation was carried out as described in “Materials and Methods” from at least five gels for each seed sample.
Table I.
Arabidopsis proteins whose abundance significantly increased in dry mature seeds according to controlled deterioration time
AGI, Arabidopsis Genome Initiative.
| No.a | Experimental Molecular Mass | Experimental pI | Arabidopsis Protein Name | Theoretical Molecular Mass | Theoretical pI | AGI No. | Coverage | Relative Abundanceb | T1/2 |
|---|---|---|---|---|---|---|---|---|---|
| kD | kD | ratio 7 d:0 d | d | ||||||
| 7c | 37.55 | 5.09 | 60S acidic ribosomal protein | 34.37 | 5.08 | At3g11250 | 24% | 3.4 ± 0.1 | 2.2 ± 0.3 |
| 212d | 42.57 | 5.18 | Actin 2 | 41.21 | 5.43 | At3g18780 | 33% | 2.5 ± 0.4 | 5.2 ± 2.7 |
| 253d | 40.29 | 5.84 | Glyceraldehyde-3-P dehydrogenase, cytosolic | 36.99 | 6.34 | At3g04120 | 18% | >16 | >7 |
| 293d | 96.69 | 5.61 | Peptidase M1 family protein | 103.40 | 6.09 | At1g63770 | 4% | >5.1 | >7 |
| 302d | 40.34 | 6.03 | Glyceraldehyde-3-P dehydrogenase | 36.90 | 7.21 | At1g13440 | 30% | 7.7 ± 1.4 | 6.1 ± 2.0 |
| 305d | 68.25 | 5.70 | Phosphoglucomutase | 63.46 | 5.57 | At1g70730 | 34% | 2.2 ± 0.1 | 1.3 ± 0.4 |
| 308c | 28.52 | 5.89 | α-Cruciferin 12S (seed storage protein fragment) | 50.56 | 6.53 | At1g03880 | 10% | 3.4 ± 0.5 | 4.8 ± 1.7 |
| 312c | 14.21 | 3.82 | NADP-dependent glyceraldehyde-3-P dehydrogenase (fragment) | 53.04 | 6.61 | At2g24270 | 8% | 6.0 ± 0.3 | 1.8 ± 0.3 |
| 320c | 57.36 | 5.82 | Ribulose bisphosphate carboxylase large chain | 53.47 | 6.25 | AtCg00490 | 24% | >2.7 | >7 |
| 321c | 57.36 | 5.84 | Ribulose bisphosphate carboxylase large chain | 53.47 | 6.25 | AtCg00490 | 28% | >2.8 | >7 |
| 322c | 35.21 | 5.20 | 60S acidic ribosomal protein | 33.65 | 4.93 | At2g40010 | 15% | >4.4 | >7 |
| 376d | 57.35 | 4.92 | Tubulin β-8 chain | 50.59 | 4.46 | At5g23860 | 9% | 5.0 ± 0.2 | 1.5 ± 0.2 |
Protein numbering following Arabidopsis seed protein reference maps available at http://www.seed-proteome.com.
Data obtained from densitometric analysis of individual spots from proteins on 2D gels stained with silver nitrate (see Fig. 2A for an example of a 2D gel): normalized spot volume in the deteriorated seeds (7 d of CDT) divided by the normalized spot volume in the nondeteriorated control seeds (0 d of CDT), from five different gels and three independent extractions.
Listed proteins correspond to previously identified proteins (Gallardo et al., 2001, 2002a; Rajjou et al., 2004, 2006a; Job et al., 2005).
Listed proteins correspond to proteins identified during this work; the peptide sequences determined are available in Supplemental Table S2.
Table II.
Arabidopsis proteins whose abundance significantly decreased in dry mature seeds according to controlled deterioration time
| No.a | Experimental Molecular Mass | Experimental pI | Arabidopsis Protein Name | Theoretical Molecular Mass | Theoretical pI | AGI No. | Coverage | Relative Abundanceb | T1/2 |
|---|---|---|---|---|---|---|---|---|---|
| kD | kD | ratio 7 d:0 d | d | ||||||
| 4c | 57.35 | 4.89 | Tubulin β2β3 chain | 50.73 | 4.7 | At5g62700 | 35% | 0.26 ± 0.01 | 0.17 ± 0.05 |
| 146c | 37.67 | 5.03 | MST | 41.89 | 5.95 | At1g79230 | 35% | 0.33 ± 0.05 | 1.12 ± 0.33 |
| 254c | 22.1 | 4.96 | Dehydrin | 18.44 | 7.95 | At5g66400 | 6% | 0.13 ± 0.04 | 0.93 ± 0.15 |
| 255c | 21.65 | 5.18 | Dehydrin | 18.44 | 7.95 | At5g66400 | 6% | <0.22 | >7 |
| 304c | 69.3 | 5.56 | Phosphoglucomutase | 63.44 | 5.73 | At1g70730 | 37% | 0.30 ± 0.04 | 0.37 ± 0.12 |
| 311c | 14.1 | 3.2 | β-Cruciferin 12S (seed storage protein fragment) | 21.20 | 6.19 | At4g28520 | 11% | 0.19 ± 0.01 | 0.56 ± 0.04 |
Protein numbering following Arabidopsis seed protein reference maps available at http://www.seed-proteome.com.
Data obtained from densitometric analysis of individual spots from proteins on 2D gels stained with silver nitrate (see Fig. 2A for an example of a 2D gel): normalized spot volume in the deteriorated seeds (7 d of CDT) divided by the normalized spot volume in the nondeteriorated control seeds (0 d of CDT), from five different gels and three independent extractions.
Listed proteins correspond to previously identified proteins (Gallardo et al., 2001, 2002a; Rajjou et al., 2004, 2006a; Job et al., 2005).
Figure 3.
Dynamic evolution of protein spot abundance in Arabidopsis seeds submitted to CDT. The relative abundance of protein spots was calculated by dividing the normalized spot volumes in the deteriorated seeds (4 and 16 h, and 1, 2, 3, 5, and 7 d) by the corresponding normalized spot volumes in the nondeteriorated seeds (0 d). A, Time courses of relative abundance increase in deteriorated seeds during CDT for the two protein spots (spots 253 and 312). B, Time courses of relative abundance decrease in deteriorated seeds during CDT for the two protein spots (spots 146 and 311). Protein spot quantitation was carried out as described in “Materials and Methods” from at least five gels for each seed sample. Solid lines were obtained by nonlinear regression analysis using the following equations: relative abundance of spot = a − b.exp(−t/T1/2) and relative abundance of spot = a + b.exp(−t/T1/2), for increase and decrease in relative abundance, respectively, where a and b are constant parameters, T1/2 is the time (days) needed to reach half variation in spot volume during the CDT, and t is the time of CDT (days). T1/2 values are listed in Tables I, II, IV, and V.
Our results clearly reveal that, despite an expected metabolic quiescent state and a relatively low water content characteristic of the seeds, proteome variations can occur under the low hydration conditions of CDT, as CDT only increases the seed water content from 5.8% to 10.5% (Tesnier et al., 2002). It remains to be determined whether these changes arose from de novo transcription, translation of stored mRNAs, or nonenzymatic modifications of the seed proteins, altering their migration on 2-D gels.
The Glycolytic Pathway Is Affected during Seed Aging
Among the 12 proteins that were more abundant in deteriorated seeds than in control seeds, four of them belonged to the glycolytic pathway (Table I). Thus, the abundance of these protein spots, corresponding to glyceraldehyde-3-P dehydrogenase (EC 1.2.1.12; spots 253 and 302; and EC 1.2.1.13; spot 312) and to phosphoglucomutase (EC 2.7.5.1; spot 305), significantly increased in seeds submitted to CDT. Interestingly, a recent study demonstrated that Arabidopsis cells exposed to oxidative stress react by substantially increasing the levels of hexose phosphates, Glc-6-P and Fru-6-P, as well as 3-phosphoglycerate (Baxter et al., 2007). Our results thereby confirmed that seeds underwent an oxidative stress during CDT (Goel et al., 2003; Bailly, 2004; Sattler et al., 2004; Job et al., 2005; Kibinza et al., 2006) and mounted a protective response through modification of the glycolytic pathway.
The β-Mercaptopyruvate Sulfurtransferase Exhibits Varying Accumulation Levels during Seed Aging
The protein β-mercaptopyruvate sulfurtransferase (MST; EC 2.8.1.2; spot 146; Figs. 2 and 3; Table II) was abundant in control high-vigor seeds (0 d). However, during CDT, the level of this protein showed an important decline. The MST catalyzes the transfer of sulfur from mercaptopyruvate to sulfur acceptors such as thiols and cyanide (Papenbrock and Schmidt, 2000a, 2000b), presumably contributing to cyanide detoxification (Cipollone et al., 2007). Despite the fact that at low concentrations cyanide is beneficial for releasing seed dormancy and improving germination (Taylorson and Hendricks, 1973; Bethke et al., 2006), its production is often associated with deleterious mechanisms and therefore must be controlled. Also, cyanide can inhibit the activity of heme proteins such as peroxidases (Ellis and Dunford, 1968; Job and Ricard, 1975) and catalases (Tejera García et al., 2007). Furthermore, this molecule is a potent inhibitor of mitochondrial ascorbate synthesis in plants (Bartoli et al., 2000), thus potentially impeding plant defense against ROS attack. Most frequently, cyanide production in plants results from the catabolism of cyanogenic glycosides or during cyanolipid hydrolysis (Poulton, 1990). It has been shown that the cyanide potential of sorghum (Sorghum spp.), a plant containing high levels of hydrogen cyanide, shows a rapid transient increase during germination and early seedling formation (Busk and Møller, 2002). Similarly, a recent work showed that imbibition of dormant and nondormant sunflower (Helianthus annuus) embryos entails a substantial rise in cyanide content (Oracz et al., 2008). However, no data are available on the metabolism of cyanogenic compounds in dry and germinating Arabidopsis seeds. In plants, cyanide can also be released as a by-product of ethylene biosynthesis (Adams and Yang, 1981; Peiser et al., 1984). However, temporal patterns of accumulation of enzymes involved in Met and S-adenosyl-Met synthesis in seeds are consistent with an essential role of endogenous ethylene in Arabidopsis only after radicle protrusion (Gallardo et al., 2002b), findings that do not favor the hypothesis that cyanide accumulates to high toxic levels through the ethylene pathway during germination sensu stricto. Finally, the decomposition of glucosinolates has also been suggested as a possible source of cyanide production in brassicaceous plants (Cipollini and Gruner, 2007). The glucosinolates of Arabidopsis seeds are distinguished by their high concentration, unique aliphatic constituents, and low level of indole compounds (Brown et al., 2003). However, the cyanide production from stored-seed glucosinolates has never been observed but should be investigated in the context of seed biology. Besides cyanide detoxification, proposed roles for sulfurtransferases are sulfur metabolism (Donadio et al., 1990) and the mobilization of sulfur for iron-sulfur cluster biosynthesis or repair (Pagani et al., 1984; Bonomi et al., 1985). Also, MST plays a physiological role in the protection against oxidative stress, and it particularly contributes to the maintenance of cellular redox homeostasis via the metabolic regulation of Cys degradation (Nagahara and Katayama, 2005). In plants, the mobilization of sulfur for transport processes in older leaves was also proposed (Papenbrock and Schmidt, 2000a, 2000b).
Our results reveal, for the first time to our knowledge, that a loss in seed vigor is associated with a decreased level of MST, highlighting further the importance of sulfur metabolism and homeostasis in seeds (Gallardo et al., 2002b). Furthermore, our data suggest as yet unknown important role(s) of this enzyme in seed physiology and quality.
The Dehydrin/RAB Group of LEA Proteins Contributes to Seed Vigor
Our proteomic analysis revealed two protein spots (spots 254 and 255) progressively disappearing in seeds according to the time of CDT. These spots correspond to two isoforms of the RAB18 (Responsive to ABA18) dehydrin, belonging to the LEA group 2 (D11) protein family. This result was unexpected, because a previous work showed an absence of correlation between accumulation of the dehydrin/RAB group of LEA proteins and seed longevity (Wechsberg et al., 1994). These proteins are inducible by dehydration and abscisic acid (Skriver and Mundy, 1990) and have been suggested to play a role in desiccation tolerance, particularly during seed development (Dure, 1993; Close, 1996, 1997). Expression of the RAB18 gene is high in Arabidopsis dry mature seeds at both the mRNA and protein levels (Lång and Palva, 1992; Parcy et al., 1994; Rajjou et al., 2004). Such a high expression could correspond to a remnant accumulation during the maturation program of seed development, in response to the acquisition of desiccation tolerance. However, it is also likely that RAB18 expression is needed to prevent environmental stress that may occur at the start of the germination process (Lopez-Molina et al., 2002; Rajjou et al., 2004, 2006a). The high hydrophilicity and thermostability of dehydrins suggest their involvement in large-scale hydrophobic interactions, such as those of membrane structures or hydrophobic patches of proteins with chaperone-like properties (Ismail et al., 1999; Borovskii et al., 2002). Interestingly, in seeds from cabbage (Brassica oleracea), a related cruciferous species, mRNA levels of the RAB18 homolog are correlated with seed stress tolerance (Soeda et al., 2005). Thus, its transcript level increased during seed maturation and declined during priming (an invigoration treatment of seeds based upon their controlled hydration; Heydecker et al., 1973) and germination. Furthermore, the mRNA can be reinduced during a slow and warm drying treatment of primed seeds to increase their storability (Soeda et al., 2005). As in cabbage, our results suggest that RAB18 protein abundance in dry mature Arabidopsis seeds is strongly correlated with seed aging. One possible explanation is that the decreased level of this protein in deteriorated seeds is associated with membrane destabilization and/or alterations in protein structure. It is noted that RAB18 protein displays both cytosolic and nuclear localization, suggesting multiple functionalities as yet unclear. In this proteomic study, we have not identified dehydrins other than the RAB18 dehydrin. This could mean that only this member of the large LEA protein family (Bies-Ethève et al., 2008) plays a role in seed vigor. Alternatively, we cannot exclude the possibility that other LEA proteins are also involved in seed vigor but are present at too low levels, so their detection escaped our analysis.
Seed Deterioration Entails a Massive Increase in Carbonylated Proteins
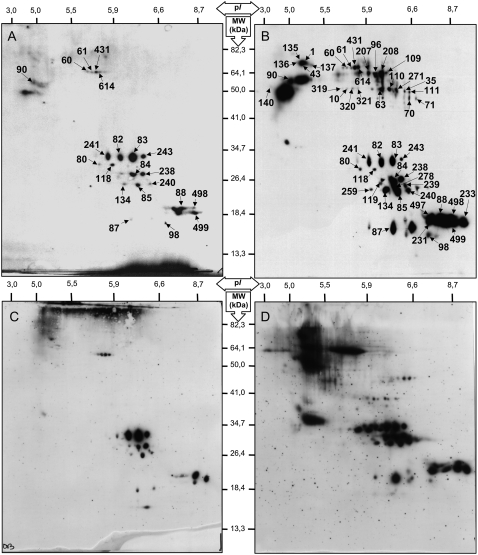
In all organisms, oxidative stress has been postulated to be a causal factor in aging processes (Harman, 1956). The extent of oxidative damage to nucleic acids, lipids, and proteins has been found to increase with age, providing support for this basic tenet (Levine and Stadtman, 2001). Indeed, a progressive accumulation of oxidative damage of these macromolecules in aged tissues is thought to contribute to the decline in biological functions characteristic of the aged phenotype (Stadtman, 2001, 2004). There is strong evidence that proteins are the most important targets for oxidants (Davies, 2005). Protein carbonylation has been widely used as an indicator of oxidative damage in several organisms and has been shown to increase in aged tissues (Nyström, 2005; Møller et al., 2007). It results from oxidative attack on Arg, Lys, Pro, or Thr residues of proteins (Levine et al., 1990), which can affect enzyme activities or alter susceptibility of the modified proteins to proteolysis (Berlett and Stadtman, 1997; Davies, 2005). We characterized the influence of CDT on the oxidized proteome of Arabidopsis seeds (Fig. 4). Carbonylated proteins were then identified by matching the 2,4-dinitrophenylhydrazone (DNP)-derivatized protein spots to master gel maps of Arabidopsis seed proteins (Gallardo et al., 2001, 2002a; Rajjou et al., 2004, 2006a; Job et al., 2005; http://www.seed-proteome.com). The results revealed that protein carbonylation strongly increased in deteriorated seeds, indicating the occurrence of ROS during CDT. In agreement with previous data (Job et al., 2005; Rajjou et al., 2006a, 2007a, 2007b), several polypeptides corresponding to the α- and β-subunits of the 12S cruciferins (legumin-type seed storage proteins) were strongly carbonylated in deteriorated seeds compared with control seeds. However, the present work also revealed that several other proteins ought to be oxidized in Arabidopsis seeds submitted to CDT (Fig. 4; Table III). Among them, several isoforms of the Rubisco large subunit (spots 10, 319, 320, and 321) proved to be oxidized. Rubisco catalyzes the first step in net photosynthetic CO2 assimilation and photorespiratory carbon oxidation. This protein has already been shown to be a preferential target of ROS in Arabidopsis (Johansson et al., 2004; Job et al., 2005). Also, previous work demonstrated that one of the first apparent symptoms of leaf senescence is the aggregation and the deterioration of Rubisco (Feller and Fischer, 1994). Finally, deterioration of Rubisco has also been observed during oxidative stress or ozone treatment (Pell et al., 1997). All of these observations point out the hypersensitivity to oxidative stress of this protein, which was also found in our study.
Figure 4.
Protein carbonyl patterns in Arabidopsis seeds following CDT or natural aging. Protein extracts were prepared as described in “Materials and Methods” from the dry mature seeds and analyzed by 2D PAGE. Carbonylated proteins were characterized in nondeteriorated seeds (0 d, control seeds; A), 7-d deteriorated seeds (B), freshly harvested seeds (C), and 11-year-old dry mature seeds (D). Proteins were separated by 2D gel electrophoresis as shown in Figure 2A. Following transfer to nitrocellulose, the appearance of carbonyl groups in proteins was analyzed by immunodetection of protein-bound DNP after derivatization with the corresponding hydrazine, as described (Job et al., 2005). Proteins undergoing carbonylation are labeled with black arrows; they are listed in Table III. These findings show that both CDT and natural aging induce an oxidative stress on specific proteins, presumably through the generation of ROS.
Table III.
Identification and relative carbonyl content of oxidized proteins during Arabidopsis seed aging
| No.a | Experimental Molecular Mass | Experimental pI | Arabidopsis Protein Name | Theoretical Molecular Mass | Theoretical pI | AGI No. | Coverage | Relative Abundanceb |
|---|---|---|---|---|---|---|---|---|
| kD | kD | ratio 7 d:0 d | ||||||
| Protein metabolism | ||||||||
| 140c | 60.26 | 4.15 | Calreticulin 1 (precursor) | 48.51 | 4.2 | At1g56340 | 15% | 2.3 |
| 1c | 79.36 | 5.08 | dnaK-type molecular chaperone BiP | 71.17 | 5.08 | At5g42020 | 20% | >100 |
| 135c | 80.5 | 5.04 | dnaK-type molecular chaperone BiP | 71.17 | 5.08 | At5g42020 | 20% | >100 |
| 136c | 77.87 | 4.87 | Heat shock cognate 70-kD protein 1 | 71.37 | 5.03 | At5g02500 | 19% | >100 |
| 43c | 78.9 | 5.06 | Heat shock cognate 70-kD protein 3 | 71.13 | 4.69 | At3g09440 | 15% | >100 |
| 137c | 76.06 | 5.07 | Heat shock protein 70 | 71.37 | 5.03 | At3g12580 | 38% | >100 |
| 614d | 68.68 | 5.83 | Aspartyl-tRNA synthetase | 62.90 | 6.12 | At4g31180 | 26% | 3.8 |
| 90c | 64.11 | 4.81 | Protein disulfide isomerase | 55.60 | 4.81 | At1g21750 | 39% | 2.1 |
| Energy metabolism and photosynthesis | ||||||||
| 109c | 64.79 | 6.35 | Malate oxidoreductase or malic enzyme | 64.28 | 6.32 | At2g19900 | 14% | >100 |
| 63c | 57.09 | 6.11 | Isocitrate lyase | 50.42 | 6.29 | At3g21720 | 30% | >100 |
| 10c | 57.36 | 5.77 | Ribulose bisphosphate carboxylase large chain | 53.47 | 6.25 | AtCg00490 | 30% | >100 |
| 319c | 57.36 | 5.57 | Ribulose bisphosphate carboxylase large chain | 53.47 | 6.25 | AtCg00490 | 26% | >100 |
| 320c | 57.36 | 5.82 | Ribulose bisphosphate carboxylase large chain | 53.47 | 6.25 | AtCg00490 | 21% | >100 |
| 321c | 57.36 | 5.84 | Ribulose bisphosphate carboxylase large chain | 53.47 | 6.25 | AtCg00490 | 30% | >100 |
| Stress response | ||||||||
| 60c | 66.68 | 5.74 | Late embryogenesis abundant (LEA) | 67.19 | 5.78 | At2g42560 | 25% | 1.2 |
| 61c | 66.68 | 5.78 | Late embryogenesis abundant (LEA) | 67.19 | 5.78 | At2g42560 | 43% | 1.5 |
| 431c | 66.68 | 5.79 | Late embryogenesis abundant (LEA) | 67.19 | 5.78 | At2g42560 | 32% | 2.2 |
| Hydrolase | ||||||||
| 96c | 64.62 | 6.08 | Glycosyl hydrolase family 1 protein | 57.83 | 6.02 | At3g21370 | 11% | >100 |
| 207c | 64.96 | 5.92 | Glycosyl hydrolase family 1 protein | 57.83 | 6.02 | At3g21370 | 21% | >100 |
| 208c | 64.96 | 6.23 | Glycosyl hydrolase family 1 protein | 57.83 | 6.02 | At3g21370 | 14% | >100 |
| Seed storage proteins | ||||||||
| 35c | 56.47 | 6.84 | 12S seed storage protein (precursor) | 55.86 | 6.36 | At4g28520 | 16% | >100 |
| 70c | 49.43 | 7.19 | 12S seed storage protein (precursor) | 52.59 | 7.68 | At5g44120 | 22% | >100 |
| 71c | 50.44 | 7.67 | 12S seed storage protein (precursor) | 52.59 | 7.68 | At5g44120 | 34% | >100 |
| 110c | 57 | 6.4 | 12S seed storage protein (precursor) | 55.86 | 6.36 | At4g28520 | 19% | >100 |
| 111c | 56.89 | 7.19 | 12S seed storage protein (precursor) | 55.86 | 6.36 | At4g28520 | 16% | >100 |
| 271d | 59.96 | 6.52 | 12S seed storage protein (precursor) | 55.86 | 6.36 | At4g28520 | 30% | >100 |
| 239c | 28.95 | 6.64 | α-Cruciferin 12S (seed storage protein fragment) | 48.03 | 6.56 | At1g03880 | 18% | >100 |
| 80c | 32.64 | 5.85 | α-Cruciferin 12S (seed storage protein fragment) | 34.68 | 6.42 | At4g28520 | 33% | 3.29 |
| 82c | 33.89 | 6.24 | α-Cruciferin 12S (seed storage protein fragment) | 34.68 | 6.42 | At4g28520 | 42% | 1.6 |
| 83c | 34.35 | 6.42 | α-Cruciferin 12S (seed storage protein fragment) | 34.68 | 6.42 | At4g28520 | 33% | 1 |
| 84c | 30.46 | 6.61 | α-Cruciferin 12S (seed storage protein fragment) | 31.75 | 6.49 | At5g44120 | 42% | 2.1 |
| 85c | 27.2 | 6.5 | α-Cruciferin 12S (seed storage protein fragment) | 27.24 | 6.34 | At1g03880 | 32% | 2.9 |
| 118c | 29.21 | 6.04 | α-Cruciferin 12S (seed storage protein fragment) | 34.68 | 6.42 | At4g28520 | 26% | 1 |
| 119c | 30.66 | 6.16 | α-Cruciferin 12S (seed storage protein fragment) | 31.75 | 6.49 | At5g44120 | 28% | 2.9 |
| 134c | 26.35 | 6.34 | α-Cruciferin 12S (seed storage protein fragment) | 27.24 | 6.34 | At1g03880 | 21% | 3.9 |
| 238c | 29.42 | 6.5 | α-Cruciferin 12S (seed storage protein fragment) | 31.75 | 6.49 | At5g44120 | 28% | 2.1 |
| 240c | 27.28 | 6.74 | α-Cruciferin 12S (seed storage protein fragment) | 27.24 | 6.34 | At1g03880 | 16% | 3.57 |
| 241c | 34.11 | 5.87 | α-Cruciferin 12S (seed storage protein fragment) | 34.68 | 6.42 | At4g28520 | 32% | 1.2 |
| 243c | 34.92 | 6.62 | α-Cruciferin 12S (seed storage protein fragment) | 34.68 | 6.42 | At4g28520 | 19% | 1 |
| 278c | 27.24 | 6.44 | α-Cruciferin 12S (seed storage protein fragment) | 31.75 | 6.49 | At5g44120 | 24% | >100 |
| 98c | 18.08 | 6.12 | β-Cruciferin 12S (seed storage protein fragment) | 20.8 | 7.03 | At1g03880 | 45% | 4.9 |
| 498c | 22.82 | 8.85 | β-Cruciferin 12S (seed storage protein fragment) | 21.2 | 6.19 | At4g28520 | 79% | 12.4 |
| 87c | 18.43 | 6.36 | β-Cruciferin 12S (seed storage protein fragment) | 20.8 | 7.03 | At1g03880 | 33% | >100 |
| 88c | 22.82 | 8.68 | β-Cruciferin 12S (seed storage protein fragment) | 21.2 | 6.19 | At4g28520 | 44% | 1.8 |
| 231c | 19.71 | 8.82 | β-Cruciferin 12S (seed storage protein fragment) | 21.2 | 6.19 | At4g28520 | 39% | >100 |
| 233c | 18.45 | 9.1 | β-Cruciferin 12S (seed storage protein fragment) | 20.84 | 9.06 | At5g44120 | 52% | >100 |
| 497c | 18.45 | 6.12 | β-Cruciferin 12S (seed storage protein fragment) | 21.2 | 6.19 | At4g28520 | 64% | >100 |
| 499c | 20.67 | 8.85 | β-Cruciferin 12S (seed storage protein fragment) | 20.84 | 9.06 | At5g44120 | 53% | 5.3 |
| Other processes | ||||||||
| 259c | 27.01 | 6.1 | Expressed protein | 27.28 | 6.7 | At1g05510 | 21% | 5.7 |
Protein numbering following Arabidopsis seed protein reference maps available at http://www.seed-proteome.com.
Data obtained from densitometric analysis of individual spots from carbonylated proteins on 2D gels revealed by anti-DNP immunoassay (examples are shown in Fig. 4): normalized spot volume in the deteriorated seeds (7 d of CDT) divided by the normalized spot volume in the nondeteriorated control seeds (0 d of CDT; ratio of carbonylation in deteriorated dry seeds [7 d of CDT] over carbonylation in nondeteriorated dry seeds [0 d of CDT]) from three different and independent protein extractions. >100 means that the accumulation level of the corresponding carbonylated protein in the nondeteriorated dry seeds (0 d of CDT) was close to background.
Listed proteins correspond to previously identified proteins (Gallardo et al., 2001, 2002a; Rajjou et al., 2004, 2006a; Job et al., 2005).
Listed proteins correspond to proteins identified during this work; the peptide sequences determined are available in Supplemental Table S2.
Many proteins with chaperone activities were also favored targets for oxidation (spots 1, 43, 90, 135, 136, 137, and 140; Table III). Among them, three HSP70 proteins (spots 43, 136, and 137) are described as being abundant in dry and imbibed seeds (Gallardo et al., 2001; Rajjou et al., 2006a; http://www.seed-proteome.com). Molecular chaperones are known to be targets of carbonylation in yeast and bacteria challenged by oxidative stress (Tamarit et al., 1998; Cabiscol et al., 2000), presumably because they act as shields protecting other proteins against ROS damage (Cabiscol et al., 2000). Other chaperone proteins associated with the endoplasmic reticulum, such as the luminal binding protein BiP (spots 1 and 135), calreticulin (spot 140), and protein disulfide isomerase (spot 90; Laboissière et al., 1997; Freedman et al., 1998; Wilkinson and Gilbert, 2004), are also oxidized in deteriorated dry seeds (Table III). The fundamental role of the ER and associated proteins in the stress response and aging was recently reviewed in human (Yoshida, 2007). Our results support this hypothesis in plants.
It is worth noting that three LEA protein isoforms (spots 60, 61, and 431), encoded by the At2g42560 gene, appeared to be more oxidized in deteriorated seeds than in control seeds (Table III). The protein sequence of this LEA protein displays strong homology with that of previously described seed biotinylated proteins from pea (Pisum sativum; Duval et al., 1994b; Job et al., 2001), soybean (Glycine max; Franca Neto et al., 1997; Shatters et al., 1997; Hsing et al., 1998), and barley (Hordeum vulgare; March et al., 2007). This type of LEA protein exhibits a characteristic biochemical feature in that it binds in vivo the vitamin biotin (Duval et al., 1994a; Alban et al., 2000; Job et al., 2001). Biotin (vitamin B7), also known as vitamin H, is a fundamental molecule for all living organisms, being the cofactor of housekeeping enzymes involved in carboxylation, decarboxylation, and transcarboxylation reactions (Patton et al., 1998; Alban et al., 2000; Nikolau et al., 2003). It has been proposed that the seed biotinylated LEA proteins may play a role in sequestering this vitamin late in embryogenesis for subsequent use during germination and/or to maintain a metabolic quiescent state in dry seeds by biotin deprivation (Alban et al., 2000; Job et al., 2001). It is also important to note the existence of an ortholog of this biotinylated LEA protein in Medicago truncatula seeds, whose accumulation level is strongly associated with the reinduction of desiccation tolerance in radicles (Boudet et al., 2006). Hence, a specific carbonylation of this biotinylated LEA protein caused by CDT could entail an alteration of Arabidopsis seed survival in the dry state.
Overall, our results document that CDT generates an oxidative stress, which in turn induces chemical modifications of proteins by carbonylation, thus providing an explanation for the decrease in seed vigor associated with this treatment.
CDT Exerts an Influence on Proteome Expression during Germination
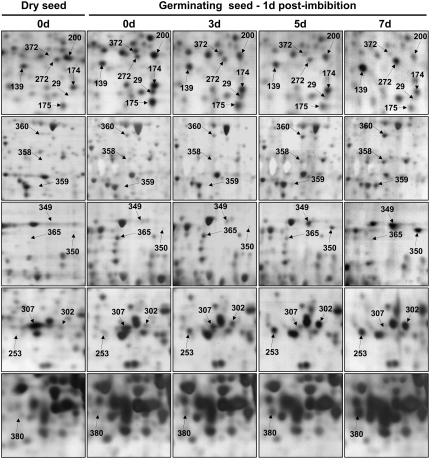
The evolution of the seed proteome during germination was also analyzed 1 d after imbibition for the control and deteriorated seeds. This stage corresponds to the germination sensu stricto of the Arabidopsis (Ler) high-vigor control seeds, as none of these seeds showed radicle protrusion at that time (Fig. 1). Of 45 protein spots presenting reproducible variations in their accumulation level, 29 protein spots were less abundant, while 16 protein spots were more abundant, in germinating deteriorated seeds than in control seeds (Fig. 5; Tables IV and V). One of the specific features observed is the maintenance of a high level of storage protein precursors (spots 151, 177, and 354) in germinating deteriorated seeds, implying that deteriorated seeds are less active than control seeds in mobilizing their storage protein reserves. In the same way, lipid storage mobilization was also strongly affected. Thus, isocitrate lyase (threo-d-isocitrate-glyoxylate lyase; EC 4.1.3.1), which is the key enzyme in seed lipid mobilization via the glyoxylate cycle (Graham, 2008), increased about 5-fold during germination sensu stricto of the control seeds (0 d), whereas its relative accumulation decreased steadily in deteriorated seeds according to the time of CDT (spot 365; Fig. 5; Table V). Isocitrate lyase plays a crucial role in the synthesis of carbohydrates from storage lipids during seed germination and seedling establishment (Eastmond and Graham, 2001). Also, it has been proposed that glyoxylate cycle activity is a good indicator of seedling emergence potential and seed vigor in sugar beet (Beta vulgaris) and Arabidopsis, notably under stress conditions (de los Reyes et al., 2003; Rajjou et al., 2006a). This suggests that a rapid onset of the glyoxylate pathway during germination can facilitate the mobilization of the lipid storage reserves, enabling the fast establishment of a vigorous seedling. Our results are in perfect agreement with these previous studies (de los Reyes et al., 2003; Rajjou et al., 2006a) and further document the fundamental role of the glyoxylate cycle for germination and seed vigor. Moreover, isocitrate lyase is regarded suitable to determine the transition from late embryogenesis to germination (Goldberg et al., 1989). In summary, this enzyme is a very good candidate as a diagnostic marker of seed vigor.
Figure 5.
Changes in protein accumulation patterns in deteriorated seeds during germination sensu stricto (1 d after imbibition). An equal amount (150 μg) of total soluble protein extracts was loaded on each gel. Proteins were separated by 2D gel electrophoresis. A representative silver-stained 2D gel of total soluble proteins from nondeteriorated dry mature seeds is presented in Figure 2A. The analysis was carried out on nondeteriorated seed samples (0 d, control seeds) and deteriorated seeds (3, 5, and 7 d of CDT) imbibed in water for 1 d. The 17 labeled protein spots (spots 29, 139, 174, 175, 176, 200, 253, 272, 302, 349, 350, 358, 359, 360, 365, 372, and 380) were identified by mass spectrometry and by comparison with Arabidopsis seed protein reference maps (Gallardo et al., 2001, 2002a; Rajjou et al., 2004, 2006a; Job et al., 2005; http://www.seed-proteome.com; Tables IV and V; Supplemental Table S2). Protein spot quantitation was carried out as described in “Materials and Methods” from at least five gels for each seed sample.
Table IV.
Arabidopsis proteins whose abundance was significantly greater in deteriorated seeds than in control seeds during germination sensu stricto (1 d after imbibition)
| No.a | Experimental Molecular Mass | Experimental pI | Arabidopsis Protein Name | Theoretical Molecular Mass | Theoretical pI | AGI No. | Coverage | Relative Abundanceb | T1/2 |
|---|---|---|---|---|---|---|---|---|---|
| kD | kD | ratio 7 d:0 d | d | ||||||
| Energy metabolism | |||||||||
| 253c | 40.29 | 5.84 | Glyceraldehyde-3-P dehydrogenase, cytosolic | 36.99 | 6.34 | At3g04120 | 18% | >6.2 | >7 |
| 302c | 40.34 | 6.03 | Glyceraldehyde-3-P dehydrogenase | 36.90 | 7.21 | At1g13440 | 30% | >7.6 | >7 |
| 368d | 40.36 | 6.24 | Glyceraldehyde-3-P dehydrogenase | 36.90 | 7.21 | At1g13440 | 17% | >4.5 | >7 |
| 369d | 40.33 | 6.20 | Glyceraldehyde-3-P dehydrogenase | 36.90 | 7.21 | At1g13440 | 22% | >5.0 | >7 |
| 349d | 64.85 | 6.77 | Malate oxidoreductase | 64.24 | 6.68 | At2g19900 | 12% | >32.0 | >7 |
| 350d | 64.70 | 7.21 | Malate oxidoreductase | 64.24 | 6.68 | At2g19900 | 2% | >39.0 | >7 |
| Amino acid metabolism | |||||||||
| 359c | 56.90 | 5.57 | S-Adenosyl-l-homo-Cys hydrolase | 53.36 | 5.83 | At4g13940 or At3g23810 | 13% | >20.0 | >7 |
| 370c | 41.66 | 6.50 | Glutamate dehydrogenase | 44.51 | 6.02 | At5g18170 or At3g03910 | 13% | >2.5 | >7 |
| Seed storage proteins | |||||||||
| 354d | 42.61 | 6.60 | 12S seed storage protein (precursor) | 52.56 | 8.08 | At5g44120 | 25% | >9.2 | >7 |
| 177d | 45.47 | 6.07 | 12S seed storage protein (precursor) | 48.03 | 6.56 | At1g03880 | 17% | >3.0 | >7 |
| 151d | 46.11 | 5.38 | Cupin family protein | 49.66 | 5.45 | At1g03890 | >6.0 | >7 | |
| Hydrolase and protease | |||||||||
| 378c | 80.28 | 5.77 | Subtilisin-like Ser proteinase | 81.80 | 6.76 | At3g14067 | 19% | >3 | >7 |
| 360c | 64.96 | 5.75 | Glycosyl hydrolase family | 84.29 | 7.71 | At5g64570 | 14% | >3.0 | >7 |
| Other processes | |||||||||
| 139d | 40.07 | 5.56 | Reversibly glycosylated polypeptide | 40.7 | 5.61 | At3g02230 or At5g15650 | 7% | >2.5 | >7 |
| 377d | 38.42 | 6.58 | Potassium channel β-subunit | 36.52 | 7.49 | At1g04690 | 15% | >3.8 | >7 |
| 380d | 26.31 | 5.89 | Nucleoside diphosphate kinase II, chloroplast | 25.53 | 9.30 | At5g63310 | 12% | >3.8 | >7 |
Protein numbering following Arabidopsis seed protein reference maps available at http://www.seed-proteome.com.
Data obtained from densitometric analysis of individual spots from proteins on 2D gels stained with silver nitrate (see Fig. 2A for an example of a 2D gel): normalized spot volume in the deteriorated seeds (7 d of CDT) incubated for 1 d in water divided by the normalized spot volume in the nondeteriorated control seeds (0 d of CDT) incubated for 1 d in water, from five different gels and three independent extractions.
Listed proteins correspond to proteins identified during this work; the peptide sequences determined are available in Supplemental Table S2.
Listed proteins correspond to previously identified proteins (Gallardo et al., 2001, 2002a; Rajjou et al., 2004, 2006a; Job et al., 2005).
Table V.
Arabidopsis proteins whose abundance was significantly smaller in deteriorated seeds than in control seeds during germination sensu stricto (1 d after imbibition)
| No.a | Experimental Molecular Mass | Experimental pI | Arabidopsis Protein Name | Theoretical Molecular Mass | Theoretical pI | AGI No. | Coverage | Relative Abundanceb | T1/2 |
|---|---|---|---|---|---|---|---|---|---|
| kD | kD | ratio 7 d:0 d | d | ||||||
| Translation and protein metabolism | |||||||||
| 128c | 47.45 | 5.45 | Elongation factor 1B-γ | 46.66 | 5.36 | At1g09640 | 26% | <0.28 | >7 |
| 129c | 47.20 | 5.50 | Eukaryotic initiation factor 4A-1 | 46.70 | 5.47 | At3g13920 | 28% | <0.49 | >7 |
| 105c | 48.63 | 5.61 | Elongation factor 1-γ2 | 46.4 | 5.55 | At1g57720 | 41% | <0.5 | >7 |
| 314d | 37.54 | 5.22 | 60S acidic ribosomal protein P0-C | 34.37 | 4.78 | At2g40010 | 7% | <0.25 | >7 |
| 200c | 40.57 | 5.72 | Protein disulfide isomerase-like (PDIL) | 39.50 | 5.80 | At2g47470 | 20% | <0.44 | >7 |
| 364d | 65.22 | 5.56 | T-complex protein 1, θ-subunit (TCP-1-θ) | 58.92 | 5.01 | At3g03960 | 2% | <0.30 | >7 |
| Energy metabolism | |||||||||
| 39c | 38.52 | 6.23 | Glyceraldehyde-3-P dehydrogenase | 36.91 | 6.62 | At3g04120 | 26% | <0.48 | >7 |
| 40c | 38.55 | 6.26 | Glyceraldehyde-3-P dehydrogenase | 36.91 | 6.62 | At3g04120 | 27% | <0.44 | >7 |
| 307c | 40.30 | 5.87 | Glyceraldehyde-3-P dehydrogenase | 36.91 | 6.62 | At3g04120 | 19% | <0.21 | >7 |
| 365c | 62.77 | 6.30 | Isocitrate lyase | 50.42 | 6.29 | At3g21720 | 16% | <0.21 | >7 |
| 126c | 27.16 | 5.51 | Triose phosphate isomerase, cytosolic | 27.15 | 5.24 | At3g55440 | 10% | <0.36 | >7 |
| 353c | 55.70 | 6.60 | Phosphoglucomutase | 63.44 | 5.73 | At1g70730 | 6% | <0.10 | >7 |
| 372c | 41.08 | 5.71 | Phosphoglycerate kinase | 42.11 | 5.49 | At1g79550 | 21% | <0.35 | >7 |
| 194c | 45.5 | 6.15 | 3-Oxoacyl-[acyl-carrier-protein] synthase I | 50.41 | 8.29 | At5g46290 | 44% | <0.49 | >7 |
| Cell detoxification and stress response | |||||||||
| 23c | 56.48 | 6.64 | Catalase | 56.93 | 6.63 | At4g35090 | 16% | <0.25 | >7 |
| 146c | 37.67 | 5.03 | MST | 41.89 | 5.95 | At1g79230 | 35% | <0.30 | >7 |
| 29c | 38.35 | 5.72 | 2-Alkenal reductase | 38.13 | 5.81 | At5g16970 | 14% | <0.26 | >7 |
| 284d | 25.34 | 5.43 | Reduced glutathione-dependent dehydroascorbate reductase | 23.62 | 5.79 | At1g19570 | 57% | <0.21 | >7 |
| 94c | 34.92 | 5.21 | Late embryogenesis abundant (LEA) | 31.31 | 5.21 | At3g17520 | 21% | <0.27 | >7 |
| Amino acid metabolism | |||||||||
| 196c | 41.46 | 6.57 | Asp aminotransferase | 44.3 | 6.8 | At5g19550 | 28% | <0.31 | >7 |
| 174c | 38.7 | 5.73 | O-Acetyl-Ser-thiol-lyase | 33.8 | 5.9 | At4g14880 | 9% | <0.26 | >7 |
| 175c | 37.85 | 5.71 | O-Acetyl-Ser-thiol-lyase | 33.8 | 5.9 | At4g14880 | 29% | <0.22 | >7 |
| Cell division | |||||||||
| 24c | 42.94 | 5.06 | Actin 7 | 41.73 | 5.31 | At5g09810 | 23% | <0.43 | >7 |
| 4c | 57.35 | 4.89 | Tubulin β2β3 chain | 50.73 | 4.70 | At5g62700 | 35% | <0.32 | >7 |
| Seed storage proteins | |||||||||
| 504c | 60.87 | 6.20 | 12S seed storage protein (precursor) | 55.86 | 6.36 | At4g28520 | 7% | <0.18 | >7 |
| 311c | 14.1 | 3.2 | β-Cruciferin 12S (seed storage protein fragment) | 58.23 | 6.53 | At4g28520 | 11% | <0.21 | >7 |
| Other processes | |||||||||
| 182c | 50.76 | 5.53 | DEAD box RNA helicase | 48.38 | 5.49 | At5g11200 | 32% | <0.42 | >7 |
| 361c | 54.65 | 5.22 | 4-Methyl-5(b-hydroxyethyl)-thiazole monophosphate biosynthesis protein | 41.84 | 5.08 | At3g14990 | 6% | <0.16 | >7 |
| 116c | 30.24 | 5.77 | Expressed protein | 28.78 | 5.92 | At5g45690 | 40% | <0.35 | >7 |
Protein numbering following Arabidopsis seed protein reference maps available at http://www.seed-proteome.com.
Data obtained from densitometric analysis of individual spots from proteins on 2D gels stained with silver nitrate (see Fig. 2A for an example of a 2D gel): normalized spot volume in the deteriorated seeds (7 d of CDT) incubated for 1 d in water divided by the normalized spot volume in the nondeteriorated control seeds (0 d of CDT) incubated for 1 d in water, from five different gels and three independent extractions.
Listed proteins correspond to previously identified proteins (Gallardo et al., 2001, 2002a; Rajjou et al., 2004, 2006a; Job et al., 2005).
Listed proteins correspond to proteins identified during this work; the peptide sequences determined are available in Supplemental Table S2.
Among proteins accumulating to lower amounts in germinating deteriorated seeds than in corresponding control seeds, two isoforms of the cytosolic O-acetyl-Ser(thiol)lyase (EC 2.5.1.47), encoded by the At4g14880 gene, were identified (spots 174 and 175; Fig. 5; Table V). This enzyme forms a complex with Ser acetyltransferase to catalyze the last step of Cys synthesis (Droux et al., 1998). It is noted that in Arabidopsis, the cytosolic form encoded by At4g14880 is the major contributor to the total O-acetyl-Ser(thiol)lyase activity of the plant (Lopez-Martin et al., 2008). Our observations, therefore, suggest that Cys synthesis is an important feature of germination potential. Cys is the essential precursor of all organic molecules containing reduced sulfur, ranging from the amino acid Met to peptides such as glutathione or phytochelatins, proteins, vitamins, cofactors such as S-adenosyl-Met, and hormones (Höfgen et al., 2001). All of these sulfur compounds play fundamental roles in plant metabolism (Ravanel et al., 1998). Furthermore, Met and/or Met derivatives have been shown to play an important promotive role in Arabidopsis seed germination and early seedling growth (Gallardo et al., 2002b). Finally, knocking out of the At4g14880 gene showed that Cys is an important determinant of the antioxidative capacity of the cytosol in Arabidopsis, as the resulting mutant appeared to be oxidatively stressed, accumulated ROS, and exhibited lesions characteristic of spontaneous cell death in the leaves (Lopez-Martin et al., 2008). We conclude that the reduced efficiency of deteriorated seeds in synthesizing Cys can induce an irreparable loss of seed vigor, owing to its general implication in metabolism and antioxidative potential in plants.
2-Alkenal reductase (EC 1.3.1.74), encoded by the At5g16970 gene, also showed a strongly depressed accumulation in germinating deteriorated seeds (spot 29; Table V). Interestingly, conditional overexpression of the 2-alkenal reductase gene in Arabidopsis results in increased salt tolerance during germination (Papdi et al., 2008). The 2-alkenal reductase possesses NADPH-dependent oxidoreductase activity, which has been shown to play a key role in the detoxification of reactive carbonyls occurring during the degradation of lipid peroxides and, hence, in the protection of cells against oxidative stress (Mano et al., 2005). As lipid peroxides accumulate during seed aging (Devaiah et al., 2007), our data highlight the role of this enzyme in seed vigor.
Protein Metabolism and Translation Are Major Components of Seed Vigor
It has been shown that seed germination has an absolute requirement for protein synthesis. Thus, cycloheximide, an inhibitor of protein translation, induces a complete inhibition of Arabidopsis seed germination (Rajjou et al., 2004). In this study, an interesting feature supports these previous observations and concerns the apparent correlation between protein metabolism and translation and the reduction of seed vigor induced by CDT (Table V). Indeed, several proteins associated with translation, such as initiation factor 4A-1, elongation factor 1-γ2, elongation factor 1B-γ, and ribosomal protein 60S (spots 129, 128, 105, and 314), are less abundant in deteriorated seeds during germination sensu stricto (Table V). Moreover, we already observed in this study that many other proteins involved in protein metabolism are altered by carbonylation in deteriorated seeds (spots 1, 43, 90, 135, 136, 137, and 140). Protein metabolism regroups several biological functions, such as protein folding, protein translocation, thermotolerance, oligomeric assembly, and switching between active and inactive protein conformations. Our results demonstrate that simultaneous impairment of these functions is closely linked with the loss of seed vigor. Our conclusion is supported by a recent study showing that transgenic seeds overaccumulating a heat stress transcription factor, which enhances the accumulation of HSPs, exhibit improved seed resistance to CDT (Prieto-Dapena et al., 2006). Moreover, the preservation of a robust stress response and protein disposal by the action of HSPs is indispensable for health and longevity in all organisms (for review, see Söti and Csermely, 2007).
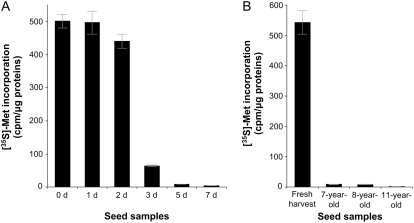
To get direct insight on the importance of protein synthesis activity in seed vigor, proteins that were neosynthesized in vivo following seed imbibition were labeled in the presence of radioactive [35S]Met. The control seeds (0 d), which had a maximum germination of 100%, exhibited a very high extent of [35S]Met incorporation, testifying to a high translational activity during germination sensu stricto. As shown in Figure 6, the extent of [35S]Met incorporation declined dramatically in the deteriorated seed samples. For example, seeds deteriorated for 3 d of CDT presented an 8-fold decrease in [35S]Met incorporation compared with control seeds, although under these conditions the aged seeds still kept good vigor, with a maximum germination of about 80% (Fig. 1). This result disclosed that translational capacity can be an excellent feature for the estimation of seed vigor, a finding that is in good agreement with previous work demonstrating a loss in translational capacity during seed aging in soybean (Pillay, 1977). The seed samples deteriorated for 5 and 7 d had germination maxima of about 28% and 2%, respectively, and showed almost no translational activity. The consequences of the observed reduction of protein synthesis can be diverse, for example, affecting the systems necessary for the maintenance, repair, and normal resumption of metabolism and cell cycle activity, the efficiency of detoxification, the efficiency of the signaling pathways, and/or the production and secretion of several metabolites and plant hormones like gibberellins.
Figure 6.
Influence of CDT and natural aging on de novo protein synthesis during germination sensu stricto (1 d after imbibition). Arabidopsis seeds were incubated in water containing [35S]Met for 1 d. Protein synthesis was measured by TCA precipitation of aliquots of reaction mixtures spotted on Whatmann GF/C filters; after 10 washing steps in cold 5% TCA and 0.04 m sodium pyrophosphate and two washing steps in absolute ethanol, filters were dried and counted for radioactivity in a liquid scintillation counter. A, Incorporation of [35S]Met in proteins synthesized de novo during germination sensu stricto of deteriorated seeds (0, 1, 2, 3, 5, and 7 d of CDT). B, Incorporation of [35S]Met in proteins synthesized de novo during germination sensu stricto of naturally aged seeds (freshly harvested seeds as well as 7-, 8-, and 11-year-old seeds).
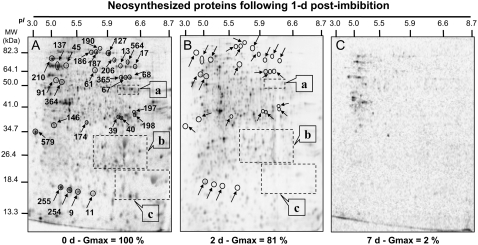
To investigate more closely this question and to reveal seed proteins whose neosynthesis during germination sensu stricto was altered by CDT, we characterized the neosynthesized proteome of three seed samples submitted to this treatment for 0, 2, and 7 d. Radiolabeled proteins were separated by 2D electrophoresis and reveled by autoradiography (Fig. 7). Translational activity of the nondeteriorated seeds (0 d) was high, as shown previously (Fig. 6). The analysis of this neosynthesized protein pattern revealed 1,272 protein spots (Fig. 7A), of which 217 proteins could be identified using our Arabidopsis seed reference maps (Supplemental Table S1). These proteins are involved in a large number of plant metabolic processes, in cell division, and in translation and protein metabolism, and interestingly, 28 protein spots match with 12S and 7S storage proteins. These seed storage proteins neosynthesized during germination sensu stricto are likely translated from stored mRNA. Indeed, it has been shown that in dry seeds, a large amount of stored mRNAs are translated during germination sensu stricto and play a fundamental role for the metabolic restart in the initialization of the germination program (Aspart et al., 1984; Rajjou et al., 2004). Deteriorated seeds submitted to 2 d of CDT displayed reduced translational activity during germination sensu stricto (Fig. 6A). Autoradiography analysis revealed 836 neosynthesized proteins (Fig. 7B), of which a large number were also neosynthesized in the nondeteriorated seeds (0 d). Yet, some proteins were specifically translated in these seeds deteriorated for 2 d, although unfortunately, they could not be identified from our Arabidopsis seed protein reference maps, because of their very low abundance. In contrast, de novo synthesis of many proteins as seen in the nondeteriorated control seeds (0 d) was abolished in 2-d deteriorated seeds. Some of them could be identified by comparison with Arabidopsis reference maps (Table VI). Interestingly, a large number of these proteins are generally associated with the end of the seed maturation program and not with the germination program, such as 12S seed storage proteins, LEA proteins, or dehydrins (Finkelstein, 1993; Bewley and Black, 1994; Cuming, 1999). It has been shown that the seed maturation program can be reinduced during early steps of seed germination (Lane, 1991; Lopez-Molina et al., 2002; Rajjou et al., 2006a, 2006b). This recruitment of the late maturation program either by de novo transcription (Lopez-Molina et al., 2002) or by translation of stored mRNAs (Rajjou et al., 2004, 2006a) is a strategy to mount appropriate defense mechanisms in response to the vagaries of nature's water supply and to protect the embryo during the transition from a metabolic quiescent state to active metabolism. For a longer time of CDT (7 d), seeds became almost unable to support de novo protein synthesis (Fig. 7C). In summary, our results clearly indicate that seed vigor is closely associated with the ability of the seeds to reinduce the late maturation program during early stages of germination.
Figure 7.
Comparison of de novo protein synthesis patterns during germination sensu stricto (1 d after imbibition) of deteriorated seeds. A, Protein profiles of de novo synthesized proteins in nondeteriorated seeds (0 d, control seeds). B, Protein profiles of de novo synthesized proteins in 3-d deteriorated seeds (3 d of CDT). C, Protein profiles of de novo synthesized proteins in 7-d deteriorated seeds (7 d of CDT). Radiolabeling of proteins was carried out by introducing [35S]Met in the germination assays, as described in “Materials and Methods.” Soluble proteins were extracted after 1 d of imbibition and submitted to 2D gel electrophoresis, and the radiolabeled proteins were revealed as described in “Materials and Methods.” The 33 labeled protein spots were identified by mass spectrometry and by comparison with Arabidopsis seed protein reference maps (Gallardo et al., 2001, 2002a; Rajjou et al., 2004, 2006a; Job et al., 2005; http://www.seed-proteome.com; Table VI; Supplemental Table S2). a, 12S seed storage protein precursor; b, α-subunit of 12S seed storage protein; c, β-subunit of 12S seed storage protein. Several radiolabeled spots exhibiting contrasting specific accumulation in deteriorated seeds could not be identified because they had no match with proteins detected in Arabidopsis seed protein reference maps (http://www.seed-proteome.com).
Table VI.
Arabidopsis proteins whose de novo synthesis was inhibited during germination sensu stricto (1 d after imbibition) of deteriorated seeds submitted to CDT for 2 d compared with nondeteriorated control seeds
| No.a | Experimental Molecular Mass | Experimental pI | Arabidopsis Protein Name | Theoretical Molecular Mass | Theoretical pI | AGI No. | Coverage |
|---|---|---|---|---|---|---|---|
| kD | kD | ||||||
| Translation and protein metabolism | |||||||
| 127b | 93.92 | 5.89 | Elongation factor EF-2 | 94.25 | 5.89 | At1g56070 | 24% |
| 9b | 17.94 | 5.50 | HSP17.6 | 17.83 | 5.22 | At5g12030 | 14% |
| 45b | 76.38 | 5.24 | HSP70 | 70.91 | 5.30 | At1g16030 | 24% |
| 137b | 76.06 | 5.07 | HSP70 | 71.37 | 5.03 | At3g12580 | 19% |
| 190b | 101.62 | 5.82 | HSP101 | 101.27 | 5.99 | At1g74310 | 16% |
| 364c | 65.22 | 5.56 | T-complex protein 1, θ-subunit (TCP-1-θ) | 58.92 | 5.01 | At3g03960 | 2% |
| 579b | 36.05 | 3.98 | Gly-rich protein similar to P23 cochaperone | 25.47 | 4.17 | At4g02450 | 24% |
| Energy metabolism | |||||||
| 17b | 73.40 | 6.58 | Phosphoenolpyruvate carboxykinase | 73.40 | 6.61 | At4g37870 | 17% |
| 197b | 40.91 | 6.77 | Fru-bisphosphate aldolase | 38.37 | 7.46 | At2g36460 | 23% |
| 198b | 40.72 | 6.89 | Fru-bisphosphate aldolase | 38.37 | 7.46 | At2g36460 | 41% |
| 39b | 38.52 | 6.23 | Glyceraldehyde-3-P dehydrogenase, cytosolic | 36.99 | 6.34 | At3g04120 | 26% |
| 40b | 38.55 | 6.26 | Glyceraldehyde-3-P dehydrogenase, cytosolic | 36.99 | 6.34 | At3g04120 | 27% |
| 365b | 62.77 | 6.30 | Isocitrate lyase | 50.42 | 6.29 | At3g21720 | 16% |
| 188b | 96.69 | 5.83 | Aconitate hydratase, cytoplasmic | 98.13 | 6.36 | At4g35830 | 10% |
| Cell detoxification and stress response | |||||||
| 11b | 19.61 | 5.75 | Em-like protein GEA1 | 16.61 | 5.75 | At3g51810 | 26% |
| 61b | 66.68 | 5.78 | Late embryogenesis abundant (LEA) | 67.19 | 5.78 | At2g42560 | 43% |
| 91b | 61.88 | 5.06 | Late embryogenesis abundant (LEA) | 52.08 | 5.29 | At3g53040 | 17% |
| 102b | 60.51 | 5.22 | Late embryogenesis abundant (LEA) | 48.49 | 5.43 | At2g36640 | 16% |
| 210b | 88.93 | 4.93 | Low-temperature-induced 65-kD protein | 65.95 | 4.81 | At5g52300 | 20% |
| 146b | 37.67 | 5.03 | MST | 41.89 | 5.95 | At1g79230 | 35% |
| 13b | 72.97 | 6.30 | MLO-like protein | 67.21 | 10.06 | At1g61560 | 15% |
| 254b | 17.96 | 5.20 | Dehydrin | 18.44 | 7.95 | At5g66400 | 6% |
| 255b | 17.96 | 5.37 | Dehydrin | 18.44 | 7.95 | At5g66401 | 6% |
| DNA metabolism | |||||||
| 186b | 96.47 | 5.78 | DNA topoisomerase I | 102.78 | 10.05 | At5g55300 | 8% |
| 187b | 96.47 | 5.81 | DNA topoisomerase I | 102.78 | 10.05 | At5g55300 | 8% |
| Amino acid metabolism | |||||||
| 174b | 38.71 | 5.73 | O-Acetyl-Ser-thiol-lyase | 33.79 | 5.95 | At4g14880 | 9% |
| 206b | 82.05 | 6.09 | 5-Methyl-tetra-hydropteroyl-tri-Glu-homo-Cys methyltransferase | 84.34 | 6.47 | At5g17920 | 17% |
| Hydrolase and protease | |||||||
| 67b | 63.25 | 6.47 | Glycosyl hydrolase family 1 protein | 60.26 | 6.91 | At3g09260 | 46% |
| 68b | 63.47 | 6.56 | Glycosyl hydrolase family 1 protein | 60.26 | 6.91 | At3g09260 | 27% |
| 564c | 87.84 | 6.40 | Cucumisin-like Ser protease | 78.28 | 5.91 | At5g67360 | 14% |
| Seed storage proteins | |||||||
| 69b | 43.67 | 6.29 | 12S seed storage protein (precursor) | 48.03 | 6.56 | At1g03880 | 18% |
| 70b | 49.43 | 7.19 | 12S seed storage protein (precursor) | 52.59 | 7.68 | At5g44120 | 22% |
| 71b | 50.44 | 7.67 | 12S seed storage protein (precursor) | 52.59 | 7.68 | At5g44120 | 34% |
| 78b | 27.26 | 6.61 | 12S seed storage protein (precursor) | 52.59 | 7.68 | At5g44120 | 13% |
| 110b | 57.97 | 6.40 | 12S seed storage protein (precursor) | 55.86 | 6.36 | At4g28520 | 19% |
| 245b | 27.22 | 6.64 | 12S seed storage protein (precursor) | 50.61 | 6.77 | At1g03880 | 25% |
| 271c | 59.96 | 6.52 | 12S seed storage protein (precursor) | 55.86 | 6.36 | At4g28520 | 9% |
| 80b | 32.64 | 5.85 | α-Cruciferin 12S (seed storage protein fragment) | 34.68 | 6.42 | At4g28520 | 33% |
| 83b | 34.35 | 6.42 | α-Cruciferin 12S (seed storage protein fragment) | 34.68 | 6.42 | At4g28520 | 33% |
| 84b | 30.46 | 6.61 | α-Cruciferin 12S (seed storage protein fragment) | 31.75 | 6.49 | At5g44120 | 42% |
| 85b | 27.20 | 6.50 | α-Cruciferin 12S (seed storage protein fragment) | 27.24 | 6.34 | At1g03880 | 32% |
| 97b | 28.89 | 7.90 | α-Cruciferin 12S (seed storage protein fragment) | 31.75 | 6.49 | At5g44120 | 40% |
| 159b | 23.54 | 5.33 | α-Cruciferin 12S (seed storage protein fragment) | 27.24 | 6.34 | At4g28520 | 15% |
| 241b | 34.11 | 5.87 | α-Cruciferin 12S (seed storage protein fragment) | 34.68 | 6.42 | At4g28520 | 32% |
| 392c | 13.31 | 5.89 | α-Cruciferin 12S (seed storage protein fragment) | 31.75 | 6.49 | At5g44120 | 14% |
| 739c | 31.04 | 5.81 | α-Cruciferin 12S (seed storage protein fragment) | 31.75 | 6.49 | At5g44120 | 47% |
| 1,154c | 11.78 | 9.05 | α-Cruciferin 12S (seed storage protein fragment) | 31.75 | 6.49 | At5g44120 | 5% |
| 1,155c | 10.43 | 9.02 | α-Cruciferin 12S (seed storage protein fragment) | 31.64 | 9.80 | At5g44120 | 17% |
| 25b | 23.32 | 6.89 | β-Cruciferin 12S (seed storage protein fragment) | 21.20 | 6.19 | At4g28520 | 27% |
| 44b | 16.14 | 7.42 | β-Cruciferin 12S (seed storage protein fragment) | 20.80 | 7.03 | At1g03880 | 7% |
| 87b | 18.43 | 6.36 | β-Cruciferin 12S (seed storage protein fragment) | 20.80 | 7.03 | At1g03880 | 33% |
| 88b | 22.82 | 8.68 | β-Cruciferin 12S (seed storage protein fragment) | 21.20 | 6.19 | At4g28520 | 44% |
| 287b | 23.44 | 5.58 | β-Cruciferin 12S (seed storage protein fragment) | 20.84 | 9.06 | At5g44120 | 22% |
| 705b | 11.75 | 8.90 | β-Cruciferin 12S (seed storage protein fragment) | 21.20 | 6.19 | At1g03880 | 44% |
| 1,153c | 11.51 | 9.02 | β-Cruciferin 12S (seed storage protein fragment) | 21.20 | 6.19 | At4g28520 | 39% |
| 1,156c | 10.68 | 9.05 | β-Cruciferin 12S (seed storage protein fragment) | 20.84 | 9.06 | At5g44120 | 41% |
| 246b | 29.45 | 6.70 | Storage proteins 7S | 55.05 | 7.15 | At3g22640 | 28% |
| 247b | 34.02 | 6.77 | Storage proteins 7S | 55.05 | 7.15 | At3g22640 | 18% |
Protein numbering following Arabidopsis seed protein reference maps available at http://www.seed-proteome.com.
Listed proteins correspond to previously identified proteins (Gallardo et al., 2001, 2002a; Rajjou et al., 2004, 2006a; Job et al., 2005).
Listed proteins correspond to proteins identified during this work; the peptide sequences determined are available in Supplemental Table S2.
Similar Events Occur during Accelerated and Natural Aging
There is still uncertainty regarding whether CDT mimics natural aging. This is a major concern of seed companies, because, for practical reasons, they rely on CDT and germination assays to predict seed storability (Delouche and Baskin, 1973). Therefore, it was important to compare the biochemical behavior of seeds submitted to CDT and of seeds that have been naturally aged, namely, seeds that have been stored for several years in tubes in a refrigerator regulated at 5°C. For that purpose, three naturally aged Arabidopsis seed samples were examined. Two of them were 7 and 8 years old and presented maximum germination of about 45% and 23%, respectively. A third sample was 11 years old and did not germinate, even at 30 d after imbibition (Table VII).
Table VII.
The effect of natural aging on the germination of Arabidopsis seeds
| Seed Samples | Maximum Germination | sd |
|---|---|---|
| % | ||
| Freshly harvested seeds | 99.00 | 1 |
| Seven-year-old seeds | 45.33 | 2.96 |
| Eight-year-old seeds | 23.00 | 2.52 |
| Eleven-year-old seeds | 0.00 | 0 |
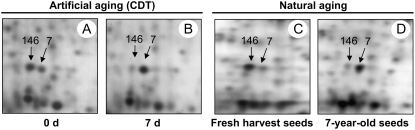
Our present proteome analysis revealed common features between the artificially and naturally aged seeds. Indeed, the evolution of the dry seed proteome during natural aging and during CDT displayed common changes, as shown in Figure 8 for two protein spots (spots 146 and 7). Spot 146 (Figs. 2, 3, and 8; Tables II and V), corresponding to MST, was abundant in nondeteriorated seeds (0 d) and in freshly harvested seeds. However, during both CDT and natural aging, the abundance of this protein was strongly reduced in the dry seeds. An opposite behavior was observed for protein spot 7, corresponding to the 60S ribosomal protein, whose level strongly increased during both artificial and natural aging (Figs. 2, 3, and 8; Tables II and V).
Figure 8.
Similar protein pattern evolution during CDT or natural aging. An equal amount (150 μg) of total protein extracts was loaded on each gel. A representative silver-stained 2D gel of total proteins from nondeteriorated seeds (0 d, control seeds) is presented Figure 2A. A, Enlarged window of 2D gels as shown in Figure 2Ab for nondeteriorated seeds. B, Enlarged window of 2D gels as shown in Figure 2Ab for deteriorated seeds (7 d of CDT). C, Enlarged window of 2D gels as shown in Figure 2Ab for freshly harvested seeds. D, Enlarged window of 2D gels as shown in Figure 2Ab for 7-year-old seeds (natural aging).
Another spectacular similarity observed between natural and artificial aging concerned the oxidation patterns of the seed proteome. As depicted in Figure 4, almost identical protein carbonylation events occurred during natural and artificial aging. In both cases, the extent of protein carbonylation was strongly increased and the protein targets of carbonylation were nearly the same. In particular, these results confirm our previous finding that protein oxidation is not a random process but targets very specific proteins (Job et al., 2005).
Finally, it is remarkable that translational capacity was strongly repressed in naturally aged seeds (Fig. 6), a specific feature also observed with CDT (Figs. 6 and 7).
Overall, our data provide the first molecular indication supporting the usefulness of CDT for the prediction of seed storability.
A Reduction in Amino Acid Pools during Aging Is Not the Cause of Seed Vigor Loss
Aging of the seeds, either by CDT or natural aging, caused large reductions in protein synthesis during the first day of imbibition. One of the hypotheses is that this can be caused by preferential use of amino acids as alternative energy sources, thereby limiting the substrate for protein synthesis. This hypothesis was tested by incubating CDT and control seeds in solutions of different amino acids, pyruvate, or Glc.
We found that Asp, Glu, or Met at 1 m could not stimulate the germination of seeds submitted to CDT. Also, neither Glc nor pyruvate could stimulate the germination of CDT seeds (data not shown).
A Reduction in Template Activity of Stored mRNAs Is Not the Cause of Seed Vigor Loss
As documented above, the potential of de novo protein synthesis was severely reduced during both artificial and natural aging. A possible explanation could be that the translational machinery was damaged, which is supported by our present data (Fig. 6). However, a different explanation to account for this behavior could be that the stored mRNA pool is damaged in seeds challenged by CDT or following natural aging. To further explore this question, stored mRNAs were extracted from nondeteriorated and aged seeds and the translation potential of these mRNAs was evaluated by in vitro translation assays using a commercially available wheat germ translation system, as described in “Materials and Methods.” For all seed samples, we found that stored mRNAs can be used as templates in this system (data not shown). It should be noted, however, that this conclusion is based on the use of an in vitro heterologous translation system that might not reproduce all facets of the in vivo situation. Furthermore, this assay allowed only global estimation of the template activity of the extracted pool of stored mRNAs, and at present we cannot exclude the possibility that particular stored mRNAs playing a role in seed quality could be damaged by aging. Nevertheless, our data strongly indicate that reduced activity of the translational machinery is one of the main factors involved in seed longevity integrity, either due to reduced integrity or to an inhibition of this machinery.
CONCLUSION
In conclusion, proteomics provided an innovative and powerful tool for investigating the molecular mechanisms of seed vigor and seed viability during aging. From a methodological point of view, it is worth noting that the proteins analyzed here could be readily identified from our previous studies establishing reference protein maps of Arabidopsis seeds (http://www.seed-proteome.com). On the one hand, this illustrates the robustness of the proteomic approach, notably concerning the reproducibility of protein patterns on 2D gels. On the other hand, this shows the usefulness of establishing such protein maps, especially considering the cost and effort needed for protein identification. From this work, it appears that changes in the regulation of protein synthesis, posttranslational modifications, and protein turnover are crucial determinants of the age-related decline in the maintenance, repair, and survival of the seed. The CDT used to mimic natural seed aging was efficient to alter germinative ability, as indicated by germination behavior. A decrease of seed vigor was observed in relation to the duration of treatment. This experimental protocol allowed comparing a differentially deteriorated seed proteome in order to gain a better understanding of complex mechanisms controlling seed aging. In particular, our proteomic analyses revealed that the loss in seed vigor induced by aging can be accounted for both by protein changes in the dry seeds and by an inability of the low-vigor seeds to display a normal proteome during germination. We characterized several proteins whose accumulation levels varied as a consequence of CDT and the loss of the ability to germinate. Therefore, these proteins should play an important role in the expression of seed vigor. In this context, our results revealed essential mechanisms for seed vigor, such as translational capacity, mobilization of seed storage reserves, and detoxification efficiency. Furthermore, the observed increase in protein oxidation, both in artificially and naturally aged seeds, lends support to the finding that oxidative stress accompanies seed aging. The accumulation of oxidative damage in seeds was correlated with the loss of germination vigor. Increased protein oxidation (carbonylation) might induce a loss of functional properties of target seed proteins and enzymes and/or enhance their susceptibility toward proteolysis. Since protein oxidation mainly results from attack by ROS, this suggests an important role of antioxidant systems through detoxification and protection of upstream mechanisms to maintain seed vigor.
Another fundamental feature shown by our study was the dramatic reduction of the protein neosynthesis capacity of the aged seeds. Their translational activity was strongly reduced during the first day of imbibition, corresponding to germination sensu stricto. It may be strongly impaired by aging, through protein oxidation and/or degradation. The 3-d CDT seeds, although having a considerable reduction in translation potential during the first day of imbibition, still kept a high vigor, as 80% of these seeds could germinate under the experimental conditions. Since these seeds would require a functional translational machinery, we assume that their translational machinery is both damaged and repaired, or that its activity is temporary halted, or a combination of both. It can be hypothesized that during germination, the seeds have feedback mechanisms inhibiting mRNA translation until DNA damage is repaired. Induction of DNA damage during seed aging has been demonstrated for a long time (Osborne et al., 1980/1981).
Our results are in agreement with previous experiments showing that Arabidopsis seeds are unable to germinate in the presence of the translational inhibitor cycloheximide, thereby implying that seed germination requires de novo protein synthesis (Rajjou et al., 2004). This study on the model plant Arabidopsis enables a better understanding of the molecular mechanisms underlying loss of germination vigor during seed aging. Finally, for the first time, to our knowledge, we demonstrate that the CDT protocol, extensively used by the seed industry to control seed quality, mimics truly molecular and biochemical events occurring during natural seed aging. Further work is in progress in our laboratories to validate the role of the characterized proteins in seed vigor by reverse genetics.
MATERIALS AND METHODS
Plant Material and Germination Experiments
Nondormant seeds of Arabidopsis (Arabidopsis thaliana), accession Ler, were used in all experiments. Germination assays were carried out at 25°C, with 16 h of light and 8 h of dark daily, as described (Rajjou et al., 2004, 2006a). Naturally aged seeds had been stored for several years in tubes in a refrigerator regulated at 5°C.
CDT
CDT was performed according to Tesnier et al. (2002). Briefly, seeds were equilibrated at 85% relative humidity (20°C) in an electronically controlled environment cabinet (van den Berg Klimaattechniek), and day-0 controls were immediately dried back at 32% relative humidity. Treatment was done by storing the seeds (after equilibration at 85% relative humidity) for various times (0, 4, and 16 h, and 1, 2, 3, 5, and 7 d) at 40°C. Then, seeds were dried back at 32% relative humidity (20°C) and stored at 4°C.
Preparation of Total Protein Extracts
Total protein extracts were prepared from dry mature seeds and from seeds at 1 d after imbibition as described previously (Rajjou et al., 2006a). Following grinding of 100 mg of seeds using mortar and pestle in liquid nitrogen, total soluble proteins were extracted at 4°C in 1.2 mL of thiourea/urea lysis buffer (Harder et al., 1999) containing 7 m urea (GE Healthcare), 2 m thiourea (Merck), 4% (w/v) CHAPS (Amersham Biosciences), and 1% (v/v) Pharmalyte pH 3 to 10 carrier ampholytes (GE Healthcare). This extraction buffer also contained 18 mm Tris-HCl (Trizma HCl; Sigma), 14 mm Trizma base (Sigma), the protease inhibitor cocktail Complete Mini from Roche Diagnostics, 53 units/mL DNase I (Roche Diagnostics), 4.9 Kunitz units/mL RNase A (Sigma), and 0.2% (v/v) Triton X-100. After 10 min at 4°C, 14 mm dithiothreitol (DTT; GE Healthcare) was added and the protein extracts were stirred for 20 min at 4°C, then centrifuged (35,000g, 10 min) at 4°C. The supernatant was submitted to a second clarifying centrifugation as above. The final supernatant corresponded to the total soluble protein extract. Protein concentrations in the various extracts were measured according to Bradford (1976). Bovine serum albumin was used as a standard.
Two-Dimensional Electrophoresis
Proteins were first separated by electrophoresis according to charge. Isoelectric focusing was realized with protein samples with an equivalent to an extract of 100 seeds, corresponding to about 150 mg of protein for all samples. Proteins from the various extracts were separated using gel strips forming an immobilized nonlinear pH gradient from 3 to 10 (Immobiline DryStrip pH 3–10 NL, 18 cm; GE Healthcare). Strips were rehydrated for 14 h at 20°C with the thiourea/urea lysis buffer containing 2% (v/v) Triton X-100, 20 mm DTT, and the protein extracts. Isoelectric focusing was performed at 20°C in the Multiphor II system (Amersham Biosciences) for 1 h at 300 V and for 7 h at 3,500 V. Proteins were then separated according to size. Prior to the second dimension, the gel strips were equilibrated twice for 30 min each in 2 × 100 mL of equilibration solution containing 6 m urea, 30% (v/v) glycerol, 2.5% (w/v) SDS, 0.15 m BisTris, and 0.1 m HCl (Görg et al., 1987; Harder et al., 1999). DTT (50 mm) was added to the first equilibration solution, and iodoacetamide (4% [w/v]) was added to the second (Harder et al., 1999). Equilibrated gel strips were placed on top of vertical polyacrylamide gels (10% [v/v] acrylamide, 0.33% [w/v] piperazine diacrylamide, 0.18 m Trizma base, 0.166 m HCl, 0.07% [w/v] ammonium persulfate, and 0.035% [v/v] N,N,N′,N′-tetramethylethylenediamine [TEMED]). A denaturing solution (1% [w/v] low-melting agarose [Gibco BRL], 0.4% [w/v] SDS, 0.15 m BisTris, and 0.1 m HCl) was loaded onto gel strips. After agarose solidification, electrophoresis was performed at 10°C in a buffer (pH 8.3) containing 25 mm Trizma base, 200 mm taurine, and 0.1% (w/v) SDS for 1 h at 35 V and for 14 h at 100 V. Ten gels (200 × 250 × 1.0 mm) were run in parallel (Isodalt system from Amersham Biosciences). For each condition analyzed, 2D gels were made at least in triplicate and from three independent protein extractions; kinetic data shown in Figure 2 and Tables I, II, IV, and V were obtained from at least five gels for each seed sample (nondeteriorated and deteriorated seeds). 2D gels were stained with silver nitrate according to Blum et al. (1987) for densitometric analyses. Image analysis was carried out with ImageMaster 2D Elite version 4.01 software (Amersham Biosciences). The kinetics of protein accumulation were analyzed by nonlinear regression analysis using exponential relationships and KaleidaGraph software (Synergy Software).
Detection of Oxidized Proteins and Western Blotting
Detection of oxidized proteins by carbonylation was performed by derivatization of protein extracts with 2,4-dinitrophenylhydrazine and immunological detection of the DNP adducts with monoclonal anti-DNP antibody (OxyBlot Oxidized Protein Detection Kit; Chemicon) as described previously (Korolainen et al., 2002; Johansson et al., 2004; Job et al., 2005).
De Novo Protein Synthesis
Labeled proteins were synthesized in vivo by imbibing seeds on water for 1 d in the presence of [35S]Met (1.85 MBq; ICN Biomedicals). Protein synthesis was measured by TCA precipitation of aliquots of reaction mixtures spotted on Whatmann GF/C filters; after 10 washing steps in cold 5% TCA and 0.04 m sodium pyrophosphate and two washing steps in absolute ethanol, filters were dried and counted for radioactivity in a liquid scintillation counter (Rajjou et al., 2004).
Protein Identification
Proteins of interest correspond to previously identified seed proteins from 2D electrophoresis experiments and localized on 2D electrophoresis reference maps (Gallardo et al., 2001, 2002a; Rajjou et al., 2004, 2006a; Job et al., 2005; http://www.seed-proteome.com). In this work, the protein identification of each spot of interest was verified by mass spectrometry as described by Rajjou et al. (2006a).
RNA Integrity
Stored RNAs were extracted using hot borate (Wan and Wilkins, 1994). The integrity of the rRNA and mRNA pools was analyzed, respectively, by gel electrophoresis and using a wheat germ extract in vitro translation assay (Promega) with 35S-labeled Met (1.85 MBq; ICN Biochemicals).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. Arabidopsis proteins whose de novo synthesis occurs during germination sensu stricto (1 d after imbibition) of nondeteriorated control seeds (0 d).
Supplemental Table S2. Peptide sequences identified by tandem mass spectrometry sequencing and corresponding to novel proteins in Arabidopsis seeds identified in this work.
Supplementary Material
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Loïc Rajjou (loic.rajjou@agroparistech.fr).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Abdalla FH, Roberts EH (1968) The effects of temperature and moisture on the induction of chromosome damage in seeds of barley, broad bean and pea during storage. Ann Bot (Lond) 32 119–136 [Google Scholar]
- Adams DO, Yang SF (1981) Ethylene, the gaseous plant hormone: mechanism and regulation of biosynthesis. Trends Biochem Sci 6 161–164 [Google Scholar]
- Agrawal GK, Yonekura M, Iwahashi Y, Iwahashi H, Rakwal R (2005. a) Systems, trends and perspectives of proteomics in dicot plants Part I: Technologies in proteome establishment. J Chromatogr B Analyt Technol Biomed Life Sci 815 109–123 [DOI] [PubMed] [Google Scholar]
- Agrawal GK, Yonekura M, Iwahashi Y, Iwahashi H, Rakwal R (2005. b) Systems, trends and perspectives of proteomics in dicot plants Part II: Proteomes of the complex developmental stages. J Chromatogr B Analyt Technol Biomed Life Sci 815 125–136 [DOI] [PubMed] [Google Scholar]
- Agrawal GK, Yonekura M, Iwahashi Y, Iwahashi H, Rakwal R (2005. c) Systems, trends and perspectives of proteomics in dicot plants Part III: Unraveling the proteomes influenced by the environment, and at the levels of functions and genetic relationships. J Chromatogr B Analyt Technol Biomed Life Sci 815 137–145 [DOI] [PubMed] [Google Scholar]
- Alban C, Job D, Douce R (2000) Biotin metabolism in plant. Annu Rev Plant Physiol Plant Mol Biol 51 17–47 [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408 796–815 [DOI] [PubMed] [Google Scholar]
- Aspart L, Meyer Y, Laroche M, Penon P (1984) Developmental regulation of the synthesis of proteins encoded by stored mRNA in radish embryos. Plant Physiol 76 664–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly C (2004) Active oxygen species and antioxidants in seed biology. Seed Sci Res 14 93–107 [Google Scholar]
- Bartoli CG, Pastori GM, Foyer CH (2000) Ascorbate biosynthesis in mitochondria is linked to the electron transport chain between complexes III and IV. Plant Physiol 123 335–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter CJ, Redestig H, Schauer N, Repsilber D, Patil KR, Nielsen J, Selbig J, Liu J, Fernie AR, Sweetlove LJ (2007) The metabolic response of heterotrophic Arabidopsis cells to oxidative stress. Plant Physiol 143 312–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentsink L, Alonso-Blanco C, Vreugdenhil D, Tesnier K, Groot SPC, Koornneef M (2000) Genetic analysis of seed-soluble oligosaccharides in relation to seed storability of Arabidopsis. Plant Physiol 124 1595–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentsink L, Jowett J, Hanhart CJ, Koornneef M (2006) Cloning of DOG1, a quantitative trait locus controlling seed dormancy in Arabidopsis. Proc Natl Acad Sci USA 103 17042–17047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlett BS, Stadtman ER (1997) Protein oxidation in aging, disease, and oxidative stress. J Biol Chem 272 20313–20316 [DOI] [PubMed] [Google Scholar]
- Bethke PC, Libourel IG, Reinohl V, Jones RL (2006) Sodium nitroprusside, cyanide, nitrite, and nitrate break Arabidopsis seed dormancy in a nitric oxide-dependent manner. Planta 223 805–812 [DOI] [PubMed] [Google Scholar]
- Bewley JD, Black M (1994) Seeds: Physiology of Development and Germination. Plenum Press, New York
- Bies-Ethève N, Gaubier-Comella P, Debures A, Lasserre E, Jobet E, Raynal M, Cooke R, Delseny M (2008) Inventory, evolution and expression profiling diversity of the LEA (late embryogenesis abundant) protein gene family in Arabidopsis thaliana. Plant Mol Biol 67 107–124 [DOI] [PubMed] [Google Scholar]
- Blum H, Beier H, Gross HJ (1987) Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis 8 93–99 [Google Scholar]
- Bonomi F, Pagani S, Kurtz DM Jr (1985) Enzymic synthesis of the 4Fe-4S clusters of Clostridium pasteurianum ferredoxin. Eur J Biochem 148 67–73 [DOI] [PubMed] [Google Scholar]
- Borovskii GB, Stupnikova IV, Antipina AI, Vladimirova SV, Voinikov VK (2002) Accumulation of dehydrin-like proteins in the mitochondria of cereals in response to cold, freezing, drought and ABA treatment. BMC Plant Biol 2 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudet J, Buitink J, Hoekstra FA, Rogniaux H, Larré C, Satour P, Leprince O (2006) Comparative analysis of the heat stable proteome of radicles of Medicago truncatula seeds during germination identifies late embryogenesis abundant proteins associated with desiccation tolerance. Plant Physiol 140 1418–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein using the principle of protein-dye binding. Anal Biochem 72 248–254 [DOI] [PubMed] [Google Scholar]
- Brown PD, Tokuhisa JG, Reichelt M, Gershenzon J (2003) Variation of glucosinolate accumulation among different organs and developmental stages of Arabidopsis thaliana. Phytochemistry 62 471–481 [DOI] [PubMed] [Google Scholar]
- Busk PK, Møller BL (2002) Dhurrin synthesis in sorghum is regulated at the transcriptional level and induced by nitrogen fertilization in older plants. Plant Physiol 129 1222–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabiscol E, Piulats E, Echave P, Herrero E, Ros J (2000) Oxidative stress promotes specific protein damage in Saccharomyces cerevisiae. J Biol Chem 275 27393–27398 [DOI] [PubMed] [Google Scholar]
- Cánovas FM, Dumas-Gaudot E, Recorbet G, Jorrín J, Mock HP, Rossignol M (2004) Plant proteome analysis. Proteomics 4 285–298 [DOI] [PubMed] [Google Scholar]
- Cipollini D, Gruner B (2007) Cyanide in the chemical arsenal of garlic mustard, Alliaria petiolata. J Chem Ecol 33 85–94 [DOI] [PubMed] [Google Scholar]
- Cipollone R, Ascenzi P, Visca P (2007) Common themes and variations in the rhodanese superfamily. IUBMB Life 59 51–59 [DOI] [PubMed] [Google Scholar]
- Clarke S (2003) Aging as war between chemical and biochemical processes: protein methylation and the recognition of age-damaged proteins for repair. Ageing Res Rev 2 263–285 [DOI] [PubMed] [Google Scholar]
- Clerkx EJ, Blankestijn-De Vries H, Ruys GJ, Groot SPC, Koornneef M (2004. a) Genetic differences in seed longevity of various Arabidopsis mutants. Physiol Plant 121 448–461 [Google Scholar]
- Clerkx EJ, El-Lithy ME, Vierling E, Ruys GJ, Blankestijn-De Vries H, Groot SPC, Vreugdenhil D, Koornneef M (2004. b) Analysis of natural allelic variation of Arabidopsis seed germination and seed longevity traits between the accessions Landsberg erecta and Shakdara, using a new recombinant inbred line population. Plant Physiol 135 432–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Close TJ (1996) Dehydrins: emergence of a biochemical role of a family of plant dehydration proteins. Physiol Plant 97 795–803 [Google Scholar]
- Close TJ (1997) Dehydrins: a commonality in the response of plants to dehydration and low temperature. Physiol Plant 100 291–296 [Google Scholar]
- Cuming AC (1999) LEA proteins. In PR Shewry, R Casey, eds, Seed Proteins. Kluwer Academic Press, Dordrecht, The Netherlands, pp 753–780
- Davies MJ (2005) The oxidative environment and protein damage. Biochim Biophys Acta 1703 93–109 [DOI] [PubMed] [Google Scholar]
- Debeaujon I, Léon-Kloosterziel KM, Koornneef M (2000) Influence of the testa on seed dormancy, germination, and longevity in Arabidopsis. Plant Physiol 122 403–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de los Reyes BG, Myers SJ, McGrath JM (2003) Differential induction of glyoxylate cycle enzymes by stress as a marker for seedling vigor in sugar beet (Beta vulgaris). Mol Genet Genomics 269 692–698 [DOI] [PubMed] [Google Scholar]
- Delouche JC, Baskin CC (1973) Accelerated aging techniques for predicting the relative storability of seed lots. Seed Sci Technol 1 427–452 [Google Scholar]
- Devaiah SP, Pan X, Hong Y, Roth M, Welti R, Wang X (2007) Enhancing seed quality and viability by suppressing phospholipase D in Arabidopsis. Plant J 50 950–957 [DOI] [PubMed] [Google Scholar]
- Donadio S, Shafiee A, Hutchinson CR (1990) Disruption of a rhodanese like gene results in cysteine auxotrophy in Saccharopolyspora erythraea. J Bacteriol 172 350–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Droux M, Ruffet ML, Douce R, Job D (1998) Interactions between serine acetyltransferase and O-acetylserine (thiol) lyase in higher plants: structural and kinetic properties of the free and bound enzymes. Eur J Biochem 255 235–245 [DOI] [PubMed] [Google Scholar]
- Dure L III (1993) Structural motifs in LEA proteins of higher plants. In TJ Close, EA Bray, eds, Response of Plants to Cellular Dehydration during Environmental Stress. American Society of Plant Physiologists, Rockville, MD, pp 91–103
- Duval M, DeRose RT, Job C, Faucher D, Douce R, Job D (1994. a) The major biotinyl protein from Pisum sativum seeds covalently binds biotin at a novel site. Plant Mol Biol 26 265–273 [DOI] [PubMed] [Google Scholar]
- Duval M, Job C, Alban C, Douce R, Job D (1994. b) Developmental patterns of free and protein-bound biotin during maturation and germination of seeds of Pisum sativum: characterization of a novel seed-specific biotinylated protein. Biochem J 299 141–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastmond PJ, Graham IA (2001) Re-examining the role of the glyoxylate cycle in oilseeds. Trends Plant Sci 6 72–78 [DOI] [PubMed] [Google Scholar]
- Ellis WD, Dunford HB (1968) The kinetics of cyanide and fluoride binding by ferric horseradish peroxidase. Biochemistry 7 2054–2062 [DOI] [PubMed] [Google Scholar]
- Feller U, Fischer A (1994) Nitrogen metabolism in senescing leaves. Crit Rev Plant Sci 13 241–273 [Google Scholar]
- Finkelstein RR (1993) Abscisic acid-insensitive mutations provide evidence for stage-specific signal pathways regulating expression of an Arabidopsis late embryogenesis-abundant (lea) gene. Mol Gen Genet 238 401–408 [DOI] [PubMed] [Google Scholar]
- Franca Neto JB, Shatters RG, West SH (1997) Developmental pattern of biotinylated proteins during embryogenesis and maturation of soybean seed. Seed Sci Res 7 377–384 [Google Scholar]
- Freedman RB, Dunn AD, Ruddock LW (1998) Protein folding: a missing redox link in the endoplasmic reticulum. Curr Biol 8 R468–R470 [DOI] [PubMed] [Google Scholar]
- Gallardo K, Job C, Groot SPC, Puype M, Demol H, Vandekerckhove J, Job D (2001) Proteomic analysis of Arabidopsis seed germination and priming. Plant Physiol 126 835–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo K, Job C, Groot SPC, Puype M, Demol H, Vandekerckhove J, Job D (2002. a) Proteomics analysis of Arabidopsis seed germination: a comparative study of wild-type and GA-deficient seeds. Plant Physiol 129 823–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo K, Job C, Groot SPC, Puype M, Demol H, Vandekerckhove J, Job D (2002. b) Importance of methionine biosynthesis for Arabidopsis seed germination and seedling growth. Physiol Plant 116 238–247 [DOI] [PubMed] [Google Scholar]
- Goel A, Goel AK, Sheoran IS (2003) Changes in oxidative stress enzymes during artificial ageing in cotton (Gossypium hirsutum L.) seeds. J Plant Physiol 160 1093–1100 [DOI] [PubMed] [Google Scholar]
- Goldberg RB, Barker SJ, Perez-Grau L (1989) Regulation of gene expression during plant embryogenesis. Cell 56 149–160 [DOI] [PubMed] [Google Scholar]
- Görg A, Postel W, Weser J, Günther S, Strahler JR, Hanash SM, Somerlot L (1987) Elimination of point streaking on silver stained two-dimensional gels by addition of iodoacetamide to the equilibration buffer. Electrophoresis 8 122–124 [Google Scholar]
- Graham IA (2008) Seed storage oil mobilization. Annu Rev Plant Biol 59 115–142 [DOI] [PubMed] [Google Scholar]
- Halmer P (2000) Commercial seed treatment technology. In M Black, JD Bewley, eds, Seed Technology and Its Biological Basis. Sheffield Academic Press, Sheffield, England, pp 257–286
- Harder A, Wildgruber R, Nawrocki A, Fey SJ, Larsen PM, Görg A (1999) Comparison of yeast cell protein solubilization procedures for two-dimensional electrophoresis. Electrophoresis 20 826–829 [DOI] [PubMed] [Google Scholar]
- Harman D (1956) Aging: a theory based on free radical and radiation chemistry. J Gerontol 11 298–300 [DOI] [PubMed] [Google Scholar]
- Heydecker W, Higgins J, Gulliver RL (1973) Accelerated germination by osmotic seed treatment. Nature 246 42–44 [Google Scholar]
- Höfgen R, Kreft O, Willmitzer L, Hesse H (2001) Manipulation of thiol contents in plants. Amino Acids 20 291–299 [DOI] [PubMed] [Google Scholar]
- Holdsworth MJ, Finch-Savage WE, Grappin P, Job D (2008) Post-genomics dissection of seed dormancy and germination. Trends Plant Sci 13 7–13 [DOI] [PubMed] [Google Scholar]
- Hsing YC, Tsou CH, Hsu TF, Chen ZY, Hsieh KL, Hsieh JS, Chow TY (1998) Tissue- and stage-specific expression of a soybean (Glycine max L.) seed-maturation, biotinylated protein. Plant Mol Biol 38 481–490 [DOI] [PubMed] [Google Scholar]
- Ingrosso D, Cimmino A, D'Angelo S, Alfinito F, Zappia V, Galletti P (2002) Protein methylation as a marker of aspartate damage in glucose-6-phosphate dehydrogenase-deficient erythrocytes: role of oxidative stress. Eur J Biochem 269 2032–2039 [DOI] [PubMed] [Google Scholar]
- Ismail AM, Hall AE, Close TJ (1999) Purification and partial characterization of a dehydrin involved in chilling tolerance during seedling emergence of cowpea. Plant Physiol 120 237–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Job C, Laugel S, Duval M, Gallardo K, Job D (2001) Biochemical characterization of atypical biotinylation domains in seed proteins. Seed Sci Res 11 149–161 [Google Scholar]
- Job C, Rajjou L, Lovigny Y, Belghazi M, Job D (2005) Patterns of protein oxidation in Arabidopsis seeds and during germination. Plant Physiol 138 790–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Job D, Ricard J (1975) Kinetic and equilibrium studies of cyanide and fluoride binding to turnip peroxidases. Arch Biochem Biophys 170 427–437 [DOI] [PubMed] [Google Scholar]
- Johansson E, Olsson O, Nyström T (2004) Progression and specificity of protein oxidation in the life cycle of Arabidopsis thaliana. J Biol Chem 279 22204–22208 [DOI] [PubMed] [Google Scholar]
- Jorrín JV, Maldonado AM, Castillejo MA (2007) Plant proteome analysis: a 2006 update. Proteomics 7 2947–2962 [DOI] [PubMed] [Google Scholar]
- Kibinza S, Vinel D, Côme D, Bailly C, Corbineau F (2006) Sunflower seed deterioration as related to moisture content during ageing, energy metabolism and active oxygen species scavenging. Physiol Plant 128 496–506 [Google Scholar]
- Korolainen MA, Goldsteins G, Alafuzoff I, Koistinaho J, Pirttila T (2002) Proteomic analysis of protein oxidation in Alzheimer's disease brain. Electrophoresis 23 3428–3433 [DOI] [PubMed] [Google Scholar]
- Laboissière MC, Sturley SL, Raines RT (1997) Protein disulfide isomerase in spore germination and cell division. Biol Chem 378 431–437 [DOI] [PubMed] [Google Scholar]
- Lane BG (1991) Cellular desiccation and hydration: developmentally regulated proteins, and the maturation and germination of seed embryos. FASEB J 5 2893–2901 [DOI] [PubMed] [Google Scholar]
- Lång V, Palva ET (1992) The expression of a rab-related gene, rab18, is induced by abscisic acid during the cold acclimation process of Arabidopsis thaliana (L.) Heynh. Plant Mol Biol 20 951–962 [DOI] [PubMed] [Google Scholar]
- Lanteri S, Nada E, Belletti P, Quagliotti L, Bino RJ (1996) Effects of controlled deterioration and osmoconditioning on germination and nuclear replication in seeds of pepper (Capsicum annuum L.). Ann Bot (Lond) 77 591–597 [Google Scholar]
- Levine RL, Garland D, Oliver CN, Amici A, Climent I, Lenz AG, Ahn BW, Shaltiel S, Stadtman ER (1990) Determination of carbonyl content in oxidatively modified proteins. Methods Enzymol 186 464–478 [DOI] [PubMed] [Google Scholar]
- Levine RL, Stadtman ER (2001) Oxidative modification of proteins during aging. Exp Gerontol 36 1495–1502 [DOI] [PubMed] [Google Scholar]
- Lopez-Martin MC, Becana M, Romero LC, Gotor C (2008) Knocking out cytosolic cysteine synthesis compromises the antioxidant capacity of the cytosol to maintain discrete concentrations of hydrogen peroxide in Arabidopsis. Plant Physiol 147 562–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, McLachlin DT, Chait BT, Chua NH (2002) ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J 32 317–328 [DOI] [PubMed] [Google Scholar]
- Mano J, Belles-Boix E, Babiychuk E, Inzè D, Torii Y, Hiraoka E, Takimoto K, Slooten L, Asada K, Kushnir S (2005) Protection against photooxidative injury of tobacco leaves by 2-alkenal reductase: detoxication of lipid peroxide-derived reactive carbonyls. Plant Physiol 139 1773–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- March TJ, Able JA, Schultz CJ, Able AJ (2007) A novel late embryogenesis abundant protein and peroxidase associated with black point in barley grains. Proteomics 7 3800–3808 [DOI] [PubMed] [Google Scholar]
- McDonald MB (1999) Seed deterioration: physiology, repair and assessment. Seed Sci Technol 27 177–237 [Google Scholar]
- Miura K, Lin Y, Yano M, Nagamine T (2002) Mapping quantitative trait loci controlling seed longevity in rice (Oryza sativa L.). Theor Appl Genet 104 981–986 [DOI] [PubMed] [Google Scholar]
- Møller IM, Jensen PE, Hansson A (2007) Oxidative modifications to cellular components in plants. Annu Rev Plant Biol 58 459–481 [DOI] [PubMed] [Google Scholar]
- Nagahara N, Katayama A (2005) Post-translational regulation of mercaptopyruvate sulfurtransferase via a low redox potential cysteine-sulfenate in the maintenance of redox homeostasis. J Biol Chem 280 34569–34576 [DOI] [PubMed] [Google Scholar]
- Nakabayashi K, Okamoto M, Koshiba T, Kamiya Y, Nambara E (2005) Genome-wide profiling of stored mRNA in Arabidopsis thaliana seed germination: epigenetic and genetic regulation of transcription in seed. Plant J 41 697–709 [DOI] [PubMed] [Google Scholar]
- Nikolau BJ, Ohlrogge JB, Wurtele ES (2003) Plant biotin-containing carboxylases. Arch Biochem Biophys 414 211–222 [DOI] [PubMed] [Google Scholar]
- Nyström T (2005) Role of oxidative carbonylation in protein quality control and senescence. EMBO J 24 1311–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M, Hanada A, Yamauchi Y, Kuwahara A, Kamiya Y, Yamaguchi S (2003) Gibberellin biosynthesis and response during Arabidopsis seed germination. Plant Cell 15 1591–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oracz K, El-Maarouf-Bouteau H, Bogatek R, Corbineau F, Bailly C (2008) Release of sunflower seed dormancy by cyanide: cross-talk with ethylene signalling pathway. J Exp Bot 59 2241–2251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne DJ, Sharon R, Ben-Ishai R (1980/1981) DNA integrity and repair. Isr J Bot 29 259–272 [Google Scholar]
- Pagani S, Bonomi F, Cerletti P (1984) Enzymic synthesis of the iron-sulfur cluster of spinach ferredoxin. Eur J Biochem 142 361–366 [DOI] [PubMed] [Google Scholar]
- Papdi C, Abraham E, Joseph MP, Popescu C, Koncz C, Szabados L (2008) Functional identification of Arabidopsis stress regulatory genes using the controlled cDNA overexpression system. Plant Physiol 147 528–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papenbrock J, Schmidt A (2000. a) Characterization of a sulfurtransferase from Arabidopsis thaliana. Eur J Biochem 267 145–154 [DOI] [PubMed] [Google Scholar]
- Papenbrock J, Schmidt A (2000. b) Characterization of two sulfurtransferase isozymes from Arabidopsis thaliana. Eur J Biochem 267 5571–5579 [DOI] [PubMed] [Google Scholar]
- Parcy F, Valon C, Raynal M, Gaubier-Comella P, Delseny M, Giraudat J (1994) Regulation of gene expression programs during Arabidopsis seed development: roles of the ABI3 locus and of endogenous abscisic acid. Plant Cell 6 1567–1582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park OK (2004) Proteomic studies in plants. J Biochem Mol Biol 37 133–138 [DOI] [PubMed] [Google Scholar]
- Patton DA, Schetter AL, Franzmann LH, Nelson K, Ward ER, Meinke DW (1998) An embryo-defective mutant of Arabidopsis disrupted in the final step of biotin synthesis. Plant Physiol 116 935–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiser GD, Wang TT, Hoffman NE, Yang SF, Liu HW, Walsh CT (1984) Formation of cyanide from carbon 1 of 1-aminocyclopropane-1-carboxylic acid during its conversion to ethylene. Proc Natl Acad Sci USA 81 3059–3063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pell EJ, Schlagnhaufer CD, Arteca RN (1997) Ozone-induced oxidative stress: mechanisms of action and reaction. Physiol Plant 100 264–273 [Google Scholar]
- Pillay DTN (1977) Protein synthesis in aging soybean cotyledons: loss in translational capacity. Biochem Biophys Res Commun 79 796–804 [DOI] [PubMed] [Google Scholar]
- Poulton JE (1990) Cyanogenesis in plants. Plant Physiol 94 401–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell AA (1995) The controlled deterioration test. In HA van de Venter, ed, Seed Vigour Testing Seminar. International Seed Testing Association, Zurich, pp 73–87
- Prieto-Dapena P, Castaño R, Almoguera C, Jordano J (2006) Improved resistance to controlled deterioration in transgenic seeds. Plant Physiol 142 1102–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajjou L, Belghazi M, Huguet R, Robin C, Moreau A, Job C, Job D (2006. a) Proteomic investigation of the effect of salicylic acid on Arabidopsis seed germination and establishment of early defense mechanisms. Plant Physiol 141 910–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajjou L, Gallardo K, Debeaujon I, Vandekerckhove J, Job C, Job D (2004) The effect of alpha-amanitin on the Arabidopsis seed proteome highlights the distinct roles of stored and neosynthesized mRNAs during germination. Plant Physiol 134 1598–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajjou L, Gallardo K, Job C, Job D (2006. b) Proteome analysis for the study of developmental processes in plants. In C Finnie, ed, Plant Proteomics. Annual Plant Reviews 28. Blackwell Publishing, Oxford, pp 151–184
- Rajjou L, Lovigny Y, Job C, Belghazi M, Groot S, Job D (2007. a) Seed quality and germination. In S Navie, S Adkins, S Ashmore, eds, Seeds: Biology, Development and Ecology. CAB International Publishing, Cambridge, MA, pp 324–332
- Rajjou L, Miché L, Huguet R, Job C, Job D (2007. b) Proteome and transcriptome profiling to understand seed germination and identify intrinsic markers determining seed quality, germination efficiency and early seedling vigor. In S Navie, S Adkins, S Ashmore, eds, Seeds: Biology, Development and Ecology. CAB International Publishing, Cambridge, MA, pp 149–158
- Ravanel S, Gakière B, Job D, Douce R (1998) The specific features of methionine biosynthesis and metabolism in plants. Proc Natl Acad Sci USA 95 7805–7812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossignol M, Peltier JB, Mock HP, Matros A, Maldonado AM, Jorrín JV (2006) Plant proteome analysis: a 2004-2006 update. Proteomics 6 5529–5548 [DOI] [PubMed] [Google Scholar]
- Sattler SE, Gilliland LU, Magallanes-Lundback M, Pollard M, DellaPenna D (2004) Vitamin E is essential for seed longevity and for preventing lipid peroxidation during germination. Plant Cell 16 1419–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shatters RG, Boo SP, Franca Neto JB, West SH (1997) Identification of biotinylated proteins in soybean [Glycine max (L.) Merrill] seeds and their characterization during germination and seedling growth. Seed Sci Res 7 373–376 [Google Scholar]
- Shen-Miller J (2002) Sacred lotus, the long-living fruits of China Antique. Seed Sci Res 12 131–143 [Google Scholar]
- Shen-Miller J, Mudgett MB, Schopf JW, Clarke S, Berger R (1995) Exceptional seed longevity and robust growth: ancient sacred lotus from China. Am J Bot 82 1367–1380 [Google Scholar]
- Skriver K, Mundy J (1990) Gene expression in response to abscisic acid and osmotic stress. Plant Cell 2 503–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soeda Y, Konings MCJM, Vorst O, van Houwelingen AMML, Stoopen GM, Maliepaard CA, Kodde J, Bino RJ, Groot SPC, van der Geest AHM (2005) Gene expression programs during Brassica oleracea seed maturation, osmopriming and germination are indicators of progression of the germination process and the stress tolerance level. Plant Physiol 137 354–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville C, Koornneef M (2002) A fortunate choice: the history of Arabidopsis as a model plant. Nat Rev Genet 3 883–889 [DOI] [PubMed] [Google Scholar]
- Söti C, Csermely P (2007) Protein stress and stress proteins: implications in aging and disease. J Biosci 32 511–515 [DOI] [PubMed] [Google Scholar]
- Stadtman ER (2001) Protein oxidation in aging and age-related diseases. Ann N Y Acad Sci 928 22–38 [DOI] [PubMed] [Google Scholar]
- Stadtman ER (2004) Role of oxidant species in aging. Curr Med Chem 11 1105–1112 [DOI] [PubMed] [Google Scholar]
- Tamarit J, Cabiscol E, Ros J (1998) Identification of the major oxidatively damaged proteins in Escherichia coli cells exposed to oxidative stress. J Biol Chem 273 3027–3032 [DOI] [PubMed] [Google Scholar]
- Taylorson RB, Hendricks SB (1973) Promotion of seed germination by cyanide. Plant Physiol 52 23–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tejera García NA, Iribarne C, Palma F, Lluch C (2007) Inhibition of the catalase activity from Phaseolus vulgaris and Medicago sativa by sodium chloride. Plant Physiol Biochem 45 535–541 [DOI] [PubMed] [Google Scholar]
- TeKrony DM (1995) Accelerated aging. In HA van de Venter, ed, Seed Vigour Testing Seminar. International Seed Testing Association, Zurich, pp 53–73
- Terskikh VV, Zeng Y, Feurtado JA, Giblin M, Abrams SR, Kermode AR (2008) Deterioration of western redcedar (Thuja plicata Donn ex D. Don) seeds: protein oxidation and in vivo NMR monitoring of storage oils. J Exp Bot 59 765–777 [DOI] [PubMed] [Google Scholar]
- Tesnier K, Strookman-Donkers HM, van Pijlen JG, van der Geest AHM, Bino RJ, Groot SPC (2002) A controlled deterioration test of Arabidopsis thaliana reveals genetic variation in seed quality. Seed Sci Technol 30 149–165 [Google Scholar]
- Walters C (2004) Temperature dependency of molecular mobility in preserved seeds. Biophys J 86 1253–1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters C, Wheeler LM, Grotenhuis JM (2005) Longevity of seeds stored in a genebank: species characteristics. Seed Sci Res 15 1–20 [Google Scholar]
- Walters C, Wheeler LM, Stanwood PC (2004) Longevity of cryogenically stored seeds. Cryobiology 48 229–244 [DOI] [PubMed] [Google Scholar]
- Wan CY, Wilkins TA (1994) A modified hot borate method significantly enhances the yield of high-quality RNA from cotton (Gossypium hirsutum L.). Anal Biochem 223 7–12 [DOI] [PubMed] [Google Scholar]
- Wechsberg GE, Probert RJ, Bray CM (1994) The relationship between “dehydrin-like” proteins and seed longevity in Ranunculus sceleratus L. J Exp Bot 45 1027–1030 [Google Scholar]
- Wilkinson B, Gilbert HF (2004) Protein disulfide isomerase. Biochim Biophys Acta 1699 35–44 [DOI] [PubMed] [Google Scholar]
- Xu Q, Belcastro MP, Villa ST, Dinkins RD, Clarke SG, Downie AB (2004) A second protein L-isoaspartyl methyltransferase gene in Arabidopsis produces two transcripts whose products are sequestered in the nucleus. Plant Physiol 136 2652–2664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H (2007) ER stress and diseases. FEBS J 274 630–658 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.