Abstract
Small 10-kD acyl-coenzyme A-binding proteins (ACBPs) are highly conserved proteins that are prevalent in eukaryotes. In Arabidopsis (Arabidopsis thaliana), other than the 10-kD ACBP homolog (designated Arabidopsis ACBP6), there are five larger forms of ACBPs ranging from 37.5 to 73.1 kD. In this study, the cytosolic subcellular localization of Arabidopsis ACBP6 was confirmed by analyses of transgenic Arabidopsis expressing autofluorescence-tagged ACBP6 and western-blot analysis of subcellular fractions using ACBP6-specific antibodies. The expression of Arabidopsis ACBP6 was noticeably induced at 48 h after 4°C treatment by northern-blot analysis and western-blot analysis. Furthermore, an acbp6 T-DNA insertional mutant that lacked ACBP6 mRNA and protein displayed increased sensitivity to freezing temperature (−8°C), while ACBP6-overexpressing transgenic Arabidopsis plants were conferred enhanced freezing tolerance. Northern-blot analysis indicated that ACBP6-associated freezing tolerance was not dependent on the induction of cold-regulated COLD-RESPONSIVE gene expression. Instead, ACBP6 overexpressors showed increased expression of mRNA encoding phospholipase Dδ. Lipid profiling analyses of rosettes from cold-acclimated, freezing-treated (−8°C) transgenic Arabidopsis plants overexpressing ACBP6 showed a decline in phosphatidylcholine (−36% and −46%) and an elevation of phosphatidic acid (73% and 67%) in comparison with wild-type plants. From our comparison, the gain in freezing tolerance in ACBP6 overexpressors that was accompanied by decreases in phosphatidylcholine and an accumulation of phosphatidic acid is consistent with previous findings on phospholipase Dδ-overexpressing transgenic Arabidopsis. In vitro filter-binding assays indicating that histidine-tagged ACBP6 binds phosphatidylcholine, but not phosphatidic acid or lysophosphatidylcholine, further imply a role for ACBP6 in phospholipid metabolism in Arabidopsis, including the possibility of ACBP6 in the cytosolic trafficking of phosphatidylcholine.
Extensive exchange of acyl-CoA derivatives occurs between the chloroplasts and the endoplasmic reticulum (ER) via the cytosol (Ohlrogge and Browse, 1995). De novo fatty acid biosynthesis in higher plants occurs in the chloroplasts (Ohlrogge and Browse, 1995), and the majority of plastid-synthesized fatty acids are exported as palmitoyl-CoA and oleoyl-CoA to the ER for glycerolipid biosynthesis (Browse et al., 1986; Maréchal et al., 1997). However, it is not clear how these acyl-CoAs are transported from the chloroplasts to the ER and whether shuttle proteins are involved in this process. Since recombinant Arabidopsis (Arabidopsis thaliana) 10-kD acyl-CoA-binding protein (ACBP) has been shown to bind oleoyl-CoA and protect it from degradation by microsomal acyl hydrolases (Engeseth et al., 1996), we were interested to experimentally verify its subcellular localization and biological functions.
In mammals, 10-kD ACBPs have been implicated in the binding and transport of cytosolic acyl-CoA esters as well as in gene regulation (Mikkelsen and Knudsen, 1987; Black et al., 2000; Petrescu et al., 2003). The 10-kD bovine ACBP was confirmed to be a cytosolic protein by western-blot analysis using anti-ACBP antibodies on different subcellular liver fractions (Mikkelsen and Knudsen, 1987). Homologs of the 10-kD ACBP have been localized in both cytoplasm and nuclei of monkey kidney fibroblast CV-1 cells (Helledie et al., 2000) and human hepatocellular liver carcinoma cells (Nitz et al., 2005). In nuclei of rat hepatocytes, the 10-kD ACBP interacts with nuclear factor-4α, a transcriptional activator of genes associated with lipid and Glc metabolism (Elholm et al., 2000; Petrescu et al., 2003).
In Arabidopsis, other than the 10-kD ACBP (Engeseth et al., 1996), which we have designated ACBP6 (Xiao et al., 2008), there are five other forms of ACBPs ranging from 37.5 to 73.1 kD (Leung et al., 2004). These include membrane-associated ACBP1 and ACBP2, which have been subcellularly localized to the ER and plasma membrane (Chye et al., 1999; Li and Chye, 2003), extracellularly targeted ACBP3 (Leung et al., 2006), and Kelch motif-containing ACBP4 and ACBP5 (Leung et al., 2004). Only ACBP6, the smallest member of this family, has well-characterized homologs in other eukaryotes (Hills et al., 1994; Faergeman and Knudsen, 1997). Little is known of ACBP1 to ACBP5 homologs in other organisms. Conserved domains that potentially mediate protein-protein interactions occur in the larger Arabidopsis ACBPs: ankyrin repeats (ACBP1 and ACBP2) and Kelch motifs (ACBP4 and ACBP5; Leung et al., 2004; Li and Chye, 2004). The function of the acyl-CoA-binding domain in binding acyl-CoA esters has been established for ACBP1 to ACBP5 using His-tagged recombinant proteins and site-directed mutagenesis (Chye et al., 2000; Leung et al., 2004, 2006). These ACBPs bind differentially to various acyl-CoA esters, implying that they have different cellular functions.
Proteomics analysis of phloem exudates revealed that homologs of ACBP6 exist in cucumber (Cucumis sativus) and pumpkin (Cucurbita maxima; Walz et al., 2004). In rice (Oryza sativa), its homolog is also a major phloem sap protein (Suzui et al., 2006), suggesting that plant 10-kD ACBPs may be associated with long-distance transport (possibly of long-chain acyl-CoA esters) and/or in stress and defense, since phloem proteins primarily belong to these classes (Walz et al., 2004; Suzui et al., 2006). Here, we sought to investigate if Arabidopsis ACBP6 expression is responsive to abiotic and biotic stresses. We observed that ACBP6 expression is cold (4°C) inducible, that the acbp6 knockout mutant displays enhanced sensitivity to freezing treatment (−8°C), and that transgenic Arabidopsis plants overexpressing ACBP6 are conferred freezing tolerance.
RESULTS
ACBP6 Is Localized to the Cytosol
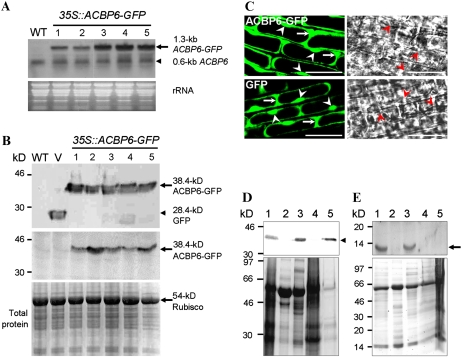
Using the PSORT Web server (http://psort.nibb.ac.jp), ACBP6 was predicted to be localized to the cytosol. To verify this, a 35S∷ACBP6-GFP construct was generated by fusing the ACBP6 coding region to the autofluorescent protein tag, eGFP, in vector pBI-eGFP (Shi et al., 2005) for expression from the cauliflower mosaic virus (CaMV) 35S promoter. Transgenic Arabidopsis plants expressing 35S∷ACBP6-GFP were generated by Agrobacterium tumefaciens-mediated transformation. The expression of the 1.3-kb ACBP6-GFP mRNA in five independent 35S∷ACBP6-GFP transformants was detected by northern-blot analysis using an ACBP6 cDNA probe that also hybridized to the endogenous 0.6-kb ACBP6 mRNA (Fig. 1A). In the 35S∷ACBP6-GFP lines, the expression of the 38.4-kD ACBP6-GFP (consisting of a 10.4-kD ACBP6 fused to a 28-kD GFP) was further confirmed in western-blot analyses using anti-GFP and anti-ACBP6 antibodies (Fig. 1B).
Figure 1.
Subcellular localization of ACBP6 in Arabidopsis. A, Northern-blot analysis with digoxigenin-labeled ACBP6 cDNA of five independent 35S∷ACBP6-GFP transgenic lines (lanes 1–5). The arrow indicates ACBP6-GFP mRNA, and the arrowhead indicates ACBP6 mRNA. An RNA gel (30 μg lane−1) stained with ethidium bromide is shown at bottom. WT, Wild type. B, Western-blot analyses using anti-GFP (top) and ACBP6-specific (bottom) antibodies on the same five independent 35S∷ACBP6-GFP transformants. ACBP6-GFP (arrows) and GFP (arrowhead) cross-reacting bands are indicated. The bottom panel shows a gel identically loaded and stained with Coomassie Blue. V, Vector-transformed control. C, Confocal microscopy of premature root cells of Arabidopsis 35S∷ACBP6-GFP line 1 (top) showing the localization of ACBP6-GFP in the cytosol (arrows) and nuclei (arrowheads). GFP vector-transformed Arabidopsis is shown at bottom. Bars = 20 μm. D, Western-blot analysis using anti-GFP antibodies on subcellular fractions of whole plant protein from transgenic Arabidopsis 35S∷ACBP6-GFP line 1. Subcellular fractions are from total whole plant protein (lane 1), membrane (lane 2), cytosol (lane 3), large particles including mitochondria, chloroplasts, and peroxisomes (lane 4), and nuclei (lane 5). The arrowhead indicates a 38.4-kD ACBP6-GFP cross-reacting band. The bottom panel shows a gel identically loaded and stained with Coomassie Blue. E, Western-blot analysis using ACBP6-specific antibodies on subcellular fractions of whole plant protein from wild-type Arabidopsis. Subcellular fractions are from total whole plant protein (lane 1), membrane (lane 2), cytosol (lane 3), large particles including mitochondria, chloroplasts, and peroxisomes (lane 4), and nuclei (lane 5). The arrow indicates a 10.4-kD ACBP6 cross-reacting band. The bottom panel shows a gel identically loaded and stained with Coomassie Blue.
When premature root cells of 2-week-old T2 transgenic Arabidopsis seedlings from 35S∷ACBP6-GFP line 1 were examined by confocal laser-scanning microscopy, fluorescence was detected primarily in the cytosol, with some signals in the nuclei (white arrowheads in Fig. 1C, top). The GFP control showed expression in both nuclei and cytosol (Fig. 1C, bottom).
Subcellular fractions of protein from rosette leaves from 35S∷ACBP6-GFP line 1, obtained following differential centrifugation, were analyzed by western-blot analysis using anti-GFP antibodies. Figure 1D shows a cross-reacting 38.4-kD ACBP6-GFP band in total protein (lane 1) as well as in the cytosolic (lane 3) and nuclear (lane 5) fractions. This band was absent in the membrane fraction (lane 2) and the fraction containing large particles, including mitochondria, chloroplasts, and peroxisomes (lane 4). Nuclear localization of ACBP6-GFP overexpressed from the 35S∷ACBP6-GFP line may have resulted from passive diffusion through nuclear pore complexes (Görlich and Mattaj, 1996; Li et al., 2006), as the size of ACBP6-GFP (38.4 kD) is smaller than the size exclusion limit (approximately 40–60 kD) for such diffusion. Hence, it was pertinent to determine the subcellular localization of native ACBP6.
To this end, subcellular fractions of protein from rosette leaves from wild-type (ecotype Columbia [Col-0]) Arabidopsis, obtained following differential centrifugation, were analyzed by western-blot analysis using ACBP6-specific antibodies. Figure 1E shows a cross-reacting 10.4-kD ACBP6 band in total protein (lane 1) and in the cytosolic (lane 3) fraction. Absence of this band in the membrane fraction (lane 2), the fraction containing large particles, including mitochondria, chloroplasts, and peroxisomes (lane 4), and the nuclear fraction (lane 5) confirmed that ACBP6 is a cytosolic protein and that ACBP6-GFP had diffused into the cell nuclei of transgenic Arabidopsis overexpressing ACBP6-GFP. Our results suggest that, unlike some mammalian 10-kD ACBPs, which interact directly with nuclear factors in the nuclei (Petrescu et al., 2003), ACBP6 seems to be confined to the cytosol, consistent with its predicted localization and its lack of a nuclear targeting signal.
The Expression of ACBP6 Is Cold Inducible
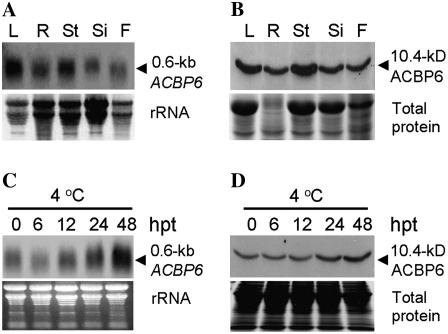
Northern-blot analyses were used to examine the spatial pattern of ACBP6 expression, using total RNAs extracted from various organs, and to analyze the response of ACBP6 expression to various forms of biotic and abiotic stresses. ACBP6 mRNA was more highly expressed in leaves and stalks compared with roots, flowers, and siliques (Fig. 2A). Western-blot analysis using ACBP6-specific antibodies reflected a similar distribution pattern of the ACBP6 protein (Fig. 2B). ACBP6 mRNA was observed to be cold inducible (Fig. 2C) but was not induced by treatments using fungal elicitor (arachidonic acid), high salt, and methyl jasmonate in whole plants (data not shown). Lack of induction with high-salt and methyl jasmonate treatments observed by northern-blot analysis is consistent with information available (www.weigelworld.org/resources/microarray) from microarray data analysis of ACBP6 (At1g31812) expression. Northern-blot analysis using total RNA from 4-week-old wild-type Arabidopsis exposed to 4°C for 0, 6, 12, 24, and 48 h indicated that ACBP6 mRNA expression increased upon cold treatment and was most significant at 48 h after treatment (Fig. 2C). Consistently, by western-blot analysis (Fig. 2D), ACBP6 protein showed its highest accumulation at 48 h following cold treatment. In comparison with microarray data (www.weigelworld.org/resources/microarray), cold induction of ACBP6 expression was not detectable in microarrays at 24 h after 4°C treatment, and no data were available for a period exceeding 24 h.
Figure 2.
Northern-blot analysis using a digoxigenin-labeled probe prepared from full-length ACBP6 cDNA and western-blot analysis using ACBP6-specific antibodies on wild-type Arabidopsis. A, Spatial expression of ACBP6 in various tissues (L, leaf; R, root; St, stalk; Si, silique; F, flower). Total RNA (30 μg well−1) was hybridized to ACBP6 cDNA. The bottom panel shows an ethidium bromide-stained gel before blotting. B, Western-blot analysis using ACBP6-specific antibodies (top). The bottom panel shows a gel identically loaded and stained with Coomassie Blue. C, Cold induction of ACBP6 expression. The northern blot shows total RNA isolated from rosettes of wild-type Arabidopsis at the indicated times after treatment (hours after treatment [hpt]) at 4°C. An RNA gel (30 μg lane−1) was stained with ethidium bromide (bottom). D, Western blot using ACBP6-specific antibodies on total protein extracted from cold-treated rosettes from wild-type Arabidopsis (top). The bottom panel shows a gel identically loaded and stained with Coomassie Blue.
Identification of an acbp6 Knockout Mutant
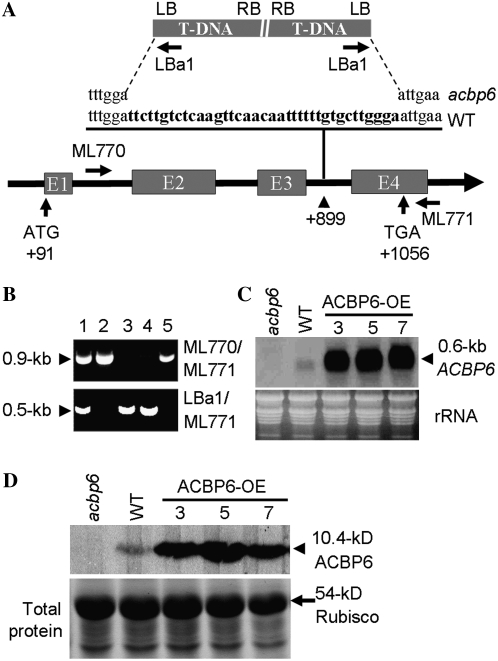
To further investigate the function of ACBP6 upon cold treatment, an acbp6 T-DNA knockout mutant (SALK_104339) was obtained from The Arabidopsis Information Resource (TAIR) and was subsequently characterized. The presence of a T-DNA insert in ACBP6 in this acbp6 homozygous mutant was confirmed by PCR using gene-specific primers (ML770 and ML771) and a T-DNA border primer, LBa1 (Fig. 3A). On PCR analysis using ML770/ML771 (Fig. 3B, top), a 0.9-kb band was amplified from wild-type Arabidopsis (lanes 2 and 5) and the acbp6 heterozygous mutant (lane 1) but not from homozygous mutants (lanes 3 and 4). When LBa1/ML771 primers were used in PCR (Fig. 3B, bottom), a 0.5-kb band was observed in the acbp6 heterozygous (lane 1) and homozygous (lanes 3 and 4) mutants but not in the wild type (lanes 2 and 5).
Figure 3.
Characterization of the acbp6 knockout mutant (SALK_104339) and 35S∷ACBP6 transgenic Arabidopsis. A, T-DNA insertion in the third intron of ACBP6 resulted in a 37-bp deletion (boldface in the wild-type [WT] sequence). The locations of primers used to genotype the acbp6 allele are shown. B, Specificity of the primer combinations ML770/ML771 (top gel) and LBa1/ML771 (bottom gel) in PCR to identify acbp6 homozygous mutants (lanes 3 and 4). The samples in lanes 2 and 5 resemble wild-type samples. Lane 1, Heterozygous mutant. C, Northern-blot analysis of wild type, acbp6 mutant, and ACBP6 overexpressor (OE-3, OE-5, and OE-7) plants using a digoxigenin-labeled ACBP6 cDNA probe. The acbp6 homozygous mutant lacked ACBP6 mRNA. ACBP6 overexpressor lines showed higher ACBP6 expression than the wild type. Total RNA (30 μg lane−1) stained with ethidium bromide before blotting is shown at bottom. D, Western-blot analysis using ACBP6-specific antibodies. Total protein (15 μg lane−1) was extracted from rosettes of wild-type, acbp6 mutant, and three independent ACBP6 overexpressor (OE-3, OE-5, and OE-7) plants. The bottom panel shows a gel identically loaded and stained with Coomassie Blue.
When the PCR products spanning the junctions between ACBP6 and the T-DNA were sequenced, results indicated that the T-DNA was inserted in the third intron of ACBP6, with a resultant 37-bp deletion in ACBP6 (Fig. 3A). Northern-blot analysis indicated that transcription of ACBP6 was disrupted in the acbp6 homozygous mutant, while a 0.6-kb mRNA was detected in wild-type Arabidopsis (Fig. 3C). On western-blot analysis, the 10.4-kD ACBP6 cross-reacting band evident in the wild type was absent in the homozygous mutant, confirming that the mutant is a knockout line (Fig. 3D).
Generation of ACBP6-Overexpressing Transgenic Arabidopsis
To test whether ACBP6 overexpression enhances cold tolerance, transgenic Arabidopsis plants overexpressing ACBP6 were generated by A. tumefaciens-mediated transformation (Clough and Bent, 1998). The ACBP6 full-length cDNA was expressed from the CaMV 35S promoter in binary vector pSMB (Mylne and Botella, 1998) for transformation of Arabidopsis (Col-0). Three independent T2 ACBP6 overexpressor lines (OE-3, OE-5, and OE-7) were identified by northern-blot analysis to overexpress the 0.6-kb ACBP6 mRNA (Fig. 3C) and by western-blot analysis to accumulate the 10.4-kD ACBP6 protein (Fig. 3D).
The acbp6 Mutant Exhibits Enhanced Sensitivity to Freezing Stress, While ACBP6 Overexpressors Are Freezing Tolerant
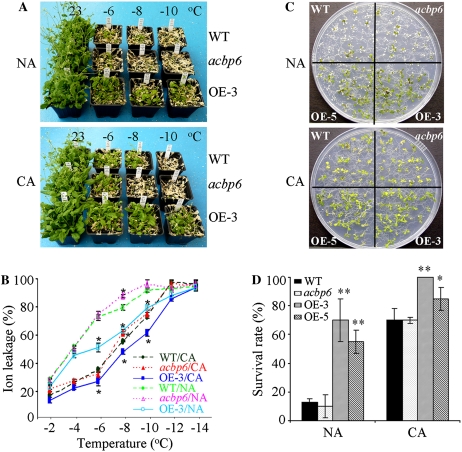
To investigate the effects of the ACBP6 mutation and ACBP6 overexpression on freezing tolerance, 5-week-old wild-type, acbp6 mutant, and ACBP6 overexpressor plants from nonacclimated (NA) and cold-acclimated (CA) sets were examined. As shown in Figure 4A, few of the wild-type and acbp6 plants tolerated freezing temperatures of −6°C, −8°C, and −10°C without cold acclimation. However, most of the ACBP6 overexpressor (OE-3) plants survived in freezing temperature as low as −6°C, and 45% (P < 0.05) of them survived even at −8°C and −10°C (Fig. 4A, top). After cold acclimation at 4°C for 3 d, freezing tolerance was enhanced in all three genotypes. More CA wild-type plants than CA mutants survived at −8°C; all CA acbp6 mutants did not survive at −8°C (Fig. 4A, bottom). In comparison, CA ACBP6 overexpressor (OE-3) plants tolerated freezing stress at −8°C and −10°C better than CA wild-type or mutant plants and NA OE-3 plants (Fig. 4A).
Figure 4.
ACBP6 overexpressors show enhanced freezing tolerance. A, Freezing tolerance of ACBP6 overexpressor (OE-3) plants tested at different temperatures below freezing. NA and CA wild-type (WT), acbp6 mutant, and ACBP6 overexpressor (OE-3) plants were photographed after 7 d of recovery under 16-h-light (23°C)/8-h-dark (21°C) cycles. A similar phenotype was observed with another ACBP6 overexpressor line (OE-5; data not shown). B, Electrolyte leakage of NA and CA wild-type, acbp6 mutant, and ACBP6 overexpressor (OE-3) plants after 1 h of treatment at temperatures below freezing, followed by thawing at 4°C overnight. For cold acclimation, plants were incubated at 4°C for 3 d. NA plants remained in a growth chamber at 23°C until measurement. Asterisks indicate significant differences from the wild type (P < 0.05). Values are means ± sd (n = 3) calculated from three independent experiments. C, Phenotypes of NA and CA 11-d-old wild-type, acbp6 mutant, and ACBP6 overexpressor (OE-3 and OE-5) seedlings after freezing treatment at −12°C for 1 h. Plates were thawed overnight at 4°C and transferred to a growth chamber (16-h-light [23°C]/8-h-dark [21°C] photoperiods) for a 7-d recovery before photography. D, Survival rate of NA and CA wild-type, acbp6 mutant, and ACBP6 overexpressor (OE-3 and OE-5) seedlings after freezing treatment at −12°C for 1 h, followed by growth at 23°C for 7 d. Asterisks indicate significant differences from the wild type (** P < 0.01, * P < 0.05). Values are means ± sd (n = 3) calculated from three independent experiments.
To evaluate freezing injury after freezing treatment, electrolyte leakage was measured using both NA and CA freezing-treated leaves from wild-type, acbp6 mutant, and ACBP6 overexpressor plants. Results showed that ionic leakage following treatment at −8°C was significantly greater in both NA and CA acbp6 mutants than in corresponding NA and CA wild-type plants (P < 0.05; Fig. 4B). In comparison, the ionic leakage at −6°C, −8°C, and −10°C of NA and CA ACBP6 overexpressor (OE-3) plants was significantly lower (P < 0.05) than in wild-type plants (Fig. 4B).
To test the effects of freezing treatment on seedling development, NA and CA 11-d-old seedlings of wild-type, acbp6 mutant, and ACBP6 overexpressor (OE-3 and OE-5) plants were grown on Murashige and Skoog (MS) medium and treated at −12°C for 1 h. As shown in Figure 4, C and D, the survival rates for NA wild-type and NA acbp6 mutant seedlings were only 13% and 10%, respectively, significantly lower than those of ACBP6-overexpressing OE-3 and OE-5 (70% and 55%, respectively). With CA seedlings, 70% of wild-type and acbp6 mutant seedlings survived, in comparison with 100% and 85% of ACBP6-overexpressing OE-3 and OE-5, respectively (Fig. 4, C and D). These results, which were averages of three replicate experiments, were significant using Student's t test (P < 0.01 or P < 0.05). Our findings suggest that knockout of ACBP6 expression led to an enhanced sensitivity to freezing, while the overexpression of ACBP6 in transgenic Arabidopsis conferred freezing tolerance.
ACBP6-Conferred Freezing Tolerance Is Independent of Induced COR Gene Expression
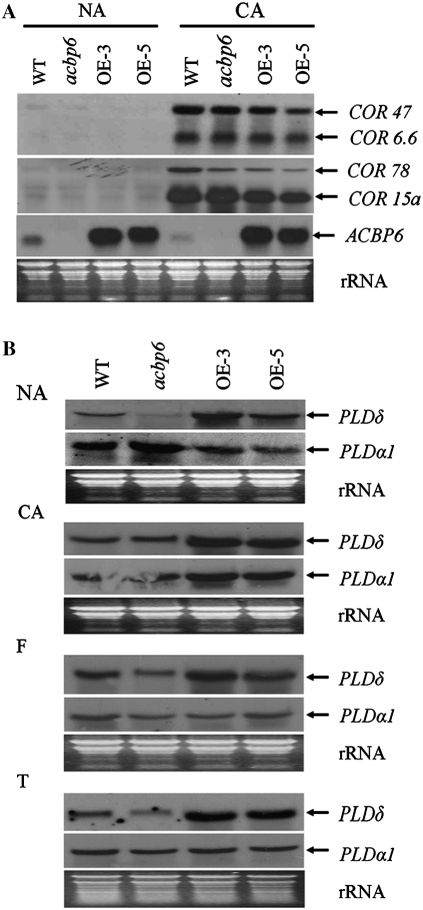
In many cases during CA, the expression of several COLD-RESPONSIVE (COR) genes is induced (Thomashow, 1999). The four major Arabidopsis COR genes, COR6.6, COR15a, COR47, and COR78, encode hydrophilic proteins that stabilize membranes during freezing-induced dehydration (Thomashow, 1999). Steponkus et al. (1998) further showed that COR15a overexpression in transgenic Arabidopsis enhanced freezing tolerance in isolated protoplasts. To determine whether ACBP6-conferred freezing tolerance is associated with induced COR gene expression, northern-blot analyses were carried out to examine the expression of these four genes using PCR-generated probes. In NA wild-type (Col-0), acbp6 mutant, and ACBP6 overexpressor (OE-3 and OE-5) plants, the COR6.6, COR15a, COR47, and COR78 transcripts were not detected. In contrast, these COR genes were induced after CA in all three genotypes (Fig. 5A). Although CA promotes ACBP6-conferred freezing tolerance, expression of these four COR genes was not further enhanced in OE-3 and OE-5 plants. It appears that ACBP6-conferred freezing tolerance is not dependent on the induction of COR gene expression.
Figure 5.
COR, PLDα1, and PLDδ expression in wild-type (WT), acbp6 mutant, and ACBP6 overexpressor (OE-3 and OE-5) plants. A, Northern-blot analysis using digoxigenin-labeled COR47, COR6.6, COR78, and COR15a PCR-generated cDNA probes. The membrane was subsequently stripped and hybridized to digoxigenin-labeled ACBP6 cDNA probe. Total RNA was extracted from rosettes of wild-type, acbp6, and transgenic Arabidopsis before (NA) and after (CA) cold acclimation for 3 d. The bottom panel shows total RNA (30 μg lane−1) stained with ethidium bromide. B, Northern-blot analysis of PLDα1 and PLDδ expression in wild-type, acbp6 mutant, and ACBP6 overexpressor (OE-3 and OE-5) plants. Total RNA (30 μg lane−1) was extracted from rosettes of wild-type, acbp6, OE-3, and OE-5 plants harvested before (NA) or after (CA) 3 d of cold acclimation, followed by freezing at −8°C for 1 h (F) and thawing at 4°C for 8 h (T).
ACBP6-Conferred Freezing Tolerance Is Related to Enhanced Phospholipase Dδ Expression
In Arabidopsis, two phospholipases, PLDα1 and PLDδ, are important in mediating freezing tolerance (Welti et al., 2002; Li et al., 2004, 2008; Rajashekar et al., 2006). PLDα1-suppressed (Welti et al., 2002; Rajashekar et al., 2006) and PLDδ-overexpressed (Li et al., 2004) Arabidopsis exhibit freezing tolerance. To investigate possible modulations in PLD expression in ACBP6-conferred freezing tolerance, the expression of PLDα1 and PLDδ in wild-type, acbp6 mutant, and ACBP6 overexpressor plants was examined by northern-blot analyses using PCR-generated digoxigenin-labeled cDNA probes. Transcript levels of PLDδ were higher in ACBP6 overexpressors (OE-3 and OE-5) than in wild-type plants at NA, CA, freezing, or thawing stages, while the acbp6 mutant showed lower expression than the overexpressor lines (Fig. 5B). In comparison, PLDα1 expression in OE-3 and OE-5 lines was lower than in wild-type and acbp6 mutant plants at NA stage, but its expression in overexpressor lines was higher than in wild-type and mutant plants at CA stage (Fig. 5B).
Changes in Lipid Molecular Species following Freezing Treatment of CA Wild-Type and ACBP6 Overexpressor Plants
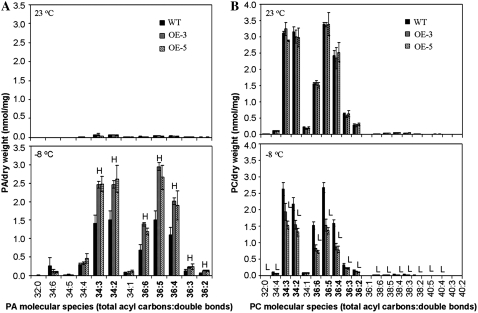
No significant changes were observed in the lipid composition between acbp6 mutant and wild-type plants before and after CA followed by freezing treatment (Table I). However, analyses of the lipid composition of wild-type and ACBP6 overexpressor (OE-3 and OE-5) plants, before and after CA followed by freezing treatment (−8°C), displayed in Figure 6, indicate several significant differences after treatment. Comparison of leaf samples from wild-type and ACBP6 overexpressor (OE-3 and OE-5) plants before treatment (grown at 23°C) showed no significant differences in the total amounts of phosphatidic acid (PA), phosphatidylcholine (PC), digalactosyldiacylglycerol (DGDG), monogalactosyldiacylglycerol (MGDG), phosphatidylethanolamine (PE), phosphatidylglycerol (PG), phosphatidylinositol (PI), lysoPG, lysoPC, and lysoPE, except for a slight decrease in phosphatidyl-Sers (PS) content in the ACBP6 overexpressors (Table II). However, following CA and freezing treatment, significant differences (P < 0.05) were observed in the total amounts of PA and PC between wild-type plants and the ACBP6 overexpressors OE-3 and OE-5 (Table II). The total amount of PA in wild-type plants increased 29-fold, while 49- and 57-fold increases occurred in OE-3 and OE-5, respectively (Table II). Hence, the ACBP6 overexpressors accumulated 73% (OE-3) and 67% (OE-5) more PA than wild-type plants. In particular, the 34:3 PA, 34:2 PA, 36:6 PA, 36:5 PA, 36:4 PA, 36:3 PA, and 36:2 PA contents in the ACBP6 overexpressors were significantly higher than in wild-type plants (P < 0.05; Fig. 6A).
Table I.
Total amount of lipid in each head group class in leaves of wild-type (Col-0) and acbp6 mutant plants grown at 23°C or CA followed by freezing at −8°C
Values are means ± sd (nmol mg−1 dry weight; n = 5). No significant differences between the wild type and the mutant were observed.
| Lipid Class | 23°C
|
−8°C
|
||
|---|---|---|---|---|
| Wild Type | acbp6 | Wild Type | acbp6 | |
| PC | 15.1 ± 2.0 | 15.7 ± 0.8 | 7.4 ± 1.2 | 6.5 ± 0.6 |
| PA | 0.21 ± 0.04 | 0.28 ± 0.07 | 9.2 ± 2.1 | 11.2 ± 1.06 |
| DGDG | 41.6 ± 6.9 | 42.7 ± 1.5 | 36.4 ± 2.7 | 33.8 ± 1.5 |
| MGDG | 160.4 ± 24.1 | 168.5 ± 9.0 | 128.7 ± 13.0 | 116.5 ± 7.8 |
| PG | 8.2 ± 1.3 | 8.8 ± 0.4 | 8.7 ± 0.6 | 8.2 ± 0.4 |
| PE | 9.9 ± 1.7 | 10.6 ± 0.7 | 4.9 ± 0.9 | 4.3 ± 0.3 |
| PI | 3.9 ± 0.7 | 4.0 ± 0.2 | 3.7 ± 0.1 | 3.7 ± 0.1 |
| PS | 0.9 ± 0.1 | 1.0 ± 0.1 | 0.5 ± 0.02 | 0.5 ± 0.02 |
| LysoPG | 0.01 ± 0.00 | 0.02 ± 0.00 | 0.05 ± 0.03 | 0.08 ± 0.01 |
| LysoPC | 0.03 ± 0.00 | 0.03 ± 0.00 | 0.18 ± 0.02 | 0.18 ± 0.03 |
| LysoPE | 0.05 ± 0.01 | 0.05 ± 0.00 | 0.07 ± 0.01 | 0.07 ± 0.00 |
Figure 6.
Freezing-induced changes in PA and PC species of Arabidopsis wild type (WT) and ACBP6 overexpressors (OE-3 and OE-5) before and after CA followed by freezing treatment. The black bar represents the wild type, and the hatched bars represent OE-3 and OE-5 (as indicated). Numbers in boldface indicate species with increases in PA that have corresponding decreases in PC. H, Value higher than the wild type (P < 0.05); L, value lower than the wild type (P < 0.05). Values are means ± sd (n = 3).
Table II.
Total amount of lipid in each head group class in leaves of wild-type (Col-0) and ACBP6-overexpressing (OE-3 and OE-5) plants grown at 23°C or CA followed by freezing at −8°C
Values are means ± sd (nmol mg−1 dry weight; n = 3). Significant differences (P < 0.05) from the wild type in the same experiment are indicated in boldface.
| Lipid Class | 23°C
|
−8°C
|
||||
|---|---|---|---|---|---|---|
| Wild Type | OE-3 | OE-5 | Wild Type | OE-3 | OE-5 | |
| PC | 15.1 ± 0.53 | 14.8 ± 0.54 | 14.7 ± 1.19 | 11.4 ± 0.80 | 7.3 ± 0.77a | 6.2 ± 0.51a |
| PA | 0.24 ± 0.02 | 0.25 ± 0.02 | 0.21 ± 0.03 | 7.06 ± 1.16 | 12.2 ± 0.4b | 11.9 ± 1.5b |
| DGDG | 40.1 ± 5.9 | 39.7 ± 1.5 | 38.4 ± 0.24 | 34.9 ± 2.5 | 35.1 ± 2.3 | 32.0 ± 1.2 |
| MGDG | 190.7 ± 29.2 | 179.0 ± 6.2 | 168.0 ± 4.7 | 113.1 ± 10.4 | 120.5 ± 10.0 | 97.3 ± 4.9 |
| PG | 8.2 ± 1.3 | 9.2 ± 0.4 | 8.1 ± 1.0 | 6.9 ± 0.8 | 6.8 ± 0.3 | 7.3 ± 1.6 |
| PE | 9.9 ± 1.7 | 11.6 ± 1.2 | 11.6 ± 2.1 | 4.9 ± 0.9 | 6.8 ± 1.5 | 6.4 ± 1.0 |
| PI | 4.5 ± 0.5 | 4.7 ± 0.2 | 4.4 ± 0.2 | 4.5 ± 0.2 | 4.5 ± 0.20 | 4.3 ± 0.3 |
| PS | 0.34 ± 0.03 | 0.25 ± 0.02a | 0.19 ± 0.00a | 0.13 ± 0.00 | 0.13 ± 0.02 | 0.13 ± 0.02 |
| LysoPG | 0.04 ± 0.01 | 0.04 ± 0.02 | 0.05 ± 0.06 | 0.13 ± 0.09 | 0.08 ± 0.04 | 0.08 ± 0.03 |
| LysoPC | 0.04 ± 0.00 | 0.05 ± 0.01 | 0.05 ± 0.01 | 0.38 ± 0.03 | 0.35 ± 0.03 | 0.30 ± 0.04 |
| LysoPE | 0.07 ± 0.00 | 0.09 ± 0.01 | 0.08 ± 0.01 | 0.25 ± 0.02 | 0.17 ± 0.01 | 0.12 ± 0.00 |
Value is lower than the wild-type value in the same experiment (P < 0.05).
Value is higher than the wild-type value in the same experiment (P < 0.05).
In contrast, the PC content decreased in both genotypes after CA followed by freezing. The total amount of PC decreased by 25% in the wild type and by 51% and 58% in the ACBP6 overexpressors OE-3 and OE-5, respectively. Furthermore, OE-3 and OE-5 accumulated 36% and 46% less PC, respectively, than wild-type plants (P < 0.05). In particular, the molecular species 32:0 PC, 34:4 PC, 34:3 PC, 34:2 PC, 36:6 PC, 36:5 PC, 36:4 PC, 36:3 PC, 36:2 PC, 38:6 PC, 38:5 PC, 38:4 PC, 38:3 PC, 38:2 PC, 40:5 PC, and 40:4 PC in the ACBP6 overexpressors OE-3 and OE-5 were significantly lower (P < 0.05) than in wild-type plants (Fig. 6B). Interestingly, the decreases in the molecular species 34:3 PC, 34:2 PC, 36:6 PC, 36:5 PC, 36:4 PC, 36:3 PC, and 36:2 PC (Fig. 6B, numbers in boldface) corresponded well to the increases in species of PA (Fig. 6A, numbers in boldface).
(His)6-ACBP6 Interacts with PC in Vitro
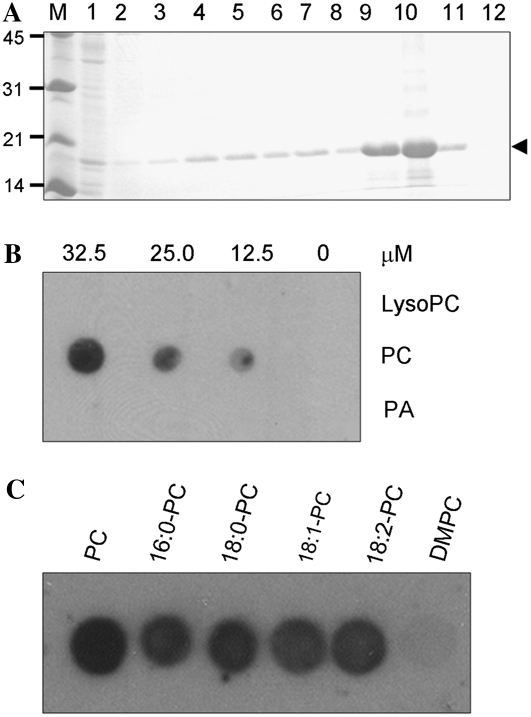
We carried out in vitro filter-binding assays to test for the interactions between ACBP6 and various phospholipids, PC, PA, and lysoPC. To this end, the 18.9-kD His-tagged ACBP6 recombinant protein was expressed and purified from Escherichia coli (Fig. 7A). Results from filter-binding assays indicated that (His)6-ACBP6 binds PC but not PA or lysoPC (Fig. 7B). As the PC used in Figure 7B is 1,2-diacyl-sn-glycero-3-phosphocholine, which consists of 33% 16:0, 13% 18:0, 31% 18:1, and 15% 18:2 fatty acids, the binding of several fatty acid species of PC to (His)6-ACBP6 was subsequently tested. Results showed that (His)6-ACBP6 binds most species of PC (16:0-PC, 18:0-PC, 18:1-PC, and 18:2-PC) tested but did not bind 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC; Fig. 7C).
Figure 7.
Purification of (His)6-ACBP6 recombinant protein and its interaction with PC. A, Purification of (His)6-ACBP6 recombinant protein. An SDS-PAGE gel shows the 18.9-kD (His)6-ACBP6 protein purified from E. coli at 3 h after isopropylthio-β-galactoside induction. M, Marker. Lane 1, Flow-through fraction; lane 2, washing fraction at pH 6.3; lanes 3 to 8, eluted fractions at pH 5.9; lanes 9 to 12, eluted fractions at pH 4.5. B, (His)6-ACBP6/lipid binding on filters. Various concentrations (0, 12.5, 25.0, and 32.5 μm) of lipids (PA, PC, and lysoPC) were spotted onto nitrocellulose and incubated with 1 μg mL−1 purified (His)6-ACBP6 protein. The (His)6-ACBP6/lipid binding was detected by immunoblotting with HRP-conjugated anti-(His)6 antibodies. C, Effect of PC acyl species on (His)6-ACBP6/lipid binding. Fifty micromolar lipid (PC, 16:0-PC, 18:0-PC, 18:1-PC, 18:2-PC, or DMPC) spotted onto nitrocellulose was incubated with 1 μg mL−1 purified (His)6-ACBP6 protein. The (His)6-ACBP6/lipid binding was detected by immunoblotting with HRP-conjugated anti-(His)6 antibodies.
DISCUSSION
Environmental factors, including cold, drought, and high salt, significantly restrict crop productivity. Together with biotic stress factors, they cause severe losses in agriculture (Vasil, 2002). Low-temperature limitations have been overcome by the identification of cold-tolerant genes for applications in genetically transformed crops. In transgenic tobacco (Nicotiana tabacum), chilling tolerance at 1°C for 7 d was achieved by the overexpression of a gene encoding chloroplast ω-3 fatty acid desaturase (Kodama et al., 1994). Furthermore, tolerance at 1°C for 11 d was conferred using a gene encoding a nonspecific cyanobacterial desaturase, and the resultant transgenic tobacco plants showed a reduction in saturated fatty acid content in membrane lipids (Ishizaki-Nishizawa et al., 1996). The overexpression of glycerol-3-phosphate acyltransferase altered the unsaturation of fatty acids and conferred chilling tolerance in transgenic plants (Ariizumi et al., 2002; Sakamoto et al., 2003; Sui et al., 2007). Hence, modifications in lipid composition that stabilize cell membranes and prevent cellular leakage lead to cold tolerance. Alternatively, the constitutive expression of COR15 protected protoplasts from leaves of NA transgenic Arabidopsis, at a range between −6°C and −8°C (Artus et al., 1996). The COR genes are regulated by transcription factors that can directly confer freezing tolerance. The constitutive expression of the transcriptional activator CBF1/DREB1B enhanced freezing tolerance (Jaglo-Ottosen et al., 1998), while DREB1A overexpression improved drought, salt, and freezing tolerance in transgenic Arabidopsis (Kasuga et al., 1999). Besides the transcriptional regulatory pathway mediated by DREB activators (for review, see Yamaguchi-Shinozaki and Shinozaki, 2005, 2006), other independent pathways leading to freezing tolerance have been proposed (Xin and Browse, 1998; Welti et al., 2002; Li et al., 2004).
In this study, our results from both northern-blot and western-blot analyses indicated that the expression of Arabidopsis ACBP6 is up-regulated by cold treatment. We further demonstrated that alterations in ACBP6 expression in the acbp6 knockout mutant and ACBP6-overexpressing transgenic Arabidopsis culminated in decreased and enhanced freezing tolerance, respectively. ACBP6-mediated freezing tolerance was not dependent on induction of COR gene expression but was accompanied by increased PLDδ expression, decreased PC content, and increased PA production, implying a role for ACBP6 in enhancing freezing tolerance via the PLDδ-mediated pathway. In wild-type Arabidopsis, freezing is accompanied by decreases in many species of PC, PE, and PG, but increases in their metabolites, PA, and lysophospholipids (Welti et al., 2002). Two phospholipases that produce PA from phospholipids and that mediate freezing tolerance are PLDδ, which plays a positive role, and PLDα1, which plays a negative role (Welti et al., 2002; Li et al., 2004; Zhang et al., 2004; Rajashekar et al., 2006; Li et al., 2008). During freezing treatment, the PLDα1 mutant displayed enhanced freezing tolerance, with decreases in freezing-induced hydrolysis of PC, and therefore generated less PA (Zhang et al., 2004; Rajashekar et al., 2006; Li et al., 2008). In contrast, the PLDδ knockout mutant showed increased freezing (−12°C) sensitivity, while transgenic Arabidopsis overexpressing PLDδ was conferred enhanced freezing tolerance with elevated PA production (Li et al., 2004). The gain in freezing tolerance accompanied by PA accumulation upon freezing treatment in ACBP6 overexpressors is consistent with previous observations of PLDδ overexpressors. Our comparison of the increases in various PA species of freezing-treated PLDδ- and ACBP6-overexpressing transgenic plants revealed that they show similarities in the elevated production of 34:2-PA, 34:3-PA, 36:5-PA, and 36:6-PA. Furthermore, all increases in PA species in the ACBP6 overexpressors were correlated with decreases in corresponding PC species, indicating that these PA accumulations were primarily derived from PC. Consistent with lipid profiling results, irrespective of NA, CA, freezing, or thawing stage, PLDδ expression was comparably higher in the ACBP6 overexpressors than in the wild type. In the acbp6 mutant, down-regulation of PLDδ expression subsequently resulted in enhanced freezing sensitivity. ACBP6-mediated freezing tolerance appears to be closely related to increased PLDδ expression and its subsequent action on PC. It has been reported that PLDδ mediates freezing tolerance by stabilizing membranes through its interaction with the cytoskeleton, and that the PA it produces (which constitutes about 20% of the total PA generated during freezing) not only promotes a nonlamellar phase membrane lipid but also inhibits phospholipase A activity (Li et al., 2004, 2008). Furthermore, we compared the expression profiles of Arabidopsis PLDδ (Katagiri et al., 2001) and ACBP6 and noticed that both genes are expressed in leaves, roots, stalks, and flowers, suggesting the feasibility of their interaction in phospholipid metabolism within these plant organs.
When we used filter-binding assays to test the binding of ACBP6 to phospholipids using His-tagged ACBP6, we observed that ACBP6 binds PC but not PA or lysoPC, suggesting a role for ACBP6 in phospholipid metabolism in Arabidopsis. One possibility of ACBP6 participation in phospholipid metabolism could be in the regulation of PLDδ expression, resembling the yeast 10-kD ACBP, which controls genes encoding proteins involved in stress responses as well as in fatty acid and phospholipid synthesis (Feddersen et al., 2007). Some of these stress-related proteins include catalase and heat shock proteins, while those associated with lipid metabolism include OLE1 (stearoyl-CoA desaturase), INO1 (myoinositol-3-phosphate synthase), PSD1 (PS decarboxylase 1), PSD2 (PS decarboxylase 2), CHO2 (PE N-methyltransferase), and OPI3 (methylene-fatty-acyl-phospholipid synthase). Feddersen et al. (2007) further suggested that the yeast ACBP-acyl-CoA ester complex can modulate gene regulation and other cellular processes by donation of acyl-CoA esters. The enhanced expression of PLDδ in ACBP6 overexpressors may be a consequence of similar sequestering action by ACBP6. Since ACBP6 binds PC (this study) and acyl-CoAs (Engeseth et al., 1996), it may possibly maintain an intracellular PC or acyl-CoA pool that can participate in regulating genes, including PLDδ, and/or their corresponding proteins. Fatty acids and their derivatives have already been demonstrated to regulate gene expression in bacteria, yeast, and mammals (Kliewer et al., 1997; Black et al., 2000).
Given the high conservation of 10-kD ACBPs among species, it would not be surprising if some of their functions, including those in the maintenance of intracellular cytosolic lipid pools and in gene regulation, are retained. Some such properties already known of the Arabidopsis homolog include the binding and protection of oleoyl-CoA from degradation by microsomal acyl hydrolases (Engeseth et al., 1996). If ACBP6 is indeed involved in gene regulation, there would still be two other predicted cytosolic ACBPs (ACBP4 and ACBP5) in this Arabidopsis ACBP family that can function in shuttling acyl-CoAs from the chloroplast to the ER. Given that the dissociation constants for recombinant (His)6-ACBP4, (His)6-ACBP5, and (His)6-ACBP6 in binding oleoyl-CoA esters are 5.0 × 10−7 m, 9.3 × 10−7 m, and 3.7 × 10−6 m, respectively, it is likely that ACBP4 and ACBP5 play more significant roles than ACBP6 in this transfer (M.-L. Chye and S. Xiao, unpublished data). The lipid analysis of rosettes presented here, showing a lack of difference in galactolipid composition under normal growth conditions at 23°C between the wild type and ACBP6 overexpressors, seems to support the probability that ACBP6 is not involved in oleoyl-CoA transport from chloroplasts to the “eukaryotic” pathway in the ER. Instead, ACBP6 has been demonstrated in this study to play a role in mediating freezing stress responses associated with phospholipid metabolism. Its ability to bind PC suggests it can transport intracellular PC within the cytosol.
MATERIALS AND METHODS
Plant Materials, Growth Conditions, and Stress Treatments
For northern-blot analysis, total RNA was extracted from rosettes of 4-week-old Arabidopsis (Arabidopsis thaliana) wild-type (Col-0) plants grown in 16-h-light (23°C)/8-h-dark (21°C) cycles. For 4°C treatment, 4-week-old Col-0 plants were transferred from a plant growth chamber (16-h-light [23°C]/8-h-dark [21°C] cycles) to a 4°C cold room under white light, and rosettes were harvested at 0, 6, 12, 24, and 48 h after treatment.
The acbp6 allele is a T-DNA insertion mutant (SALK_104339 from the SALK collection; http://signal.salk.edu/) obtained from TAIR (http://www.arabidopsis.org/). For growth on MS medium (Murashige and Skoog, 1962) supplemented with 2% Suc, seeds of Arabidopsis wild-type, acbp6 mutant, and ACBP6-overexpressing transgenic (ecotype Col-0) plants were surface sterilized and chilled at 4°C for 2 d. Subsequently, seeds were germinated and grown on MS medium supplemented with 2% Suc under 16-h-light (23°C)/8-h-dark (21°C) cycles. Soil-grown plants were also grown under 16-h-light (23°C)/8-h-dark (21°C) cycles.
Freezing treatment was carried out following Zhu et al. (2004). NA plants were grown in a growth chamber under 16-h-light (23°C)/8-h-dark (21°C) cycles until treatment, while CA plants were transferred from the growth chamber to a cold room (4°C) and grown for 3 d prior to treatment. Soil-grown plants (5 weeks old) or 11-d-old seedlings grown on MS medium plates were subjected to a temperature drop from 4°C to −2°C at 2°C h−1 in the growth chamber (Watlow series 942). When the temperature reached −2°C, ice crystals were placed on the plates or soil to induce crystallization and prevent supercooling. After 2 h at −2°C, the temperature was lowered to −12°C at 2°C h−1. After 1 h at the final temperature, the plants or seedlings were thawed at 4°C overnight. Following recovery for 7 d under 16-h-light (23°C)/8-h-dark (21°C) cycles, the plants were photographed.
Generation of 35S∷ACBP6-GFP Transgenic Lines
To investigate the subcellular localization of ACBP6, an ACBP6-GFP fusion was prepared by reverse transcription-PCR of a 369-bp ACBP6 cDNA using RNA from wild-type Arabidopsis and the ACBP6-specific primers ML750 (5′-ATATGGATCCCACGCGTTGTCCTCGTCTTCT-3′; BamHI site underlined) and ML838 (5′-CAGGATCCTGAAGCCTTGGAAGCAGCAACT-3′; BamHI site underlined). The PCR product was digested with BamHI and cloned into the BamHI restriction site on plasmid pBI121-eGFP (Shi et al., 2005) to yield plasmid pAT376, in which ACBP6 was transcribed from the CaMV 35S promoter.
The plant transformation vector was mobilized from Escherichia coli to Agrobacterium tumefaciens strain LBA4404 by triparental mating (Horsch et al., 1985). The resultant A. tumefaciens was used in plant transformation of wild-type Arabidopsis by the floral dip method (Clough and Bent, 1998). Putative transgenic plants expressing ACBP6-GFP were selected on MS medium containing kanamycin (50 μg mL−1) and verified by PCR using the CaMV 35S promoter-specific forward primer 35SB (5′-CAATCCCACTATCCTTCGCAAGACC-3′) and the gene-specific reverse primer ML838, followed by northern-blot analysis using an ACBP6 full-length cDNA and western-blot analyses using ACBP6-specific antibodies and anti-GFP antibodies. Subsequently, the T2 homozygous lines were tested on kanamycin-containing MS medium, and the resistant plants were used in further analyses.
Northern-Blot Analysis
Rosettes from 4-week-old plants grown at 23°C or 4°C were collected in liquid nitrogen at the indicated times following treatment. Total RNA was extracted using TRIzol reagent (Invitrogen) following the manufacturer's protocol. Northern-blot analysis was carried out using the Digoxigenin Nucleic Acid Detection Kit (Roche). Equal amounts of RNA (30 μg) were separated on a 1.5% agarose gel containing 6% formaldehyde and transferred to Hybond-N membranes (Amersham). The PCR Digoxigenin Probe Synthesis Kit was used to generate cDNA probes according to the manufacturer's instructions (Roche). The gene-specific primers used were ML750 and ML751 (5′-AATATATCATCTTGAATTCAACTG-3′) for ACBP6, ML880 (5′-GCTAACATGAGCTGTTCTCAC-3′) and ML881 (5′-GAATGTGACGGTGACTGTGG-3′) for COR15a, ML882 (5′-CAGAGACCAACAAGAATGCC-3′) and ML883 (5′-CGTAGTACATCTAAAGGGAG-3′) for COR6.6, ML884 (5′-CAAGATTACTCTGCTAGAGGAGC-3′) and ML885 (5′-GTATACGATGAGTGTTATGGG-3′) for COR47, ML886 (5′-CAGAGGAACCACCACTCAAC-3′) and ML887 (5′-CTCCTCTGTTTTCTCATCTC-3′) for COR78, ML921 (5′-TATGCGACGATTGATCTGCA-3′) and ML922 (5′-CTGAGAGCCTGAATCACATC-3′) for PLDα1, and ML923 (5′-AGCGACTCTAGCTCGAACAC-3′) and ML924 (5′-CAAGCATAAGAAGAACCCAG-3′) for PLDδ. Hybridization and detection were performed according to standard procedures as specified by the manufacturer (Roche).
Western-Blot Analysis
Total plant protein for western-blot analysis was extracted from 4-week-old plants of wild-type Arabidopsis, acbp6 mutant, ACBP6 overexpressors, and 35S∷ACBP6-GFP transgenic lines. Protein concentration was determined by the method of Bradford (1976). Fifteen micrograms of total protein was loaded per well for SDS-PAGE. The proteins were electrophoretically transferred to Hybond-C membranes (Amersham) using the Trans-Blot Cell (Bio-Rad). ACBP6-specific and anti-GFP antibodies (Invitrogen) were used for western-blot analyses. To generate ACBP6-specific antibodies, a synthetic peptide (VEGKSSEEAMNDY) corresponding to amino acids 63 to 75 of ACBP6 was used for the immunization of rabbits.
For analyses of subcellular fractions of plant protein by western blots, protein was extracted from 3-week-old rosettes of 35S∷ACBP6-GFP line 1 and wild-type (Col-0) Arabidopsis that had been confirmed by northern-blot analysis and western-blot analysis. Subcellular fractionation was carried out by differential centrifugation according to Smith et al. (1988).
Confocal Laser-Scanning Microscopy
A Zeiss LSM 510 inverted confocal laser-scanning microscope equipped with helium/neon lasers and multitracking was used for the analysis of ACBP6-GFP localization. GFP fluorescence was excited at 488 nm, filtered through a primary dichroic filter (UV/488/543), a secondary dichroic filter of 545 nm, and subsequently a BP505- to 530-nm emission filter to the photomultiplier tube detector. The images were processed using the LSM 510 software (Zeiss).
Identification of an acbp6 Mutant
The acbp6 T-DNA insertion mutant (SALK_104339) was screened from a T-DNA seed pool prepared by the SALK Institute Genomic Analysis Laboratory (http://signal.salk.edu/). The T-DNA insertion in the gene was identified using the T-DNA left border primer LBa1 (5′-TTTTTCGCCCTTTGACGTTGGA-3′) and the ACBP6-specific forward primer ML770 (5′-ACTGATCACGCTTTTTCTCTG-3′) and reverse primer ML771 (5′-TTCTGGTATAGCTCCTGCCTG-3′). The PCR product was sequenced and the T-DNA insertion site was confirmed. Individual homozygous T-DNA mutant plants were identified by PCR. PCR amplification was initiated with denaturation at 95°C for 3 min, followed by 30 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min, and an extension at 72°C for 10 min.
Generation of ACBP6-Overexpressing Plants
A 0.6-kb full-length cDNA of ACBP6 was amplified by reverse transcription-PCR using RNA isolated from wild-type Arabidopsis plants and the ACBP6-specific primer pair ML750 and ML751 (5′-AATATATCATCTTGAATTCAACTG-3′; EcoRI site underlined). The PCR product was cloned into pGEM-T Easy vector (Promega) to generate pAT323. The ACBP6 SpeI-EcoRI fragment from pAT323 was inserted into similar restriction sites on binary vector pSMB (Mylne and Botella, 1998) to generate plasmid pAT332. In the resultant vector, expression of the ACBP6 cDNA is under the control of the CaMV 35S promoter.
The construct was mobilized from E. coli to A. tumefaciens strain LBA4404 by triparental mating (Horsch et al., 1985). The resultant A. tumefaciens was used in plant transformation of Arabidopsis (ecotype Col-0) by the floral dip method (Clough and Bent, 1998). The T1 generation (designated ACBP6-OE) was selected using BASTA (57.8 μg mL−1 glufosinate solution) and was further verified by PCR using the CaMV 35S promoter-specific forward primer 35SB and the gene-specific reverse primer ML771. The putative positive transformants were confirmed by northern-blot analysis using an ACBP6 full-length cDNA probe and by western-blot analysis using ACBP6-specific antibodies.
Electrolyte Leakage
Ionic leakage measurements were carried out according to Welti et al. (2002). Rosettes from NA and CA plants were collected 1 h after freezing at the indicated temperatures and then incubated at 4°C for 24 h. Deionized water was added, and the conductivity of the solution was measured after gentle agitation at 23°C for 1 h. Total ionic strength was determined after heating the solution in a 100°C water bath for 10 min and cooling to 23°C. Ionic leakage was determined using a conductivity meter (YSI model 55).
Lipid Profiling
Lipid extraction was carried out according to the protocol provided by the Kansas Lipidomics Research Center (www.K-state.edu/lipid/lipidomics). Five-week-old plants were CA for 3 d at 4°C and then frozen at −8°C for 2 h, following which rosettes from two to three plants were harvested immediately. The nontreated NA plants remained in a growth chamber at 23°C until harvest. The rosettes were transferred immediately to 3 mL of isopropanol with 0.01% butylated hydroxytoluene at 75°C and incubated for 15 min. Subsequently, 1.5 mL of chloroform and 0.6 mL of water were added. The tubes were shaken for 1 h, followed by removal of the extract for lipid analysis. The tissue was reextracted with chloroform:methanol (2:1) with 0.01% butylated hydroxytoluene four to five times with 30 min of agitation each until all of the plant tissue turned white. The remaining plant tissue was heated overnight at 105°C and weighed to yield dry weight. The combined extracts were washed once with 1 mL of 1 m KCl and once with 2 mL of water, after which the solvent was evaporated under nitrogen. These samples were sent by courier service for lipid profiling at the Kansas Lipidomics Research Center.
Purification of Recombinant His-Tagged ACBP6 for Filter-Binding Assays
Expression and purification of His-tagged ACBP6 recombinant protein was carried out according to Xiao et al. (2008). Binding of (His)6-ACBP6 to various lipids on filters was carried out as described previously (Zhang et al., 2004) with minor modifications. Briefly, various concentrations of lipids were spotted onto nitrocellulose and incubated at room temperature for 1 h in dark. LysoPC, PC, PA, 18:0-PC, and 18:2-PC were purchased from Sigma, and 16:0-PC, 18:1-PC, and DMPC were purchased from Echelon Biosciences. The lipid-bound filter was blocked with Tris-buffered saline (TBS) with 1% nonfat milk for 1 h. After incubation with 1 μg mL−1 purified (His)6-ACBP6 protein in blocking buffer for 2 h, the filter was gently washed three times with TTBS (TBS plus 0.1% Tween 20), each for 10 min. Following incubation with the horseradish peroxidase (HRP)-conjugated anti-(His)6 antibodies (1:2,000; Qiagen; catalog no. 1014922) for 1 h at room temperature, the filter was again washed three times with TTBS, each for 10 min, and then detected with the ECL Western Blotting Detection Kit (Amersham) following the manufacturer's protocols.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers NM_102916 (ACBP6), NM_129815 (COR15a), NM_121602 (COR6.6), NM_101894 (COR47), NM_124610 (COR78), NM_112443 (PLDα1), and NM_119745 (PLDδ).
Acknowledgments
We thank J.R. Botella (University of Queensland) and W.C. Yang (Institute of Genetics and Developmental Biology, Chinese Academy of Science) for providing vectors pSMB and pBI-eGFP, respectively, S.F. Chen (University of Hong Kong) for providing the conductivity meter, R. Welti (Kansas Lipidomics Research Center) for her comments on the manuscript, M. Roth (Kansas Lipidomics Research Center) for lipid profiling, and TAIR for providing acbp6 knockout mutant seed pools.
This work was supported by a Croucher Senior Research Fellowship awarded to M.-L.C. and by the University of Hong Kong (grant no. 10208034). Q.-F.C. and S.X. were supported by University of Hong Kong postgraduate studentships. The Kansas Lipidomics Research Center was supported by the National Science Foundation (grant nos. EPS 0236913, MCB 0455318, and DBI 0521587), the Kansas Technology Enterprise Corporation, the Kansas IDeA Network of Biomedical Research Excellence of the National Institutes of Health (grant no. P20RR16475), and Kansas State University.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Mee-Len Chye (mlchye@hkucc.hku.hk).
Open Access articles can be viewed online without a subscription.
References
- Ariizumi T, Kishitani S, Inatsugi R, Nishida I, Murata N, Toriyama K (2002) An increase in unsaturation of fatty acids in phosphatidylglycerol from leaves improves the rates of photosynthesis and growth at low temperatures in transgenic rice seedlings. Plant Cell Physiol 43 751–758 [DOI] [PubMed] [Google Scholar]
- Artus NN, Uemura M, Steponkus PL, Gilmour SJ, Lin C, Thomashow MF (1996) Constitutive expression of the cold-regulated Arabidopsis COR15α gene affects both chloroplast and protoplast freezing tolerance. Proc Natl Acad Sci USA 93 13404–13409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black PN, Faergeman NJ, DiRusso CC (2000) Long-chain acyl-CoA-dependent regulation of gene expression in bacteria, yeast and mammals. J Nutr 130 305S–309S [DOI] [PubMed] [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Annu Rev Biochem 72 248–254 [DOI] [PubMed] [Google Scholar]
- Browse J, Warwick N, Somerville CR, Slack CR (1986) Fluxes through the prokaryotic and eukaryotic pathways of lipid-synthesis in the ‘16-3’ plant Arabidopsis. Biochem J 235 25–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chye ML, Huang BQ, Zee SY (1999) Isolation of a gene encoding Arabidopsis membrane-associated acyl-CoA binding protein and immunolocalization of its gene product. Plant J 18 205–214 [DOI] [PubMed] [Google Scholar]
- Chye ML, Li HY, Yung MH (2000) Single amino acid substitutions at the acyl-CoA-binding domain interrupt [14C]palmitoyl-CoA binding of ACBP2, an Arabidopsis acyl-CoA-binding protein with ankyrin repeats. Plant Mol Biol 44 711–721 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Elholm M, Garras A, Neve S, Tornehave D, Lund TB, Skorve J, Flatmark T, Kristiansen K, Berge RK (2000) Long-chain acyl-CoA esters and acyl-CoA binding protein are present in the nucleus of rat liver cells. J Lipid Res 41 538–545 [PubMed] [Google Scholar]
- Engeseth NJ, Pacovsky RS, Newman T, Ohlrogge JB (1996) Characterization of an acyl-CoA-binding protein from Arabidopsis. Arch Biochem Biophys 331 55–62 [DOI] [PubMed] [Google Scholar]
- Faergeman NJ, Knudsen J (1997) Role of long-chain fatty acyl-CoA esters in the regulation of metabolism and in cell signalling. Biochem J 323 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feddersen S, Neergaard TB, Knudsen J, Faergeman NJ (2007) Transcriptional regulation of phospholipid biosynthesis is linked to fatty acid metabolism by an acyl-CoA-binding-protein-dependent mechanism in Saccharomyces cerevisiae. Biochem J 407 219–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlich D, Mattaj IW (1996) Nucleocytoplasmic transport. Science 271 1513–1518 [DOI] [PubMed] [Google Scholar]
- Helledie T, Antonius M, Sørensen RV, Hertzel AV, Bernlohr DA, Kølvraa S, Kristiansen K, Mandrup S (2000) Lipid-binding proteins modulate ligand-dependent transactivation by peroxisome proliferator-activated receptors and localize to the nucleus as well as the cytoplasm. J Lipid Res 41 1740–1751 [PubMed] [Google Scholar]
- Hills MJ, Dann R, Lydiate D, Sharpe A (1994) Molecular cloning of a cDNA from Brassica napus L. for a homologue of acyl-CoA-binding protein. Plant Mol Biol 25 917–920 [DOI] [PubMed] [Google Scholar]
- Horsch RB, Fry JE, Hoffmann NL, Eichholtz D, Rogers SG, Fraley RT (1985) A simple and general method for transferring genes into plants. Science 227 1229–1231 [DOI] [PubMed] [Google Scholar]
- Ishizaki-Nishizawa O, Fuji T, Azume M, Sekiguchi K, Murata N, Ohtani T, Toguri T (1996) Low-temperature resistance of higher plants is significantly enhanced by a nonspecific cyanobacterial desaturase. Nat Biotechnol 14 1003–1006 [DOI] [PubMed] [Google Scholar]
- Jaglo-Ottosen KR, Gilmour SJ, Zarka DG, Schabenberger O, Thomashow MF (1998) Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 280 104–106 [DOI] [PubMed] [Google Scholar]
- Kasuga M, Liu Q, Miura S, Yamaguchi-Shinozaki K, Shinozaki K (1999) Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat Biotechnol 17 287–291 [DOI] [PubMed] [Google Scholar]
- Katagiri T, Takahashi S, Shinozak K (2001) Involvement of a novel Arabidopsis phospholipase D, AtPLDδ, in dehydration-inducible accumulation of phosphatidic acid in stress signaling. Plant J 26 595–605 [DOI] [PubMed] [Google Scholar]
- Kliewer SA, Sundseth SS, Jones SA, Brown PJ, Wisely GB, Koble CS, Devchand P, Wahli W, Willson TM, Lenhard JM, et al (1997) Fatty acids and eicosanoids regulate gene expression through direct interactions with peroxisome proliferator-activated receptors α and γ. Proc Natl Acad Sci USA 94 4318–4323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodama H, Hamada T, Horiguchi G, Nishimura M, Iba K (1994) Genetic enhancement of cold tolerance by expression of a gene for chloroplast ω-3 fatty acid desaturase in transgenic tobacco. Plant Physiol 105 601–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung KC, Li HY, Mishra G, Chye ML (2004) ACBP4 and ACBP5, novel Arabidopsis acyl-CoA-binding proteins with Kelch motifs that bind oleoyl-CoA. Plant Mol Biol 55 297–309 [DOI] [PubMed] [Google Scholar]
- Leung KC, Li HY, Xiao S, Tse MH, Chye ML (2006) Arabidopsis ACBP3 is an extracellularly targeted acyl-CoA-binding protein. Planta 223 871–881 [DOI] [PubMed] [Google Scholar]
- Li HY, Chye ML (2003) Membrane localization of Arabidopsis acyl-CoA binding protein ACBP2. Plant Mol Biol 51 483–492 [DOI] [PubMed] [Google Scholar]
- Li HY, Chye ML (2004) Arabidopsis acyl-CoA-binding protein ACBP2 interacts with an ethylene-responsive element-binding protein, AtEBP, via its ankyrin repeats. Plant Mol Biol 54 233–243 [DOI] [PubMed] [Google Scholar]
- Li S, Ehrhardt DW, Rhee SY (2006) Systematic analysis of Arabidopsis organelles and a protein localization database for facilitating fluorescent tagging of full-length Arabidopsis proteins. Plant Physiol 141 527–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Li M, Zhang W, Welti R, Wang X (2004) The plasma membrane-bound phospholipase Dδ enhances freezing tolerance in Arabidopsis. Nat Biotechnol 22 427–433 [DOI] [PubMed] [Google Scholar]
- Li W, Wang R, Li M, Li L, Wang C, Welti R, Wang X (2008) Differential degradation of extraplastidic and plastidic lipids during freezing and post-freezing recovery in Arabidopsis. J Biol Chem 283 461–468 [DOI] [PubMed] [Google Scholar]
- Maréchal E, Block MA, Dorne AJ, Douce R, Joyard J (1997) Lipid synthesis and metabolism in the plastid envelope. Physiol Plant 100 65–77 [Google Scholar]
- Mikkelsen J, Knudsen J (1987) Acyl-CoA-binding protein from cow. Biochem J 248 709–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15 473–497 [Google Scholar]
- Mylne J, Botella JR (1998) Binary vectors for sense and antisense expression of Arabidopsis ESTs. Plant Mol Biol Rep 16 257–262 [Google Scholar]
- Nitz I, Döring F, Schrezenmeir J, Burwinkel B (2005) Identification of new acyl-CoA binding protein transcripts in human and mouse. Int J Biochem Cell Biol 37 2395–2405 [DOI] [PubMed] [Google Scholar]
- Ohlrogge J, Browse J (1995) Lipid biosynthesis. Plant Cell 7 957–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrescu AD, Payne HR, Boedecker A, Chao H, Hertz R, Bar-Tana J, Schroeder F, Kier AB (2003) Physical and functional interaction of acyl-CoA-binding protein with hepatocyte nuclear factor-4α. J Biol Chem 278 51813–51824 [DOI] [PubMed] [Google Scholar]
- Rajashekar CB, Zhou HE, Zhang Y, Li W, Wang X (2006) Suppression of phospholipase Dα1 induces freezing tolerance in Arabidopsis: response of cold-responsive genes and osmolyte accumulation. J Plant Physiol 163 916–926 [DOI] [PubMed] [Google Scholar]
- Sakamoto A, Sulpice R, Hou CX, Kinoshita M, Higashi SI, Kanaseki T, Nonaka H, Moon BY, Murata N (2003) Genetic modification of the fatty acid unsaturation of phosphatidylglycerol in chloroplasts alters the sensitivity of tobacco plants to cold stress. Plant Cell Environ 27 99–105 [Google Scholar]
- Shi DQ, Liu J, Xiang YH, Ye D, Sundaresan V, Yang WC (2005) SLOW WALKER1, essential for gametogenesis in Arabidopsis, encodes a WD40 protein involved in 18S ribosomal RNA biogenesis. Plant Cell 17 2340–2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JA, Krauss MR, Borkird C, Sung ZR (1988) A nuclear protein associated with cell divisions in plants. Planta 174 462–472 [DOI] [PubMed] [Google Scholar]
- Steponkus PL, Uemura M, Joseph RA, Gilmour SJ, Thomashow MF (1998) Mode of action of the COR15a gene on the freezing tolerance of Arabidopsis. Proc Natl Acad Sci USA 95 14570–14575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui N, Li M, Zhao SJ, Li F, Liang H, Meng QW (2007) Overexpression of glycerol-3-phosphate acyltransferase gene improves chilling tolerance in tomato. Planta 226 1097–1108 [DOI] [PubMed] [Google Scholar]
- Suzui N, Nakamura S, Fujiwara T, Hayashi H, Yoneyama T (2006) A putative acyl-CoA-binding protein is a major phloem sap protein in rice (Oryza sativa L.). J Exp Bot 57 2571–2576 [DOI] [PubMed] [Google Scholar]
- Thomashow MF (1999) Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu Rev Plant Physiol Plant Mol Biol 50 571–599 [DOI] [PubMed] [Google Scholar]
- Vasil IK (2002) The science and politics of plant biotechnology: a personal perspective. Nat Biotechnol 21 849–851 [DOI] [PubMed] [Google Scholar]
- Walz C, Giavalisco P, Schad M, Juenger M, Klose J, Kehr J (2004) Proteomics of curcurbit phloem exudate reveals a network of defence proteins. Photochemistry 65 1795–1804 [DOI] [PubMed] [Google Scholar]
- Welti R, Li W, Li M, Sang Y, Biesiada H, Zhou HE, Rajashekar CB, Williams TD, Wang X (2002) Profiling membrane lipids in plant stress responses: role of phospholipase Dα in freezing-induced lipid changes in Arabidopsis. J Biol Chem 277 31994–32002 [DOI] [PubMed] [Google Scholar]
- Xiao S, Gao W, Chen QF, Ramalingam S, Chye ML (2008) Overexpression of membrane-associated acyl-CoA-binding protein ACBP1 enhances lead tolerance in Arabidopsis. Plant J 54 141–151 [DOI] [PubMed] [Google Scholar]
- Xin Z, Browse J (1998) eskimo1 mutants of Arabidopsis are constitutively freezing-tolerant. Proc Natl Acad Sci USA 95 7799–7804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K (2005) Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci 10 88–94 [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K (2006) Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol 57 781–803 [DOI] [PubMed] [Google Scholar]
- Zhang W, Qin C, Zhao J, Wang X (2004) Phospholipase Dα1-derived phosphatidic acid interacts with ABI1 phosphatase 2C and regulates abscisic acid signaling. Proc Natl Acad Sci USA 101 9508–9513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JH, Shi HZ, Lee BH, Damsz B, Cheng S, Stirm V, Zhu JK, Hasegawa PM, Bressan RA (2004) An Arabidopsis homeodomain transcription factor gene, HOS9, mediates cold tolerance through a CBF-independent pathway. Proc Natl Acad Sci USA 101 9873–9878 [DOI] [PMC free article] [PubMed] [Google Scholar]