Abstract
The mammalian architectural HMGB-Box transcription factor UBF is ubiquitously expressed in two variant forms as the result of a differential splicing event, that in the UBF2 deletes 37 amino acid from the second of six HMGB-boxes. Several attempts to define a function for this shorter UBF2 protein have been less than satisfactory. However, since all mammals appear to display similar levels of the longer and shorter UBF variants, it is unlikely that UBF2 is simply nonfunctional. Previously we showed that phosphorylation of UBF by the MAP-kinase ERK regulates chromatin folding and transcription elongation, explaining the rapid response of the ribosomal RNA genes to growth factors. Here we have investigated the roles the UBF variants play in the response of these genes to ERK activity. We demonstrate that the variant HMGB-box 2 of UBF2 has lost the ability to bind bent DNA and hence to induce chromatin folding. As a result it is significantly less effective than UBF1 at arresting RNAPI elongation but at the same time is more responsive to ERK phosphorylation. Thus, UBF2 functionally simulates a hemi-phosphorylated UBF whose expression may provide a means by which to tune the response of the ribosomal RNA genes to growth factor stimulation.
INTRODUCTION
A common phenomenon of higher eukaryotes is the co-expression of two or more protein variants from the same gene via differential splicing of the primary transcript. In some cases specific functions have been attributed to these variants, but in many cases their functional significance is unknown. Such is the case with the shorter splice variant of the RNA polymerase I (RNAPI) transcription factor UBF. This factor was originally identified some 19 years ago as a multi-HMGB-box protein that enhanced in vitro assembly of the RNAPI initiation complex (1). Soon after it was found to be expressed in two distinct forms via differential splicing in tetrapods as diverse as human and frog and some data has suggested that the relative levels of the splice variants are linked to changes occurring during early development (2–4). A single common differential UBF splice event occurs in all mammals studied and leads to the deletion of 37 amino acid from the second HMGB-box (Figure 1). Several attempts to define a function for the shorter UBF2 protein have been less than satisfactory. In vitro assays of initiation complex assembly showed that this form was nonfunctional (5), while reporter gene assays suggested that it was less able to enhance RNAPI transcription than full length UBF1 (6). However, since every mammalian cell studied to date has displayed similar levels of both the full length UBF1 and the shorter UBF2 variant, e.g. see (3,4,7), it seems unlikely that UBF2 is simply nonfunctional.
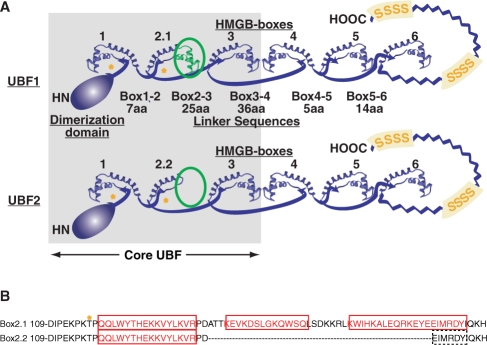
Figure 1.
(A) The domain structure of the mammalian UBF splice variants. The HMGB-Box domains are shown using HMGD as model (27). The UBF2 splice variation is shown as a hypothetical folded structure and the variant polypeptide indicated in green. The asterisk indicates the ERK phosphorylation sites and ‘S’ the serine rich segment in the acidic ‘tail’ domain (zig-zag line). (B) The protein sequence of full length HMGB-Box 2.1 and shortened Box2.2 showing the effect of differential UBF mRNA splicing. Predicted alpha-helical regions are shown in red and asterisk indicates the ERK phosphorylation site.
Recently, we demonstrated that in contrast to previous assumptions RNAPI transcription of the rRNA genes responds immediately to growth factor stimulation and ERK activation and that this response requires a regulation of the transcription elongation rate (8). We further showed that this regulation results from a direct phosphorylation of the first two HMGB-boxes of UBF (9,10). In vitro elongation studies quite surprisingly demonstrated that in its unphosphorylated state UBF was able to arrest the elongating RNAPI and that this arrest was abrogated by ERK phosphorylation of HMGB-boxes 1 and 2 (10). We further showed that ERK phosphorylation of UBF did not affect its affinity for DNA but abrogated DNA bending by the HMGB-Boxes and hence prevented the formation of the looped DNA structure we have referred to as the ribosomal enhancesome (9,11–13). These data suggested that RNAPI elongation was in fact regulated by the folding of the rRNA gene into the enhancesome structure. Consistent with this, UBF is present throughout the rRNA gene DNA and is required for to maintain rRNA genes transcriptionally active (14) and (Sanij et al., manuscript submitted).
The UBF2 splice variant has lost a major segment of HMGB-Box 2 (Figure 1B). This suggested to us that the shorter HMGB-Box2 would probably also have lost the ability to bend DNA and that this could have consequences for the regulation of RNAPI elongation. Here we demonstrate that the variant HMGB-box 2 of UBF has indeed lost the ability to bind bent DNA. More importantly, we show that UBF2 is significantly less effective at arresting RNAPI elongation and that in the case of UBF2 this arrest is more easily abrogated by ERK phosphorylation than is the case for UBF1. Thus, varying relative levels of expression of UBF2 provides a mechanism by which to tune the response of the rRNA genes to growth factors and to ERK activity.
MATERIALS AND METHODS
Expression of UBF constructs
Core mouse UBF1 and UBF2 (amino acid 2–404 and 2–367, respectively) were expressed as GST-fusion proteins in E. coli after sub-cloning the rat cDNA, mutated to encode a T at amino acid 242 as in mouse, into the BamHI/EcoRI site of pGEX2T and were purified from the soluble protein fraction on G-Sepharose (Amersham Biosciences, GE Healthcare, Little Chalfont, Buckinghamshire, UK) and recovered by thrombin cleavage (15,16) as previously described (16). Full-length rat UBF1 and 2 were expressed in Sf9 cells (ATCC, Rockville, MD, USA) using the Bac-to-Bac Baculovirus Expression System (Invitrogen, Carlsbad, CA, USA) following the manufacturer's instructions and subsequently purified on Anti-Flag M2 Affinity Resin (Sigma Aldrich, St Louis, MO, USA), also as previously described (10). Mouse UBF HMGB-Box 1 (amino acids 110–184), Box 2.1 (amino acids 194–267) and Box 2.2 (amino acids 194–230) were expressed in E. coli after sub-cloning into the BamHI/EcoRI site of pGEX2T as for core mouse UBFs. The use of full-length rat instead of mouse UBF was due to its efficient expression in the baculovirus system. It varies from the core mouse UBF at only one amino acid, amino acid 242, and as will be seen from the data the full-length rat and core mouse UBF proteins were functionally interchangeable.
In vitro transcription assays
The template used in all reactions was the 34 bp G-less template (GL34) containing the mouse rRNA gene sequences from –168 to + 297 previously described (10). It was linearized at + 320 within the cloning vector by HindIII.
Multiple initiation round reactions (25 µl) contained 30 ng of linearized template, 9 µl of DEAE 280 fraction (17), 1 µl RNAguard (Amersham Biotech), 0.5 mM ATP, CTP and GTP and 0.05 mM UTP (Amersham Biotech) and 1 µCi α-[32P]-UTP (Amersham Biotech), in 4 mM HEPES–KOH, pH 7.9, 8 mM Glycerol, 7 mM DTT, 0.05 mM EDTA, 4 mM MgCl2, 98 mM KCl, 250 μg ml−1 α-amanitin (Sigma). One to three microliters of UBF protein or buffer was added in TBS. Reactions were stopped at 60 min, or as indicated, with 180 µl TE, pH 8.3, 0.1% SDS, 0.1 mg/ml Proteinase K (Sigma), RNA resolved on 5% TBE-urea sequencing gels and analyzed on a STORM 860 (Molecular Dynamics, GE Healthcare, Little Chalfont, Buckinghamshire, UK).
In single-round reactions the initial reaction was as above but for 20 min at 30°C in the absence of UBF and GTP. After addition of UBF protein or buffer, incubation was continued for another 15 min at 30°C. Subsequently, 0.5 mM of GTP and UTP were added and incubation continued at 30°C for 20 min. The samples were resolved on a 12% TBE-urea sequencing gel and analyzed as above. 5S RNA contained in the DEAE 280 fraction was end-labeled by endogenous enzymes during the transcription reaction.
Phosphoimages were analyzed using the ImageQuant (Molecular Dynamics) software. Errors were either estimated from the Standard Error of multiple experimental series or, where the analysis of a typical experimental series is shown, from an estimate of experimental errors in combination with the data scatter in that experimental series.
In vitro phosphorylation
Active ERK2 was prepared as (18). Two to five micrograms of UBF construct was phosphorylated at 37°C in 20 µl of kinase buffer (20 mM HEPES, pH 7.3, 10 mM MgCl2, 1 mM benzamidine, 1 mM DTT, 0.5 mM ATP) with 4 µl activated ERK2. Mock reactions used heat-inactivated ERK2 (5 min at 95°C).
Cruciform DNA mobility shift assays
The cruciform DNA structure was prepared according to (19) and used as previously described (9). Each mobility shift reaction was performed in 10 μl consisting of 5 µl of 2 × Binding Buffer (16% Ficoll, 200 mM NaCl, 20 mM HEPES, pH 7.9, 10 mM KCl, 2 mM EDTA, 2 mM spermidine, 1 mM DTT), 2 µl (100 fmol) of cruciform DNA in TMS (TBS plus 10 mM MgCl2) and 3 µl of TBS (10 mM Tris–HCl pH 7.5, 100 mM NaCl) containing varying amounts of the respective HMG box proteins. After 30 min incubation on ice and the addition of 1 µl 0.1% xylene cyanol, the samples were loaded on a 6.5% PAGE (30 acrylamide: 0.8 bisacrylamide) in 0.5× TBE, separated for 4 h at 11 V/cm, dried and autoradiographed.
RESULTS
UBF2 is less effective at inhibiting RNAPI transcription than UBF1
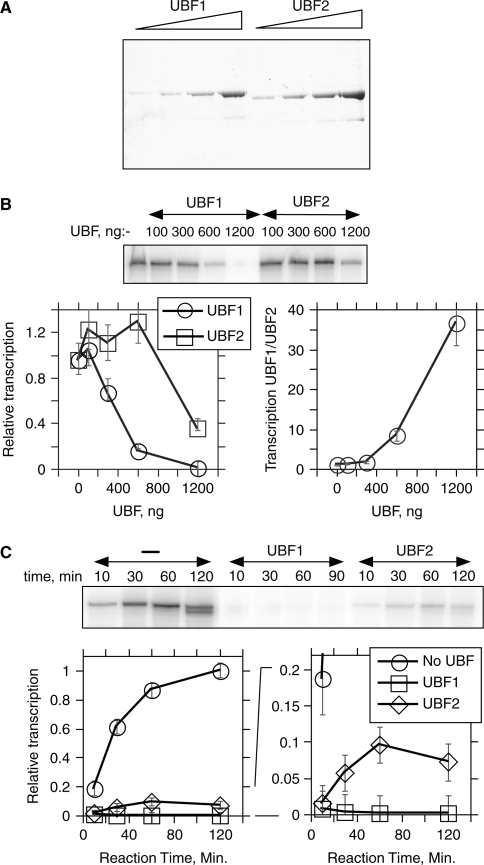
To better understand the function of UBF2, homogeneous baculovirus expressed rat UBF1 and 2 (Figure 2A) were assayed in an in vitro RNAPI transcription system able to support approximately 10 rounds of promoter-driven initiation and elongation per template. We previously demonstrated that addition of UBF1 to this system reduces transcript yield by specifically arresting RNAPI elongation complex (10). As was previously shown, levels of UBF1 sufficient to engage >90% of the template DNA (1200 ng/reaction) strongly inhibited the yield of RNAPI run-off transcripts. As will be seen from Figure 4B and as previously reported, this inhibition was not due to squelching since it required only the DNA architectural core region of UBF1 that is unable to compete for any of the basal RNAPI factors. When UBF2 was added to this system, though it significantly reduced transcript yield, at equivalent concentrations it did so significantly less well than UBF1 (Figure 2B) and this difference was evident throughout the range of UBF concentrations assayed. Accumulation of transcripts followed the same time course whether in the presence or absence of UBF2, demonstrating that UBF2 did not simply lead to a temporal inactivation of transcription (Figure 2C).
Figure 2.
UBF1 and UBF2 differ in their abilities to inhibit RNAPI transcription during multiple rounds of initiation. (A) Baculovirus expressed full-length rat UBFs. A Commassie stained gel of increasing amounts of the two proteins is shown. (B) In vitro RNAPI transcription in the presence of increasing amounts of the full-length UBFs. The phosphoimage of a typical electrophoretic gel analysis is shown and below this the quantitation of this analysis. (C) Typical time course of in vitro transcript accumulation in the presence or absence of 1200 ng per standard reaction of UBF1 or 2. As in B, quantitation of the analysis is shown below the phosphoimage of the gel, the right-hand panel shows a Y-axis expansion. Error bars indicate estimated measurement errors in B and C.
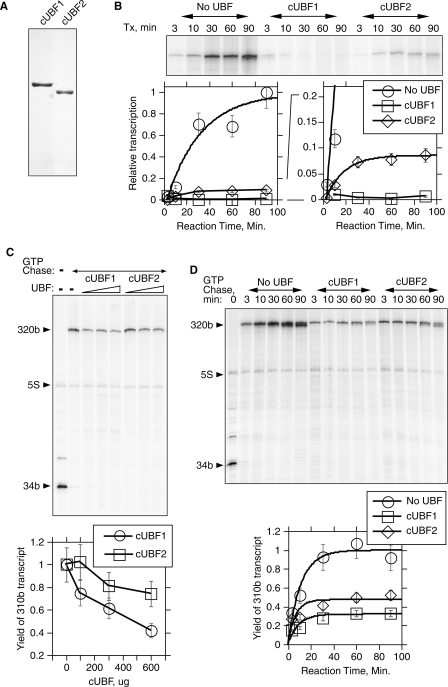
Figure 4.
The DNA architectural core regions of UBF1 and 2 emulate the activities of the full-length proteins. (A) Commassie stained gel analysis of mouse core UBF (cUBF) 1 and 2 proteins. (B) Time course of transcript yield during multi-round transcription reactions in the presence of 600 ng of cUBF1 or cUBF2. A typical analysis is shown above the quantitation. The lower right hand panel shows a Y-axis expansion of the data. (C) Elongation assays on the G-less cassette template in the presence of increasing amounts (100, 300 and 600 ng) of cUBF1 or 2. Lower panel gives the mean of three experiments and the standard errors. (D) Typical time course of elongation in the presence of 600 ng of cUBF1 or 2 for increasing GTP (+UTP) chase times. Upper panel shows the phosphoimage and lower panel the quantitation. Error bars indicate estimated measurement errors. The experimental design and nomenclature are as in Figure 3A. In B and D the curves show the best fits to the function; a + b[1−(exp−ct)], where t = time.
UBF2 is also less efficient at arresting RNAPI elongation complexes
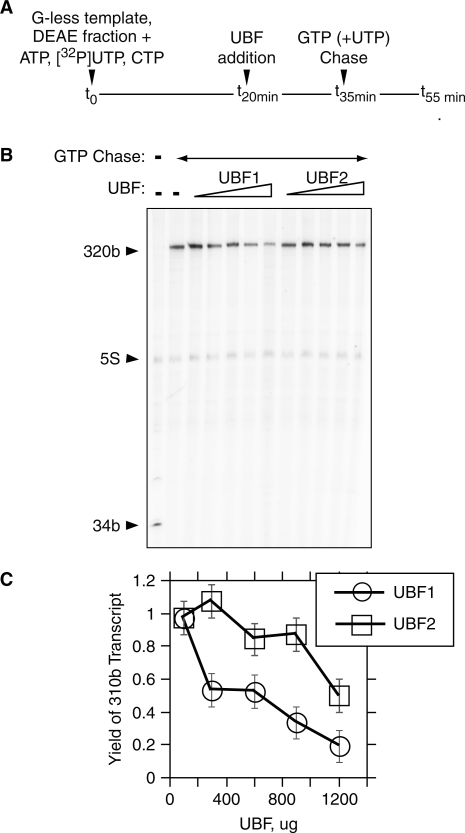
Using a G-less cassette approach it was shown that the major inhibitory effect of UBF1 is due to its ability to arrest RNAPI transcription elongation (10). We therefore used this assay to determine the relative capacity of UBF2 to arrest elongation. As can be seen from Figure 3, elongating RNAPI poised at nucleotide 34 in the absence of GTP is able to efficiently complete transcription of the G-less template on addition of GTP. As expected, the yield of full-length transcripts was severely reduced by increasing amounts of UBF1. But when UBF2 was assayed under the same conditions it was clearly less able to arrest elongation. The inhibitory effect on elongation during a single round of transcription, (maximally ∼80% for UBF1 and ∼50% for UBF2; Figure 3C), was significantly less than that seen in the multi-round transcription assay in Figure 2 (maximally 99.9% for UBF1 and 70–90% for UBF2). However, each new round of transcription in the multi-round transcription assay was subject to a similar degree of arrest, elongation complexes need only to be stably arrested for half the reaction time, the equivalent of about five rounds of transcription, to achieve the inhibition observed in the multi-round assays. Thus, the UBF catalyzed elongation arrest observed in the single round G-less cassette transcription quantitatively explains the transcription inhibition observed in the multiple round assay.
Figure 3.
UBF1 arrests elongation by RNAPI transcription complexes more efficiently than UBF2. (A) Diagram delineating the experimental procedure to measure elongation efficiency. Transcription of the RNAPI template containing a G-less cassette, bases 1 and 34, was initiated by the addition of the DEAE 280 nuclear protein fraction (DEAE fraction, see ‘Materials and methods’ section) in the presence of ATP, CTP and α[32P]-UTP. One thousand two hundred nanograms of UBF was added to the RNAPI elongation complexes arrested at +34 and finally GTP and excess UTP unlabeled was added to permit elongation to continue to the end of the template. (B) Typical phosphoimage analysis of a G-less cassette transcription assays. For tracks in which full-length rat UBF1 or 2 were added to the reactions, the amounts from left to right were 100, 300, 600, 900 and 1200 ng. ‘34b’ refers to the transcripts from elongation complexes arrested at the end of the G-less cassette and ‘320b’ to the full-length ‘run-off’ transcripts after addition of GTP (and excess UTP). Endogenous 5S RNA was also labeled during the reaction (10). (C) Quantitation of the yield of full-length (320b) transcript in the analysis shown in B as a function of UBF1 or 2 addition. Error bars indicate estimated measurement errors.
Core UBF is sufficient to arrest transcription elongation
It was previously shown that the minimal or core region of UBF (Figure 1A) is sufficient for elongation arrest and coincides with the region necessary for the formation of the enhancesome structure (10). Since the UBF splice variation falls within this region we also tested UBF1 and 2 in the context of core UBF (cUBF). At equivalent molarity, the ability of homogeneous cUBF1 and 2 to inhibit multiple round transcription was found to be very similar to the equivalent full-length proteins (Figure 4A and B). cUBF1 and 2 inhibited transcript yield respectively by >99 and 90%, in very close agreement with the data for UBF1 and 2 (compare Figure 2C with 4B). We also found that the cUBF1 and 2 forms arrested elongation in the single round transcription assay to similar degrees to their full-length counterparts (Figure 4C). Thus, the splice variants in the context of cUBF retained their differential ability to arrest RNAPI transcription elongation.
Two scenarios can be envisaged to explain the differential abilities of UBF1 and 2 to arrest RNAPI elongation. In the first, UBF1 and 2 would be equally likely to arrest RNAPI but the stability of the arrested elongation complex would be lower in the case of UBF2. This would be consistent with the partial loss of one key DNA binding domain in UBF2. In the second scenario, UBF2 would be less likely to induce RNAPI arrest in the first place, the arrested complex then may or may not be less stable than that arrested by UBF1. To determine the relative stabilities of the RNAPI elongation complexes arrested by UBF1 or 2, we formed arrested complexes on the G-less template and followed the yield of full-length transcripts with time. If RNAPI arrest was ‘leaky’ in the case of UBF2, transcript yield should continue to increase at longer reaction times. On the other hand, if arrest was stable the yield of transcripts in the presence of UBF1 or 2 should plateau. In fact regardless of the UBF variant used the yield of transcripts clearly plateaued. Thus, once arrested by either UBF variant the RNAPI complex did not resume elongation for the remaining reaction time (>60 min) (Figure 4D). The RNAPI elongation complexes were therefore stably arrested by both UBF1 and 2. The difference between UBF1 and 2 then appears to lie in the probability that arrest of RNAPI elongation will occur at any UBF–DNA complex. Given the low intrinsic stability of the UBF–DNA complex (9,16) this finding is somewhat surprising. It clearly infers that UBF must interact specifically with RNAPI to prevent its continued elongation, presumably at the same time stabilizing its own interaction with the template DNA. A possible interaction between UBF and RNAPI has been demonstrated in free solution (20–22), but it is unclear at present whether this is related to UBF's ability to arrest elongating RNAPI.
The short splice variant of HMGB-Box2 is unable to bind bent DNA
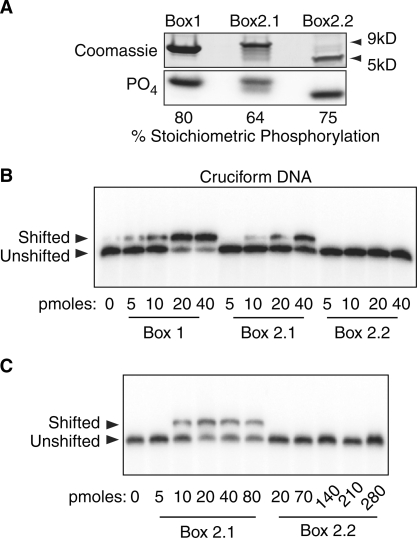
We have shown that HMGB-Boxes 1–3 are capable of significant in-phase DNA bending and in this way induce the DNA loop characteristic of the enhancesome (11–13). Furthermore, phosphorylation of HMGB-Boxes 1 and 2 by ERK abrogates this bending as well as the ability of UBF1 to arrest RNAPI elongation complexes (9,10). Given that the UBF2 splice variant is missing much of its HMGB-Box2 (Figure 1), it seemed possible that the remnant Box would no longer induce DNA bending and hence might functionally emulate constitutive ERK phosphorylation. To test this possibility, we first sought to determine if the short HMG-Box2.2 had indeed lost its ability to selectively bind bent DNA.
The ability of the HMGB-Boxes to bend DNA is reflected in their tight binding to prebent DNAs. Binding of HMGB-Boxes to cruciform DNA can be several orders of magnitude higher than for linear DNA and indeed this is the case for Boxes 1 and 2 of UBF1 (9). We, therefore, expressed the minimal HMGB-Boxes (Figure 5A) and determined their affinity for cruciform DNA. As expected, Box1 and the full-length Box 2.1 in their unphosphorylated forms both shifted cruciform DNA at very low concentrations (Figure 5B), characteristic of the estimated Kd for these Boxes of 1 and 3 μM, respectively (9). In contrast, the short unphosphorylated Box2.2 variant displayed no detectable interaction with cruciform DNA even at elevated concentrations (Figure 5C). This was consistent with the short splice variant of HMGB-Box2 being unable to bend the target DNA and predicts that UBF2 could not generate the in-phase bending required for enhancesome formation.
Figure 5.
The short HMGB-Box2.2 splice variant does not recognize prebent DNA. (A) Upper panel shows the Coomassie stained SDS–PAGE analysis of recombinant HMGB-Box1 protein, and the longer and shorter protein variants of HMGB-Box2, respectively Box2.1 and Box2.2. The lower panel shows the analysis of in vitro phosphorylation of the Boxes catalyzed by active ERK2. The percent stoichiometric levels of phosphorylation of each Box are indicated (% Stoichiometric Phosphorylation). (B) and (C) Mobility shift analyses of the binding of increasing amounts of the unphosphorylated Boxes to the standard cruciform DNA structure (9,19).
The short UBF2 splice variant displays an enhanced response to ERK phosphorylation
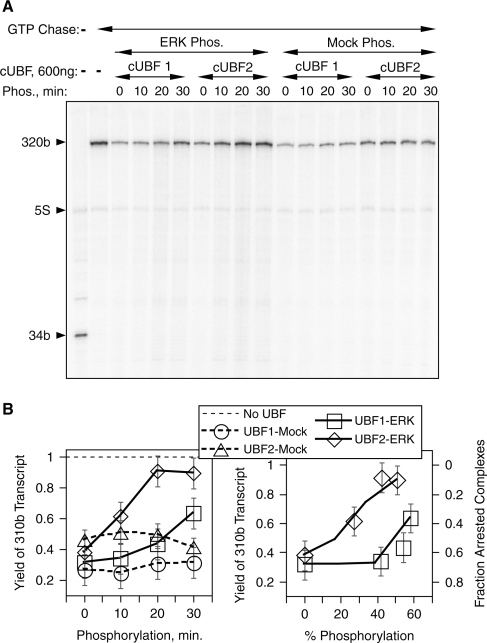
We next determined whether modulation of transcription elongation by ERK phosphorylation depended on which UBF splice variant was used. As can be seen in Figure 1B, the ERK site in HMGB-Box2 is retained in the short splice variant, and this site was found to be fully accessible to phosphorylation by activated ERK2 (Figure 5A). As previously shown (10), the ability of UBF1 to arrest the RNAPI elongation complex was significantly reduced by its ERK phosphorylation (Figure 6A). However, this effect was not directly proportional to the degree of UBF1 phosphorylation, 50% stoichiometric phosphorylation giving only a 20–25% reduction in the number of arrested elongation complexes (Figure 6B). This is consistent with efficient elongation through the UBF1–DNA complex requiring phosphorylation of both HMGB-Box1 and -Box2, since at 50% stoichiometric phosphorylation about 25% of UBF1 molecules will be phosphorylated on both Boxes 1 and 2, 50% only on one or other Box and 25% will be un-phosphorylated. In contrast, elongation arrest by UBF2 was nearly directly proportional to its level of phosphorylation, 50% stoichiometric phosphorylation reducing the number of arrested complexes by about the same factor (Figure 6B). This is consistent with the short splice variant functionally emulating constitutive phosphorylation of Box2, that is at 50% stoichiometric phosphorylation half the UBF2 molecules would have Box1 phosphorylated and in each case this will be in combination with the short Box 2.2. Thus, the short splice variant of HMGB-Box2 bestows on UBF2 the ability to be more responsive to ERK phosphorylation at the expense of being less effective in arresting RNAPI transcription. This suggests a role for UBF2 in fine-tuning the response of the rRNA genes to growth factor stimulation.
Figure 6.
UBF2 is more responsive to ERK phosphorylation. Core UBF1 and 2 (cUBF1, 2) were phosphorylated by ERK2 for increasing times or mock phosphorylated and then assayed for their abilities to arrest RNAPI elongation in the G-less cassette reaction. (A) Shows a phosphoimage of a typical gel electrophoretic analysis and (B) the quantitation of elongation efficiency plotted as full-length transcript yield as a function of time of phosphorylation and as a function of phosphorylation. Error bars indicate estimated measurement errors.
DISCUSSION
Our data demonstrate that UBF1 and 2 possess distinct abilities to regulate the transcription elongation rate of RNAPI. UBF2 is less efficient than UBF1 at arresting the RNAPI transcription complex, but is at the same time more responsive to ERK phosphorylation. This appears to be the result of the short splice variant of HMGB-Box2 (Box2.2) present in UBF2 functionally simulating the phosphorylation of this Box (Figure 1). Consistent with this, Box2.2 has lost the ability to recognize prebent DNA and hence probably also the ability to bend DNA. The data also suggest that UBF arrests the RNAPI elongation complex not by presenting a physical barrier to elongation, but rather by specifically interacting with RNAPI to inhibit its ability to elongate the transcript.
It was recently shown that the RNAPI transcription elongation rate plays a key role in regulating rRNA synthesis in response to growth factors in mammalian cell culture (10). The present data now suggest that the UBF1/2 ratio determines the responsiveness of the rRNA genes to extra-cellular signals. Changes in the UBF1/2 ratio could then tune the basal rate of RNAPI elongation and hence the rate of rRNA synthesis while at the same time determining an appropriate response to growth stimuli in different cell types or tissues. Higher levels of UBF1 would reduce the unstimulated or basal level of rRNA synthesis but also make transcription less responsive to ERK activation. In contrast, higher levels of UBF2 allow a somewhat higher basal rate of transcription, but make transcription more responsive to ERK. Differences in the UBF1:2 ratio have been observed between cell lines, e.g. (23) and during differentiating (2,4) but their significance has not been demonstrated.
Previous studies have almost exclusively tested the ability of UBF to activate transcription initiation either in vitro or in the context of a reporter gene. These studies showed that in comparison with UBF1, UBF2 essentially lacked the ability to activate rRNA transcription and suggested that it represented an essentially nonfunctional form of UBF (5,6). However, it is rather surprising that an apparently nonfunctional UBF isoform should consistently be found to represent around 50% of total UBF in actively growing cells, especially considering the abundant nature of UBF (24). More recent studies found that UBF binds throughout the transcribed region and forms a distinct rRNA gene chromatin through which RNAPI must transcribe (14). It was also shown that, without the aid of remodeling of this chromatin by the ERK-dependent phosphorylation of UBF, RNAPI could only but poorly transcribe the underlying DNA, explaining the growth factor regulation of RNAPI transcription elongation (10). The present finding that UBF2 is more permissive to transcription elongation and more responsive to the ERK pathway than UBF1 suggests that, far from it being nonfunctional, UBF2 plays an important role in tuning RNAPI elongation rates and hence rRNA transcription levels. However, an in vivo determination of the effects of changes in the UBF1:2 ratio was confounded by the observation that siRNA knock-down of UBF causes a major reduction in the numbers of active rRNA genes and induces cell cycle arrest and that UBF1 but not UBF2 is required to maintain the rRNA genes active (Sanji et al. manuscript in revision). Thus, it appears likely that UBF1 is necessary to maintain transcriptional competence of the rRNA genes, possibly by inducing a specialized rRNA chromatin, but that in so doing it presents a barrier to RNAPI elongation. Both ERK phosphorylation of UBF1 and the presence of UBF2 together enable RNAPI to surmount this barrier to elongation and provide a mechanism to regulate rRNA synthesis.
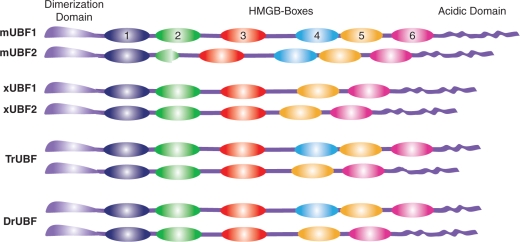
Though UBF1 and 2 are essentially identical in all mammals studied (3,7), more distantly related vertebrates display quite different UBF variants. In particular, while all mammals contain an HMGB-Box4, Xenopus UBF is missing a Box 4 homolog and what is more it is subject to differential splicing between Boxes 3 and 5 (25,26) (Figure 7 and Supplementary Data). To better understand the range of UBF variants we searched the vertebrate cDNA data bases and have identified several isoforms in both Zebrafish (Danio rerio) and Pufferfish (Takifugu rubripes). The most striking finding is that while the HMGB-Boxes of a given UBF are extremely divergent one to another, both the identity and order of the Boxes is highly conserved from fish to mammal. Further we were surprised to find that both Zebrafish and Pufferfish encode UBFs with an identifiable homologue of mammalian Box4. Thus, HMGB-Box4 is not specific to mammals and indeed probably predates the evolutionary divergence between fish and amphibia. What is more, both Zebrafish and Pufferfish express UBF variants with and without a Box4 homolog.
Figure 7.
Comparison of the domain structures of variant UBFs from four animal species. Orthologous HMGB-Box domains are indicated by a common coloring. In the case of Zebrafish (Danio rerio) the diagrams present only the major variants, in fact at least five distinct variants are expressed from two genes. In the case of Pufferfish (Takifugu rubripes) two distinct genes were identified but no cDNA data was available from which to determine possible splice variants. The diagrammatic representations are based on the following UBF sequences; Rat P25977, Xenopus laevis CAA40487 and CAA42523, Pufferfish (Takifugu rubripes) Ensembl peptides SINFRUP00000177953 and SINFRUP00000181780, and Zebrafish (Danio rerio) Ensembl peptides ENSDARP00000048724 and ENSDARP00000056633.
HMGB-Boxes 1 to 3 cooperate to induce in-phase bending in DNA, creating a nucleoprotein structure, the enhancesome, that is analogous in mass and size to the nucleosome (11,12). The mammalian UBF2 splice variant very probably disrupts this cooperativity by preventing DNA bending by Box2 and in this way probably causes a major unfolding of the enhancesome. However, it is possible that changes in the spacing between Boxes5/6 and Box3, or the addition or removal of Box4, could equally affect enhancesome structure, by interfering with the positioning of Box3 and hence disrupting structural cooperativity with Boxes 1 and 2. Though this is pure conjecture, it is a testable hypothesis that could explain the apparent contradiction between the high degree of conservation of the HMGB-Box domains and the high degree of variability in domain juxtaposition observed in the UBF protein family.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
This work was supported by an operating grant from the Canadian Institutes of Health Research (CIHR). The Research Centre of the CHUQ, in which the Cancer Research Centre is housed, is supported by a grant from the FRSQ (Québec). Funding to pay the Open Access publication charges for this article was provided by CIHR.
Conflict of interest statement. None declared.
REFERENCES
- 1.Jantzen H-M, Admon A, Bell SP, Tjian R. Nucleolar transcription factor hUBF contains a DNA-binding motif with homology to HMG proteins. Nature. 1990;344:830–836. doi: 10.1038/344830a0. [DOI] [PubMed] [Google Scholar]
- 2.Guimond A, Moss T. Variants of the Xenopus laevis ribosomal transcription factor xUBF are developmentally regulated by differential splicing. Nucleic Acids Res. 1992;20:3361–3366. doi: 10.1093/nar/20.13.3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O’Mahony DJ, Rothblum LI. Identification of two forms of the RNA polymerase I transcription factor UBF. Proc. Natl Acad. Sci. USA. 1991;88:3180–3184. doi: 10.1073/pnas.88.8.3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Larson DE, Xie W, Glibetic M, O'Mahony D, Sells BH, Rothblum LI. Coordinated decreases in rRNA gene transcription factors and rRNA synthesis during muscle cell differentiation. Proc. Natl Acad. Sci. USA. 1993;90:7933–7936. doi: 10.1073/pnas.90.17.7933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuhn A, Voit R, Stefanovsky V, Evers R, Bianchi M, Grummt I. Functional differences between the two splice variants of the nucleolar transcription factor UBF: the second HMG box determines specificity of DNA binding and transcriptional activity. EMBO J. 1994;13:416–424. doi: 10.1002/j.1460-2075.1994.tb06276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hannan R, Stefanovsky V, Arino T, Rothblum L, Moss T. Cellular regulation of ribosomal DNA transcription: both rat and Xenopus UBF1 stimulate rDNA transcription in 3T3 fibroblasts. Nucleic Acids Res. 1999;27:1205–1213. doi: 10.1093/nar/27.4.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bolívar J, Goenechea LG, Grenett H, Pendón C, Valdivia MM. Cloning and sequencing of the genes encoding the hamster ribosomal transcription factors UBF1 and UBF2. Gene. 1996;176:257–258. doi: 10.1016/0378-1119(96)00207-7. [DOI] [PubMed] [Google Scholar]
- 8.Stefanovsky VY, Pelletier G, Hannan R, Gagnon-Kugler T, Rothblum LI, Moss T. An immediate response of ribosomal transcription to growth factor stimulation in mammals is mediated by ERK phosphorylation of UBF. Mol. Cell. 2001;8:1063–1073. doi: 10.1016/s1097-2765(01)00384-7. [DOI] [PubMed] [Google Scholar]
- 9.Stefanovsky VY, Pelletier G, Bazett-Jones DP, Moss T. ERK modulates DNA bending and enhancesome structure by phosphorylating HMG1-boxes 1 and 2 of the RNA polymerase I transcription factor UBF. Biochemistry. 2006;45:3626–3634. doi: 10.1021/bi051782h. [DOI] [PubMed] [Google Scholar]
- 10.Stefanovsky VY, Langlois F, Gagnon-Kugler T, Rothblum LI, Moss T. Growth factor signaling regulates elongation of RNA polymerase I transcription in mammals via UBF phosphorylation and rchromatin remodeling. Mol. Cell. 2006;21:629–639. doi: 10.1016/j.molcel.2006.01.023. [DOI] [PubMed] [Google Scholar]
- 11.Stefanovsky VY, Pelletier G, Bazett-Jones DP, Crane-Robinson C, Moss T. DNA looping in the RNA polymerase I enhancesome is the result of non-cooperative in-phase bending by two UBF molecules. Nucleic Acids Res. 2001;29:3241–3247. doi: 10.1093/nar/29.15.3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stefanovsky VY, Bazett-Jones DP, Pelletier G, Moss T. The DNA supercoiling architecture induced by the transcription factor xUBF requires three of its five HMG-boxes. Nucleic Acids Res. 1996;24:3208–3215. doi: 10.1093/nar/24.16.3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bazett-Jones DP, Leblanc B, Herfort M, Moss T. Short-range DNA looping by the Xenopus HMG-box transcription factor, xUBF. Science. 1994;264:1134–1137. doi: 10.1126/science.8178172. [DOI] [PubMed] [Google Scholar]
- 14.O'Sullivan AC, Sullivan GJ, McStay B. UBF binding in vivo is not restricted to regulatory sequences within the vertebrate ribosomal DNA repeat. Mol. Cell. Biol. 2002;22:657–668. doi: 10.1128/MCB.22.2.657-668.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith DB, Corcoran LM. Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. New York: Greene Publishing Associates & Wiley-Interscience; 1991. Expression and purification of glutathione-S-transferase fusion proteins; pp. 16.17.11–16.17.17. [Google Scholar]
- 16.Leblanc B, Read C, Moss T. Recognition of the Xenopus ribosomal core promoter by the transcription factor xUBF involves multiple HMG box domains and leads to an xUBF interdomain interaction. EMBO J. 1993;12:513–525. doi: 10.1002/j.1460-2075.1993.tb05683.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schnapp A, Grummt I. Purification, assay, and properties of RNA polymerase I and class I-specific transcription factors in mouse. Methods Enzymol. 1996;273:233–248. doi: 10.1016/s0076-6879(96)73023-9. [DOI] [PubMed] [Google Scholar]
- 18.Wilsbacher JL, Cobb MH. Bacterial expression of activated mitogen-activated protein kinases. Methods Enzymol. 2001;332:387–400. doi: 10.1016/s0076-6879(01)32217-6. [DOI] [PubMed] [Google Scholar]
- 19.Bianchi ME. Interaction of a protein from rat liver nuclei with cruciform DNA. EMBO J. 1988;7:843–849. doi: 10.1002/j.1460-2075.1988.tb02883.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schnapp G, Santori F, Carles C, Riva M, Grummt I. The HMG box-containing nucleolar transcription factor UBF interacts with a specific subunit of RNA polymerase I. EMBO J. 1994;13:190–199. doi: 10.1002/j.1460-2075.1994.tb06248.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanada K, Song CZ, Yamamoto K, Yano K, Maeda Y, Yamaguchi K, Muramatsu M. RNA polymerase I associated factor 53 binds to the nucleolar transcription factor UBF and functions in specific rDNA transcription. EMBO J. 1996;15:2217–2226. [PMC free article] [PubMed] [Google Scholar]
- 22.Hempel WM, Cavanaugh AH, Hannan RD, Taylor L, Rothblum LI. The species-specific RNA polymerase I transcription factor SL-1 binds to upstream binding factor. Mol. Cell. Biol. 1996;16:557–563. doi: 10.1128/mcb.16.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poortinga G, Hannan KM, Snelling H, Walkley CR, Jenkins A, Sharkey K, Wall M, Brandenburger Y, Palatsides M, Pearson RB, et al. MAD1 and c-MYC regulate UBF and rDNA transcription during granulocyte differentiation. EMBO J. 2004;23:3325–3335. doi: 10.1038/sj.emboj.7600335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moss T, Langlois F, Gagnon-Kugler T, Stefanovsky V. A housekeeper with power of attorney: the rRNA genes in ribosome biogenesis. Cell. Mol. Life Sci. 2007;64:29–49. doi: 10.1007/s00018-006-6278-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bachvarov D, Moss T. The RNA polymerase I transcription factor xUBF contains 5 tandemly repeated HMG homology boxes. Nucleic Acids Res. 1991;19:2331–2335. doi: 10.1093/nar/19.9.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bachvarov D, Normandeau M, Moss T. Heterogeneity in the Xenopus ribosomal transcription factor xUBF has a molecular basis distinct from that in mammals. FEBS Lett. 1991;288:55–59. doi: 10.1016/0014-5793(91)81002-p. [DOI] [PubMed] [Google Scholar]
- 27.Murphy FV, Sweet RM, Churchill ME. The structure of a chromosomal high mobility group protein-DNA complex reveals sequence-neutral mechanisms important for non-sequence-specific DNA recognition. EMBO J. 1999;18:6610–6618. doi: 10.1093/emboj/18.23.6610. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.