Abstract
Bacteriophage terminases package DNA through the portal ring of a procapsid during phage maturation. We have probed the mechanism of the phage T4 large terminase subunit gp17 by analyzing linear DNAs that are translocated in vitro. Duplex DNAs of random sequence from 20 bp to 500 bp were efficiently packaged. Dye and short single-stranded end extensions were tolerated, whereas 20-base extensions, hairpin ends, 20 bp DNA-RNA hybrid, and 4 kb dsRNA substrates were not packaged. Molecules 60 bp long with 10 mismatched bases were translocated; substrates with 20 mismatched bases, a related D-loop structure or ones with 20 base single-strand regions were not. A single nick in 100 or 200 bp duplexes, irrespective of location, reduced translocation efficiency; but a singly nicked 500 bp molecule was packaged as effectively as an un-nicked control. A fluorescence correlation spectroscopy (FCS)-based assay further showed that a 100 bp nicked substrate did not remain stably bound by the terminase-prohead. Taken together, two unbroken DNA strands seem important for packaging, consistent with a proposed torsional compression translocation mechansim.
Keywords: Bacteriophage, Terminase, Helicase, DNA packaging, Fluorescence Correlation Spectroscopy
Introduction
The packaging of bacteriophage DNA during phage morphogenesis is driven by some of the most powerful molecular motors yet characterized 1; 2. During phage assembly a preformed prohead is filled with genomic DNA to near crystalline density by the action of the terminase complex. The packaging machinery of double-stranded DNA and RNA bacteriophages, herpesvirus, adenovirus and poxvirus families share structural and mechanistic similarities. Phage packaging systems are of considerable interest as they serve as paradigms for viral assembly pathways, chromosome condensation mechanisms, and the mechano-chemical events by which the energy of ATP hydrolysis is converted into movement at the molecular level.
In a T4 infected host cell the genomic DNA available to the packaging machinery during phage morphogenesis exists as a transcriptionally and recombinationally active, endless concatamer 3. From this dynamic, branched substrate the 170 kb linear genome molecules that ultimately appear in filled T4 proheads are generated 4. The terminases are the enzymes primarily responsible for both the generation of single genome-length molecules and the active translocation of these units into the proheads 5. The T4 terminase enzyme comprises multiple copies of a large (70 kDa) subunit gp17 - which possess the ATPase domains that couple ATP hydrolysis to DNA translocation 6; 7 - and a small (18 kDa) subunit gp16 8; 9. The other essential protein component of the T4 packaging machinery is the gp20 portal or connector protein, occupying a unique vertex in the T4 prohead 10; 11. Initiation of packaging by terminase depends on the physical interaction of gp17 with a complex of the late sigma factor gp55 and the T4 sliding clamp gp45 pre-loaded onto the endless DNA 12, as well as with the gp16 small terminase subunit 12; 13. Interaction of gp17 with the gp20-prohead is also needed to complete assembly of the packasome 14: see also Figure 7a. A free end of DNA is generated by the gp17 nuclease15; 16 and ATP-dependent translocation of DNA proceeds until the prohead is filled to capacity with one genome equivalent. A second nuclease cut is then made by gp17 on the DNA to release the filled prohead from the terminase complex, for further phage maturation events.
Figure 7. Phage DNA packaging via a torsional compression mechanism.
(a) A fully assembled packasome complex is sketched, with the empty prohead (red) carrying the gp20 portal ring (blue) shown at one vertex, and the DNA duplex drawn in black. The multimeric state of the gp17 terminase (orange) remains to be precisely determined: only two subunits are shown for clarity. The terminase subunits are sketched with a minor lobe, representing a flexible region that undergoes a conformational change (black dashed arrow) coupled with ATP binding and hydrolysis (white solid arrow). No detailed gp17 structure is implied; nor do we specify the exact steps and temporal order by which the binding and hydrolysis of ATP is coupled to movement of gp17 during the reaction cycle. (b) Directed linear motion of the flexible arm of the lower gp17 subunit engages the DNA substrate and translocates this towards the prohead. This movement coupled with interaction of the DNA with the portal region causes induced changes to the helical pitch and temporarily stores energy. (c) The stored energy is released by translocation into the prohead (green arrow), enabling a restoration of the B-form helical repeat. For clarity, only one power stroke from one gp17 subunit is shown: the model also allows for multiple power strokes occurring at this stage before translocation occurs. (d) A nick in the DNA strand disrupts the accumulation and translocation potential of energy temporarily stored in the duplex, preventing translocation. Instead, loss of torsional energy in the compressed nicked duplex might even facilitate backward release of the substrate, as suggested by the red arrow in (e).
The packaging machinery in certain well characterized phage systems seems to be relatively tolerant of structural defects in the substrate DNA. Nicks (i.e. a stretch of fully paired bases on one DNA strand that lacks a phospho-diester bond between two adjacent residues) do not significantly inhibit packaging of phage T3 DNA17 or of T5 DNA, as nicked DNA was identified within mature heads in the latter system 18. Small (19 bp) regions of heterology were tolerated by a λ packaging system 19 and the φ29 system was shown to package nicked, but not gapped DNA 20. By contrast, the T4 DNA packaging apparatus appears to be much more stringent in its substrate requirements. Nicks, gaps or branches in the dynamic T4 concatamer all seem to impede the translocation process in vivo, as deficiencies in the activities of T4 or host DNA ligases, T4 gp49 endonuclease VII or T4 topoisomerase II were all inhibitory to efficient packaging 21; 22; 23; 24.
Variations in the details of DNA packaging between different phages highlight the need for defined in vitro systems to delineate events at the molecular level, and fluorescence-based microscopy and single-molecule techniques are providing new insights to this end.
Firstly, D. E. Smith and others have shown using single molecule optical tweezers that the φ29 motor translocates 2 bp of DNA per ATP hydrolyzed and can package DNA against forces approaching 100 pN or more 25; 26; 27. Equivalent studies on the λ and T4 motors established that each could package DNA against comparable forces, although the T4 system seemed more prone to ‘slips’ on DNA during the translocation process 28. Secondly, one long-standing model for the phage DNA packaging mechanism involved portal rotation relative to the prohead as an essential requirement for translocation 29; 30; although this model has recently been severely challenged experimentally. Single molecule studies of a fluorescently-tagged subunit of the φ29 portal failed to detect any portal rotation during DNA packaging 31; corroborating separate data obtained in this laboratory showing that immobilization of the T4 portal by protein fusions caused no inhibition to packaging 32.
We have recently developed a system for studying T4 packaging events in vitro 33 utilizing a novel application of fluorescence correlation spectroscopy (FCS) 34. This is a non invasive approach by which the diffusion coefficients of fluorescent molecules can be determined. By applying FCS to a T4 in vitro packaging system we readily detected a dramatic change in DNA diffusibility during packaging as faster-moving fluorescently-tagged 100 bp DNA molecules (65 kDa) became progressively sequestered inside the more slowly diffusing 75 MDa prohead. We were able to achieve near real-time monitoring of the packaging kinetics, and the rate constants determined by FCS matched those obtained by a separate in vitro assay 12. In addition, using recombinant proheads that encapsidated free GFP 35, energy transfer between the incoming acceptor-labeled DNA and donor GFP was detected, establishing directly by novel means that the DNA was translocated fully into the prohead interior.
To gain fresh mechanistic insights into the T4 packaging reaction we extended the biochemical analyses to include several short, structurally defined DNA substrates that can be packaged by the T4 system. Specifically, we examined the packaging efficiency in the context of single-stranded ends, hairpin ends, RNA-containing duplexes, mismatches, gaps and nicks. Our results extend the characterization of DNA that is processed by gp17 and have implications for the biology of T4 maturation, the mechanism of DNA translocation, and future biophysical applications of the T4 motor.
Results
T4 in vitro packaging assays
DNA packaging can be realized in vitro using preformed, unfilled gp20-proheads; the gp17 terminase; linear DNA and ATP 12; 28; 36. (The small terminase subunit gp16 is non-essential, in fact inhibitory, to packaging of linear DNA in vitro and so was not included in any of the experiments presented here). In a standard nuclease protection assay, the phage components and DNA are mixed together and packaging typically initiated by addition of ATP. After incubation, DNase I is added to the mix, and any DNA sequestered in the prohead is thus protected from nuclease digestion. The packaged material is released by SDS-Proteinase K digestion of the prohead and the amount of translocated DNA determined by gel analysis. Alternatively the FCS-based assay allows the kinetics of packaging to be monitored in near real-time 33. Identical cofactor requirements and kinetics of packaging seen with both approaches, as well as with a T4 phage assembly assay 37, confirm that the in vitro assays are faithful reconstructions of the T4 packaging reaction.
Short DNA molecules are efficiently translocated by gp17
In the current work we explored the minimum length of DNA that could be packaged by the T4 system. Molecules as short as 20 bp, made by annealing two complementary oligonucleotides, were efficiently translocated (Figure 1a). This reaction was dependent on the presence of ATP and gp17, showing that encapsidation of 20 bp DNA arises not from passive diffusion into the prohead but from bona fide translocation by the T4 motor. In other experiments, 1-10 nM of purified proheads (along with a 10-fold molar excess of gp17 monomer) typically protected up to 50 nM or more of input DNA: a molar ratio consistent with estimates from the FCS assay 33 and by other laboratories 36. Also, in reactions where a range of prohead concentrations were incubated in parallel with 100 nM DNA substrates that ranged in size from 20 to 100 bp, the extent of packaging was independent of the length of the molecule (Figure 1b). This result implies that translocation of short molecules is not the rate-limiting step in the packaging reaction.
Figure 1. Effect of DNA length on packaging by the T4 terminase.
(a) A 20 bp DNA substrate (100 nM) was incubated in a reaction mix with gp17, ATP and ∼ 6, 12 or 24 nM proheads, as indicated. Control reactions comprised the DNA alone (Input or ‘I’ lane), or a mix with DNA and proheads but lacking gp17 (Digested or ‘D’ lane). After 60 min at 25 °C DNase I was added to all tubes except (I) and any free DNA was digested at 37 °C for 20 min. Proteinase K, EDTA and SDS were added to degrade the proheads (by incubation at 65 °C for 30 min) and products were resolved on a 7.5 % polyacrylamide gel. DNA bands were revealed by EtBr staining. Further control reactions are shown in the right panel. The 20 bp substrate (100 nM) was incubated alone (I) or with ∼ 10 nM proheads in packaging assays with all components (Packaged or ‘P’ lane) or reactions omitting gp17 or ATP. Samples were processed as above. (b) DNA substrates 20 or 40 bp in length were constructed by annealing appropriate oligos, or a 100 bp substrate was obtained by PCR. Equimolar amounts were incubated in packaging assays as above, with the same range of prohead concentrations. The relative amounts protected were determined by quantifying the amounts of DNA on polyacrylamide gels, and the data plotted as shown. Points ± error bars show the mean ± range of data from two separate experiments. (c) Similar experiments were performed with plasmid DNA 4.8 kb in length cut with EcoRI (‘R’) to leave a 4 base 5′ overhang; the same plasmid cut with PstI (‘P’) to leave a 4 base 3′ overhang; or with 48 kb lambda DNA with the 12 base 5′ overhang of the cos site. Points ± error bars give the mean ± standard deviation of three separate experiments.
Tolerance of the T4 packaging motor to structural variations in the DNA ends
Plasmid DNA molecules 5 kb in length containing 4-base 5′ extensions or 4-base 3′ extensions were packaged with similar efficiencies (Figure 1c). The 48 kb phage λ genome with the 12 base 5′ extension of the cos site was also packaged, as reported previously 36. (In these experiments, the λ DNA was heated to 65 °C for 10 min and rapidly quenched on ice-water immediately before use in packaging assays, to ensure minimal cos site re-annealing.) This latter feature suggests that 12 base single-strands regions do not inhibit initiation, arguing against the possibility that gp17 might bind sufficiently strongly to ssDNA to prevent active translocation. Furthermore, the 48 kb and 5 kb substrates were packaged to comparable extents (Figure 1c). Again this result points to translocation not being a rate-limiting step for T4 packaging. However, neither of the longer substrates was as efficiently packaged as the 20-100 bp substrates (compare the axes of traces in Figures 1b and 1c). This discrepancy may arise from some compaction of the longer molecules in the 5 % PEG, 1.5 mM spermidine environment of the assay mix, perhaps making the ends of the molecule less accessible for initiation of packaging by the terminase.
Partly for the above reason, we further explored the effects on packaging of defined DNA end structures on short molecules. Substrates carrying either a 20- or a 40-base 3′ extension were constructed by annealing a 100-mer with complementary 80- or 60-mers. A related substrate with 20 bases of single-stranded DNA internally in a 100 bp molecule was also included. Pre-treatment with S1 nuclease, which degrades DNA at nicks or single-strand regions but leaves duplex DNA largely intact, confirmed the extent of the single-strand regions in these substrates (Figure 2a). In packaging assays, neither construct with a 3′ single-strand overhang was protected under conditions where a fully intact 100 bp duplex DNA was readily packaged (Figure 2b). These results taken together with the results of Figure 1c suggest that extensions up to 12 bases at the end of the DNA do not inhibit translocation, whereas 20 or more do have a significant effect on end recognition and/or translocation by the T4 motor. In addition, the 100 bp molecule with an internal 20-base gap was consistently not protected (Figure 2b). Thus a stretch of 20 single stranded bases seems to block motor function, regardless of whether the region is located at one end or internally in the substrate.
Figure 2. Packaging of short DNA molecules with varying end structures.
(a) Partly single-stranded DNA molecules were constructed by annealing two or three oligos, with the lengths sketched above each set of lanes. Each was incubated without or with S1 nuclease (- and +), and the products resolved on a polyacrylamide gel. (b) The same substrates were incubated with ∼ 10 nM proheads in a nuclease protection assay as above. A 100 bp PCR product of fully duplex DNA was used as a control. ‘I’ represents the Input DNA (from tubes lacking proheads and DNase I digestion), ‘D’ represents DNase I digestion of reactions lacking ATP and ‘P’ represents the Packaged material remaining after DNase I and Proteinase K/SDS treatments. (c) Alkali gel analysis of hairpin-containing molecules. The main substrate was a 65-mer, sketched in the 3rd lane, with 5 cytosine residues forming the unpaired loop. Treatment with DNA polymerase and dNTP’s formed a fully duplex 40 bp hairpin (2nd lane). A 40 bp PCR product of the same sequence was also used as a control (1st lane). (d) Each substrate was used in packaging assays, and Input (I) and Packaged (P) samples are shown.
The effect of a hairpin was also assessed. A molecule with a (dC)5 hairpin, 20 bp of duplex DNA and a 20 base 3′ extension was compared with similar constructs with no extension (i.e. a hairpin molecule with 40 bp throughout) and the same 40 bp sequence as a linear control. Analysis of all three substrates on denaturing alkali gels confirmed the nature of each form (Figure 2c). A tiny proportion at best of the hairpin-containing molecules was protected under conditions where essentially all of the 40 bp duplex control substrate was translocated (Figure 2d).
Effects of structural alterations to B-form duplex DNA on T4 packaging
The necessity of the helical repeat itself was examined by using duplex molecules that contained one or two RNA strands; structures that in each case tend to adopt an A-form-like helical structure 38; 39. A 4 kb double-stranded RNA substrate was not readily packaged by the T4 system under conditions where a 5 kb linear DNA molecule was fully translocated (Figure 3a). A 20 bp hybrid DNA-RNA molecule was also not packaged (Figure 3b) whereas a control 20 bp DNA duplex of identical sequence and concentration was efficiently protected.
Figure 3. Effect of RNA-containing duplexes on packaging.
(a) Packaging assays were set up with 10 nM proheads, gp17, ATP and ∼1 nM substrates as before, but after incubation at room temperature for 1 hr the reactions were loaded directly onto a 0.8 % agarose gel, which was stained with EtBr after electrophoresis. Under these conditions the proheads alone run as a faint band indicated by side arrow (lane 1) but this vanishes upon addition of Proteinase K (‘K’ in lane 2). The rest of the lanes show packaging reactions performed in the absence (-) or presence (A) of ATP. A 5 kb DNA substrate in a reaction lacking ATP runs as indicated by side arrow (lane 3) but in the presence of ATP the substrate is sequestered into the prohead, and so the DNA co-migrates with the proheads (lane 4). This pattern is not reproduced with a 4 kb dsRNA substrate (lanes 5 and 6): instead little if any RNA is up-shifted in this way. (b) Two 20 bp substrates of identical sequence but comprised of either double stranded DNA or hybrid DNA:RNA duplexes were constructed by annealing appropriate oligos. Each duplex (100 nM) was incubated alone with no added components (I) or subjected to digestion by a mixture of both DNase I and RNase A (‘0’). In addition, increasing amounts of proheads (2-20 nM) were used in packaging assays, as indicated by the triangle. ‘M’ comprises a separate 20 bp DNA marker in each case.
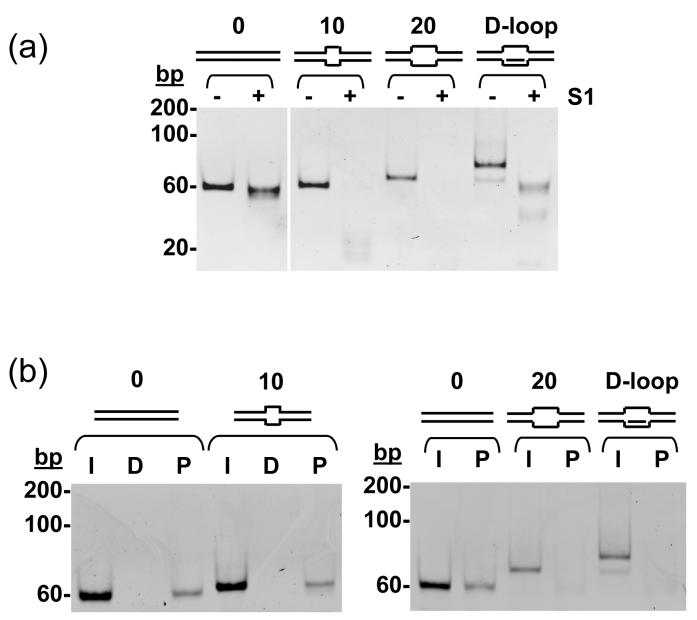
The effects of mismatched bases in a DNA duplex were also determined. Substrates with up to 20 mismatched bases were assembled by annealing appropriate 60-mer oligos. Treatment with S1 nuclease confirmed correct assembly (Figure 4a). The motor packaged the fully paired 60 bp duplex and molecules with 1, 2 or 5 (not shown) and 10-base mismatched DNAs with comparable efficiency, but the DNA bearing 20 mismatched bases was not packaged (Figure 4b). A related D-loop substrate was also constructed, with a third 20-mer complementary in sequence to the mismatched region. A small proportion (<10 %) of the molecules were present as 20 base mismatched DNA molecules rather than D-loops, due to incomplete annealing. Nevertheless, the addition of the third oligo formed a substrate much more resistant to S1 digestion (Figure 4a), as expected for this construct. In packaging assays, however, no protection of the D-loop was observed (Figure 4b).
Figure 4. Inhibition of packaging by mismatches and D-loops.
Substrates 60 bp in length were constructed by annealing the same 60-mer ‘Top’ strand with appropriate 60-mer oligos to make molecules with 0, 10 or 20 centrally located mismatched bases. A D-loop variant of the latter was assembled by including a third 20-mer oligo in the initial annealing reaction. (a) S1 nuclease digestion to confirm the integrity of the duplex, mismatched and D-loop forms. Numbers above each sketched substrate indicate the lengths of unpaired bases. (b) Each substrate (50 nM) was incubated with ∼12 nM proheads as in previous experiments. I, D and P show the Input, DNase-digested and Packaged material.
Single nicks in the DNA substrate inhibit packaging
We next examined the effect of a single nick in the sugar-phosphate backbone. In one set of experiments, appropriate substrates of 200 bp or 500 bp were treated with the modified restriction enzyme N.BsmI, which hydrolyses one DNA strand only at the recognition site. The PCR reaction was used to generate molecules that differed both in size and in position of the N.BsmI site relative to the entire molecule: thus variations in both DNA length and location of the nick could be assessed. The specific hydrolysis of one strand was confirmed by analyzing the the N.BsmI-treated samples on denaturing alkali gels (Figures 5a and 5c), and further control reactions using T4 ligase confirmed a single nick - as opposed to a more extensive gapped region - was present in the digested DNA substrates (data not shown).
Figure 5. Effect of DNA nicks on the T4 packaging reaction.
Fragments 500 or 200 bp in length containing an N.BsmI site were generated by PCR. (a) Alkali gel analysis of un-nicked (‘500-0’) and centrally nicked (‘250-250’) forms of the 500 bp substrate. (b) Extent of protection of 5 nM of both 500 bp forms in packaging assays, performed as described above. Points ± error bars show the mean ± range of values of two separate experiments. (c) Alkali gel analysis of 200 bp forms, either un-nicked (‘200-0’), centrally nicked (‘100-100’) or nicked 20 bp from one end of the molecule (‘180-20’). (d) Packaging assays performed on 15 nM of the 200 bp forms as above. In addition, 100 bp substrates were constructed by annealing oligos to generate an un-nicked control (‘100-0’), nicked molecules with a nick 20 or 50 bp from one end (‘80-20’ or ‘50-50’) or a molecule with a centrally located one-base gap (‘50-49’). (e) Alkali gel analysis of the nicked substrates and (f) packaging assays are shown.
Equimolar amounts of the nicked and un-nicked forms of the DNA were tested in the standard packaging assay. Substrates 500 bp in size were packaged at comparable levels regardless of whether one strand was nicked centrally or not (Figure 5b). Likewise in separate experiments two 5 kb molecules, without or with a nick centered in one strand, were packaged to similar extents (data not shown). Thus a single DNA nick does not seem to inhibit packaging on molecules 500 bp or greater in size. A reduction in packaging efficiency was observed, however, with nicked molecules of 200 bp compared with the un-nicked controls (Figure 5d). The degree of inhibition was comparable regardless of whether the nicks in the 200 bp DNA substrates were located either 20 bp or 100 bp from one end. This latter result argues strongly against the possibility of an unproductive initiation complex - formed by gp17 on any DNA distortion induced by the nick 20 bp from one end for example - as being an explanation for the reduction in packaging efficiency. Rather it seems that the nick itself forms an impediment to efficient DNA translocation.
The effects of nicks in 100 bp DNA molecules were also determined. Substrates for these experiments were constructed by annealing appropriate oligos; and an un-nicked control DNA, or substrates with the nick 20 or 50 bases from one end, were used (Figure 5e). In every experiment performed with nicked 100 bp DNA’s packaging was inhibited to some degree, although we tended to see some differences between the extent of inhibition and the particular batch of terminase - including a 6-His-tagged variant of full-length gp17 - that was used (data not shown). Figure 5f presents the data for a representative set of experiments where the same reaction mixes, i.e. those containing the same batch of terminase and dilutions of proheads, were used with the three 100 bp substrates. The two nicked forms of DNA inhibited packaging, and the extent of inhibition was equivalent regardless of whether the nick was 20 or 50 bases from the DNA end. This result again implies that a nick impedes packaging, independent of its location relative to the end of the molecule.
In addition, a variation of the centrally nicked substrate was built using a 49-mer, thus generating a molecule with a centrally located, one base gap in the DNA duplex. This construct inhibited packaging, to a comparable extent than the centrally nicked DNA (Figure 5f). While Figure 2B showed that a 20-base gap in a 100 bp DNA molecule was sufficient to abolish translocation, this latter result now suggests that a gap must be larger than a single base in order to fully block DNA translocation by the gp17 terminase.
FCS analysis of packaging of 100 bp substrates
The FCS packaging assay was then used to extend the analysis of the 100 bp DNA’s, utilizing rhodamine-tagged forms of the un-nicked substrate or that with the nick 20 bases from the 5′ end. Additionally, a rhodamine form of the 100 bp molecule bearing a 20-base internal gap (see Figure 2b, rightmost panel) was also used. The traces presented in Figure 6 show the time-dependent decay of the fluorescence autocorrelation function G(τ) of the substrates, a parameter which is related to the diffusion constant of the fluorescent species 34. More specifically, the G(τ) function will decay more slowly for a slower diffusing fluorophore than for a faster diffusing (i.e. smaller or apparently smaller) one. In the FCS packaging reaction, the 100 bp DNA substrates that are encapsidated move with the apparent diffusion of the 75 MDa prohead itself, causing an increase in the lag time of G(τ), compared with the G(τ) of unpackaged molecules. This is readily illustrated in Figure 6a, which shows the average FCS traces for the last five min of 30 min packaging reactions performed with ∼ 5 nM proheads and ∼ 10 nM un-nicked substrate DNA, with and without ATP. (In these experiments autocorrelation functions were collected at 20 sec intervals; thus multiple traces per reaction were obtained.) After 30 min, the G(τ) function of the molecules in the ATP-containing reaction showed a significant increase in lag time compared to those in the no ATP control, consistent with the majority of the DNA being sequestered inside proheads. By contrast, the nicked 100 bp substrate showed no change in G(τ) after 30 min even in the presence of ATP (Figure 6b), implying that the nicked substrate was largely un-translocated under these conditions. Similarly, the substrate with a 20-base gap also showed no evidence of being packaged by this assay, or of remaining terminase-bound (data not shown), consistent with the data in Figure 2b.
Figure 6. FCS analysis of packaging of nicked and un-nicked 100 bp substrates.
(a) FCS analysis of the last 5 min of a 32 min packaging reaction performed with 100 bp linear substrates carrying a 5′ rhodamine fluorophore, in the presence (+) or absence (○) of ATP. A single-species biophysical model was used to fit the data for the reaction lacking ATP, yielding a diffusion coefficient for the Rh-DNA substrate of 12.1 μm2/s and volume occupancy of 90 molecules. For reactions with ATP a two-species biophysical model was used to fit the data,yielding diffusion constants of 12.1 μm2/s and 0.91 μm2/s that arise from free DNA or prohead-encapsidated DNA respectively. The volume occupancy was determined to be 40 molecules of free DNA and 7 proheads, each containing an average of 6.7 DNAs (87 total DNAs). Note the two different y-axis scales. (b) The same analysis of reactions on a 100 bp nicked substrate, with (+) or without (○) ATP. In this panel the G(τ) values were normalized to unity. The autocorrelation curves yield diffusion coefficients of 7.3 μm2/s or 6.6 μm2/s for the nicked DNA, in reactions without and with ATP respectively.
The data of Figure 6a might initially seem to contradict that in Figure 5d, where some translocation of a 100 bp DNA with a nick 20 bp from one end was observed. However, it should be noted that differing batches of prohead and terminase preps, when assayed with nicked 100 bp substrates, showed some variations in translocation inhibition in any case. Also, the DNA substrates used for the FCS analyses necessarily had a bulky 5′ fluorophore attached. This adduct routinely caused some reduction in packaging efficiency in the standard nuclease protection assay, when compared with un-tagged DNA constructs (data not shown). By contrast, it might be argued that a molecule which blocked translocation due to a structural defect could remain associated with the prohead-terminase in a stalled complex, and so remain susceptible to DNase I attack in the nuclease assay. The diffusion coefficient of the DNA in this scenario, however, would still appear as that of the prohead complex itself in the FCS assay. In fact, no change in diffusion coefficient of the DNA over the course of the reaction was observed. Thus the result of Figure 6b implies that not only was the 100 bp nicked DNA not translocated by the T4 motor, but it also failed to remain associated with the terminase; arguing against the existence of a stalled but stable prohead-terminase-nicked DNA complex.
Discussion
In this work we have extended the characterization of small DNA molecules in packaging reaction of the T4 terminase complex. Substrates generally a few tens of nucleotides in length were assayed for their efficacy of protection by an in vitro packaging assay. Single-stranded overhangs of 12 bases at the end of DNA or 10 mismatched bases internally in a duplex were readily packaged, whereas a hairpin end or duplexes with at least one RNA strand were not tolerated. A single nick was an impediment to translocation of short substrates: an FCS analysis confirmed this and further revealed the lack of stable association of nicked DNA with the T4 motor. These results give fresh insights into the biological role of the T4 gp17 terminase subunit in phage packaging.
During the T4 infection cycle a free end of genomic DNA is not normally available to the gp17 terminase. Instead this is generated by the action of gp16 and other components of the late transcription and replication machinery acting in concert with gp17 and the endless genomic concatamer substrate 12; 15; 16. The in vitro packaging systems, by contrast, circumvent the need for these additional functions and enable the ability of gp17 to initiate packaging on a pre-formed linear DNA end to be readily assessed. Thus gp17 was relatively tolerant of varying structures for initiating the translocation reaction. Single-strand regions 4 bases long arising from restriction digests did not alter packaging efficiency, nor did the 12-base overhang of the lambda cos site. Furthermore bulky fluorescent dyes linked to a 5′ terminus formed substrates that still gave detectable - albeit reduced - levels of DNA packaging 33. In fact, the only end structures we analyzed that did prevent packaging were a 20- (or 40) base extension, as well as a closed hairpin loop at one end of a small (40 bp) substrate. Assuming productive initiation of DNA translocation has some requirement for a region of duplex DNA, these results imply that 12 to 20 bases represent the potential length of a gp17-DNA initiation footprint. It is also very interesting in this regard, and likely significant, that 20 bp duplex DNA is capable of binding to a recently identified DNA binding site in the ATPase domain of gp17 40; and in addition 20 bp molecules were readily translocated by the T4 motor (Figure 1). Indeed, this latter result now strongly implies that 20 bp can be taken as an upper bound for the length of DNA needed for productive engagement and translocation by the terminase-portal complex.
One of two central findings of this work relevant to gp17 function is that substrates which departed from duplex DNA generally had a significant inhibitory effect on DNA translocation. Thus duplex molecules containing one or two RNA strands failed to be packaged. This inhibition to translocation could arise from the alteration to the helical pitch and base stacking of the duplex induced by the ribonucleic acid bases. Alternatively, the 2′ OH group of the RNA strand(s) may inhibit binding to a region of gp17 tailored to specifically recognize deoxyribonucleic sugars of the substrate. Other structural departures from intact DNA also caused severe impediments to translocation. Substrates 100 bp in length that carried 20 base single-strand regions failed to be packaged, regardless of whether the single-strand region was at one end or internal in the molecule. There may be an absolute need for the translocation machinery to contact a substantial region of duplex DNA to function, with 20 unpaired bases of the substrate being too long a stretch for this to occur. Strikingly, however, a 10 base mismatched region within a 60 bp substrate was tolerated by the terminase, although 20 mismatched bases essentially eliminated translocation. Thus intact DNA strands per se seem more important for translocation than a requirement for fully base-paired regions.
The other central finding bearing on the packaging mechanism is that discontinuities in short duplex DNA substrates are poorly tolerated by the gp17 terminase, since gaps 1 or 20 bases in length either inhibited or essentially blocked packaging, respectively. Single nicks also inhibited packaging. Furthermore, an FCS analysis of 100 bp nicked or 20-base gapped DNAs revealed no significant steady-state association of these molecules with the terminase-prohead complex; implying these substrates were not bound the motor. At first sight these observations might appear singular to the T4 packaging machinery. However, observations in other well established phage systems are also consistent with these results. Thus although sequence-specific nicks are present in the encapsidated phage T5 DNA 18; 41, and to a lesser extent the T7 DNA 42, these nicks likely are introduced into the DNA during packaging, and so the terminase-nuclease may be tolerant of self-generated nicks. Recent work demonstrated that the φ29 system was capable of packaging nicked, but not gapped DNA 20: however these observations were of full length (19.3 kb) φ29 DNA and are thus consistent with our finding that the T4 motor can readily package nicked DNA 500 bp or longer. Additionally, while the precise lengths of the gaps in the studies on the φ29 system were not presented, the current work suggests between 1 and 20 bases represents the length capable of blocking DNA translocation by the T4 motor. The observed dependence of phage T4 concatemeric DNA packaging upon ligase activity in vivo could be due to gaps rather than nicks, or due to terminase interaction with DNA repair proteins bound to nicks that disengage the packaging motor 21, thus is not inconsistent with our in vitro observations here.
The structural demands of gp17 for the substrate are relatively strict, in that nicks, gaps, single-strand regions, hairpins and hybrid DNA-RNA duplex regions are all impediments to translocation. This is of biological significance, as the T4 genome that is a substrate for packaging in vivo is also actively undergoing replication, recombination and late transcription, and so such structures are likely to be encountered frequently by the packaging complex. Indeed, initiation of late transcription itself was shown to be dependent on the presence of discontinuities in the T4 genome 43. The necessity of intact DNA for gp17 function could allow the terminase to effectively scan the incoming genome, such that any duplex interruption is sufficient to cause a halt to packaging, at least long enough for appropriate repair enzymes to act. Consistent with this, a recent single molecule optical tweezers study of T4 packaging revealed the gp17 motor was prone to ‘slips’ and ‘pauses’ during packaging, even on a largely intact DNA substrate 28. Any structural defect in DNA could then likely extend the duration of such naturally occurring pauses, providing sufficient opportunity for the repair enzymes to restore genome integrity.
Other results from the Smith laboratory 28 established that a T4 packasome actively translocating DNA at the expense of ATP hydrolysis was a stable and highly processive entity. This is not necessarily inconsistent with the observation that the T4 system is also prone to slips and pauses. Once even a few tens of base-pairs of packaged DNA had accumulated inside the prohead and had compacted to accommodate further incoming material, the resulting DNA structure could serve to anchor the remaining DNA to the prohead-terminase complex. Even if the gp17 were to undergo spontaneous (or defect-induced) pauses in such a scenario, minimal extrusion of the DNA or dissociation of the packasome would be expected, accounting for the observed processivity. A corollary of the above model is that only very small molecules would be free to dissociate easily from a stalled gp17-prohead complex. This scenario can neatly account for the failure of short molecules with 20-base single strand overhangs or hairpin ends to be fully encapsidated; the length dependence of the inhibitory effect of a DNA nick, as well as FCS evidence that small substrates inhibitory to translocation fail to remain stably associated with the prohead. Since the T4 system readily packages small molecules, the combination of both experimental approaches presented here clearly facilitates analysis of these potentially new aspects of the phage packaging mechanism.
It is now unlikely that DNA packaging is carried out by a rotary motor 31; 32; instead the terminase mechanism may well be more closely related to that of helicases that appear to generate force through the linear motion 7; 44, as already established for the phage φ6 motor: one which translocates ssRNA 45. Consistent with this, our observations showing the importance of intact duplex strands for DNA translocation support a torsional DNA packaging mechanism, one form of which was suggested previously 46. Based on this and the current work, as well as recent studies establishing that the portal of the phage SPP1 system engages with DNA during packaging 47, we propose that the duplex is translocated by a compression and release mechanism that is also dependent upon storing torsional energy in the duplex. Figure 7 presents an overview of how this could occur. In this model energy would be introduced into the DNA duplex by a linear terminase motor operating by a helicase-like, ATP-driven flexible arm; one that engages the DNA and introduces a forward translocation force into the duplex. The central distinguishing feature of the proposed mechanism is that DNA binding by the gp20 portal coupled with the gp17-manifested power stroke induces a transient compression in the DNA through local structural changes in the dimensions of the duplex. Sufficient compression would then promote release of the translocated segment via entry into the prohead. Upon encountering discontinuities in duplex DNA, however, the gp17-generated force would be dissipated before translocation - at least on very small molecules. Conversely, if anchored within the prohead, the nick-bearing region on longer molecules could be recycled through the terminase multimer until a productive motor power step re-engages and encapsidates it.
This model thus neatly accommodates our data of the inhibitory effects of nicks and other discontinuities on the packaging of short molecules. In addition, a key prediction of the model, namely that the local dimensions of the duplex are altered by the power stroke of the terminase motor, is currently being tested by energy transfer analysis of double fluorescent tagged DNA substrates in both translocating and in stalled motor complexes. The fact that structural changes to DNA central to transcription, introduced by a motor protein via a DNA scrunching mechanism, were detected by similar experimental means 48; 49 suggests the feasibility of testing the proposed torsional mechanism in the same way.
Experimental Procedures
Enzymes and chemicals
Nicking restriction endonuclease N.Bsm I was obtained from New England Biolabs. Taq DNA polymerase, S1 nuclease, DNA ligase, DNase I, RNase A and Proteinase K were all used according to the manufacturers’ instructions. Standard cloning and electrophoresis techniques were used throughout 50.
Purification of terminase components
Unmodified full-length gp17 was purified as described 51. The concentration and purity was determined by the Bradford assay and SDS PAGE analysis. Expanded proheads were obtained from extracts of P301 E. coli cells infected for 1 hr at 37 °C with T4[16amN66 17amA465 13amE111 rIIA(ΔH88)] essentially as described 12; 52. The purity of the prohead preparations was assessed by SDS PAGE analysis, and amounts calibrated by SDS-PAGE comparison with the major capsid protein of CsCl-purified phage T4 particles.
Oligo Sequences
The following oligonucleotides were used: pET-UP (5′ GCA TCG TGG TGT CAC GCT CG), pET-DN (5′ GCT TTT TTG CAC AAC ATG GG), pET-20-DN (5′ CGA GCG TGA CAC CAC GAT GC), pET-100-DN (5′ GCT TTT TTG CAC AAC ATG GG), 60-UP (5′ CAG CTC CGG TTC CCA ACG ATC AAG GCG AGT TAC ATG ATC CCC CAT GTT GTG CAA AAA AGC), 80-UP (5′ TCG TTT GGT ATG GCT TCA TTC AGC TCC GGT TCC CAA CGA TCA AGG CGA GTT ACA TGA TCC CCC ATG TTG TGC AAA AAA GC), 100-DN (5′ GCT TTT TTG CAC AAC ATG GGG GAT CAT GTA ACT CGC CTT GAT CGT TGG GAA CCG GAG CTG AAT GAA GCC ATA CCA AAC GAC GAG CGT GAC ACC ACG ATG C), HP-UP (5′ TCG TTT GGT ATG GCT TCA TTC AGC TCC GGT TCC CAA CGA TCC CCC ATC GTT GGG AAC CGG AGC TG), pET-40-DN (5′ AAT GAA GCC ATA CCA AAC GA), 60-ss-0 (5′ GCT TTT TTG CAC AAC ATG GGG GAT CAT GTA ACT CGC CTT GAT CGT TGG GAA CCG GAG CTG), 60-ss-10 (5′ GCT TTT TTG CAC AAC ATG GGG GAT CTA CAT TGA GCC CTT GAT CGT TGG GAA CCG GAG CTG), 60-ss-20 (5′ GCT TTT TTG CAC AAC ATG GGC CTA GTA CAT TGA GCG GAA CAT CGT TGG GAA CCG GAG CTG), pET-60D-UP (5′ GTT CCG CTC AAT GTA CTA GG) and an RNA oligo r-pET-DN (5′ CGA GCG UGA CAC CAC GAU GC). Oligos used for constructing substrates were purified from acrylamide gels. Further primers were used that were designed to generate 200 or 500 bp PCR fragments encompassing the unique N.BsmI site (GAATGC) at position 1840 of pL16 9; 53. For FCS experiments pET-UP was also obtained (from Invitrogen) with a 6-carbon linker ending in a primary amine group attached at the 5′ end. The modified oligo was purified by ethanol precipitation and taken up in freshly prepared 0.2 M Na2CO3. This was mixed with an equal volume of 5-(and-6) carboxyrhodamine 6G succinimidyl ester (Invitrogen Molecular Probes) dissolved in DMF (1 mg/mL) and the tube rocked gently overnight in the dark at room temperature. Free dye was removed by passage through NAP-5 columns (GE Healthcare) and the conjugated dye-oligo eluted with water. The labeled oligo was purified from PAGE gels and stored in water at -20° C. Amounts were determined by OD260 measurements.
DNA substrates
The sequence from positions 4103 - 4202 in pL16 9; 53, adjacent to the unique PstI site, was obtained by PCR with pET-UP and pET-DN and was used as a standard 100 bp substrate. Most of the other constructs used were based on this sequence. Additionally, pL16 itself was cut with appropriate restriction enzymes. Purified λ DNA was from New England Biolabs. Linear 4063 bp M segment dsRNA of phage φ6 was gel purified from RNA extracted from highly purified phage particles that were a generous gift of Leonard Mindich, Public Health Research Institute, N.Y. Nicked DNA molecules were obtained by incubating an appropriate PCR fragment with N.BsmI followed by heat denaturation and ethanol precipitation. Other short substrates were assembled from the oligos listed above. Molecules with 20 or 40 base single-strand extensions were assembled from 100-DN + 80-UP or 100-DN + 60-UP respectively. The hairpin-containing molecules used HP-UP either alone or pre-incubated with Klenow polymerase and dNTP’s. A 20 bp DNA duplex was formed from pET-UP and pET-20-DN, while the DNA-RNA hybrid form was obtained using pET-UP and r-pET-20-DN. Other substrates were formed with 60-UP and either 60-ss-0 or 60-ss-20 to give constructs with 0 or 20 centrally located mismatches (A-A, C-C, G-G or T-T in all cases). The D-loop substrate was assembled from 60-UP, 60-ss-20 and pET-60D-UP. The 100 bp nicked DNA came from pET-UP, 80 UP and 100-DN: the 20 base gapped variant was obtained by substituting 60-UP for 80-UP. All these substrates were assembled by mixing equimolar amounts of the relevant oligos together in a solution comprising 10 mM Tris-HCl pH 8.0, 50 mM NaCl, 0.5 mM EDTA. This was heated to 95 °C and al lowed to cool slowly. The annealed constructs were stored at -20 °C.
Packaging Assays and Gel Analyses
The reaction buffer comprised 50 mM Tris-HCl pH 7.5, 6 mM MgCl2, 100 mM NaCl, 1.5 mM spermidine, 0.1 mg/ml BSA, 1.5 mM DTT, 1.5 mM ATP, 5 % PEG (Fluka, 20k), 50 nM gp17 (monomer), 10-20 nM DNA substrate and 1-10 nM purified proheads: typically in 16 μL volumes. (Phage λ DNA was pre-heated to 65 °C and rapidly quenched on ice-water immediately before use, to ensure complete melting of the cos site ends.) Reaction tubes were incubated 1 hr at room temperature: 1 μL of a 5 mg/ml DNase I was then added and tubes incubated a further 30 min at 37°C. A 1:1:1 mix of 0.5 M EDTA, 5 mg/ml proteinase K and 10 % SDS (3μL) was added, and tubes incubated at 65° C for 30 min. Reactions were resolved on agarose gels or on 7.5 % (37.5:1) polyacrylamide gels developed in TBE buffer. Bands were detected by EtBr staining and images obtained and quantitated on a UVP Epichem3 darkroom apparatus. Denaturing acrylamide gels were prepared in 50 mM NaOH, 1 mM EDTA: these components also comprised the running solution. Samples were prepared by addition of an equal volume of 0.1 M NaOH, loaded directly onto the gels, and electrophoresed at low voltage (<5 V/cm) to prevent overheating. Gels were washed 2-3 times in 0.1 M Tris-HCl pH 7.5 and stained with 0.1 μg/ml EtBr dissolved in the same.
FCS Analysis
Correlation spectroscopy was performed with an Alba FCS system (ISS, Il) coupled to an Olympus IX71 microscope. R6G end-labeled DNAs were excited using 5 μW of 514.5 nm light from an Ar+ laser, which was expanded then reflected off a Z514RDC dichroic mirror (Chroma, Vt) to fill the back aperture of a 40× 1.2 N.A. water immersion objective. R6G fluorescence was focused through a 50 μm pinhole, filtered with a 530lp longpass filter (Chroma) and imaged onto an avalanche photodiode. Raw intensity data were collected with a sampling frequency of 50 kHz, and autocorrelated with software provided by the manufacturer. Packaging reactions were monitored in real-time by taking FCS data at 20 s intervals for 30 min. For curve fitting the time-dependent autocorrelation function G(τ), the following biophysical model was used:
where GD(τ)=(1+4 ·D·τ/w0)-1 ·(1+4·D·τ/z0)-1/2. In the above expression, the first term describes the fluorophore blinking kinetics (i.e., triplet state formation and collisional quenching), with characteristic time τT and fraction T (less than 0.2 with 5 μW power). The second term represents the variation in intensity fluctuations as m fluorescent species with diffusion coefficient D traverse the 3D Gaussian confocal volume, defined by radius w0 and half-axis height z0. The average number and brightness of each species is denoted by n and b, respectively. A two-species model (free DNA and DNA inside proheads) was used to fit the FCS curves for packaging reactions (+ATP). During fitting procedures, a negative FCS control lacking ATP was used to determine the number of R6G-labeled molecules: this number was then used to constrain the fitting parameters for FCS data obtained from ATP reactions; such that the total number of DNAs (n1b1 + n2b2) was conserved. All fitting was done with software code written in Igor Pro (Wavemetrics).
Acknowledgements
We thank Diana Oram and Julienne Mullaney for critical reading of the manuscript and helpful suggestions, and Douglas Smith for discussions and communication of results prior to publication. We thank the staff at the University of Maryland Center for Fluorescence Studies for support and for access to equipment, and Leonard Mindich for the generous gift of phage φ6. This work was supported by NIH grant AI11676.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Feiss M, Catalano C. Bacteriophage Lambda Terminase and the Mechanisms of Viral DNA Packaging. In: Catalano C, editor. Viral Genome Packaging Machines: Genetics, Structure, and Mechanism. Springer Press; New York: 2005. pp. 5–39. [Google Scholar]
- 2.Guo P, Lee TJ. Viral nanomotors for packaging of dsDNA and dsRNA. Mol Microbiol. 2007;64:886–903. doi: 10.1111/j.1365-2958.2007.05706.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kreuzer KN. Interplay between DNA replication and recombination in prokaryotes. Annu Rev Microbiol. 2005;59:43–67. doi: 10.1146/annurev.micro.59.030804.121255. [DOI] [PubMed] [Google Scholar]
- 4.Black LW. DNA packaging in dsDNA bacteriophages. Annu Rev Microbiol. 1989;43:267–92. doi: 10.1146/annurev.mi.43.100189.001411. [DOI] [PubMed] [Google Scholar]
- 5.Fujisawa H, Morita M. Phage DNA packaging. Genes Cells. 1997;2:537–45. doi: 10.1046/j.1365-2443.1997.1450343.x. [DOI] [PubMed] [Google Scholar]
- 6.Sun S, Kondabagil K, Gentz PM, Rossmann MG, Rao VB. The structure of the ATPase that powers DNA packaging into bacteriophage T4 procapsids. Mol Cell. 2007;25:943–9. doi: 10.1016/j.molcel.2007.02.013. [DOI] [PubMed] [Google Scholar]
- 7.Draper B, Rao VB. An ATP hydrolysis sensor in the DNA packaging motor from bacteriophage T4 suggests an inchworm-type translocation mechanism. J Mol Biol. 2007;369:79–94. doi: 10.1016/j.jmb.2007.03.019. [DOI] [PubMed] [Google Scholar]
- 8.Rao VB, Black LW. Cloning, overexpression and purification of the terminase proteins gp16 and gp17 of bacteriophage T4. Construction of a defined in-vitro DNA packaging system using purified terminase proteins. J Mol Biol. 1988;200:475–88. doi: 10.1016/0022-2836(88)90537-2. [DOI] [PubMed] [Google Scholar]
- 9.Lin H, Simon MN, Black LW. Purification and characterization of the small subunit of phage T4 terminase, gp16, required for DNA packaging. J Biol Chem. 1997;272:3495–501. doi: 10.1074/jbc.272.6.3495. [DOI] [PubMed] [Google Scholar]
- 10.Black LW. DNA packaging and cutting by phage terminases: control in phage T4 by a synaptic mechanism. Bioessays. 1995;17:1025–30. doi: 10.1002/bies.950171206. [DOI] [PubMed] [Google Scholar]
- 11.Valpuesta JM, Carrascosa JL. Structure of viral connectors and their function in bacteriophage assembly and DNA packaging. Q Rev Biophys. 1994;27:107–155. doi: 10.1017/s0033583500004510. [DOI] [PubMed] [Google Scholar]
- 12.Black LW, Peng G. Mechanistic coupling of bacteriophage T4 DNA packaging to components of the replication-dependent late transcription machinery. J Biol Chem. 2006;281:25635–43. doi: 10.1074/jbc.M602093200. [DOI] [PubMed] [Google Scholar]
- 13.Kondabagil KR, Rao VB. A critical coiled coil motif in the small terminase, gp16, from bacteriophage T4: insights into DNA packaging initiation and assembly of packaging motor. J Mol Biol. 2006;358:67–82. doi: 10.1016/j.jmb.2006.01.078. [DOI] [PubMed] [Google Scholar]
- 14.Lin H, Rao VB, Black LW. Analysis of capsid portal protein and terminase functional domains: interaction sites required for DNA packaging in bacteriophage T4. J Mol Biol. 1999;289:249–60. doi: 10.1006/jmbi.1999.2781. [DOI] [PubMed] [Google Scholar]
- 15.Kuebler D, Rao VB. Functional analysis of the DNA-packaging/terminase protein gp17 from bacteriophage T4. J Mol Biol. 1998;281:803–14. doi: 10.1006/jmbi.1998.1952. [DOI] [PubMed] [Google Scholar]
- 16.Rentas FJ, Rao VB. Defining the bacteriophage T4 DNA packaging machine: evidence for a C-terminal DNA cleavage domain in the large terminase/packaging protein gp17. J Mol Biol. 2003;334:37–52. doi: 10.1016/j.jmb.2003.09.028. [DOI] [PubMed] [Google Scholar]
- 17.Fujisawa H, Hamada K, Shibata H, Minagawa T. On the molecular mechanism of DNA translocation during in vitro packaging of bacteriophage T3 DNA. Virology. 1987;161:228–33. doi: 10.1016/0042-6822(87)90189-9. [DOI] [PubMed] [Google Scholar]
- 18.Hayward GS, Smith MG. The chromosome of bacteriophage T5. II. Arrangement of the single-stranded DNA fragments in the T5 + and T5st(O) chromosomes. J Mol Biol. 1972;63:397–407. doi: 10.1016/0022-2836(72)90436-6. [DOI] [PubMed] [Google Scholar]
- 19.Pearson RK, Fox MS. Effects of DNA heterologies on bacteriophage lambda packaging. Genetics. 1988;118:5–12. doi: 10.1093/genetics/118.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moll WD, Guo P. Translocation of nicked but not gapped DNA by the packaging motor of bacteriophage phi29. J Mol Biol. 2005;351:100–7. doi: 10.1016/j.jmb.2005.05.038. [DOI] [PubMed] [Google Scholar]
- 21.Zachary A, Black LW. DNA ligase is required for encapsidation of bacteriophage T4 DNA. J Mol Biol. 1981;149:641–58. doi: 10.1016/0022-2836(81)90351-x. [DOI] [PubMed] [Google Scholar]
- 22.Kemper B, Janz E. Function of gene 49 of bacteriophage T4. I. Isolation and biochemical characterization of very fast-sedimenting DNA. J Virol. 1976;18:992–9. doi: 10.1128/jvi.18.3.992-999.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hsiao CL, Black LW. DNA packaging and the pathway of bacteriophage T4 head assembly. Proc Natl Acad Sci U S A. 1977;74:3652–6. doi: 10.1073/pnas.74.9.3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zachary A, Black LW. Topoisomerase II and other DNA-delay and DNA-arrest mutations impair bacteriophage T4 DNA packaging in vivo and in vitro. J Virol. 1986;60:97–104. doi: 10.1128/jvi.60.1.97-104.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chemla YR, Aathavan K, Michaelis J, Grimes S, Jardine PJ, Anderson DL, Bustamante C. Mechanism of force generation of a viral DNA packaging motor. Cell. 2005;122:683–92. doi: 10.1016/j.cell.2005.06.024. [DOI] [PubMed] [Google Scholar]
- 26.Rickgauer JP, Fuller DN, Grimes S, Jardine PJ, Anderson DL, Smith DE. Portal motor velocity and internal force resisting viral DNA packaging in bacteriophage phi29. Biophys J. 2008;94:159–67. doi: 10.1529/biophysj.107.104612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fuller DN, Raymer DM, Rickgauer JP, Robertson RM, Catalano CE, Anderson DL, Grimes S, Smith DE. Measurements of single DNA molecule packaging dynamics in bacteriophage lambda reveal high forces, high motor processivity, and capsid transformations. J Mol Biol. 2007;373:1113–22. doi: 10.1016/j.jmb.2007.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fuller DN, Raymer DM, Kottadiel VI, Rao VB, Smith DE. Single phage T4 DNA packaging motors exhibit large force generation, high velocity, and dynamic variability. Proc Natl Acad Sci U S A. 2007;104:16868–73. doi: 10.1073/pnas.0704008104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hendrix RW. Symmetry mismatch and DNA packaging in large bacteriophages. Proc Natl Acad Sci U S A. 1978;75:4779–83. doi: 10.1073/pnas.75.10.4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simpson AA, Tao Y, Leiman PG, Badasso MO, He Y, Jardine PJ, Olson NH, Morais MC, Grimes S, Anderson DL, Baker TS, Rossmann MG. Structure of the bacteriophage phi29 DNA packaging motor. Nature. 2000;408:745–50. doi: 10.1038/35047129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hugel T, Michaelis J, Hetherington CL, Jardine PJ, Grimes S, Walter JM, Falk W, Anderson DL, Bustamante C. Experimental Test of Connector Rotation during DNA Packaging into Bacteriophage phi29 Capsids. PLoS Biol. 2007;5:e59. doi: 10.1371/journal.pbio.0050059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baumann RG, Mullaney J, Black LW. Portal fusion protein constraints on function in DNA packaging of bacteriophage T4. Mol Microbiol. 2006;61:16–32. doi: 10.1111/j.1365-2958.2006.05203.x. [DOI] [PubMed] [Google Scholar]
- 33.Sabanayagam CR, Oram M, Lakowicz JR, Black LW. Viral DNA Packaging Studied by Fluorescence Correlation Spectroscopy. Biophys J. 2007;93:L17–L19. doi: 10.1529/biophysj.107.111526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maiti S, Haupts U, Webb WW. Fluorescence correlation spectroscopy: diagnostics for sparse molecules. Proc Natl Acad Sci U S A. 1997;94:11753–7. doi: 10.1073/pnas.94.22.11753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mullaney JM, Thompson RB, Gryczynski Z, Black LW. Green fluorescent protein as a probe of rotational mobility within bacteriophage T4. J Virol Methods. 2000;88:35–40. doi: 10.1016/s0166-0934(00)00166-x. [DOI] [PubMed] [Google Scholar]
- 36.Kondabagil KR, Zhang Z, Rao VB. The DNA translocating ATPase of bacteriophage T4 packaging motor. J Mol Biol. 2006;363:786–99. doi: 10.1016/j.jmb.2006.08.054. [DOI] [PubMed] [Google Scholar]
- 37.Malys N, Chang DY, Baumann RG, Xie D, Black LW. A bipartite bacteriophage T4 SOC and HOC randomized peptide display library: detection and analysis of phage T4 terminase (gp17) and late sigma factor (gp55) interaction. J Mol Biol. 2002;319:289–304. doi: 10.1016/S0022-2836(02)00298-X. [DOI] [PubMed] [Google Scholar]
- 38.Arnott S, Chandrasekaran R, Millane RP, Park HS. DNA-RNA hybrid secondary structures. J Mol Biol. 1986;188:631–40. doi: 10.1016/s0022-2836(86)80011-0. [DOI] [PubMed] [Google Scholar]
- 39.Kallenbach NR, Berman HM. RNA structure. Q Rev Biophys. 1977;10:138–236. doi: 10.1017/s0033583500000202. [DOI] [PubMed] [Google Scholar]
- 40.Alam TI, Rao VB. The ATPase Domain of the Large Terminase Protein, gp17, from Bacteriophage T4 Binds DNA: Implications to the DNA Packaging Mechanism. J Mol Biol. 2007 doi: 10.1016/j.jmb.2007.12.041. [DOI] [PubMed] [Google Scholar]
- 41.Ponchon L, Boulanger P, Labesse G, Letellier L. The endonuclease domain of bacteriophage terminases belongs to the resolvase/integrase/ribonuclease H superfamily: a bioinformatics analysis validated by a functional study on bacteriophage T5. J Biol Chem. 2006;281:5829–36. doi: 10.1074/jbc.M511817200. [DOI] [PubMed] [Google Scholar]
- 42.Khan SA, Hayes SJ, Wright ET, Watson RH, Serwer P. Specific single-stranded breaks in mature bacteriophage T7 DNA. Virology. 1995;211:329–31. doi: 10.1006/viro.1995.1411. [DOI] [PubMed] [Google Scholar]
- 43.Williams KP, Kassavetis GA, Herendeen DR, Geiduschek EP. Regulation of Late Gene Expression. In: Karam JD, editor. Molecular Biology of Bacteriophage T4. ASM Press; Washington DC: 1994. pp. 161–175. [Google Scholar]
- 44.Velankar SS, Soultanas P, Dillingham MS, Subramanya HS, Wigley DB. Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell. 1999;97:75–84. doi: 10.1016/s0092-8674(00)80716-3. [DOI] [PubMed] [Google Scholar]
- 45.Mancini EJ, Kainov DE, Grimes JM, Tuma R, Bamford DH, Stuart DI. Atomic snapshots of an RNA packaging motor reveal conformational changes linking ATP hydrolysis to RNA translocation. Cell. 2004;118:743–55. doi: 10.1016/j.cell.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 46.Black LW, Silverman DJ. Model for DNA packaging into bacteriophage T4 heads. J Virol. 1978;28:643–55. doi: 10.1128/jvi.28.2.643-655.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cuervo A, Vaney MC, Antson AA, Tavares P, Oliveira L. Structural rearrangements between portal protein subunits are essential for viral DNA translocation. J Biol Chem. 2007;282:18907–13. doi: 10.1074/jbc.M701808200. [DOI] [PubMed] [Google Scholar]
- 48.Kapanidis AN, Margeat E, Ho SO, Kortkhonjia E, Weiss S, Ebright RH. Initial transcription by RNA polymerase proceeds through a DNA-scrunching mechanism. Science. 2006;314:1144–7. doi: 10.1126/science.1131399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Revyakin A, Liu C, Ebright RH, Strick TR. Abortive initiation and productive initiation by RNA polymerase involve DNA scrunching. Science. 2006;314:1139–43. doi: 10.1126/science.1131398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Edition. Cold Spring Harbor Laboratory Press; 1989. edit. [Google Scholar]
- 51.Baumann RG, Black LW. Isolation and characterization of T4 bacteriophage gp17 terminase, a large subunit multimer with enhanced ATPase activity. J Biol Chem. 2003;278:4618–27. doi: 10.1074/jbc.M208574200. [DOI] [PubMed] [Google Scholar]
- 52.Rao VB, Black LW. DNA packaging of bacteriophage T4 proheads in vitro. Evidence that prohead expansion is not coupled to DNA packaging. J Mol Biol. 1985;185:565–78. doi: 10.1016/0022-2836(85)90072-5. [DOI] [PubMed] [Google Scholar]
- 53.Lin H, Black LW. DNA requirements in vivo for phage T4 packaging. Virology. 1998;242:118–27. doi: 10.1006/viro.1997.9019. [DOI] [PubMed] [Google Scholar]