Abstract
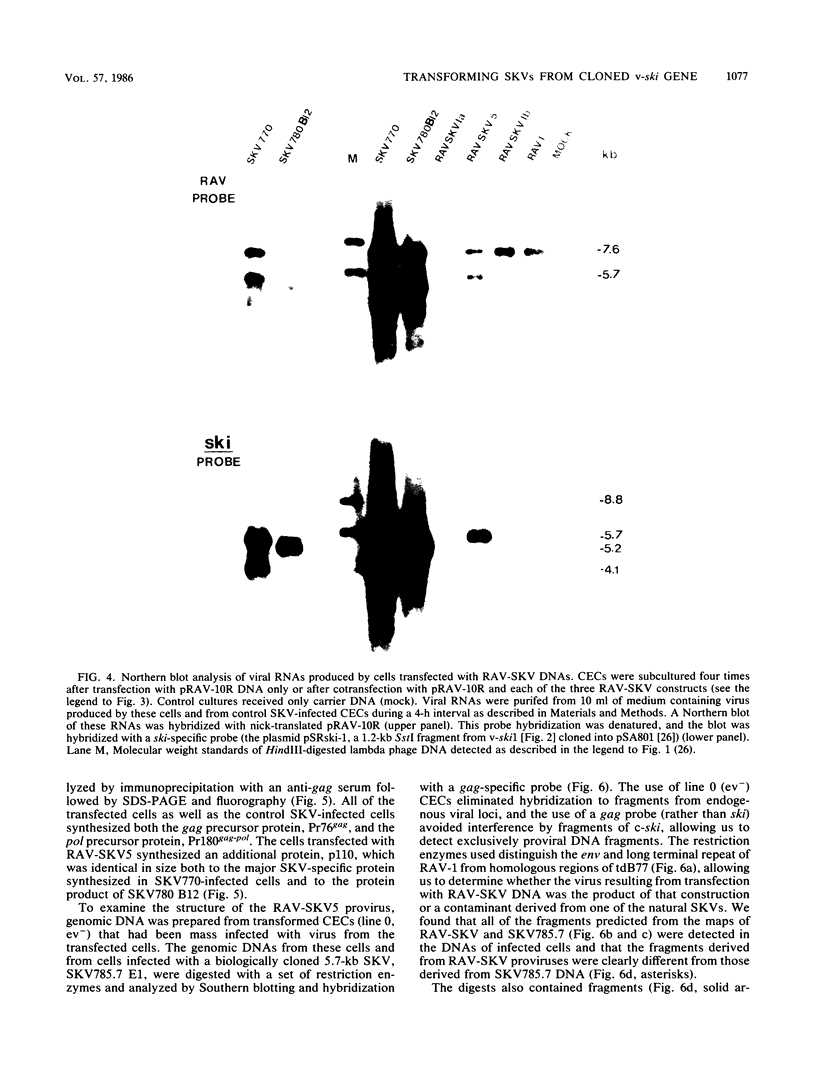
The Sloan-Kettering viruses (SKVs) are replication-defective retroviruses that transform avian cells in vitro. Each of the three SKV isolates is a mixture of viruses with genomes ranging in size from 4.1 to 8.9 kilobases (kb) with a predominant genome of 5.7 kb. Using a cDNA representing a sequence, v-ski, that is SKV specific and held in common by the multiple SKV genomes, we generated a restriction map of the 5.7-kb SKV genome and molecularly cloned a ski-containing fragment from SKV proviral DNA. Southern hybridization and sequence analysis showed that the cloned DNA fragment consisted of the 1.3-kb ski sequence embedded in the p19gag sequence and followed by the remaining 5' half of the gag gene and small portions of both the pol and env genes. A large deletion encompassing the 3' half of gag and the 5' 80% of pol was mapped to a position about 1 kb downstream from the 3' ski-gag junction. To determine whether the cloned ski sequence had transforming activity, the ski-containing fragment and a cloned Rous-associated virus 1 (RAV-1) genome were used to construct an analog of the 5.7-kb SKV genome, RAV-SKV. Cotransfection of chicken embryo cells with RAV-SKV and RAV-1 yielded foci of transformed cells whose morphology was identical to that induced by the natural SKVs. The transformed transfected cells produced transforming virus with a 5.7-kb ski-containing genome and synthesized a gag-containing polyprotein of 110 kilodaltons (kDa). Several nonproducer clones of RAV-SKV-transformed cells were analyzed, and most were found to synthesize a 5.7-kb SKV RNA and a 110-kDa polyprotein. One clone was found to contain an 8.9-kb SKV RNA, and this clone synthesized a 125-kDa polyprotein. Since both the 5.7- and 8.9-kb genomes and the 110- and 125-kDa polyproteins had been identified in studies on the natural SKVs, the present results not only demonstrate the transforming activity of these individual SKVs but also suggest mechanisms for their generation.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Astrin S. M., Robinson H. L., Crittenden L. B., Buss E. G., Wyban J., Hayward W. S. Ten genetic loci in the chicken that contain structural genes for endogenous avian leukosis viruses. Cold Spring Harb Symp Quant Biol. 1980;44(Pt 2):1105–1109. doi: 10.1101/sqb.1980.044.01.119. [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay P. K., Watanabe S., Temin H. M. Recombination of transfected DNAs in vertebrate cells in culture. Proc Natl Acad Sci U S A. 1984 Jun;81(11):3476–3480. doi: 10.1073/pnas.81.11.3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton W. D., Davis R. W. Screening lambdagt recombinant clones by hybridization to single plaques in situ. Science. 1977 Apr 8;196(4286):180–182. doi: 10.1126/science.322279. [DOI] [PubMed] [Google Scholar]
- Bishop J. M. Cellular oncogenes and retroviruses. Annu Rev Biochem. 1983;52:301–354. doi: 10.1146/annurev.bi.52.070183.001505. [DOI] [PubMed] [Google Scholar]
- Calos M. P., Miller J. H. Transposable elements. Cell. 1980 Jul;20(3):579–595. doi: 10.1016/0092-8674(80)90305-0. [DOI] [PubMed] [Google Scholar]
- Coffin J. M. Structure, replication, and recombination of retrovirus genomes: some unifying hypotheses. J Gen Virol. 1979 Jan;42(1):1–26. doi: 10.1099/0022-1317-42-1-1. [DOI] [PubMed] [Google Scholar]
- Corsaro C. M., Pearson M. L. Enhancing the efficiency of DNA-mediated gene transfer in mammalian cells. Somatic Cell Genet. 1981 Sep;7(5):603–616. doi: 10.1007/BF01549662. [DOI] [PubMed] [Google Scholar]
- DeLorbe W. J., Luciw P. A., Goodman H. M., Varmus H. E., Bishop J. M. Molecular cloning and characterization of avian sarcoma virus circular DNA molecules. J Virol. 1980 Oct;36(1):50–61. doi: 10.1128/jvi.36.1.50-61.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denhardt D. T. A membrane-filter technique for the detection of complementary DNA. Biochem Biophys Res Commun. 1966 Jun 13;23(5):641–646. doi: 10.1016/0006-291x(66)90447-5. [DOI] [PubMed] [Google Scholar]
- Fincham V. J., Neiman P. E., Wyke J. A. Novel nonconditional mutants in the src gene of Rous sarcoma virus: isolation and preliminary characterization. Virology. 1980 May;103(1):99–111. doi: 10.1016/0042-6822(80)90129-4. [DOI] [PubMed] [Google Scholar]
- Fleissner E. Virus-specific antigens in hamster cells transformed by Rous sarcoma virus. J Virol. 1970 Jan;5(1):14–21. doi: 10.1128/jvi.5.1.14-21.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff S. P., Witte O. N., Gilboa E., Rosenberg N., Baltimore D. Genome structure of Abelson murine leukemia virus variants: proviruses in fibroblasts and lymphoid cells. J Virol. 1981 May;38(2):460–468. doi: 10.1128/jvi.38.2.460-468.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes S. H., Shank P. R., Spector D. H., Kung H. J., Bishop J. M., Varmus H. E., Vogt P. K., Breitman M. L. Proviruses of avian sarcoma virus are terminally redundant, co-extensive with unintegrated linear DNA and integrated at many sites. Cell. 1978 Dec;15(4):1397–1410. doi: 10.1016/0092-8674(78)90064-8. [DOI] [PubMed] [Google Scholar]
- Hunter E. The mechanism for genetic recombination in the avian retroviruses. Curr Top Microbiol Immunol. 1978;79:295–309. doi: 10.1007/978-3-642-66853-1_7. [DOI] [PubMed] [Google Scholar]
- Li Y., Turck C. M., Teumer J. K., Stavnezer E. Unique sequence, ski, in Sloan-Kettering avian retroviruses with properties of a new cell-derived oncogene. J Virol. 1986 Mar;57(3):1065–1072. doi: 10.1128/jvi.57.3.1065-1072.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F. L., Sperle K., Sternberg N. Model for homologous recombination during transfer of DNA into mouse L cells: role for DNA ends in the recombination process. Mol Cell Biol. 1984 Jun;4(6):1020–1034. doi: 10.1128/mcb.4.6.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loenen W. A., Brammar W. J. A bacteriophage lambda vector for cloning large DNA fragments made with several restriction enzymes. Gene. 1980 Aug;10(3):249–259. doi: 10.1016/0378-1119(80)90054-2. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McMaster G. K., Carmichael G. G. Analysis of single- and double-stranded nucleic acids on polyacrylamide and agarose gels by using glyoxal and acridine orange. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4835–4838. doi: 10.1073/pnas.74.11.4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purrello M., Balazs I. Direct hybridization of labeled DNA to DNA in agarose gels. Anal Biochem. 1983 Feb 1;128(2):393–397. doi: 10.1016/0003-2697(83)90391-3. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Schwartz D. E., Tizard R., Gilbert W. Nucleotide sequence of Rous sarcoma virus. Cell. 1983 Mar;32(3):853–869. doi: 10.1016/0092-8674(83)90071-5. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Stavnezer E., Gerhard D. S., Binari R. C., Balazs I. Generation of transforming viruses in cultures of chicken fibroblasts infected with an avian leukosis virus. J Virol. 1981 Sep;39(3):920–934. doi: 10.1128/jvi.39.3.920-934.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanstrom R., Varmus H. E., Bishop J. M. Nucleotide sequence of the 5' noncoding region and part of the gag gene of Rous sarcoma virus. J Virol. 1982 Feb;41(2):535–541. doi: 10.1128/jvi.41.2.535-541.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temin H. M. Origin of retroviruses from cellular moveable genetic elements. Cell. 1980 Oct;21(3):599–600. doi: 10.1016/0092-8674(80)90420-1. [DOI] [PubMed] [Google Scholar]
- Thomas P. S. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5201–5205. doi: 10.1073/pnas.77.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varmus H. E. Form and function of retroviral proviruses. Science. 1982 May 21;216(4548):812–820. doi: 10.1126/science.6177038. [DOI] [PubMed] [Google Scholar]
- Varmus H. E., Quintrell N., Ortiz S. Retroviruses as mutagens: insertion and excision of a nontransforming provirus alter expression of a resident transforming provirus. Cell. 1981 Jul;25(1):23–36. doi: 10.1016/0092-8674(81)90228-2. [DOI] [PubMed] [Google Scholar]
- Weiss S. R., Varmus H. E., Bishop J. M. The size and genetic composition of virus-specific RNAs in the cytoplasm of cells producing avian sarcoma-leukosis viruses. Cell. 1977 Dec;12(4):983–992. doi: 10.1016/0092-8674(77)90163-5. [DOI] [PubMed] [Google Scholar]