Abstract
Recently, we found an interaction between adenomatous polyposis coli (APC) and DNA polymerase β (pol-β) and showed that APC blocks strand-displacement synthesis of long-patch base excision repair (LP-BER) however, the mechanism is not clear. Using an in vivo LP-BER assay system, we now show that the LP-BER is higher in APC−/− cells than in APC+/+ cells. In addition to pol-β, the pull-down experiments showed that the full-length APC also interacted with flap endonuclease 1 (Fen-1). To further characterize the interaction of APC with pol-β and Fen-1, we performed a domain-mapping of APC and found that both pol-β and Fen-1 interact with a 138-amino acids peptide from the APC at the DRI-domain. Our functional assays showed that APC blocks pol-β-mediated 1-nucleotide (1-nt) as well as strand-displacement synthesis of reduced abasic, nicked-, or 1-nt gapped-DNA substrates. Our further studies demonstrated that APC blocks 5′-flap endonuclease as well as 5′-3′ exonuclease activity of Fen-1 resulting in the blockage of LP-BER. From these results we concluded that APC can have three different effects in the LP-BER pathway. First, APC can block pol-β-mediated 1-nt incorporation and strand-displacement synthesis. Second, APC can block LP-BER by blocking coordinated formation and removal of the strand-displaced flap. Third, APC can block LP-BER by blocking “Hit and Run” synthesis. These studies will have important implications of APC in DNA damage-induced carcinogenesis and chemoprevention.
The genomic stability of an organism is dependent upon numerous DNA metabolic proteins. These proteins coordinate in a very orderly fashion to ensure that DNA repair, replication and recombination occur with high fidelity. However, many proteins involved in DNA metabolism have been linked with human diseases including cancer (1), premature aging syndrome (2), Huntington's disease, Friederich's ataxia and myotonic dystrophy (3). The modification or loss of DNA bases can alter the coding specificity leading to mutations, which are a vital source for genetic variations and major cause of human diseases. To deal with this type of situation, biological systems have evolved DNA repair mechanisms to protect genetic stability and integrity for the survival of organisms. DNA repair systems efficiently remove damaged DNA via several different pathways. Abasic DNA lesions account for a large proportion of the total damage and are mainly corrected by the base excision repair (BER) pathway. BER is mediated through two sub-pathways depending upon the size of the repair gap and the enzymes involved. In mammalian cells, single base lesions are repaired by “single-nucleotide base excision repair” also referred to as short-patch (SP)-BER (4) and “multi-nucleotide” also referred to as long-patch (LP)-BER (5, 6).
There are two types of DNA glycosylases – monofunctional and bifunctional. The bifunctional DNA glycosylases have additional AP lyase activity. Monofunctional DNA glycosylases cleave only the glycosidic bond between N and C′ and then protect the abasic site until AP-endonuclease (APE-1) cleaves the DNA backbone at the 5′-end of the AP-site (7). The BER process is initiated by a DNA glycosylase that recognizes a damaged base and cleaves a glycosidic bond between the sugar and the base to establish an apurinic/apyrimidinc (AP) or abasic-site. Subsequently, AP endonuclease-1 (APE1) cleaves the DNA backbone at the 5′-end of the AP-site. Depending upon the type of DNA damage, the abasic-site is repaired either by the SP- or LP-BER. During SP-BER, DNA polymerase β (pol-β) removes the 5′-deoxyribose phosphate intermediate by deoxyribose phosphate lyase (dRPase) activity to yield a 5′-phosphorylated gapped DNA strand (4). Pol-β then incorporates the correct base at the site of the damaged base and DNA ligase-I seals the gap (8). If the AP-site is oxidized or reduced, then the repair is completed by LP-BER. In this case, pol-β incorporates 1 nucleotide (1-nt). The 5′-overhang is removed by flap endonuclease-1 (Fen-1), and finally the nick is sealed by DNA ligase I (5, 9, 10). Without the removal of the flap, LP-BER cannot be accomplished. Thus, Fen-1 is a critical enzyme in the LP-BER pathway (11, 12). Fen-1 also has a 5′-3′ exonuclease activity which is required for the removal of displaced RNA-DNA primers synthesized by DNA polymerase α-primase during discontinuous lagging-strand replication. Fen-1 is also required for the removal of damaged DNA fragments during various DNA-repair pathways (5, 9, 10, 13). In vitro, Fen-1 can remove abasic-sites (14), as well as a diverse group of flap adducts such as cisplatin derivatives (15). Fen-1 interacts with several DNA repair proteins such as proliferating cell nuclear antigen (PCNA), replication protein A (RPA), replication factor-C (RF-C), DNA polymerase δ and ε (pol-δ and ε) and DNA ligase I (12). The role of Fen-1 has been implicated in maintaining genomic stability (11, 16-19), and as a novel tumor suppressor gene (20).
Mutations in adenomatous polyposis coli (APC) gene are the earliest events in the development of colorectal carcinogenesis. The APC gene contains 8535 nucleotides which encodes for 2843 amino acids or 312 kD protein. Most of the somatic mutations are clustered between codons 1284 and 1580, also known as the mutation cluster region (MCR) (21-24). APC plays a diversified role in a broad spectrum of functions ranging from cell adhesion to cell migration, Wnt/β-catenin signaling (24, 25), cell cycle control (24), apoptosis regulation (26-28), and chromosomal segregation (29). APC is present in both cytosol and nucleus (30). One of the roles of APC in the nucleus is to regulate β-catenin levels, which bind to Tcf/Lef transcription factors and regulate transcriptional activity of number of target genes (30, 31). Our recent findings implicate another role of nuclear APC in the regulation of DNA repair (32, 33). We have previously shown that APC gene expression is induced upon exposure to DNA-damaging agents in colon cancer cells (34, 35), suggesting a possibility of the interaction of APC with DNA repair machinery. Recently, we suggested that APC may interfere with pol-β-mediated strand-displacement synthesis of LP-BER (32). We described that increased and decreased levels of APC in different breast cancer cell lines was associated with a decrease or increase in LP-BER activity. Our studies suggested a role of increased level of APC in compromised LP-BER in benzo[a]pyrene (B[a]P) and cigarette smoke condensate-induced transformation of pre-malignant breast epithelial cells (33). However, the mechanism by which APC blocks LP-BER is still not clear. Previous studies have shown that pol-β-mediated strand-displacement synthesis of LP-BER is coordinated and stimulated by Fen-1 (36, 37). Whether APC has any effect on Fen-1-coordinated strand-displacement synthesis is not known. In the present investigation we have examined the mechanism by which APC plays a role in LP-BER. We hypothesize that APC plays a distinct role in LP-BER through its interaction with pol-β and Fen-1. When APC interacts with pol-β, it blocks pol-β-mediated 1-nt as well as strand-displacement synthesis. We have further shown that the interaction of APC with Fen-1 blocks 5′-flap endonuclease activity of Fen-1 and thus blocks strand-displacement synthesis of LP-BER. Our results also showed that APC blocks 5′-3′ exonuclease activity of Fen-1 and thus blocks the “Hit and Run” mechanism of LP-BER, in which Fen-1 excises a nucleotide at the ′ side of a nick and creates a gap that is filled by pol-β (37).
EXPERIMENTAL PROCEDURES
Maintenance of cells
Human colon cancer cell lines HCT-116 (expressing wild-type APC), LS411N and LoVo (expressing mutant APC) were grown in McCoy's 5a medium at 37°C under a humidified atmosphere of 5% CO2. In each case, the medium was supplemented with 10% fetal bovine serum, 100 U/ml of penicillin, and 100 μg/ml of streptomycin.
APC peptides
An APC peptide of 20-amino acids in length (1250-KVSSINQETIQTYCVEDTPI-1269) was synthesized at the Protein Chemistry and Biomarkers core facility at the ICBR of the University of Florida. They represent the DRI-domain of the wild-type APC (APCwt), or a mutated form of the APC, APC(Q-A,I-A,Y-A) or APC(I-A,Y-A) in which amino acids Q1256, I1259 and Y1262 were replaced with alanine (A) (1250-KVSSINAETAQTACVEDTPI-1269 or1250-KVSSINQETAQTACVEDTPI-1269).
Stable APC-knockdown HCT116 cell line
A vector-based overexpression of the double-stranded RNA (dsRNA) that is homologous to the APC gene was used in these studies. The oligonucleotides designed to contain a nucleotide sequence specific for the APC mRNA (oligonucleotides 5′-GATCCGCAACAGAAGCAGAGAGGTTTCAAGAGAACCTCTCTGCTTCTGTTGCTTTTTTGGAAA-′ and 5′-AGCTTTTCCAAAAAAGCAACAGAAGCAGAGAGGTTCTCTTGAAACCTCTCTGCTTCTGTTG CG-′, respectively) were annealed and subcloned into the BamH1 and HindIII sites of the pSilencer 2.1 vector system (Ambion Inc., Austin, TX) to generate pSiRNA-APC (38). A jumbled sequence of this oligonucleotide was used to generate a mutant APC plasmid. The plasmid harboring the insert, named pSiRNA-APC or pSiRNA-APCmut, was transfected into HCT-116 cells using Lipofectamine™ reagent as described by the manufacturer (Invitrogen, Carlsbad, CA). Briefly, HCT-116 cells were plated in 60 mm tissue culture dishes at 60% confluence. The DNA•lipid complex was assembled by mixing 14 μl of Lipofectamine with 300 μl of serum-free medium and 4 μg of pSiRNA plasmids. The DNA•lipid complex was allowed to form at 22°C for 45 min, and then the complex was gently added to the plates in a serum and antibiotics-free environment. About 24 h post-transfection, the medium was replaced with a fresh medium containing 0.6 g/L of G418. The cells were grown for 4-7 days in a G418 selection medium. The stably-transfected clones were grown and confirmed for APC protein levels by Western blot analysis. The HCT-116 cells expressing stable pSiRNA-APC and pSiRNA-APCmut plasmids were named as HCT-116(APC−/−) and HCT-116(APC+/+), respectively.
In vivo BER assay
The pGL2-p21, a closed circular DNA of p21(Waf-1/Cip1) promoter down stream of the luciferase-reporter gene, containing random C-residues was deaminated by 3 M sodium bisulfite in the presence of 50 mM hydroquinone (39, 40). This reaction modifies cytosine into uracil-residues (U-p21P) for SP-BER substrate. The resulting U-p21P was treated with uracil-DNA glycosylase (UDG) and then reduced with 0.1 M sodium borohydride to generate reduced AP-sites (R-p21P) for LP-BER substrate. We transiently transfected R-p21P plasmid using Lipofectamine reagent (Invitrogen, Carlsbad, CA) in stable HCT-116(APC+/+) and HCT-116(APC−/−) (clone-19) cell lines. Briefly, cells were grown to 60-70% confluence in 35 mm tissue culture dishes and transfected with 2.0 μg/ml of R-p21P DNA and 0.5 μg pCMV-β-galactoside (β-gal) plasmid using 7 μl/ml of Lipofectamine reagent. The pCMV-β-gal served as an internal control to correct the differences in the transfection efficiency. After 5 h of transfection once the cells were acclimatized, one set of cells was harvested, and the promoter activity determined at this time point, was considered as zero time point. The medium of the remaining dishes were aspirated and replaced with complete medium supplemented with 10% FBS. Cells were harvested at different time intervals and LP-BER activities were measured by determining the luciferase gene-reporter activity of cellular lysates using a Moonlight™ 3010 Illuminometer (Promega, San Diego, CA).
Immunoprecipitation
Protein-protein interaction studies were carried out using nuclear extract preparations from different colon cancer cell lines. For immunoprecipitation experiments, 150-200 μg nuclear extract protein was pre-cleared by incubating with 2 μg of normal rabbit IgG at 4°C for 2 h. The mixture was rocked for 2 h at 4°C followed by the addition of 40 μl of BSA-blocked protein A-Sepharose 4B beads. Beads were removed by centrifugation, and pre-cleared supernatant was incubated with anti-APC rabbit polyclonal antibody (Bio-Synthesis Inc., Lewisville, TX) at 4°C for 4 h. The immunocomplex was captured on BSA-blocked protein A-Sepharose 4B beads by incubating with the mixture for further 3-4 h at 4°C. Beads containing immunocomplex were washed five-times with a washing buffer containing 20 mM Hepes, pH 7.9, 100 mM KCl, 5% (v/v) glycerol, 0.2 mM EDTA, 0.2 mM EGTA, 2 mM DTT, 1 mM PMSF and 0.05% (v/v) NP40, to remove non-specifically bound proteins. Beads were resuspended in a SDS-sample buffer, boiled for 5 min, and the soluble proteins were resolved on 10% SDS-PAGE for Western blot analysis.
Western blot analysis
The protein levels of APC, Fen-1, and a-tubulin were determined by Western blot analysis with our previously described procedure (34). The antibodies were procured from the following sources – Fen-1 from Novus Biologicals, Littleton, CO; APC from Oncogene Research Products, Cambridge, MA; and α-tubulin from Sigma-Aldrich Chem. Co., St. Louis, MO.
Far-Western blot analysis
APCwt and APC(Q-A,I-A,Y-A) peptides, GST-p21, and PCNA were slot-blotted (0-10 μg) onto a polyvinylidene difluoride membrane (Amersham Biosciences, Piscatway, NJ) in a binding buffer containing 20 mM Tris-Cl, pH 7.4, 100 mM phosphate buffer, pH 7.4, 60 mM KCl, 0.25% (v/v) Nonidet P-40. After blotting, the membrane was blocked with 5% (w/v) bovine serum albumin and washed three times with Tris-buffered saline with 0.025% (v/v) Tween 20 prior to incubation with purified human Fen-1 protein. Binding was detected using anti-Fen-1 antibody. The signals were detected using the enhanced chemiluminescence technique (Amersham Biosciences, Piscatway, NJ).
Yeast two-hybrid assay
The yeast two-hybrid system was used to determine the functional interaction of APC with Fen-1 in vivo. The APC DNA fragments containing the wild type (amino acids 1190-1328) or the mutant DRI-domain (amino acids 1200-1324, in which amino acids Q1256, I1259, and Y1262 were replaced with alanine) for protein-protein interaction studies were fused to the yeast Gal4 DNA-binding domain (BD) in plasmid pGBDU-C3. Single mutant APC cDNA fragments of the DRI-domain of APC(Q1256-A), APC(I1259-A), or APC(Y1262-A) were used to further characterize the pol-β and Fen-1-interactions and also fused to the yeast Gal4-BD in plasmid pGBDU-C3. The interacting proteins such as full length pol-β and Fen-1 were fused to the yeast Gal4 activation domain (AD) in plasmid pGAD-C3. Adapters were included as needed for in-frame insertion of APC, pol-β and Fen-1 DNA sequences relative to Gal4-BD or Gal4-AD plasmids. The yeast strain S. cerevisiae PJ69-4A was co-transformed with PGBDU-C3 and pGAD-C3 derived plasmids and spread on plates containing yeast synthetic dropout (SD) – UL medium lacking only vector markers Ura for pGBDU-C3 derived plasmids and Leu for pGAD-C3 derived plasmids. To test for potential protein-protein interactions, transformed cells were screened for growth on yeast SD-ULH medium which lacked Ura, Leu, and His but contained 5 mM His3 inhibitor, 3-amino-1,2,4-triazole, to prevent His3-reporter gene autoactivation.
Synthesis and labeling of in vitro BER substrates
To examine LP-BER activity, an AP site analog (3-hydroxy-2-hydroxymethyltetrahydrofuran, noted as F) was introduced at the 24th position of the 63-mer DNA (5′-CTAGATGCCTGCAGCTGATGCGCFGTACGGATCCACGTGTACGGTACCGAGGGCGGGTCGA CA-′) called as F-DNA (32). A 23-mer (5′-CTAGATGCCTGCAGCTGATGCGC-′) and 40-mer oligonucleotides (5′-TGTACGGATCCACGTGTACGGTACCGAGGGCGGGTCGACA-′) were annealed with a 63-mer complimentary oliogonucleotide to prepare a nicked-DNA substrate. Again, the same 23-mer and 39-mer oligonucleotides (5′-GTACGGATCCACGTGTACGGTACCGAGGGCGGGTCGACA-′) were annealed to a 63-mer complementary template to prepare 1-nt gapped-DNA substrate. The 23-mer oligonucleotide and the 63-mer oligonucleotide (F-residue at 24th position) were radiolabeled at the 5′-end with [γ-32P] ATP and T4 polynucleotide kinase (New England Bio Lab, Woburn, MA). The labeled probes were purified by using nick-column (GE Healthcare, Piscataway, NJ).
Strand-displacement synthesis
The BER assay was reconstituted using purified proteins under the following conditions. The reaction mixture for strand-displacement synthesis contained 30 mM Hepes, pH 7.5, 30 mM KCl, 8 mM MgCl2, 1 mM DTT, 100 μg/ml BSA, 0.01 % (v/v) Nonidet P-40, 0.5 mM ATP, and 10 μM each of dATP, dCTP, dGTP, dTTP in a final volume of 25 μl. The BER reaction mixture was assembled on ice by the addition of 1 nM APE, 2.5 nM pol-β and Fen-1. This mixture was incubated for 5 min at 22°C. The amounts of the APCwt and APC(I-A,Y-A) peptides and Fen-1 used in each experiment are given in respective figure legends. The strand-displacement synthesis was initiated by the addition of 2.5 nM of 32P-labeled F-DNA and further incubated for 45 min at 37°C. The reaction was terminated by the addition of 0.4% (w/v) SDS, 5 mM EDTA, 1 μg of proteinase K and 10 μg of carrier RNA. After incubation for an additional 20 min at 37°C, the DNA was recovered by phenol/chloroform extraction followed by ethanol precipitation. The strand-displacement products were separated on a 15% acrylamide and 7 M urea gel. The bands were quantitated by electronic autoradiography (InstantImager™; Packard Instrument Co., Meriden, CT).
Synthesis and labeling of Fen-1 substrate
Fen-1 substrate for 5′-flap endonuclease activity was made by annealing an upstream 23-mer (5′-CTAGATGCCTGCAGCTGATGCGC-′) and a downstream 45-mer oligonucleotide (5′-FTTTTTGTACGGATCCACGTGTACGGTACCGAGGGCGGGTCGACA-′) to a 63-mer complementary template as described before (40). The 45-mer downstream oligonucleotide has a flap of 5-nt (with a teterahydrofuran, F, residue at the 5′-end), which is cleaved by Fen-1. The Fen-1 substrate for 5′-3′ exonuclease activity was made similarly except that the 5-nt flap from the 45-mer oligonucleotide was removed. We made two substrates for 5′-3′ exonuclease activity of Fen-1. One of the substrates had an F residue at the 5′-end of the 45-mer downstream oligonucleotide. After annealing with a 63-mer complementary template as described above, these oligonucleotides created a nick in the DNA suitable for Fen-1 exonuclease activity. The 45- and 40-mer downstream oligonucleotides were radiolabeled at the 5′-end with [γ-32P]ATP and T4 polynucleotide kinase (New England Bio Lab, Woburn, MA). The labeled probe was purified by using nick column (GE Healthcare, Piscataway, NJ). All the three oligonucleotides were annealed at a molar ratio of 1:1:1.
In vitro Fen-1 activity
The assays for 5′-flap endonuclease and 5′-3′ exonuclease activities for Fen-1 were performed in a final volume of 25 μl. The reaction mixture contained 30 mM Hepes, pH 7.5, 8 mM MgCl2, 1 mM DTT, 200 μg/ml BSA, 1 ng of DNA substrate and indicated amounts of Fen-1 and APC peptides. After addition to the APC peptides, the reaction mixture was incubated at room temperature for 5 min. Then 2.5 nM of the 32P-labeled flapped-DNA or nicked-DNA substrates were added in the mixture and further incubated at 37°C for 30 min. Reactions were terminated with stop solution containing 0.4% (w/v) SDS and 5 mM EDTA. The DNA was recovered by phenol/chloroform extraction followed by ethanol precipitation. The 5- and 6-nt DNA products from the 5′-flap endonuclease activity and 1-nt product from the 5′-3′ exonuclease activity were separated on a 15% acrylamide and 7 M urea gel and quantitated by electronic autoradiography (InstantImager™; Packard Instrument Co., Meriden, CT).
RESULTS
Knockdown ofAPC improves LP-BER in vivo
To understand the mechanism by which APC plays a role in BER, we first established that APC interacts with BER machinery in vivo. Today, most DNA repair assays are performed using a reconstituted system with purified proteins. Some of the in vivo DNA repair assays, which are routinely used in laboratories such as single-cell gel electrophoresis or comet assay, are non-specific (41). The comet assay, although extremely useful in the assessment of genotoxicity of DNA-damaging agents (42), does not distinguish among the type of DNA damage and repair pathways involved. Best to our knowledge, there is no suitable in vivo BER assay system available. Thus, we developed a reporter plasmid-based in vivo BER assay system (33). We randomly modified multiple cytosine (C) residues of the p21(Waf1/Cip1)-luciferase promoter DNA into uracil (U) residues (U-p21P; a substrate for SP-BER). This plasmid DNA was further treated with uracil-DNA glycosylase (UDG) and then with sodium borohydride to create reduced abasic p21P (R-p21P) substrate for LP-BER (Figure 1A). Modification of DNA by this technique is described in our earlier studies (39). The principle behind this assay is that the modified p21P plasmid when transfected into cells should show poor promoter activity as compared to the unmodified p21P plasmid. However, the promoter activity can be restored if the modified DNA is allowed to go through DNA repair process(es) in the cell. The assay is quick, sensitive and quantitative. The disadvantage with the assay is that it does not provide information whether the damage and repair is more efficient on the transcribed or non-transcribed strand of DNA. Furthermore, it also does not provide information about the number of C-residues modified and repaired within the cells in a given time period. The treatment with DNA-damaging agents will inhibit the expression of the gene by interfering with the promoter versus inhibiting expression by messing up the coding sequence which cannot be assessed by this assay. Nonetheless, it provides useful information about the DNA repair capacity of the cell.
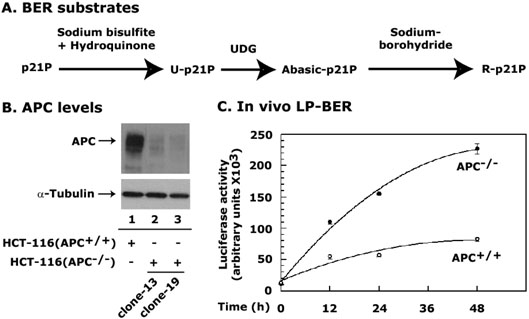
Figure 1. APC blocks BER in vivo.
Panel A describes the protocol for the modification of p21P for long-patch BER (R-p21P) DNA substrate. Panel B is a Western blot analysis of APC protein in HCT-116 stable cell lines. Clones of HCT-116(APC−/−) and HCT-116(APC+/+) cell lines show decreased levels of APC in pSiRNA-APC or pSiRNA-APCmut transfected plasmids, respectively. Clone-19 of the HCT-116(APC−/−) cell line was used in our studies. Panel C shows a Luciferase gene reporter assay of the R-p21P DNA transfected in HCT-116(APC+/+) or HCT-116(APC−/−) cell lines. Data are the mean ± SE of three different experiments.
Since APC affects the strand-displacement synthesis of LP-BER in vitro (32), we examined whether APC has similar effects on LP-BER in vivo. To test this idea, we used the p21P-luciferase promoter-based assay system mentioned above. We established an APC-knockdown HCT-116 cell line by using the SiRNA techniques as described in “Experimental Procedures”. HCT-116 cells were stably transfected with pSiRNA-APC and pSiRNA-APCmut plasmids which are referred to as HCT-116(APC−/−) and HCT-116(APC+/+), respectively. Among the several clones we screened for APC knockdown levels, data from two clones 13 and 19 are shown in Figure 1B. We used clone-19 because more than 90% of the APC protein was knocked down in this clone (Figure 1B, compare lane 1 with 3). Cell lines were transfected with R-p21P plasmid, a substrate for LP-BER, and the luciferase reporter activity was determined at different periods after transfection. In our experiments, the promoter activity of the p21P (unmodified) was the same in both HCT-116(APC+/+) and HCT-116(APC−/−) cell lines (data not shown). The R-p21P (modified) promoter activity was significantly higher in HCT-116(APC−/−) cells than in HCT-116(APC+/+) cells (Figure 1C). From these results it appears that the absence of APC caused an increase in LP-BER and in turn an increase in R-p21P promoter activity. Thus, our assay system directly tests the role of APC in LP-BER in vivo and can be a useful tool for future BER studies.
Physical interaction of APC with Fen-1
To examine the mechanism by which APC plays a role in BER, we determined its interaction with BER proteins. Since pol-β-mediated strand-displacement synthesis is known to be coordinated by Fen-1 (36, 37), we determined whether APC along with its interaction with pol-β also has a role in Fen-1-coordinated strand-displacement synthesis. First, we performed an in vitro pull-down experiment of anti-APC antibody-antigen complex with protein A-Sepharose 4B beads using cell nuclear extracts of three different human colon cancer cell lines – HCT-116, LoVo, and LS411N. The HCT-116 cell line expresses 310 kDa wild-type APC. Like the LoVo cell line, the LS411N cell line also expresses a mutant APC protein (87 kDa), which lacks the pol-β interaction domain. As shown in Figure 2A, the results indicate a physical interaction between wild-type APC and Fen-1 (lanes 1-3), but not with the truncated APC (lanes 4-9).
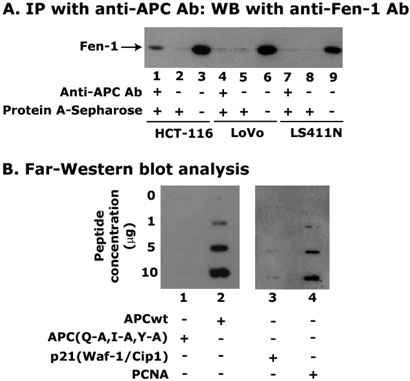
Figure 2. Interaction of APC with Fen-1.
Panel A shows an immunoblot of Fen-1. APC protein complexes from HCT-116, LoVo, and LS411N cell nuclear extracts (200 μg) were immunoprecipitated with anti-APC antibody and then immunoblotted with anti-Fen-1 antibody to detect Fen-1 protein levels. One-fifth of the nuclear extracts without antibody was run as a control in lanes 3, 6, and 9, respectively. Panel B depicts the Far-Western blot analysis of APC and Fen-1 interaction in which a 20-amino acids peptide either mutant APC(Q-A,I-A,Y-A) (lane 1) or wild-type APCwt (lane 2) was used. The interaction of Fen-1 with p21(Waf-1/Cip1) (lane 3) and PCNA (lane 4) are also shown as negative and positive controls, respectively.
Earlier we have indicated that APC may have a proliferating cell nuclear antigen (PCNA) interacting protein (PIP)-like box in the N-terminal region, which might be involved in the binding with pol-β (32). In the present study, we examined whether this region is also involved in the binding with Fen-1. We performed a Far-western analysis with synthetic APC peptide containing the PIP-like box. A 20-amino acids-long peptide, APCwt(1250-KVSSINQETIQTYCVEDTPI-1269 amino acids), with the PIP-like box of the wild-type APC protein was synthesized. To determine the specificity of the interaction, amino acid residues such as Q1256,I1259, and Y1262 in the PIP-like box motif were replaced with alanine (A). The mutant peptide was named as APC(Q-A,I-A,Y-A). We observed a dose-dependent increase in the binding of Fen-1 with APCwt but not with APC(Q-A,I-A,Y-A) peptide (Figure 2B, compare lane 1 with lane 2). As a positive control, we used the interaction of Fen-1 with PCNA (Figure 2B, lane 4), which was reported in earlier studies (43, 44), and as a negative control, we used the interaction of Fen-1 with p21(Waf-1/Cip1) (Figure 2B, lane 3). These results support that the PIP-like box region of the APC is important for the physical interaction with Fen-1.
Determination ofDNA repair inhibitory (DRI)-domain of APC
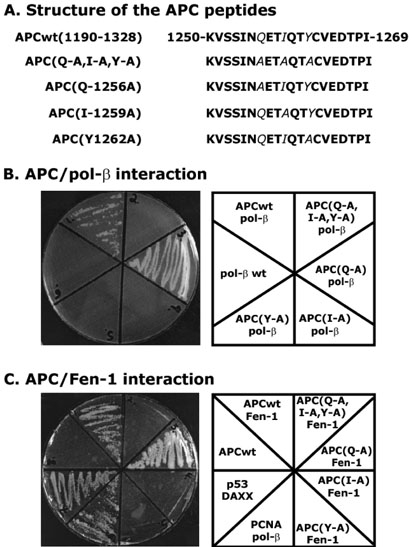
Next, we determined whether all the three amino acids Q1256, I1259, and Y1262 in the PIP-like box were important for interaction with pol-β and Fen-1 by using yeast two-hybrid analysis. We made different yeast two-hybrid constructs of APC (amino acids 1190-1328) and replaced the Q1256, I1259, and Y1262 amino acids one by one with alanine (A) by site-directed mutagenesis (Figure 3A). Plasmids were co-transfected into S. cerevisiae PJ69-4A yeast strain, and protein-protein interactions were determined by the growth of yeast cells in a histidine-selection medium as described in Materials and Methods. Results show that the Q1256A mutation has no effect on the interaction with both pol-β and Fen-1 (Figure 3B and C). However, either I1259A or Y1262A mutations abolished the ability of yeast to grow on histidine-selection medium, indicating that the interaction with pol-β and Fen-1 was not occurring with APC (Figure 3B and C). These results suggest that the APC protein has a unique site at which pol-β and Fen-1 interact. Furthermore, this establishes that either I1259 or Y1262 amino acid of the DRI-domain of APC is important for its interaction. To validate our assay system, we used well characterized interactions of PCNA/pol-β (45) and p53/DAXX (46). The plasmid containing only APCwt(1190-1324)-binding domain, that served as a background control, did not show any growth on the medium. From these results, we concluded that the pol-β and Fen-1 interaction site of APC is quite different than the known PCNA-interacting domain of QXX(h)XX(a)(a), in which “h” represents amino acids with moderately hydrophobic side chains such as Leu, Ile, or Met, “a” represents residues with highly hydrophobic residues such as Phe and Tyr, and “X” is any residue (47). However, the pol-β and Fen-1 interacting domain of APC is restricted to only two amino acids Ile (I1256) and Tyr (Y1262). Furthermore, mutations on either I1256 or Y1262 are sufficient to abolish the interaction with pol-β and Fen-1. Due to the role of APC in the inhibition of DNA repair, we renamed the pol-β and Fen-1 interacting domain of APC the DNA repair inhibitory (DRI)-domain.
Figure 3. Interaction of APC with pol-β and Fen-1 by yeast two-hybrid analysis.
The yeast two-hybrid constructs are described under Materials and Methods. Panel A shows the structure of the APC peptides used in the yeast two-hybrid analysis. Panels B and C show the interaction of APC with pol-β and Fen-1, respectively. The yeast PJ69-4A cells were co-transformed with pGBDU-C3-APCwt (amino acids 1190-1328) or pGBDU-C3-APC(TripleMut) (amino acids 1200-1324; Q1256-A, I1259-A, Y1262-A) plasmids with either pGAD-C3-pol-β or pGAD-C3-Fen-1 plasmids. For a positive control, PCNA/pol-β and p53/DAXX interactions are shown. Transformation with pGBDU-C3-APCwt-BD (amino acids 1190-1324) alone served as a negative control for background colonies.
APC blocks 1-nucleotide synthesis activity of pol-β
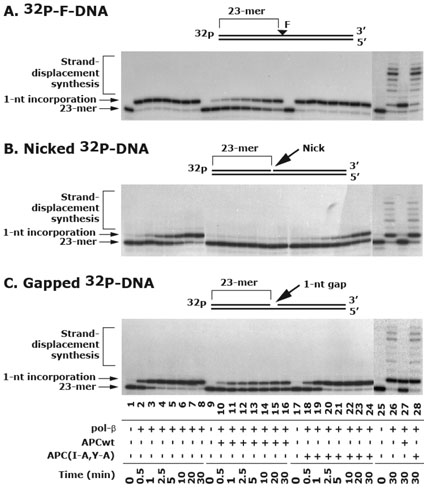
Our earlier findings suggest that APC blocks pol-β-mediated strand-displacement synthesis (32). In the present study, we determined whether APC also affects 1-nt synthesis activity of pol-β. In our experiments, we used 32P-labeled F-, nicked- and gapped-DNA substrates in a reconstituted in vitro BER assay system with purified proteins. The 32P-labeled F-DNA was pre-incubated with APE for 10 min at 22°C. Then, pol-β APCwt or APC(I-A,Y-A) peptides were added in the reaction mixture containing different DNA substrates and dCTP to allow 1-nt synthesis at the damaged site. A time course for 1-nt synthesis was followed. The 1-nt synthesis was rapid and completed in 0.5 min with F-DNA (Figure-4A, compare lane 1 with 2-8), which was partially blocked by APCwt peptide (Figure 4A, compare lane 9 with 16). The APC(I-A,Y-A) peptide did not have any effect on pol-β-mediated 1-nt synthesis with F-DNA (Figure 4A, compare lane 17 with 18-24). The kinetics of 1-nt synthesis with nicked-DNA was much slower than F-DNA. It required 30 min to achieve 90% activity (Figure 4B, compare lane 1 with 8). However, the APCwt peptide completely blocked 1-nt synthesis activity of pol-β with nicked-DNA (Figure 4B, compare lane 9 with 10-16) as compared to APC(I-A,Y-A) peptide (Figure 4B, compare lane 17 with 18-24). Next, we extended our experiments to determine whether APC also affects 1-nt synthesis activity of pol-β with gapped-DNA. In these experiments we used 1-nt 32P-labeled gapped-substrate. We found an efficient 1-nt synthesis into gapped-DNA substrate, which was completed in about 2.5 min (Figure 4C, compare lane 1 with 2-8). However, it was partially inhibited by APCwt (Figure 4C, compare lane 9 with 10-16) and uninhibited by APC(I-A,Y-A) peptide (Figure 4C, compare lane 17 with 18-24). These results suggest that pol-β-mediated 1-nt synthesis has different kinetics with different DNA substrates, which is differentially inhibited by APCwt peptide in an order of nicked-DNA > F-DNA >gapped-DNA. To further determine whether F-, nicked- and gapped-DNA can be processed by long-patch (LP)-BER pathway and whether APC will have inhibitory effect on them, we added all the four dNTPs in the reaction mixture and incubated for 30 min. Results showed a pol-β-mediated strand-displacement synthesis with all the three tested DNA substrates (Figure 4A, B and C, compare lane 25 with 26), which were blocked by APCwt (Figure 4A, B and C, compare lane 26 with 27), but not by APC(I-A,Y-A) peptide (Figure 4A, B and C, compare lane 26 with 28). Interestingly, in the presence of all the dNTPs, the 1-nt synthesis was not blocked by APCwt peptide with F-DNA (Figure 4A, compare lane 25 with 27), partially blocked with gapped-DNA (Figure 4C, compare lane 25 with 27) and completely blocked with nicked DNA (Figure 4B, compare lane 25 with 27). These results suggest that APC blocks pol-β-mediated 1-nt synthesis as well as strand-displacement synthesis and ultimately influences the LP-BER pathway.
Figure 4. APC differentially blocks 1-nt incorporation activity of pol-β.
The 32P-F-DNA was incubated with 1 nM of APE for 10 min to introduce an incision at the 5′ end of AP site. In a test tube 2 nM of pol-β and 4 μM of either APCwt or APC(I-A,Y-A) peptides were added, followed by the addition of precut 32P-labeled F-, nicked- and 1-nt gapped-DNA and dCTP. A time course was followed to determine the effect of APC on pol-β-mediated kinetics of 1-nt synthesis (lanes 1-24). In a second set of experiment, all the dNTPs were added in the reaction mixture to determine the effect of APC on the strand-displacement synthesis with F-, nicked-, and gapped-DNA substrates (lanes 25-28). The data are representative of three different experiments.
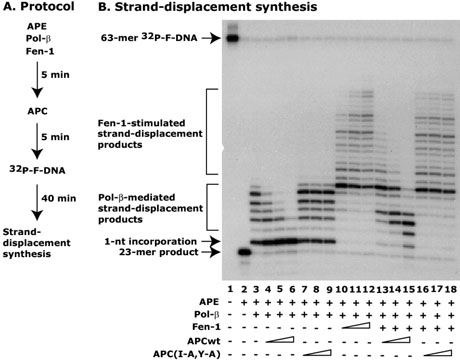
APC blocks Fen-1 activity and thus blocks strand-displacement synthesis
Once we established the interaction of APC with Fen-1, we determined its functional role in strand-displacement synthesis by using a reconstituted in vitro BER assay system with purified proteins. The LP-BER can be distinguished in two parts – pol-β-mediated strand-displacement synthesis (8) and the “Hit-and Run” mechanism supported by pol-β and Fen-1 (36, 37). We set up an in vitro BER assay system in which both of these activities were distinguished and the role of APC was determined. APE and pol-β were incubated for 5 min before adding APC peptides and then incubation was continued for an additional 5 min. DNA synthesis was initiated by 32P-labeled F-DNA at 37°C (Figure 5A). The results showed a 7-nt product of pol-β-mediated strand-displacement synthesis (Figure 5B, compare lane 2 with 3), which was blocked in a dose-dependent manner by 20-amino acid wild-type APC (APCwt, amino acids 1250-1269) peptide containing the DRI-domain (Figure 5B, compare lane 3 with 4-6). As a control, we used a DRI-domain mutant 20-amino acids peptide in which the I1259 and Y1262 were replaced with alanine (A). The mutant peptide was named as APC(I-A,Y-A). We found that the APC(I-A,Y-A) peptide did not affect the pol-β-mediated strand-displacement synthesis (Figure 5B, compare lane 3 with 7-9). The 1-nt incorporation by pol-β was not affected by APC. These results were consistent with our previous findings (32).
Figure 5. APC blocks Fen-1-coordinated strand-displacement synthesis.
The APE (1 nM), pol-β (2 nM) and either different concentrations (0, 5, 10, 15 nM) or fixed concentration (15 nM) of Fen-1 were mixed and preincubated for 5 min on ice. Then different concentrations (2, 4, 6 μM) of either APCwt or APC(I-A,Y-A) mutant peptides were added and further incubated for 5 min on ice. The stranddisplacement synthesis was initiated with 32P-F-DNA. Panel A shows a schematic representation of the protocol. Panel B shows a representative autoradiogram of the three different experiments. Arrows show different BER products including 23-mer incision product, 1-nt incorporation product, and 2–7-nt pol-β-initiated and more than 7-nt by Fen-1-coordinated strand-displacement products. Lane 1 shows the position of the 63-mer 32P-F-DNA.
Pol-β-mediated strand-displacement synthesis is also coordinated by Fen-1 (36). Therefore, we next examined whether APC plays a role in Fen-1-coordinated strand-displacement synthesis. The strand-displacement synthesis assay was set up similarly as above, except Fen-1 was added along with APE and pol-β. After 5 min of incubation, the APC peptides were added and then incubated for an additional 5 min. The DNA synthesis was initiated by 32P-labeled F-DNA (Figure 5A). We observed strong Fen-1-coordinated strand-displacement synthesis in a dose-dependent manner (more than 7-nt), which was clearly distinguished from the one mediated by pol-β alone (7-nt) (Figure 5B, compare lane 3 with 10-12). The addition of the APCwt peptide in the mixture showed a dose-dependent block of Fen-1-coordinated strand-displacement synthesis (Figure 5B, compare 10-12 with 13-15, respectively). Although the effect of APCwt on Fen-1-coordinated strand-displacement synthesis was drastic, it only partially blocked pol-β-mediated strand-displacement synthesis (Figure 5B, compare lane 6 with 15). On the other hand, the APC(I-A,Y-A) peptide did not inhibit blockage of Fen-1 coordinated strand-displacement synthesis (Figure 5B, compare lane 10-12 with 16-18). These results suggest that pol-β can synthesize up to 4-nt without a challenge or hindrance, but cannot proceed for the elongation phase of strand-displacement synthesis once it complexes with APC. Alternatively, the Fen-1 cleavage activity may be necessary to remove the flapped-DNA and allow pol-β to proceed for synthesis which might have been blocked by APC.
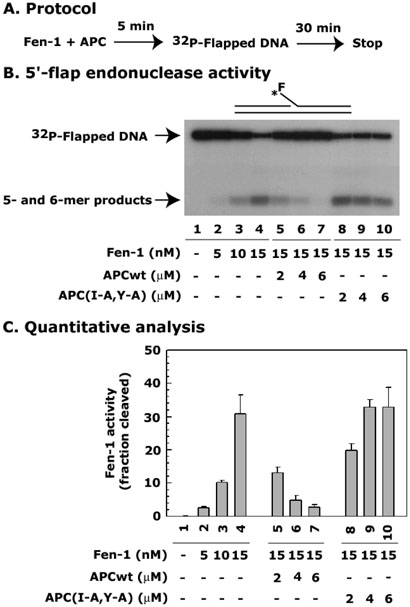
APC blocks 5′-flap endonuclease activity of Fen-1
Fen-1 has two types of cleavage activity - 5′-flap endonuclease activity and 5′-3′ exonuclease activity (13, 48). The 5′-flap endonuclease activity requires a 5′-flap structure, and the activation and position of the cleavage efficiency is increased by the presence of upstream primers adjacent to the bifurcation (49). The 5′-flap endonuclease activity may be important for strand-displacement synthesis of LP-BER in which the cleavage of the flap is necessary for DNA ligase to seal the nick. Since we found that APC interacts with Fen-1, we tested whether APC affects both activities of Fen-1. First, to test the possibility of APC blocking Fen-1 's 5′-flap endonuclease activity, we used an in vitro 5′-flap endonuclease assay of Fen-1 in the presence or absence of APC peptide using 32P-labeled nicked-flapped DNA substrate (Figure 6A).
Figure 6. APC blocks 5′-flap endonuclease activity of Fen-1.
Panel A shows a schematic representation of the protocol. Panel B shows an autoradiogram of a representative experiment. Reaction mixtures in a 25 μl final volume, contained either different (0, 5, 10, 15 nM) or fixed (15 nM) concentrations of Fen-1 coupled with different concentrations (2, 4, 6 μM) of either APCwt or mutant APC(I-A,Y-A) peptides. The mixture was incubated for 5 min at 22°C, and then 2.5 nM of 32P-labeled flapped-DNA substrate was added. The cleavage reaction was carried out at 37°C for 30 min. X-ray film autoradiography determined the 5-nt cleaved product, and the electronic autoradiography measured the radioactivity (InstantImager™; Packard Instrument Co., Meriden, CT). Panel C shows the quantitative analysis of the 5-mer cleaved product. The fraction of cleavage was calculated as a percent radioactivity present in the cleaved product as follows: % cleavage = [5-nt/(45-mer + 5-nt)] × 100. Data are the mean ± SE of three different experiments.
Fen-1 showed a 5- and 6-nt cleavage product of the substrate in a dose-dependent manner (Figure 6B, compare lane 1 with 2-4). In the presence of different concentrations of APCwt, but not mutant APC(I-A,Y-A), the cleavage of the substrate was blocked (Figure 6B, compare lane 4 with 5-7, and 8-10, respectively). A quantitative analysis of the data is shown in Figure 6C. There is about a 10-fold decrease in 5′-flap endonuclease activity of Fen-1 at the highest APCwt concentration used. Our results demonstrate that APC interacts with Fen-1 and blocks its 5′-flap endonuclease activity. Since the removal of the flap is a critical step in LP-BER, the blockage of 5′-flap endonuclease activity of Fen-1 by APC will block LP-BER.
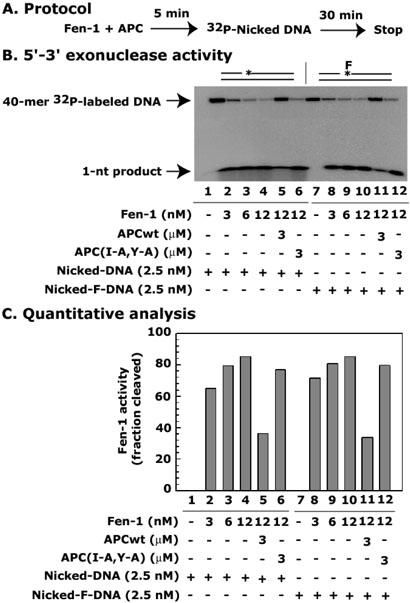
APC blocks “Hit and Run” synthesis of LP-BER by blocking 5′-3′ exonuclease activity of Fen-1
The 5′-3′ exonucleolytic cleavage by Fen-1 is influenced by the presence of the upstream primer and is reduced if it is missing or a gap is present between the upstream and downstream primers (13). In our assays, after the removal of the flap by 5′-flap endonuclease activity of Fen-1, a gap will be generated in the DNA. The gap will be filled by pol-β leaving a nick, which will further become a substrate for the 5′-3′ exonuclease activity of Fen-1 (Figure 7). The 5′-flap endonuclease and 5′-3′ exonuclease activities of Fen-1 reside in the same catalytic domain of the protein, which has a positively charged groove containing the active center and a H3TH motif proposed to mediate binding of ssDNA and dsDNA portions of the flap substrates, respectively (12, 50). Thus, it is likely that APC affects both these activities of Fen-1. In addition to removing the flap by 5′-flap endonuclease activity, the 5′-3′ exonuclease activity of Fen-1 can also be critical in LP-BER by removing 1-nt and creating a gap. The gapped-DNA will provide an opportunity for pol-β to fill up the gap by synthesizing 1-nt until DNA ligase binds and seals the nick. This is another mechanism of LP-BER which is described as the “Hit and Run” mechanism. In this mechanism, an alternating 1-nt gap is created by Fen-1, and gap-filling is completed by pol-β, rather than through coordinated formation and removal of a strand-displaced flap (37). However, if the interaction of APC with Fen-1 blocks its 5′-3′ exonuclease activity, then there will be no gap formation and no “Hit and Run” synthesis of LP-BER.
Figure 7. APC blocks 5′-3′ exonuclease activity of Fen-1.
Exonuclease activity reaction mixture was assembled in a total of 25 μl final volume and contained either different (0, 3, 6, 12 nM) or fixed (12 nM) concentrations of Fen-1 along with 3 μM of either APCwt or APC(I-A,Y-A) peptides. The mixture was incubated for 5 min at 22°C and then 2.5 nM of 63-mer oligonucleotide substrates containing either 32P-labeled 40-mer or F-40-mer downstream oligonucleotides were added. The exonuclease reaction was carried out at 37°C for 30 min. X-ray autoradiography determined the 1-nt cleaved product (Panel A) and the electronic autoradiography measured the radioactivity (Panel B) (InstantImager™; Packard Instrument Co., Meriden, CT). The fraction of cleavage was calculated as a percent radioactivity present in the cleaved product as follows: % cleavage = [1-nt/(40-mer + 1-nt)] X 100. Data are the representative of two different experiments.
To explore this possibility, we first determined the effect of APC on the 5′-3′ exonuclease activity of Fen-1. We designed nicked DNA substrates, instead of a flapped DNA substrate as used for 5′-flap endonuclease activity of Fen-1. In the nicked DNA substrates, a 23-mer upstream and 40-mer downstream oligonucleotides were annealed with a 63-mer complimentary template oligonucleotide as described in Materials and Methods. At the 5′-end of one of the downstream oligonucleotides, a tetrahydrofuran (F) was incorporated to determine whether the 5′-3′ exonuclease activity of Fen-1 has a preference upon the reduced abasic site. The 40-mer oligonucleotides were labeled with [γ-32P]ATP at the 5′-end before annealing and used in the exonuclease assays. If the Fen-1 has a 5′-3′ exonuclease activity, then we should see a cleavage of 1-nt from the 32P-labeled 40-mer oligonucleotide. Results show a very efficient cleavage of 1-nt from both the substrates in a dose-dependent manner (Figure 7, compare lane 1 with 2-4 for Nicked-DNA and lane 7 with 8-10 for Nicked-F-DNA, respectively), suggesting that the 5′-3′ exonuclease activity of Fen-1 is dependent upon the nick instead of the modifications on the nick of the DNA, which is consistent with previous findings (13). Then we incubated APC with Fen-1 to determine whether APC can interact with Fen-1 and block its 5′-3′ exonuclease activity. Results show a 64% decrease in the 5′-3′ exonuclease activity of Fen-1 when incubated with the wild-type APC but not with the mutant APC(I-A,Y-A) peptides (Figure 7, compare lane 4 with 5 and 6 for Nicked-DNA; and lane 10 with 11 and 12 for Nicked-F-DNA, respectively). These results indicate that APC blocks 5′-3′ exonuclease activity of Fen-1 and thus can block “Hit and Run” synthesis of LP-BER.
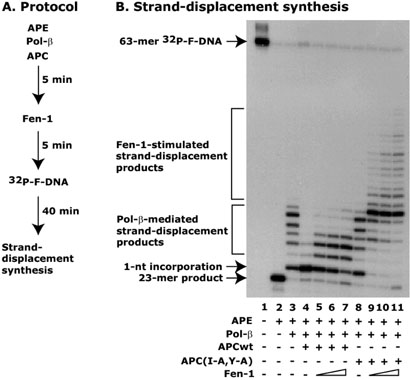
To determine whether APC plays a role in “Hit and Run” synthesis, we set up an in vitro strand-displacement synthesis assay with some modifications (Figure 8A). In this protocol to ensure the binding of APC and pol-β, the APC was first incubated with pol-β and then Fen-1 was added. Finally, the addition of 32P-labeled F-DNA initiated the reaction (Figure 8A). As we expected, the results show that APCwt peptide blocked pol-β-mediated strand-displacement synthesis, but accumulated 1-nt product (Figure 8B, compare lane 3 with 4). In the presence of Fen-1, 1-nt product was extended up to 4-nt but could not be extended further. However, in the presence of the mutant APC(I-A,Y-A) peptide, a Fen-1-coordinated LP-BER was clearly observed in a dose-dependent manner and extended synthesis beyond 4-nt (Figure 8B, compare lane 8 with 9-11, respectively). This result suggests that APC blocks LP-BER at two separate stages. First, APC inhibits strand displacement synthesis by pol-β with F-, nicked-, or 1-nt gapped-DNA; and second, APC inhibits Fen-1-mediated 5′-3 exonuclease activity. Taken together, this indicates that APC blocks LP-BER that occurs either by a strand-displacement or a “Hit and Run” mechanism.
Figure 8. APC blocks Fen-1-coordinated LP-BER.
In this experiment, the conditions were the same as described in Figure 5, except that the APC peptides were incubated with APE and pol-β before the addition of Fen-1 as shown in Panel A. Panel B shows a representative autoradiogram of the three different experiments. Arrows show different BER products including 23-mer incision product, 1-nt incorporation product, and 2-7-nt of pol-β-initiated and more than 7-nt by Fen-1-coordinated stranddisplacement products. Lane 1 shows the position of the 63-mer 32P-F-DNA.
DISCUSSION
We have recently described a role of APC in LP-BER in which it interferes with pol-β-mediated strand-displacement synthesis (32). The strand-displacement synthesis is an important step in LP-BER, and Fen-1 plays a major role in this pathway (8, 36, 37, 51). In the present study, we extended our findings to examine whether APC interacts with Fen-1 to inhibit its activity and thus to provide a second mechanism for blocking LP-BER. In our earlier study, a physical interaction of APC with pol-β was described (32); however, the interaction of APC with Fen-1 is not yet known and is being examined in the present study. Our amino acid fine mapping experiments suggest that APC has a unique DNA repair inhibitory (DRI)-domain. Since APC is a 310 kDa protein, we expect that a group of amino acids in the DRI-domain may participate in the binding with pol-β or Fen-1. However, we were intrigued to see that only one amino acid, i.e., either I1259 or Y1262 was important for the binding. These results suggested that the interaction of pol-β and Fen-1 may be at the surface instead of in the groove of the APC protein. Alternatively, other surrounding amino acid residues of the DRI-domain may be important for the interaction, but only I1259 or Y1262 are in direct contact with pol-β and Fen-1. These possibilities can be better understood with crystal structure modeling of APC with pol-β and Fen-1, respectively. However, the crystal structure of APC is not yet available to work on these predictions.
LP-BER can occur via the following two mechanisms, strand-displacement and “Hit and Run” synthesis. Fen-1 has distinct roles in each. In strand-displacement, Fen-1 cleaves the flap generated by pol-β-mediated strand displacement synthesis. In “Hit and Run” synthesis, Fen-1 excises a nucleotide at the ′ side of a nick to create a gap to be filled by pol-β (5, 14, 17, 51-53). Since the removal of the flapped-DNA by Fen-1 is a major step in the completion of LP-BER, the interaction of APC with Fen-1 suggested its possible involvement in LP-BER. Once we established the interaction of APC with Fen-1, we hypothesized that it may block Fen-1 cleavage activity and thus blocks the LP-BER. Furthermore, since APC interacts with both pol-β and Fen-1, it is likely that APC may target three distinct activities required for LP-BER. First, APC inhibits pol-β's 1-nt synthesis and strand-displacement activity. Second, APC can interact with Fen-1 and block its 5′-flap endonuclease activity. The combination of these interactions effectively blocks LP-BER that occurs by strand-displacement synthesis. Third, the interaction of APC and Fen-1 also inhibit the 5′-3′ exonuclease activity of Fen-1 and thus block LP-BER synthesis by a “Hit and Run” mechanism.
Once pol-β is recruited on an oxidized or reduced abasic site after incision by APE, it continues strand-displacement synthesis until Fen-1 binds and cleaves the flap and DNA ligase seals the nick. The interaction of APC with pol-β blocks its 1-nt incorporation and strand-displacement synthesis. However, the strand-displacement synthesis can still resume if Fen-1 is accessible to the system. In this case, a “Hit and Run” type of LP-BER can take place (37). According to this model, there are sequential events involved in LP-BER. First, pol-β recognizes and binds to the nicked-DNA which is generated by Fen-1 from the removal of the flapped DNA, extends the ′-OH displacing dRP moiety, fills the 1-nt gap, leaves a nick, and dissociates from the DNA. Then Fen-1 binds to the nicked DNA, removes 1-nt by its 5′-3′ exonuclease activity, creates a gap, and dissociates from DNA. This cycle of LP-BER is repeated several times until DNA ligase seals the nick. Once APC interacts with Fen-1, it blocks both the 5′-flap endonuclease as well as the 5′-3′ exonuclease activities of Fen-1, and affects the overall LP-BER process. More precisely, when APC interacts with Fen-1 and blocks 5′-flap endonuclease activity of Fen-1, then APC blocks the pol-β-mediated strand-displacement synthesis of LP-BER. On the other hand, when APC interacts with Fen-1 and blocks 5′-3′ exonuclease activity of Fen-1, then APC blocks “Hit and Run” synthesis of LP-BER. From these studies, it is clear that both pol-β and Fen-1 interact with the DRI-domain of APC and affect LP-BER. In our experiments, the assays were performed in vitro using purified proteins; therefore, it is not clear how pol-β and Fen-1 compete for binding with APC in vivo. Nonetheless, the interaction of APC with pol-β alone, Fen-1 alone, or pol-β and Fen-1 can block LP-BER. Another limitation of our in vitro studies is the use of synthetic APC peptide instead of the purified protein to block LP-BER. A several-fold higher concentration of APC peptide (4 μM) is needed to block the activity of 2 nM of pol-β and 12 nM of Fen-1. One may assume that the stoicheometry of interaction of APC with pol-β and Fen-1 would be in a close proximity. In our future studies, we will use purified APC protein to address this concern more precisely. Nonetheless, we have backed up our findings by using in vivo gene-reporter and comet assays to describe the involvement of APC in LP-BER (32, 33).
It is widely believed that APC is a tumor suppressor and its mutation plays a role in the impairment of cell-cell adhesion (54, 55), cell migration (56, 57), regulation of β-catenin levels (58, 59), and chromosomal stability (60, 61). However, there is no information about the role of normal APC in cells which are challenged with DNA-damaging agents. In earlier studies, we have shown that APC levels are increased in colon cancer cells after treatment with DNA-damaging agents (34, 35). The consequence of DNA damage-induced levels of APC in cells is currently not well understood. Our finding that APC blocks LP-BER is the beginning of understanding the role of APC in DNA damage-induced fate of cells. It is well established that DNA damage is an initiating event of carcinogenesis. If the DNA damage of a cell is not efficiently repaired, then cells can adapt either programmed cell death or proceed through the cell cycle with damaged DNA and cause carcinogenesis (62), which suggests a paradoxical role for APC. In fact, recently, we have shown a link of APC with carcinogenesis in which cigarette smoke condensate-induced levels of APC was associated with the blockage of LP-BER and transformation of normal breast epithelial cells (33). Thus, depending upon the environment of the cell, APC may act as a tumor suppressor by inducing cell death or causing carcinogenesis by helping to accumulate mutations. Since the DRI-domain is located toward the N-terminal region of the APC protein, which is spared by the MCR region, both the wild-type and the mutant APC proteins (containing the DRI-domain) may block DNA damage-induced LP-BER. In such cases, if the chemopreventive agents that produce DNA damage can increase APC levels in the target cell, then the decreased LP-BER and increased DNA damage may induce cell death (63), suggesting a tumor suppressor role of APC. Recently, the BER inhibitors have been suggested as possible therapeutic targets in colon cancer cells (64). Thus, the link between APC and DNA repair may provide an important clinical implication in future drug development strategies.
ACKNOWLEDGEMENTS
We are grateful to the following individuals for providing us the valuable reagents – Linda Bloom (University of Florida, Gainesville, FL) for human APE-1, Samuel H. Wilson and Rajendra Prasad (NIEHS, Research Triangle Park, NC) for human DNA polymerase β and Fen-1, and Daiqing Liao (University of Florida, Gainesville, FL) for his assistance in yeast two-hybrid analysis. We sincerely thank Linda Bloom for critically reading the scientific content and Mary Wall and Nirupama Gupta for proofreading the manuscript.
FOOT NOTES
The financial support for these studies was provided to Satya Narayan by the grants from NCI-NIH (CA-097031-01 and CA-100247-01) and Flight Attendant Medical Research Institute, Miami, FL.
Abbreviations – APC, adenomatous polyposis coli; APE, apurinic/apyrimidinic endonuclease; BER, base excision repair; DRI-domain, DNA repair inhibitory-domain; Fen-1, flap endonuclease 1; LPBER, long-patch base excision repair; pol-β, DNA polymerase β
REFERENCES
- 1.Fan J, Wilson DM. Protein-protein interactions and post-translational modifications in mammalian base excision repair. Free Radic. Biol. Med. 2005;38:1121–1138. doi: 10.1016/j.freeradbiomed.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 2.Chakraverty RK, Hickson ID. Defending genome integrity during DNA replication: a proposed role for RecQ family helicases. Bioessays. 1999;21:286–294. doi: 10.1002/(SICI)1521-1878(199904)21:4<286::AID-BIES4>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 3.Lenzmeier BA, Freudenreich CH. Trinucleotide repeat instability: a hairpin curve at the crossroads of replication, recombination, and repair. Cytogenet. Genome Res. 2003;100:7–24. doi: 10.1159/000072836. [DOI] [PubMed] [Google Scholar]
- 4.Matsumoto Y, Kim K. Excision of deoxyribose phosphate residues by DNA polymerase beta during DNA repair. Science. 1995;269:699–702. doi: 10.1126/science.7624801. [DOI] [PubMed] [Google Scholar]
- 5.Klungland A, Lindahl T. Second pathway for completion of human DNA base excision-repair: reconstitution with purified proteins and requirement for DNase IV (FEN1) EMBO J. 1997;16:3341–3348. doi: 10.1093/emboj/16.11.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gary R, Kim K, Cornelius HL, Park MS, Matsumoto Y. Proliferating cell nuclear antigen facilitates excision in long-patch base excision repair. J. Biol. Chem. 1999;274:4354–4363. doi: 10.1074/jbc.274.7.4354. [DOI] [PubMed] [Google Scholar]
- 7.Huffman JL, Sundheim O, Tainer JA. DNA base damage recognition and removal: new twists and grooves. Mutat. Res. 2005;577(12):55–76. doi: 10.1016/j.mrfmmm.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 8.Podlutsky AJ, Dianova II, Podust VN, Bohr VA, Dianov GL. Human DNA polymerase beta initiates DNA synthesis during long-patch repair of reduced AP sites in DNA. EMBO J. 2001;20:1477–1482. doi: 10.1093/emboj/20.6.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bambara RA, Murante RS, Henricksen LA. Enzymes and reactions at the eukaryotic DNA replication fork. J. Biol. Chem. 1997;272:4647–4650. doi: 10.1074/jbc.272.8.4647. [DOI] [PubMed] [Google Scholar]
- 10.Lieber MR. The FEN-1 family of structure-specific nucleases in eukaryotic DNA replication, recombination and repair. Bioessays. 1997;19:233–240. doi: 10.1002/bies.950190309. [DOI] [PubMed] [Google Scholar]
- 11.Liu Y, Kao HI, Bambara RA. Flap endonuclease 1: a central component of DNA metabolism. Annu. Rev. Biochem. 2004;73:589–615. doi: 10.1146/annurev.biochem.73.012803.092453. [DOI] [PubMed] [Google Scholar]
- 12.Shen B, Singh P, Liu R, Qiu J, Zheng L, Finger LD, Alas S. Multiple but dissectible functions of FEN-1 nucleases in nucleic acid processing, genome stability and diseases. Bioessays. 2005;27:717–729. doi: 10.1002/bies.20255. [DOI] [PubMed] [Google Scholar]
- 13.Murante RS, Huang L, Turchi JJ, Bambara RA. The calf 5′- to 3′-exonuclease is also an endonuclease with both activities dependent on primers annealed upstream of the point of cleavage. J. Biol. Chem. 1994;269:1191–1196. [PubMed] [Google Scholar]
- 14.DeMott MS, Shen B, Park MS, Bambara RA, Zigman S. Human RAD2 homolog 1 5′- to 3′-exo/endonuclease can efficiently excise a displaced DNA fragment containing a 5′-terminal abasic lesion by endonuclease activity. J. Biol. Chem. 1996;271:30068–30076. doi: 10.1074/jbc.271.47.30068. [DOI] [PubMed] [Google Scholar]
- 15.Barnes CJ, Wahl AF, Shen B, Park MS, Bambara RA. Mechanism of tracking and cleavage of adduct-damaged DNA substrates by the mammalian 5′- to 3′- exonuclease/endonuclease RAD2 homologue 1 or flap endonuclease 1. J. Biol. Chem. 1996;271:29624–29631. doi: 10.1074/jbc.271.47.29624. [DOI] [PubMed] [Google Scholar]
- 16.Murray JM, Tavassoli M, al-Harithy R, Sheldrick KS, Lehmann AR, Carr AM, Watts FZ. Structural and functional conservation of the human homolog of the Schizosaccharomyces pombe rad2 gene, which is required for chromosome segregation and recovery from DNA damage. Mol. Cell. Biol. 1994;14:4878–4888. doi: 10.1128/mcb.14.7.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reagan MS, Pittenger C, Siede W, Friedberg EC. Characterization of a mutant strain of Saccharomyces cerevisiae with a deletion of the RAD27 gene, a structural homolog of the RAD2 nucleotide excision repair gene. J. Bacteriol. 1995;177:364–371. doi: 10.1128/jb.177.2.364-371.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Storici F, Henneke G, Ferrari E, Gordenin DA, Hubscher U, Resnick MA. The flexible loop of human FEN1 endonuclease is required for flap cleavage during DNA replication and repair. EMBO J. 2002;21:5930–5942. doi: 10.1093/emboj/cdf587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vallen EA, Cross FR. Mutations in RAD27 define a potential link between G1 cyclins and DNA replication. Mol. Cell. Biol. 1995;15:4291–4302. doi: 10.1128/mcb.15.8.4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henneke G, Friedrich-Heineken E, Hubscher U. Flap endonuclease 1: a novel tumour suppresser protein. Trends Biochem. Sci. 2003;28:384–390. doi: 10.1016/S0968-0004(03)00138-5. [DOI] [PubMed] [Google Scholar]
- 21.Bodmer WF, Bailey CJ, Bodmer J, Bussey HJ, Ellis A, Gorman P, Lucibello FC, Murday VA, Rider SH, Scambler P, et al. Localization of the gene for familial adenomatous polyposis on chromosome 5. Nature. 1987;328:614–616. doi: 10.1038/328614a0. [DOI] [PubMed] [Google Scholar]
- 22.Su LK, Kinzler KW, Vogelstein B, Preisinger AC, Moser AR, Luongo C, Gould KA, Dove WF. Multiple intestinal neoplasia caused by a mutation in the murine homolog of the APC gene. Science. 1992;256:668–670. doi: 10.1126/science.1350108. [DOI] [PubMed] [Google Scholar]
- 23.Powell SM, Zilz N, Beazer-Barclay Y, Bryan TM, Hamilton SR, Thibodeau SN, Vogelstein B, Kinzler KW. APC mutations occur early during colorectal tumorigenesis. Nature. 1992;359:235–237. doi: 10.1038/359235a0. [DOI] [PubMed] [Google Scholar]
- 24.Fearnhead NS, Britton MP, Bodmer WF. The ABC of APC. Hum Mol Genet. 2001;10:721–733. doi: 10.1093/hmg/10.7.721. [DOI] [PubMed] [Google Scholar]
- 25.Polakis P. Wnt signaling and cancer. Genes Dev. 2000;14:1837–1851. [PubMed] [Google Scholar]
- 26.Hall PA, Coates PJ, Ansari B, Hopwood D. Regulation of cell number in the mammalian gastrointestinal tract: the importance of apoptosis. J Cell Sci. 1994;107:3569–3577. doi: 10.1242/jcs.107.12.3569. [DOI] [PubMed] [Google Scholar]
- 27.Hasegawa S, Sato T, Akazawa H, Okada H, Maeno A, Ito M, Sugitani Y, Shibata H, Miyazaki Ji. J., Katsuki M, Yamauchi Y, Yamamura Ki K., Katamine S, Noda T. Apoptosis in neural crest cells by functional loss of APC tumor suppressor gene. Proc. Natl. Acad. Sci. USA. 2002;99:297–302. doi: 10.1073/pnas.012264999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jaiswal AS, Narayan S. Reduced levels of the adenomatous polyposis coli (APC) protein are associated with ceramide-induced apoptosis of colon cancer cells. J. Cancer Res. Clin. Oncol. 2004;130:695–703. doi: 10.1007/s00432-004-0591-6. [DOI] [PubMed] [Google Scholar]
- 29.Kaplan KB, Burds AA, Swedlow JR, Bekir S,S, Sorger PK, Nathke IS. A role for the Adenomatous Polyposis Coli protein in chromosome segregation. Nat. Cell Biol. 2001;3:429–432. doi: 10.1038/35070123. [DOI] [PubMed] [Google Scholar]
- 30.Neufeld KL, Nix DA, Bogerd H, Kang Y, Beckerle MC, Cullen BR, White RL. Adenomatous polyposis coli protein contains two nuclear export signals and shuttles between the nucleus and cytoplasm. Proc. Natl. Acad. Sci. USA. 2000;97:12085–12090. doi: 10.1073/pnas.220401797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jaiswal AS, Balusu R, Narayan S. Involvement of adenomatous polyposis coli in colorectal tumorigenesis. Front. Biosci. 2005;10:1118–1134. doi: 10.2741/1605. [DOI] [PubMed] [Google Scholar]
- 32.Narayan S, Jaiswal AS, Balusu R. Tumor suppressor APC blocks DNA polymerase beta-dependent strand displacement synthesis during long patch but not short patch base excision repair and increases sensitivity to methylmethane sulfonate. J. Biol. Chem. 2005;280:6942–6949. doi: 10.1074/jbc.M409200200. [DOI] [PubMed] [Google Scholar]
- 33.Kundu CN, Balusu R, Jaiswal AS, Gairola CG, Narayan S. Cigarette smoke condensate-induced level of adenomatous polyposis coli blocks long-patch base excision repair in breast epithelial cells. Oncogene. 2006. (in press; DOI: 10.1038/sj.onc.1209925) [DOI] [PubMed]
- 34.Narayan S, Jaiswal AS. Activation of adenomatous polyposis coli (APC) gene expression by the DNA-alkylating agent N-methyl-N′-nitro-N-nitrosoguanidine requires p53. J. Biol. Chem. 1997;272:30619–30622. doi: 10.1074/jbc.272.49.30619. [DOI] [PubMed] [Google Scholar]
- 35.Jaiswal AS, Narayan S. p53-dependent transcriptional regulation of the APC promoter in colon cancer cells treated with DNA alkylating agents. J. Biol. Chem. 2001;276:18193–18199. doi: 10.1074/jbc.M101298200. [DOI] [PubMed] [Google Scholar]
- 36.Prasad R, Dianov GL, Bohr VA, Wilson SH. FEN1 stimulation of DNA polymerase beta mediates an excision step in mammalian long patch base excision repair. J. Biol. Chem. 2000;275:4460–4466. doi: 10.1074/jbc.275.6.4460. [DOI] [PubMed] [Google Scholar]
- 37.Liu Y, Beard WA, Shock DD, Prasad R, Hou EW, Wilson SH. DNA polymerase beta and flap endonuclease 1 enzymatic specificities sustain DNA synthesis for long patch base excision repair. J. Biol. Chem. 2005;280:3665–3674. doi: 10.1074/jbc.M412922200. [DOI] [PubMed] [Google Scholar]
- 38.Verma UN, Surabhi RM, Schmaltieg A, Becerra C, Gaynor RB. Small interfering RNAs directed against beta-catenin inhibit the in vitro and in vivo growth of colon cancer cells. Clin. Cancer Res. 2003;9:1291–1300. [PubMed] [Google Scholar]
- 39.Shortle D, Botstein D. Directed mutagenesis with sodium bisulfite. Methods Enzymol. 1983;100:457–468. doi: 10.1016/0076-6879(83)00073-7. [DOI] [PubMed] [Google Scholar]
- 40.Jaiswal AS, Bloom LB, Narayan S. Long-patch base excision repair of apurinic/apyrimidinic site DNA is decreased in mouse embryonic fibroblast cell lines treated with plumbagin: involvement of cyclin-dependent kinase inhibitor p21Waf-1/Cip-1. Oncogene. 2002;21:5912–5922. doi: 10.1038/sj.onc.1205789. [DOI] [PubMed] [Google Scholar]
- 41.Collins AR. The comet assay for DNA damage and repair: principles, applications, and limitations. Mol. Biotechnol. 2004;26:249–261. doi: 10.1385/MB:26:3:249. [DOI] [PubMed] [Google Scholar]
- 42.Brendler-Schwaab S, Hartmann A, Pfuhler S, Speit G. The in vivo comet assay: use and status in genotoxicity testing. Mutagenesis. 2005;20:245–254. doi: 10.1093/mutage/gei033. [DOI] [PubMed] [Google Scholar]
- 43.Stucki M, Jonsson ZO, Hubscher U. In eukaryotic flap endonuclease 1, the C terminus is essential for substrate binding. J. Biol. Chem. 2001;276:7843–7849. doi: 10.1074/jbc.M008829200. [DOI] [PubMed] [Google Scholar]
- 44.Sakurai S, Kitano K, Yamaguchi H, Hamada K, Okada K, Fukuda K, Uchida M, Ohtsuka E, Morioka H, Hakoshima T. Structural eukaryotic flap endonuclease 1, the C terminus is essential for substrate binding. EMBO J. 2005;24:683–693. doi: 10.1038/sj.emboj.7600519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kedar PS, Kim SJ, Robertson A, Hou E, Prasad R, Horton JK, Wilson SH. Direct interaction between mammalian DNA polymerase beta and proliferating cell nuclear antigen. J. Biol. Chem. 2002;277:31115–31123. doi: 10.1074/jbc.M201497200. [DOI] [PubMed] [Google Scholar]
- 46.Zhao LY, Liu J, Sidhu GS, Niu Y, Liu Y, Wang R, Liao D. Negative regulation of p53 functions by Daxx and the involvement of MDM2. J. Biol. Chem. 2004;279:50566–50579. doi: 10.1074/jbc.M406743200. [DOI] [PubMed] [Google Scholar]
- 47.Matsumoto Y. Molecular mechanism of PCNA-dependent base excision repair. Prog. Nucleic Acids Res. Mol. Biol. 2001;68:129–138. doi: 10.1016/s0079-6603(01)68095-4. [DOI] [PubMed] [Google Scholar]
- 48.Harrington JJ, Lieber MR. Functional domains within FEN-1 and RAD2 define a family of structure-specific endonucleases: implications for nucleotide excision repair. Genes Dev. 1994;8:1344–1355. doi: 10.1101/gad.8.11.1344. [DOI] [PubMed] [Google Scholar]
- 49.Kaiser MW, Lyamicheva N, Ma W, Miller C, Neri B, Fors L, Lyamichev VI. A comparison of eubacterial and archaeal structure-specific 5′-exonucleases. J. Biol. Chem. 1999;274:21387–21394. doi: 10.1074/jbc.274.30.21387. [DOI] [PubMed] [Google Scholar]
- 50.Friedrich-Heineken E, Henneke G, Ferrari E, Hubscher U. The Acetylatable Lysines of Human Fen1 are Important for Endo- and Exonuclease Activities. J. Mol. Biol. 2003;328:73–84. doi: 10.1016/s0022-2836(03)00270-5. [DOI] [PubMed] [Google Scholar]
- 51.Prasad R, Lavrik OI, Kim SJ, Kedar P, Yang XP, Vande Berg BJ, Wilson SH. DNA polymerase beta -mediated long patch base excision repair. Poly(ADP-ribose) polymerase-1 stimulates strand displacement DNA synthesis. J. Biol. Chem. 2001;276:32411–32414. doi: 10.1074/jbc.C100292200. [DOI] [PubMed] [Google Scholar]
- 52.Qiu J, Li X, Frank G, Shen B. Cell cycle-dependent and DNA damage-inducible nuclear localization of FEN-1 nuclease is consistent with its dual functions in DNA replication and repair. J. Biol. Chem. 2001;276:4901–4908. doi: 10.1074/jbc.M007825200. [DOI] [PubMed] [Google Scholar]
- 53.Shibata Y, Nakamura T. Defective flap endonuclease 1 activity in mammalian cells is associated with impaired DNA repair and prolonged S phase delay. J. Biol. Chem. 2002;277:746–754. doi: 10.1074/jbc.M109461200. [DOI] [PubMed] [Google Scholar]
- 54.Kawasaki Y, Senda T, Ishidate T, Koyama R, Morishita T, Iwayama Y, Higuchi O, Akiyama T. Asef, a link between the tumor suppressor APC and G-protein signaling. Science. 2000;289:1194–1197. doi: 10.1126/science.289.5482.1194. [DOI] [PubMed] [Google Scholar]
- 55.Nathke IS, Adams CL, Polakis P, Sellin JH, Nelson WJ. The adenomatous polyposis coli tumor suppressor protein localizes to plasma membrane sites involved in active cell migration. J. Cell. Biol. 1996;134:165–179. doi: 10.1083/jcb.134.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kawasaki Y, Sato R, Akiyama T. Mutated APC and Asef are involved in the migration of colorectal tumour cells. Nat. Cell Biol. 2003;5:211–215. doi: 10.1038/ncb937. [DOI] [PubMed] [Google Scholar]
- 57.Moss SF, Liu TC, Petrotos A, Hsu TM, Gold LI, Holt PR. Inward growth of colonic adenomatous polyps. Gastroenterology. 1996;111:1425–1432. doi: 10.1016/s0016-5085(96)70003-3. [DOI] [PubMed] [Google Scholar]
- 58.Hamada F, Bienz M. A Drosophila APC tumour suppressor homologue functions in cellular adhesion. Nat. Cell Biol. 2002;4:208–213. doi: 10.1038/ncb755. [DOI] [PubMed] [Google Scholar]
- 59.Peifer M, Polakis P. Wnt signaling in oncogenesis and embryogenesis-a look outside the nucleus. Science. 2000;287:1606–1609. doi: 10.1126/science.287.5458.1606. [DOI] [PubMed] [Google Scholar]
- 60.Fodde R, Kuipers J, Rosenberg C, Smits R, Kielman M, Gaspar C, van Es JH, Breukel C, Wiegant J, Giles RH, Clevers H. Mutations in the APC tumour suppressor gene cause chromosomal instability. Nat. Cell Biol. 2001;3:433–438. doi: 10.1038/35070129. [DOI] [PubMed] [Google Scholar]
- 61.Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 62.Dixon K, Kopras E. Genetic alterations and DNA repair in human carcinogenesis. Semin. Cancer Biol. 2004;14:441–448. doi: 10.1016/j.semcancer.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 63.Taverna P, Hwang HS, Schupp JE, Radivoyevitch T, Session NN, Reddy G, Zarling DA, Kinsella TJ. Inhibition alterations and DNA repair in human carcinogenesis. Cancer Res. 2003;63:838–846. [PubMed] [Google Scholar]
- 64.Liu L, Nakatsuru Y, Gerson SL. Base excision repair as a therapeutic target in colon cancer. Clin. Cancer Res. 2002;8:2985–2991. [PubMed] [Google Scholar]