Abstract
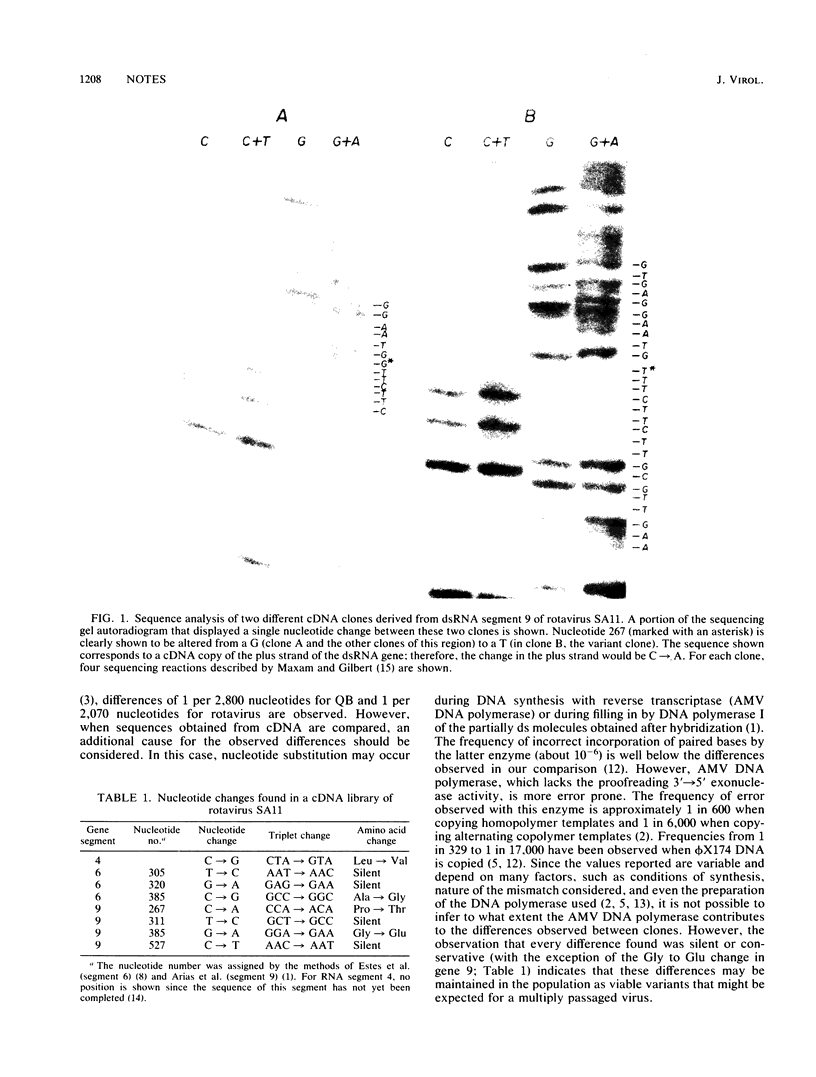
The nucleotide sequences for several complementary DNA clones of the rotavirus genome were determined. When the sequences obtained from different clones for the same regions (16,000 bases) were compared, differences in eight base positions were observed. These discrepancies, approximately 1 in 2,000 bases, may be due to differences in individual RNA genomes resulting from multiple passages; infidelity of DNA synthesis in the cloning procedure; or both factors. Whatever the cause, this frequency of base substitution found in sequences of complementary DNA obtained from the same isolate should be considered when comparing DNA sequences obtained from independent isolates. On the other hand, the frequency of base changes observed suggests that the rotavirus genome is very conserved since the virus used for cDNA synthesis has been continuously passaged for 6 years without plaque purification.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arias C. F., López S., Bell J. R., Strauss J. H. Primary structure of the neutralization antigen of simian rotavirus SA11 as deduced from cDNA sequence. J Virol. 1984 May;50(2):657–661. doi: 10.1128/jvi.50.2.657-661.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battula N., Loeb L. A. The infidelity of avian myeloblastosis virus deoxyribonucleic acid polymerase in polynucleotide replication. J Biol Chem. 1974 Jul 10;249(13):4086–4093. [PubMed] [Google Scholar]
- Both G. W., Bellamy A. R., Street J. E., Siegman L. J. A general strategy for cloning double-stranded RNA: nucleotide sequence of the Simian-11 rotavirus gene 8. Nucleic Acids Res. 1982 Nov 25;10(22):7075–7088. doi: 10.1093/nar/10.22.7075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Both G. W., Mattick J. S., Bellamy A. R. Serotype-specific glycoprotein of simian 11 rotavirus: coding assignment and gene sequence. Proc Natl Acad Sci U S A. 1983 May;80(10):3091–3095. doi: 10.1073/pnas.80.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingo E., Sabo D., Taniguchi T., Weissmann C. Nucleotide sequence heterogeneity of an RNA phage population. Cell. 1978 Apr;13(4):735–744. doi: 10.1016/0092-8674(78)90223-4. [DOI] [PubMed] [Google Scholar]
- Elleman T. C., Hoyne P. A., Dyall-Smith M. L., Holmes I. H., Azad A. A. Nucleotide sequence of the gene encoding the serotype-specific glycoprotein of UK bovine rotavirus. Nucleic Acids Res. 1983 Jul 25;11(14):4689–4701. doi: 10.1093/nar/11.14.4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estes M. K., Mason B. B., Crawford S., Cohen J. Cloning and nucleotide sequence of the simian rotavirus gene 6 that codes for the major inner capsid protein. Nucleic Acids Res. 1984 Feb 24;12(4):1875–1887. doi: 10.1093/nar/12.4.1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopinathan K. P., Weymouth L. A., Kunkel T. A., Loeb L. A. Mutagenesis in vitro by DNA polymerase from an RNA tumour virus. Nature. 1979 Apr 26;278(5707):857–859. doi: 10.1038/278857a0. [DOI] [PubMed] [Google Scholar]
- Holland J., Spindler K., Horodyski F., Grabau E., Nichol S., VandePol S. Rapid evolution of RNA genomes. Science. 1982 Mar 26;215(4540):1577–1585. doi: 10.1126/science.7041255. [DOI] [PubMed] [Google Scholar]
- Imai M., Akatani K., Ikegami N., Furuichi Y. Capped and conserved terminal structures in human rotavirus genome double-stranded RNA segments. J Virol. 1983 Jul;47(1):125–136. doi: 10.1128/jvi.47.1.125-136.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A., Eckstein F., Mildvan A. S., Koplitz R. M., Loeb L. A. Deoxynucleoside [1-thio]triphosphates prevent proofreading during in vitro DNA synthesis. Proc Natl Acad Sci U S A. 1981 Nov;78(11):6734–6738. doi: 10.1073/pnas.78.11.6734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A., Loeb L. A. On the fidelity of DNA replication. Effect of divalent metal ion activators and deoxyrionucleoside triphosphate pools on in vitro mutagenesis. J Biol Chem. 1979 Jul 10;254(13):5718–5725. [PubMed] [Google Scholar]
- López S., Arias C. F., Bell J. R., Strauss J. H., Espejo R. T. Primary structure of the cleavage site associated with trypsin enhancement of rotavirus SA11 infectivity. Virology. 1985 Jul 15;144(1):11–19. doi: 10.1016/0042-6822(85)90300-9. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Schubert M., Harmison G. G., Meier E. Primary structure of the vesicular stomatitis virus polymerase (L) gene: evidence for a high frequency of mutations. J Virol. 1984 Aug;51(2):505–514. doi: 10.1128/jvi.51.2.505-514.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]