Abstract
The molecular chaperone, Hsp90, facilitates the maturation and/or activation of over 100 ‘client proteins’ involved in signal transduction and transcriptional regulation. Largely an enigma among the families of heat shock proteins, Hsp90 is central to processes broadly ranging from cell cycle regulation to cellular transformation. Here we review the contemporary body of knowledge regarding the biochemical mechanisms of Hsp90 and update the most current paradigms defining its involvement in both normal and pathological cell physiology.
Keywords: Hsp90, heat shock protein, molecular chaperone, ATPase, genetic capacitor
Hsp90 defines a family of molecular chaperones that are highly conserved from prokaryotes to eukaryotes [1-5]. Nonessential for normal growth in most bacteria, Hsp90 is abundantly expressed in higher eukaryotes where it has been shown to be necessary for viability [6, 7]. It functions as a homodimer that associates with co-chaperones to catalyze the maturation and/or activation of over 100 substrate proteins that are known to be involved in cell regulatory pathways [5]. These ‘client proteins’ include protein kinases, nuclear hormone receptors, transcription factors, and an array of other essential proteins [8]. While much is known regarding the ATPase-driven conformational cycling of Hsp90, the precise physical effects imparted by this chaperone that serve to activate its substrates are still poorly understood [5].
Hsp90 architecture
Three highly conserved domains comprise the structure of Hsp90. These include the N-terminal domain, responsible for ATP-binding, a proteolytically resistant core domain, and the C-terminal domain that facilitates homodimerization (Fig. 1a) [9]. In eukaryotes, a more variable charged region links the N-terminal domain to the core domain. The length and composition of this linker region is highly divergent among organisms [10]. As no atomic resolution structure for full-length Hsp90 is yet available, the most thorough structural analyses for Hsp90, to date, have been based on crystallographic studies of its individual domains.
Figure 1. The structure of Hsp90 and its ATP-dependent molecular clamp.
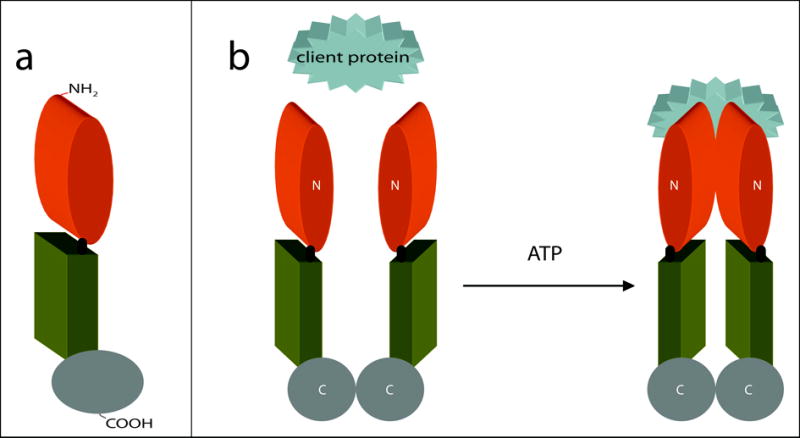
(a) Schematic representation of Hsp90. The Hsp90 monomer is comprised of three domains: the N-terminal domain responsible for ATP-binding (orange), a core domain (green), and a C-terminal domain that facilitates homodimerization (gray). In eukaryotes, a short charged region links the N-terminal and core domains (black). (b) ATP-driven molecular clamp cycle of Hsp90. In the absence of bound nucleotide, the C-termini (C) of two Hsp90 monomers interact to maintain an antiparallel dimer (left). Concurrently, the N-termini (N) of the Hsp90 homodimer preserve an open-state, facilitating the capture of client proteins (left). On the right, association with ATP induces modest changes in the conformation of Hsp90 that permit a transitory interaction between the opposing N-terminal domains. This produces the closed-form of Hsp90 where clamping of the substrate protein occurs.
A mechanistic understanding of Hsp90 was nebulous until partial sequence homology was recognized between its N-terminal domain and two types of ATP-dependent proteins. These included the type II topoisomerases and the MutL DNA mismatch repair enzymes [11]. Structural analyses of the N-terminal domain of Hsp90 revealed that this domain contains an ATP-binding site [12]. Additionally, biochemical studies suggest that transitory interactions between two N-terminal domains of the Hsp90 homodimer occur in an ATP-dependent manner, and this provides the mechanistic basis for an ATPase-driven molecular clamp [13]. Mutations in this region that impair the ability of Hsp90 to either bind or hydrolyze ATP eliminate its chaperone activity [14]. The discovery that the antibiotics, radicicol and geldanamycin, inhibit the Hsp90-dependent activation of numerous regulatory and signal transduction proteins by occupying their ATP-binding sites was a revelation in antitumor research [15]. Indeed, that was the origin of almost a decade of intense efforts focusing on Hsp90 as a therapeutic target for the treatment of cancer.
Biochemical and structural analyses of the core domain of Hsp90 determined that this domain contains a catalytic loop that accepts the γ-phosphate of ATP [16]. This led to the characterization of Hsp90 as a “split ATPase” [16]. Structural and mechanistic similarities shared between Hsp90 and DNA gyrase B serve as the basis for suggesting that the core domain is also involved in the interface of Hsp90 with its client proteins. Strengthening this position, several studies now implicate the core of Hsp90 in its interactions with p53, eNOS, and Akt [17-19].
The advent of the C-terminal crystal structure provided further evidence for the antiparallel dimeric architecture of Hsp90 that had been previously predicted by electron microscopy [20-22]. C-terminal truncations of Hsp90 abolish its ability to hydrolyze ATP, indicating that its dimeric nature is essential for its activity [13]. A highly conserved pentapeptide (MEEVD), present in the C-terminus of eukaryotic Hsp90, is recognized by co-chaperones containing tetratricopeptide repeats [23-25]. Thus, the C-terminal domain is also involved in the formation of active Hsp90 multiprotein complexes.
The structural mechanism for the chaperone activity of Hsp90 has been likened to a ‘molecular clamp.’ In the absence of bound nucleotide, the N-termini of the Hsp90 homodimer maintain an open-state, facilitating the ‘capture’ of client proteins (Fig. 1b) [13]. Association with ATP induces modest changes in the conformation of Hsp90 that permit a transitory interaction between the opposing N-terminal domains. This produces the closed-form of Hsp90 where clamping of the substrate protein occurs [13]. It is through this ATPase-driven cycle that Hsp90, with the assistance of several co-chaperones, induces the activation of its ‘clientelle’ [26].
Hsp90 co-chaperones
Hsp90 is not capable of autonomously functioning as a protein chaperone. Instead, it serves at the core of various multiprotein complexes that incorporate other chaperones, such as Hsp70, and an assortment of co-chaperones [5, 27]. The broadest class of Hsp90 co-chaperones are those containing one or more tetratricopeptide repeat (TPR) motifs that interact with the C-terminal domain of Hsp90 [28]. Beyond the conservation of their TPR motifs, these proteins are remarkably diverse, possessing few overlapping biochemical characteristics [28]. Hop/Sti1, for example, facilitates the interaction between Hsp70 and Hsp90 [29], while WISp39 serves as a client protein specificity factor [30]. A number of TPR-containing co-chaperones even convey their own catalytic activities [5]. These include such enzymes as the E3/E4-ubiquitin ligase, CHIP [31], the protein phosphatase, PP5 [32], and several prolyl isomerases [33, 34]. It is known that CHIP functions in the targeting of Hsp90 client proteins for proteasome-mediated degradation [31]. However, the biological functions associated with the recruitment of other enzymes to Hsp90 chaperone complexes are still unclear.
Several Hsp90 co-chaperones have been shown to regulate the ATPase-driven molecular clamp cycle associated with its N-terminal domain. While Hop/Sti1, p23, and Cdc37 impair the progression of this cycle [24, 35, 36], Aha1 and Cpr6 function to enhance it [35, 37]. Because Hop/Sti1 and Cdc37 are both involved with the recruitment of Hsp90 client proteins, their inhibition of the ATPase cycle is thought to permit the loading of client proteins by maintaining the open clamp conformation of Hsp90 [36, 38]. Cpr6 is known to subsequently displace Hop/Sti1 by competing for the C-terminal TPR-recognition motif of Hsp90, thereby permitting progression of the clamp cycle [24]. The Hsp90 activation potential of Aha1 is achieved through extensive associations along the core domain of Hsp90 that induce conformational changes within its catalytic loop. These adjustments place the active site of the Hsp90 loop in better proximity for the acceptance of the γ-phosphate of ATP [39].
Hsp90 client proteins
The most detailed understanding of the effects of Hsp90 on its client proteins has been gleaned from its involvement with the maturation of steroid hormone receptors. Steroid receptors must be maintained in a labile conformation that allows them to be rapidly activated in the presence of their cognate ligand [40]. Hop1/Sti1, by virtue of its ability to bind Hsp70 and Hsp90 in tandem, facilitates the transfer of Hsp70-bound receptors to the open form of Hsp90. The Hsp90 system then induces subtle alterations in the conformation of the bound steroid receptor that enhances its affinity toward its respective ligand [41].
Protein kinases comprise the most prevalent group of Hsp90 client proteins. The co-chaperone Cdc37 is known to interact both with protein kinases and Hsp90, thereby delivering client kinases to the Hsp90 chaperone complex [42, 43]. Bound to Hsp90, the client kinases are stabilized and remain in a receptive but inactive state while awaiting appropriate signals [42]. The details of the Hsp90-protein kinase chaperone system are still under investigation.
Beyond its specific in vivo role in chaperoning authentic client proteins, Hsp90 has long been noted for its capacity to impede the in vitro aggregation of a broad range of non-specific proteins induced to express in E. coli [44]. Table 1 lists proteins for which we have used strains of E. coli that over-express Hsp90 (Plus90α™; Plus90β™; Expression Technologies Inc., San Diego, CA) to prevent aggregation during expression [45-52]. This illustrates the structural and functional disparity among the in vitro clientele of Hsp90. Figure 2 demonstrates the dramatic increase in soluble protein product rendered in Hsp90-over-expressing strains of E. coli. While the selectivity of Hsp90 for its in vivo client proteins points to a highly specific mechanism, the architectural variation among its substrates, both in vivo and in vitro, implicate a far more general mechanism. In this way, Hsp90 remains an enigma among heat shock proteins.
Table 1.
Partial list of recently established Hsp90 in vitro client proteins
| Protein | Function |
|---|---|
| Smyd2 | catalyzes methylation of p53 (K370) and histone H3 (K36) [45, 48] |
| Smyd3 | catalyzes methylation of histone H3 (K4) [47] |
| RNA polymerase II | RNA polymerase [46] |
| FKBP38 | calcineurin inhibitor [49] |
| MICAL | cytoskeletal regulation [50] |
| skNAC | muscle-specific transcription factor [51] |
| Bcl2 | regulator of apoptosis [52] |
Hsp90 stabilizes over 100 client proteins [5]. Proteins shown in this table are recently established in vitro clients that we have observed in our own laboratory, based on their enhanced solubility and yields in the Hsp90 over-expressing strains of E. coli. Figure 2 illustrates representative results.
Figure 2. Hsp90α and Hsp90β enhance the solubility of proteins expressed in E. coli.
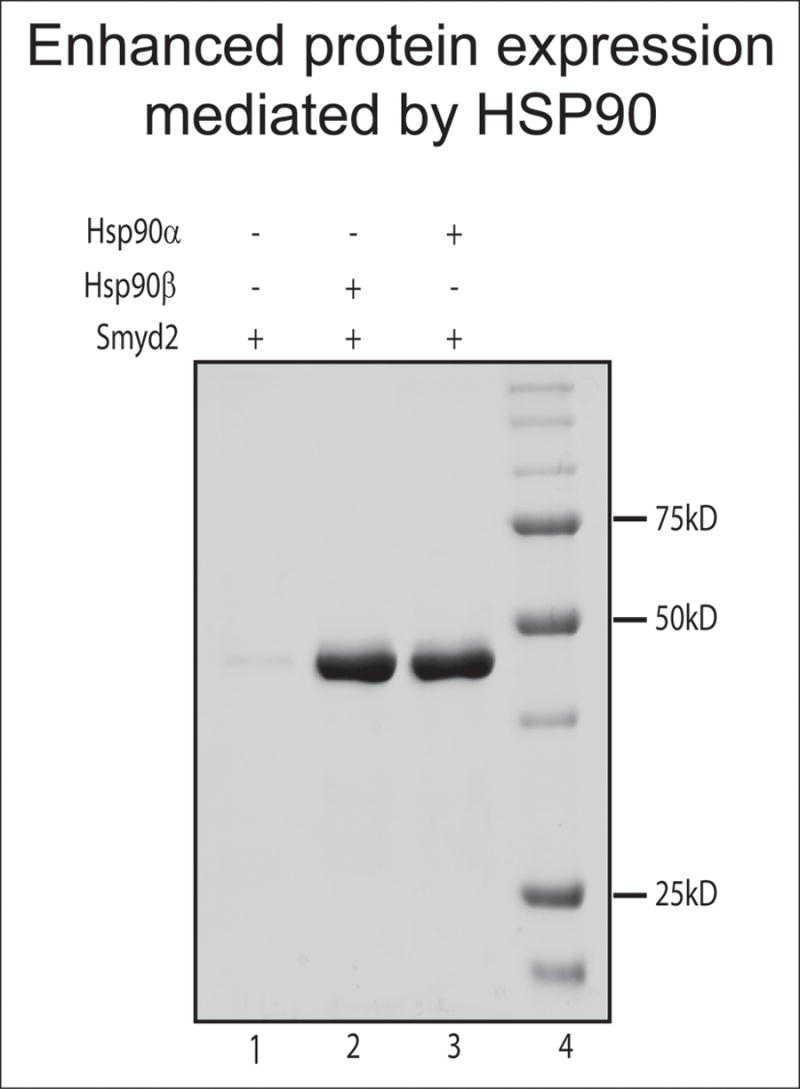
There are two isoforms of Hsp90 (Hsp90α and Hsp90β) which share 85% identity and maintain virtually indistinguishable functional properties [59]. Here, Hsp90 isoform-expressing Escherichia coli strains produce higher yields of soluble Smyd2. Wild type (lane1) and Hsp90 isoform-expressing (lanes 2 and 3) E. coli strains were transformed with a Smyd2 expression construct [45]. Equal amounts (2.5 g wet cell paste) of cultured E. coli cells were used for isolation and purification of Smyd2. The migration of molecular weight markers (in kiloDaltons) is indicated. The depicted results are representative of the enhanced solubility achieved with numerous proteins (Table 1) in Hsp90+ strains.
Chaperoning tumorigenesis
The essential roles that Hsp90 fulfills in the normal physiology of healthy cells are even more critical for the viability of transformed cells. Hsp90 is absolutely essential for the stabilization/maturation of nuclear hormone receptors, transcription factors, and protein kinases that are commonly misregulated during tumorigenesis [8]. It also serves to buffer the effects of transformation by preventing the aggregation of aberrantly expressed proteins, whose accumulation would otherwise result in toxic stress signals and progression to programmed cell death [53]. As many of the client proteins of Hsp90 are linked to growth signal pathways, Hsp90 is viewed as key player in the subversion of normal cells toward unrestrained proliferation. Amplifying the corruptive potential of Hsp90 is its ability to facilitate the evolution of neoplastic clones by stabilizing many of the mutated proteins that are often associated with cancerous lesions, including p53, Bcr-Abl, and v-Src [4, 53]. For this reason, Hsp90 is thought to be especially crucial in the development of tumors that result from the inactivation of DNA repair pathways, in which there are extensive pools of diversely mutated proteins (Fig. 3).
Figure 3. Role of Hsp90 in chaperoning tumorigenesis.
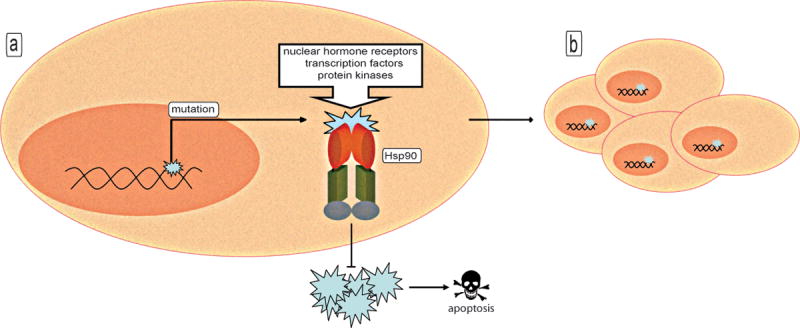
(a) Hsp90 stabilizes many mutated proteins that mediate cell transformation and prevents the aggregation of aberrantly expressed proteins (blue), which would otherwise result in toxic stress signals leading to the progression of apoptosis. (b) The chaperoning capacity of Hsp90 facilitates the evolution of neoplastic clones.
The earliest studies highlighting the antitumor capacities of geldanamycin and radicicol credited their abilities to impair the activity of oncogenic protein kinases such as ErbB-2 and v-Src [54]. It was later shown that the biological target of these drugs is actually Hsp90, and that their use blocks the Hsp90-dependent activity of Raf-1, Cdk4, Src-family kinases, and many other oncogenic targets [53, 54]. Since then, immense progress has been made in the development of pharmacological agents that act as inhibitors of Hsp90. In addition to their role in cancer therapy, these drugs will undoubtedly reveal new insights into the involvement of Hsp90 in diverse physiological processes.
Hsp90 as a ‘genetic capacitor’
By chaperoning mutated clients, Hsp90 facilitates the accumulation of mutant proteins [53, 55]. Beyond the implications this may have in tumorigenesis, the stabilization of proteins that may otherwise be degraded allows Hsp90 to act as a buffer for phenotypic change [53, 55]. Studies in Drosophila and Arabidopsis have revealed that Hsp90 curbs phenotypic variations under ordinary conditions, allowing their manifestation only when Hsp90 is functionally inert [56, 57]. As a chaperone for many proteins that lie along broad-reaching signal cascades, the function of Hsp90 is central to key developmental processes [4, 5]. Therefore, when the activity of Hsp90 is compromised due to environmental stress or the application of Hsp90 inhibitors, the effects are often pleiotropic [58]. In this capacity, Hsp90 has been portrayed as a ‘capacitor for evolution’ [57]. Since a significant portion of mutated proteins stabilized by Hsp90 likely result from genetic mutations, Hsp90 has also been described as a ‘genetic capacitor’ [58]. This adds to the ever-increasing convolution involved with the translation of genotype to phenotype.
Conclusions
From its crucial roles in signal transduction to transformation to genetic capacitance, Hsp90 is a ubiquitous molecular chaperone that influences an expansive array of cellular events through its broad range of protein clientele. Hsp90 has been the focus of intense research for the past 20 years, resulting in the establishment of several overlapping paradigms stemming from the ATP-dependent chaperoning cycle of Hsp90. In spite of this immense progress, many challenges remain. For example, while much is known regarding the ATPase-driven conformational cycling of Hsp90, the precise physical effects imparted by this chaperone that serve to activate its substrates are still poorly understood. In addition, the currently known repertoire of Hsp90-dependent proteins is far from complete. A more comprehensive listing and characterization of its clients will undoubtedly reveal the vast-reaching governance wielded by Hsp90 as an intermediate custodian of far-reaching physiological processes. As the target for several promising lines of cancer therapeutics, Hsp90 is certain to remain the focus of intense research for many years to come.
Acknowledgments
PWT thankfully acknowledges support from the Marie Betzner Morrow Endowment and the NIH (AI47209 and HL071160). MAB is supported by the Short Memorial Endowment. We thank Ms. Chhaya Das for excellent technical assistance and Dr. Chuan Li (Expression Technologies, Inc., San Diego, CA.) for providing Hsp90-expressing E. coli strains and protocols.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Finkelstein DB, Strausberg S. Identification and expression of a cloned yeast heat shock gene. J Biol Chem. 1983;258:1908–1913. [PubMed] [Google Scholar]
- 2.Koyasu S, Nishida E, Kadowaki T, Matsuzaki F, Iida K, Harada F, Kasuga M, Sakai H, Yahara I. Two mammalian heat shock proteins, HSP90 and HSP100, are actin-binding proteins. Proc Natl Acad Sci U S A. 1986;83:8054–8058. doi: 10.1073/pnas.83.21.8054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meng X, Jerome V, Devin J, Baulieu EE, Catelli MG. Cloning of chicken hsp90 beta: the only vertebrate hsp90 insensitive to heat shock. Biochem Biophys Res Commun. 1993;190:630–636. doi: 10.1006/bbrc.1993.1095. [DOI] [PubMed] [Google Scholar]
- 4.Pearl LH, Prodromou C. Structure and in vivo function of Hsp90. Curr Opin Struct Biol. 2000;10:46–51. doi: 10.1016/s0959-440x(99)00047-0. [DOI] [PubMed] [Google Scholar]
- 5.Pearl LH, Prodromou C. Structure and mechanism of the Hsp90 molecular chaperone machinery. Annu Rev Biochem. 2006;75:271–294. doi: 10.1146/annurev.biochem.75.103004.142738. [DOI] [PubMed] [Google Scholar]
- 6.Borkovich KA, Farrelly FW, Finkelstein DB, Taulien J, Lindquist S. hsp82 is an essential protein that is required in higher concentrations for growth of cells at higher temperatures. Mol Cell Biol. 1989;9:3919–3930. doi: 10.1128/mcb.9.9.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Versteeg S, Mogk A, Schumann W. The Bacillus subtilis htpG gene is not involved in thermal stress management. Mol Gen Genet. 1999;261:582–588. doi: 10.1007/s004380051004. [DOI] [PubMed] [Google Scholar]
- 8.Whitesell L, Lindquist SL. HSP90 and the chaperoning of cancer. Nat Rev Cancer. 2005;5:761–772. doi: 10.1038/nrc1716. [DOI] [PubMed] [Google Scholar]
- 9.Terasawa K, Minami M, Minami Y. Constantly updated knowledge of Hsp90. J Biochem (Tokyo) 2005;137:443–447. doi: 10.1093/jb/mvi056. [DOI] [PubMed] [Google Scholar]
- 10.Scheibel T, Siegmund HI, Jaenicke R, Ganz P, Lilie H, Buchner J. The charged region of Hsp90 modulates the function of the N-terminal domain. Proc Natl Acad Sci U S A. 1999;96:1297–1302. doi: 10.1073/pnas.96.4.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bergerat A, de Massy B, Gadelle D, Varoutas PC, Nicolas A, Forterre P. An atypical topoisomerase II from Archaea with implications for meiotic recombination. Nature. 1997;386:414–417. doi: 10.1038/386414a0. [DOI] [PubMed] [Google Scholar]
- 12.Prodromou C, Roe SM, Piper PW, Pearl LH. A molecular clamp in the crystal structure of the N-terminal domain of the yeast Hsp90 chaperone. Nat Struct Biol. 1997;4:477–482. doi: 10.1038/nsb0697-477. [DOI] [PubMed] [Google Scholar]
- 13.Prodromou C, Panaretou B, Chohan S, Siligardi G, O’Brien R, Ladbury JE, Roe SM, Piper PW, Pearl LH. The ATPase cycle of Hsp90 drives a molecular ‘clamp’ via transient dimerization of the N-terminal domains. Embo J. 2000;19:4383–4392. doi: 10.1093/emboj/19.16.4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Panaretou B, Prodromou C, Roe SM, O’Brien R, Ladbury JE, Piper PW, Pearl LH. ATP binding and hydrolysis are essential to the function of the Hsp90 molecular chaperone in vivo. Embo J. 1998;17:4829–4836. doi: 10.1093/emboj/17.16.4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roe SM, Prodromou C, O’Brien R, Ladbury JE, Piper PW, Pearl LH. Structural basis for inhibition of the Hsp90 molecular chaperone by the antitumor antibiotics radicicol and geldanamycin. J Med Chem. 1999;42:260–266. doi: 10.1021/jm980403y. [DOI] [PubMed] [Google Scholar]
- 16.Meyer P, Prodromou C, Hu B, Vaughan C, Roe SM, Panaretou B, Piper PW, Pearl LH. Structural and functional analysis of the middle segment of hsp90: implications for ATP hydrolysis and client protein and cochaperone interactions. Mol Cell. 2003;11:647–658. doi: 10.1016/s1097-2765(03)00065-0. [DOI] [PubMed] [Google Scholar]
- 17.Fontana J, Fulton D, Chen Y, Fairchild TA, McCabe TJ, Fujita N, Tsuruo T, Sessa WC. Domain mapping studies reveal that the M domain of hsp90 serves as a molecular scaffold to regulate Akt-dependent phosphorylation of endothelial nitric oxide synthase and NO release. Circ Res. 2002;90:866–873. doi: 10.1161/01.res.0000016837.26733.be. [DOI] [PubMed] [Google Scholar]
- 18.Muller L, Schaupp A, Walerych D, Wegele H, Buchner J. Hsp90 regulates the activity of wild type p53 under physiological and elevated temperatures. J Biol Chem. 2004;279:48846–48854. doi: 10.1074/jbc.M407687200. [DOI] [PubMed] [Google Scholar]
- 19.Sato S, Fujita N, Tsuruo T. Modulation of Akt kinase activity by binding to Hsp90. Proc Natl Acad Sci U S A. 2000;97:10832–10837. doi: 10.1073/pnas.170276797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harris SF, Shiau AK, Agard DA. The crystal structure of the carboxy-terminal dimerization domain of htpG, the Escherichia coli Hsp90, reveals a potential substrate binding site. Structure. 2004;12:1087–1097. doi: 10.1016/j.str.2004.03.020. [DOI] [PubMed] [Google Scholar]
- 21.Maruya M, Sameshima M, Nemoto T, Yahara I. Monomer arrangement in HSP90 dimer as determined by decoration with N and C-terminal region specific antibodies. J Mol Biol. 1999;285:903–907. doi: 10.1006/jmbi.1998.2349. [DOI] [PubMed] [Google Scholar]
- 22.Shiau AK, Harris SF, Southworth DR, Agard DA. Structural Analysis of E. coli hsp90 reveals dramatic nucleotide-dependent conformational rearrangements. Cell. 2006;127:329–340. doi: 10.1016/j.cell.2006.09.027. [DOI] [PubMed] [Google Scholar]
- 23.Chen S, Sullivan WP, Toft DO, Smith DF. Differential interactions of p23 and the TPR-containing proteins Hop, Cyp40, FKBP52 and FKBP51 with Hsp90 mutants. Cell Stress Chaperones. 1998;3:118–129. doi: 10.1379/1466-1268(1998)003<0118:diopat>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prodromou C, Siligardi G, O’Brien R, Woolfson DN, Regan L, Panaretou B, Ladbury JE, Piper PW, Pearl LH. Regulation of Hsp90 ATPase activity by tetratricopeptide repeat (TPR)-domain co-chaperones. Embo J. 1999;18:754–762. doi: 10.1093/emboj/18.3.754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Young JC, Obermann WM, Hartl FU. Specific binding of tetratricopeptide repeat proteins to the C-terminal 12-kDa domain of hsp90. J Biol Chem. 1998;273:18007–18010. doi: 10.1074/jbc.273.29.18007. [DOI] [PubMed] [Google Scholar]
- 26.Voellmy R, Boellmann F. Chaperone regulation of the heat shock protein response. Adv Exp Med Biol. 2007;594:89–99. doi: 10.1007/978-0-387-39975-1_9. [DOI] [PubMed] [Google Scholar]
- 27.Otaka M, Odashima M, Watanabe S. Role of heat shock proteins (molecular chaperones) in intestinal mucosal protection. Biochem Biophys Res Commun. 2006;348:1–5. doi: 10.1016/j.bbrc.2006.07.028. [DOI] [PubMed] [Google Scholar]
- 28.Travers SA, Fares MA. Functional coevolutionary networks of the Hsp70-Hop-Hsp90 system revealed through computational analyses. Mol Biol Evol. 2007;24:1032–1044. doi: 10.1093/molbev/msm022. [DOI] [PubMed] [Google Scholar]
- 29.Scheufler C, Brinker A, Bourenkov G, Pegoraro S, Moroder L, Bartunik H, Hartl FU, Moarefi I. Structure of TPR domain-peptide complexes: critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine. Cell. 2000;101:199–210. doi: 10.1016/S0092-8674(00)80830-2. [DOI] [PubMed] [Google Scholar]
- 30.Jascur T, Brickner H, Salles-Passador I, Barbier V, El Khissiin A, Smith B, Fotedar R, Fotedar A. Regulation of p21(WAF1/CIP1) stability by WISp39, a Hsp90 binding TPR protein. Mol Cell. 2005;17:237–249. doi: 10.1016/j.molcel.2004.11.049. [DOI] [PubMed] [Google Scholar]
- 31.Jiang J, Ballinger CA, Wu Y, Dai Q, Cyr DM, Hohfeld J, Patterson C. CHIP is a U-box-dependent E3 ubiquitin ligase: identification of Hsc70 as a target for ubiquitylation. J Biol Chem. 2001;276:42938–42944. doi: 10.1074/jbc.M101968200. [DOI] [PubMed] [Google Scholar]
- 32.Silverstein AM, Galigniana MD, Chen MS, Owens-Grillo JK, Chinkers M, Pratt WB. Protein phosphatase 5 is a major component of glucocorticoid receptor.hsp90 complexes with properties of an FK506-binding immunophilin. J Biol Chem. 1997;272:16224–16230. doi: 10.1074/jbc.272.26.16224. [DOI] [PubMed] [Google Scholar]
- 33.Dolinski K, Muir S, Cardenas M, Heitman J. All cyclophilins and FK506 binding proteins are, individually and collectively, dispensable for viability in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1997;94:13093–13098. doi: 10.1073/pnas.94.24.13093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith DF, Baggenstoss BA, Marion TN, Rimerman RA. Two FKBP-related proteins are associated with progesterone receptor complexes. J Biol Chem. 1993;268:18365–18371. [PubMed] [Google Scholar]
- 35.Panaretou B, Siligardi G, Meyer P, Maloney A, Sullivan JK, Singh S, Millson SH, Clarke PA, Naaby-Hansen S, Stein R, Cramer R, Mollapour M, Workman P, Piper PW, Pearl LH, Prodromou C. Activation of the ATPase activity of hsp90 by the stress-regulated cochaperone aha1. Mol Cell. 2002;10:1307–1318. doi: 10.1016/s1097-2765(02)00785-2. [DOI] [PubMed] [Google Scholar]
- 36.Siligardi G, Panaretou B, Meyer P, Singh S, Woolfson DN, Piper PW, Pearl LH, Prodromou C. Regulation of Hsp90 ATPase activity by the co-chaperone Cdc37p/p50cdc37. J Biol Chem. 2002;277:20151–20159. doi: 10.1074/jbc.M201287200. [DOI] [PubMed] [Google Scholar]
- 37.McLaughlin SH, Smith HW, Jackson SE. Stimulation of the weak ATPase activity of human hsp90 by a client protein. J Mol Biol. 2002;315:787–798. doi: 10.1006/jmbi.2001.5245. [DOI] [PubMed] [Google Scholar]
- 38.Richter K, Muschler P, Hainzl O, Reinstein J, Buchner J. Sti1 is a non-competitive inhibitor of the Hsp90 ATPase. Binding prevents the N-terminal dimerization reaction during the atpase cycle. J Biol Chem. 2003;278:10328–10333. doi: 10.1074/jbc.M213094200. [DOI] [PubMed] [Google Scholar]
- 39.Meyer P, Prodromou C, Liao B, Hu S, Mark Roe S, Vaughan CK, Vlasic I, Panaretou B, Piper PW, Pearl LH. Structural basis for recruitment of the ATPase activator Aha1 to the Hsp90 chaperone machinery. Embo J. 2004;23:511–519. doi: 10.1038/sj.emboj.7600060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pratt WB, Galigniana MD, Morishima Y, Murphy PJ. Role of molecular chaperones in steroid receptor action. Essays Biochem. 2004;40:41–58. doi: 10.1042/bse0400041. [DOI] [PubMed] [Google Scholar]
- 41.Steinmetz AC, Renaud JP, Moras D. Binding of ligands and activation of transcription by nuclear receptors. Annu Rev Biophys Biomol Struct. 2001;30:329–359. doi: 10.1146/annurev.biophys.30.1.329. [DOI] [PubMed] [Google Scholar]
- 42.Pearl LH. Hsp90 and Cdc37 -- a chaperone cancer conspiracy. Curr Opin Genet Dev. 2005;15:55–61. doi: 10.1016/j.gde.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 43.Shao J, Irwin A, Hartson SD, Matts RL. Functional dissection of cdc37: characterization of domain structure and amino acid residues critical for protein kinase binding. Biochemistry. 2003;42:12577–12588. doi: 10.1021/bi035138j. [DOI] [PubMed] [Google Scholar]
- 44.Baneyx F, Mujacic M. Recombinant protein folding and misfolding in Escherichia coli. Nat Biotechnol. 2004;22:1399–1408. doi: 10.1038/nbt1029. [DOI] [PubMed] [Google Scholar]
- 45.Brown MA, Sims RJ, 3rd, Gottlieb PD, Tucker PW. Identification and characterization of Smyd2: a split SET/MYND domain-containing histone H3 lysine 36-specific methyltransferase that interacts with the Sin3 histone deacetylase complex. Mol Cancer. 2006;5:26. doi: 10.1186/1476-4598-5-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ducasse M, Brown MA. Epigenetic aberrations and cancer. Mol Cancer. 2006;5:60. doi: 10.1186/1476-4598-5-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hamamoto R, Furukawa Y, Morita M, Iimura Y, Silva FP, Li M, Yagyu R, Nakamura Y. SMYD3 encodes a histone methyltransferase involved in the proliferation of cancer cells. Nat Cell Biol. 2004;6:731–740. doi: 10.1038/ncb1151. [DOI] [PubMed] [Google Scholar]
- 48.Huang J, Perez-Burgos L, Placek BJ, Sengupta R, Richter M, Dorsey JA, Kubicek S, Opravil S, Jenuwein T, Berger SL. Repression of p53 activity by Smyd2-mediated methylation. Nature. 2006;444:629–632. doi: 10.1038/nature05287. [DOI] [PubMed] [Google Scholar]
- 49.Shirane M, Nakayama KI. Inherent calcineurin inhibitor FKBP38 targets Bcl-2 to mitochondria and inhibits apoptosis. Nat Cell Biol. 2003;5:28–37. doi: 10.1038/ncb894. [DOI] [PubMed] [Google Scholar]
- 50.Suzuki T, Nakamoto T, Ogawa S, Seo S, Matsumura T, Tachibana K, Morimoto C, Hirai H. MICAL, a novel CasL interacting molecule, associates with vimentin. J Biol Chem. 2002;277:14933–14941. doi: 10.1074/jbc.M111842200. [DOI] [PubMed] [Google Scholar]
- 51.Yotov WV, St-Arnaud R. Differential splicing-in of a proline-rich exon converts alphaNAC into a muscle-specific transcription factor. Genes Dev. 1996;10:1763–1772. doi: 10.1101/gad.10.14.1763. [DOI] [PubMed] [Google Scholar]
- 52.Zinkel S, Gross A, Yang E. BCL2 family in DNA damage and cell cycle control. Cell Death Differ. 2006;13:1351–1359. doi: 10.1038/sj.cdd.4401987. [DOI] [PubMed] [Google Scholar]
- 53.Calderwood SK, Khaleque MA, Sawyer DB, Ciocca DR. Heat shock proteins in cancer: chaperones of tumorigenesis. Trends Biochem Sci. 2006;31:164–172. doi: 10.1016/j.tibs.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 54.Chiosis G, Neckers L. Tumor selectivity of Hsp90 inhibitors: the explanation remains elusive. ACS Chem Biol. 2006;1:279–284. doi: 10.1021/cb600224w. [DOI] [PubMed] [Google Scholar]
- 55.Bergman A, Siegal ML. Evolutionary capacitance as a general feature of complex gene networks. Nature. 2003;424:549–552. doi: 10.1038/nature01765. [DOI] [PubMed] [Google Scholar]
- 56.Queitsch C, Sangster TA, Lindquist S. Hsp90 as a capacitor of phenotypic variation. Nature. 2002;417:618–624. doi: 10.1038/nature749. [DOI] [PubMed] [Google Scholar]
- 57.Rutherford SL, Lindquist S. Hsp90 as a capacitor for morphological evolution. Nature. 1998;396:336–342. doi: 10.1038/24550. [DOI] [PubMed] [Google Scholar]
- 58.Sangster TA, Lindquist S, Queitsch C. Under cover: causes, effects and implications of Hsp90-mediated genetic capacitance. Bioessays. 2004;26:348–362. doi: 10.1002/bies.20020. [DOI] [PubMed] [Google Scholar]
- 59.Voss AK, Thomas T, Gruss P. Mice lacking HSP90beta fail to develop a placental labyrinth. Development. 2000;127:1–11. doi: 10.1242/dev.127.1.1. [DOI] [PubMed] [Google Scholar]


