Abstract
ATP synthase uses a unique rotary mechanism to couple ATP synthesis and hydrolysis to transmembrane proton translocation. The F1 subcomplex has three catalytic nucleotide binding sites, one on each β subunit, at the interface to the adjacent α subunit. In the x-ray structure of F1 (Abrahams, J. P., Leslie, A. G. W., Lutter, R., and Walker, J. E. (1994) Nature 370, 621–628), the three catalytic β/α interfaces differ in the extent of inter-subunit interactions between the C termini of the β and α subunits. At the closed βDP/αDP interface, a hydrogen-bonding network is formed between both subunits, which is absent at the more open βTP/αTP interface and at the wide open βE/αE interface. The hydrogen-bonding network reaches from βL328 (Escherichia coli numbering) and βQ441 via αQ399, βR398, and αE402 to βR394, and ends in a cation/π interaction between βR394 and αF406. Using mutational analysis in E. coli ATP synthase, the functional importance of the βDP/αDP hydrogen-bonding network is demonstrated. Its elimination results in a severely impaired enzyme but has no pronounced effect on the binding affinities of the catalytic sites. A possible role for the hydrogen-bonding network in coupling of ATP synthesis/hydrolysis and rotation will be discussed.
F1F0-ATP synthase catalyzes the final step of oxidative phosphorylation and photophosphorylation, the synthesis of ATP from ADP and inorganic phosphate. F1F0-ATP synthase consists of the membrane embedded F0 subcomplex with, in Escherichia coli, a subunit composition of ab2c10, and the peripheral F1 subcomplex, with a subunit composition of α3β3γδε. The energy necessary for ATP synthesis is derived from an electrochemical transmembrane proton (or, in some organisms, sodium ion) gradient. Proton flow, down the gradient, through F0 is coupled to ATP synthesis on F1 by a unique rotary mechanism. The protons flow through (half) channels at the interface of a and c subunits, which drives rotation of the ring of c subunits. The c10 ring, together with F1 subunits γ and ε, forms the rotor. Rotation of γ leads to conformational changes in the catalytic nucleotide binding sites on the β subunits, where ADP and Pi are bound. The conformational changes result in formation and release of ATP. Thus, ATP synthase converts electrochemical energy, the proton gradient, into mechanical energy in form of subunit rotation and back into chemical energy as ATP. In bacteria, under certain physiological conditions, the process runs in reverse. ATP is hydrolyzed to generate a transmembrane proton gradient, which the bacterium requires for such functions as nutrient import and locomotion (for reviews, see Refs. 1–6).
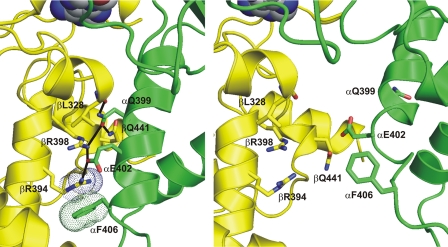
F1 (or “F1-ATPase”) has three catalytic nucleotide binding sites, with pronounced differences in their nucleotide binding affinity. The catalytic nucleotide binding sites are located on the β subunits, at the interface to the adjacent α subunit. In the original x-ray structure of bovine mitochondrial F1 (7), one of the three catalytic sites was filled with the ATP analog AMP-PNP,2 a second one with ADP (plus azide; see ref. 8), and the third site was empty. Hence, the β subunits are referred to βTP, βDP, and βE, and the β/α subunit pairs as βTP/αTP, βDP/αDP, and βE/αE, respectively. The three catalytic β/α interfaces differ in their degree of interaction between both subunits. The βDP/αDP interface has the most extensive contacts between β and α, the βTP/αTP interface has less, and the βE/αE interface has the least. These differences are most pronounced at the C-terminal domains of β and α. Unique to the βDP/αDP interface is an inter-subunit hydrogen-bonding network, reaching from the main-chain oxygen of βL3283 and the side chain of βQ441 via the side chains of αQ399, βR398, and αE402 to the side chain of βR394. The guanidino group of βR394 forms a cation/π interaction with the phenyl ring of αF406 (Fig. 1, left-hand panel). At the βTP/αTP interface, the α-helix carrying residues αQ399, αE402, and αF406 is rotated and has moved away from β by 3–4 Å, preventing all these interactions (Fig. 1, right-hand panel). At the βE/αE interface, the C-terminal domains of two subunits are even further apart.
FIGURE 1.
The βDP/αDP hydrogen-bonding network. The βDP/αDP hydrogen-bonding network is shown in the left-hand panel. Protein backbone and selected side-chain carbon atoms of the β subunit are depicted in yellow;inthe α subunits, these elements are shown in green. Oxygen atoms are in red, nitrogen atoms in blue. The catalytic-site-bound nucleotide is visible at the top of the figure in “space-fill” representation. Possible inter-subunit hydrogen bonds are indicated by black lines. The “dots” represent the van der Waals radii of the guanidino function of βR394 and the phenyl ring of αF406, showing how these groups form a cation/π interaction. For comparison, the right-hand panel shows the same residues at the βTP/αTP interface, which lacks the hydrogen-bonding network. The figure was generated using PyMOL (W. L. DeLano (2002) PyMOL, DeLano Scientific, San Carlos, CA).
The amino acids that make up the hydrogen-bonding network are conserved between bovine mitochondrial ATP synthase and the enzyme from E. coli. In the report presented here, we investigate the functional importance of this hydrogen-bonding network at the βDP/αDP interface of E. coli ATP synthase by perturbing it to varying degrees by site-directed mutagenesis. Single conservative amino acid substitutions were used to change charges and/or geometry of individual hydrogen bonds, single alanine substitutions removed one, two, or three possible hydrogen bonds, and a triple alanine mutant, βR394A/βR398A/βQ441A, prevented formation of all hydrogen bonds except one, as well as formation of the cation/π interaction. Analysis of the mutants demonstrated the functional importance of the hydrogen-bonding network. A possible role for the network in energy coupling will be discussed.
EXPERIMENTAL PROCEDURES
E. coli Strains and Plasmids—The source of wild-type F1 was strain SWM1 (9), the source of βY331W mutant F1 was strain pSWM4/JP17 (10). The template for mutagenesis was plasmid pSN6 (11); thus, all new mutants would carry in addition the βY331W mutation, to allow fluorescence-based nucleotide binding measurements. Mutagenesis was performed using the QuikChange II XL kit (Stratagene). The mutagenic oligonucleotides were designed in such a way that, in addition to the desired mutation, a restriction site would be eliminated or generated, to facilitate screening: Plasmids containing the desired mutation were transformed into strain DK8 (12).
Preparation of Membranes and Enzymes and Functional Analysis of Mutant Strains and Enzymes—Growth yields of E. coli strains in limiting glucose and growth on succinate plates were measured as described previously (13). E. coli membranes were prepared as described (14). F1 was isolated as described previously (15). NADH- and ATP-driven proton pumping in membranes was measured using acridine orange fluorescence quenching (16). The amount of F1 in membrane preparations was estimated via Western blots, using an anti-β antibody (Agrisera, Vännäs, Sweden) or an anti-α/anti-β antibody (kindly provided by Dr. Bill Brusilow, Wayne State University). The staining intensity was quantified using a Photodyne imaging system and the program Image J (National Institutes of Health). ATPase activities were measured in 50 mm Tris/H2SO4, 10 mm ATP, 4 mm MgSO4, pH 8.0, at 23 °C. Released phosphate was determined as described (17, 18).
Fluorescence Measurements—A general outline of nucleotide binding experiments using βTrp331 fluorescence has been described previously (15). The experiments were performed on a spectrofluorometer type Fluorolog 3 (HORIBA Jovin Yvon, Edison, NJ), at 23 °C. The buffer was 50 mm Tris/H2SO4, 2.5 mm MgSO4, pH 8.0. F1 concentration was 30–60 nm. ATP and ADP were added as indicated. To determine MgADP binding in the presence of scandium fluoride, the enzyme was incubated with 0.3 mm ScCl3, 10 mm NaF, 2.5 mm MgSO4, plus the indicated concentration of ADP (19). To correct for dilution and inner filter effects, parallel titrations with wild-type F1 were performed. Kd values were determined from the titration curves as described previously (15).
Modeling—Homology modeling, including energy minimization refinement, was performed using the program PRIME (Schroedinger Inc.). Templates were Protein Data Bank entries 1BMF and 1H8E, the structures of bovine mitochondrial F1 with two (7) and three (20) catalytic sites occupied, respectively. Due to program restrictions regarding the number of amino acids, the N-terminal domains of the α and β subunits were removed.
RESULTS
Assaying the Mutants: An Overview—With the exception of residue βL328, which participates in the βDP/αDP hydrogen-bonding network via its main-chain carbonyl oxygen, the function of the other residues involved in the network was studied by mutational analysis. In addition, residue αF406, which forms a cation/π interaction with one of the terminal residues of the network, βR394, was included in the analysis. The results of the mutagenesis experiments are summarized in Table 1. To characterize the functional effect of each mutation on oxidative phosphorylation in vivo, we determined growth yields in 3 mm glucose and growth on plates containing succinate as the sole carbon source. After preparation of membranes from the mutant strains, we analyzed NADH- and ATP-driven proton gradient formation by measuring quenching of acridine orange fluorescence, and we measured ATPase activity. A low percentage of NADH-induced quenching is indicative of proton permeability of the membranes, which can be caused by oligomeric instability of the mutant ATP synthase complex; of the mutations tested here, only the αQ399C mutant showed a pronounced reduction in NADH-induced acridine orange quenching. The degree of quenching by ATP depends on the balance between ATP-driven proton pumping and proton permeability of the membranes. To determine if variations in enzymatic activities were direct functional consequences of the respective mutation or due to changes in the amount of enzyme on the membrane, we quantified the amount of F1 present in the membrane preparations by Western blots. For most mutants, the amount of F1 was within ±25% of that observed for the parental strain pSN6/DK8. Exceptions were the βQ441A mutant, which had ∼2/3 of the F1 content of the control, the βQ441C mutant, which contained ∼50% more F1 than the control, and the αQ399A mutant, where no F1 was found on the membrane.
TABLE 1.
Functional properties of strains and membranes containing mutations perturbing the βDP/αDP hydrogen-bonding network Growth yield in limiting (3 mm) glucose and growth on succinate plates were determined as in (13). Growth yield data were measured via the turbidity (A590) and are expressed as percentage of the value for the positive control. Quenching of acridine orange fluorescence by NADH and ATP was measured as in (16). The determination of the amount of F1 on the membranes is based on the quantitative evaluation of western blots by densitometric analysis (see “Experimental Procedures”). ATPase activities were determined in 50 mm Tris/H2SO4, 10 mm ATP, 4 mm MgSO4, pH 8.0, at 23 °C. The relative ATPase activity in the last column is expressed as percentage of the activity of the positive control, corrected for the different amounts of F1 on the membrane. Strain pSN6/DK8 served as positive control; it expresses ATP synthase containing a βY331W mutation. βY331W mutant ATP synthase is a normal, active enzyme (10, 15, 46). All mutant strains described in the table were derived from strain pSN6/DK8. Strain pUC118/DK8 does not express ATP synthase and served as negative control.
|
Strain/mutation |
Growth yield in limiting glucose |
Growth on succinate |
Acridine orange quenching |
Amount of F1 on membranes |
Membrane ATPase activity |
||
|---|---|---|---|---|---|---|---|
| NADH-induced | ATP-induced | ||||||
| % | % | % | % | Units/mg | % | ||
| A) Controls | |||||||
| pSN6/DK8 | 100 | ++++ | 94 | 85 | 100 | 0.55 | 100 |
|
pUC118/DK8 (unc–)
|
42
|
–
|
93
|
0
|
0
|
<0.01
|
NDa |
| B) Single mutations | |||||||
| αQ399N | 88 | +++ | 84 | 81 | 112 | 0.45 | 73 |
| αQ399C | 46 | + | 68 | 0 | 82 | 0.02 | 4 |
| αQ399A | 38 | – | 84 | 0 | 0 | <0.01 | ND |
| αE402D | 76 | ++ | 90 | 54 | 111 | 0.31 | 50 |
| αE402Q | 94 | +++ | 92 | 75 | 122 | 0.49 | 72 |
| αE402A | 51 | ++ | 90 | 15 | 121 | 0.06 | 9 |
| αF406Wb | 101b | ||||||
| αF406Cb | 77b | ||||||
| αF406A | 48 | + | 93 | 6 | 104 | 0.18 | 31 |
| βR394K | 58 | ++ | 93 | 21 | 93 | 0.13 | 25 |
| βR394Q | 54 | ++ | 93 | 19 | 88 | 0.13 | 27 |
| βR394A | 52 | ++ | 87 | 22 | 107 | 0.10 | 17 |
| βR398K | 96 | +++ | 91 | 80 | 90 | 0.44 | 89 |
| βR398Hc | 92c | +++ | 71c | ||||
| βR398Q | 92 | +++ | 93 | 66 | 107 | 0.45 | 76 |
| βR398Wc | 102c | ++++ | 91c | 89c | 100c | ||
| βR398Cc | 100c | ++++ | 88c | 85c | 88c | ||
| βR398A | 89 | +++ | 92 | 68 | 88 | 0.37 | 76 |
| βQ441N | 94 | +++ | 92 | 79 | 115 | 0.40 | 63 |
| βQ441C | 68 | ++ | 93 | 53 | 151 | 0.35 | 42 |
|
βQ441A
|
50
|
+
|
88
|
4
|
68
|
0.10
|
27
|
| C) Triple mutation | |||||||
| βR394A/βR398A/βQ441A | 46 | + | 91 | 11 | 104 | 0.06 | 10 |
| S.D.d | 5 | 5 | 10 | 20 | 0.05 | ||
ND, not determinable (correction for amount of F1 leads to division by 0)
Mutations were generated previously.4 The plasmid containing the αF406W mutation was derived from plasmid pBOW1 (47), encoding ATP synthase with Trp-free F1. The plasmid containing the αF406C mutation was derived from pBWU13.4 (48), encoding wild-type ATP synthase. Growth yields are given as percentage of the growth yield of the wild-type control (pBWU13.4/DK8). From both mutant strains an active F1 could be isolated
Mutations were described previously (34, 35). In all cases, wild-type enzyme was used as background. Growth yields and membrane ATPase activities are expressed as percentage of the respective values for the wild-type control. βR398W mutant F1 was isolated and showed normal function (49)
Maximum standard deviation for the values in the respective column. All assays were run at least in triplicate, except for the determination of the amount of F1 on the membrane which was done at least in duplicate
The results of the experiments listed in Table 1 show that the hydrogen-bonding network plays an important role in the function of the enzyme. Many of the mutants showed impaired activity. In most cases, the effect of the mutations on ATP synthesis and hydrolysis was comparable. Although, in general, conservative substitutions had less influence on the enzymatic function less than an alanine replacement, in some cases these differences were smaller than expected. In the following, the results of the mutagenesis experiments will be analyzed in detail.
Mutation of βQ441—The side chain of residue βQ441 can form a hydrogen bond with the side chain of residue αQ399. The mutagenesis results suggest that this hydrogen bond can still be formed when the side chain of βQ441 is shortened by one methylene group to asparagine, because the βQ441N mutant has close-to-normal functional properties. Even the βQ441C mutant seems to have preserved some hydrogen-bonding capabilities. Abolishing the possibility for hydrogen bond formation in the βQ441A mutant impairs the enzyme significantly.
Mutation of αQ399—The side chain of residue αQ399 has a central position in the βDP/αDP hydrogen-bonding network. It can form bonds to the side chains of βQ441 and βR398 as well as to the main-chain carbonyl oxygen of βL328. Like for βQ441, the results of the functional assays suggest that shortening the side chain of αQ399 in the αQ399N mutant leaves the functionally important hydrogen bonds largely unperturbed. Modeling (not shown) of the αQ399N mutation suggested that the hydrogen bonds to the side chains of βQ441 and βR398 might be preserved; in addition, a new hydrogen bond to the hydroxyl group of βY444 seemed possible. Eliminating all potential hydrogen bonds in which the side chain of αQ399 is involved, in the αQ399A mutant, appears to prevent assembly of the enzyme. Visual inspection of the βDP/αDP interaction site indicates that the αQ399C mutant should be capable to form at least one intersubunit hydrogen bond, most likely to βQ441. However, this residual hydrogen-bonding capability does not seem enough to overcome the loss of the other hydrogen bonds; the αQ399C mutant is very strongly impaired.
Mutation of βR398—The guanidino group of βR398 can form hydrogen bonds with the side chains of αQ399 and αE402, plus an intra-subunit hydrogen bond to the main-chain carbonyl oxygen of βQ441. These hydrogen bonds seem of less importance for the functionality of the enzyme. Even a complete removal in the βR398A mutant gives an enzyme with considerable activity.
Mutation of αE402—The carboxylate group of αE402 can form hydrogen bonds with the guanidino functions of βR398 and βR394. Loss of the negative charge in the αE402Q mutant has only a minor effect on enzymatic function. Shortening the side chain by removal of one methylene group in the αE402D mutant reduces the activity by about a half, possibly by preventing formation of one of the hydrogen bonds. Loss of both hydrogen bonds in the αE402A mutant leaves an enzyme with overall <20% residual activity.
Mutation of βR394—The guanidino group of βR394 can form a hydrogen bond with the carboxylate group of αE402, plus a cation/π interaction with the phenyl ring of αF406. In addition, formation of intra-subunit hydrogen bonds with the hydroxyl function of βY367 and the carboxylate group of βE440 seems likely. Complete removal of the hydrogen-bonding capability in the βR394A mutant reduces the activity severely. Interestingly, in the more conservative mutants, βR394K and βR394Q, a similar decrease in activity was observed. A possible explanation is that the lysine and glutamine side chains preferentially form intra-subunit hydrogen bonds, which are not required for the activity of the enzyme, but fail to provide the functionally important interactions with the α subunit.
Mutation of αF406—The phenyl ring of αF406 can make a cation/π interaction with the guanidino group of βR394. Elimination of this interaction, in the αF406A mutant, results in a significant impairment of the enzyme. Earlier studies4 had shown that the phenyl ring can be replaced by an indole ring system, in the αF406W mutant, without loss of activity. The αF406C mutant shows considerably higher activity the αF406A mutant, which might suggest that the cation/π interaction between the side chains of αF406 and βR394 has been substituted by a hydrogen bond between the sulfhydryl group of the Cys and the guanidino group of the Arg, with partial preservation of functionality.
The βR394A/βR398A/βQ441A Triple Mutant—In an attempt to remove the βDP/αDP hydrogen-bonding network as completely as possible, without preventing assembly of the enzyme, we constructed a βR394A/βR398A/βQ441A triple mutant. (It is actually a quadruple mutant as the plasmid used to generate the mutations described here contained the βY331W mutation, to allow fluorescence-based nucleotide binding measurements. As described earlier (10), the βY331W mutation gives a normal, fully functional enzyme.) The βR394A/βR398A/βQ441A mutations would eliminate all possible hydrogen bonds plus the cation/π interaction, with the exception of one hydrogen bond between αQ399 and the backbone carbonyl oxygen of βL328. As expected, the data in Table 1 did not give any indication of assembly problems. This is also supported by the fact that we were successful in preparing F1 from this strain (see below). As can be seen from Table 1, the βR394A/βR398A/βQ441A triple mutant has ∼10% residual activity.
F1 was prepared from the strain containing the βR394A/βR398A/βQ441A triple mutation by the standard procedure (15). The elution profile of the gel chromatography column used as the last purification step indicated that the isolated F1 subcomplex had normal size and shape. SDS-PAGE of the final product revealed a normal subunit composition (data not shown). The isolated F1 exhibited ATPase activity of ∼10% of that of the parental βY331W enzyme (0.56 unit/mg for βR394A/βR398A/βQ441A+βY331W F1, as compared with 5.9 units/mg for βY331W mutant F1; see Refs. 10 and 21).
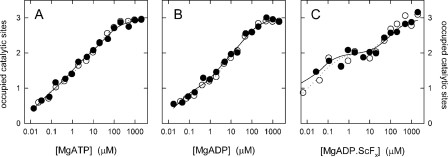
Nucleotide Binding to the βR394A/βR398A/βQ441A Mutant—The fluorescence of the tryptophan in position β331 was used to determine the affinities for binding of MgATP and MgADP to the three catalytic sites (10, 15). The results of the titrations are shown in Fig. 2 (A and B). As can be seen, the affinities of βR394A/βR398A/βQ441A+βY331W mutant F1 do not differ significantly from those of the βY331W control enzyme. For MgATP, the following Kd values were determined: for βR394A/βR398A/βQ441A+βY331W mutant F1, Kd1 = 21 nm, Kd2 = 0.9 μm, Kd3 = 39 μm; for βY331W mutant F1, Kd1 = 20 nm, Kd2 = 1.5 μm, Kd3 = 35 μm. For MgADP, the values were as follows: for βR394A/βR398A/βQ441A+βY331W mutant F1, Kd1 = 24 nm, Kd2 = 2.4 μm, Kd3 = 53 μm; for βY331W mutant F1, Kd1 = 28 nm, Kd2 = 3.6 μm, Kd3 = 41 μm. These data show that the hydrogen-bonding network, which is present in the βY331W control F1, but reduced to one possible hydrogen bond in the βR394A/βR398A/βQ441A+βY331W enzyme, does not contribute to the affinity of any of the three catalytic sites. Specifically, it does not increase the affinity of the medium-affinity site 2, which was recently shown to be located on βDP (22).
FIGURE 2.
Nucleotide binding to the catalytic sites of βR394A/βR398A/βQ441A mutant F1. Nucleotide binding was measured by the decrease in fluorescence of the insertedβW331 residue. Closed symbols, βR394A/βR398A/βQ441A+βY331W F1; open symbols, βY331W control. A, binding of MgATP; B, binding of MgADP; C, binding of the transition state analog MgADP·ScFx. The lines are fits of theoretical curves to the measured data points, assuming three independent sites. For the resulting Kd values and further details, see text.
MgADP·AlFx and MgADP·ScFx are analogs of the transition state that is formed when MgATP is hydrolyzed to MgADP and Pi. Interestingly, although in F1 only the high affinity catalytic site 1 is catalytically active (23), a transition-state-like complex forms with MgADP·AlFx and MgADP·ScFx also at the medium affinity site; formation of this complex manifests itself as a significant increase in nucleotide binding affinity (19, 24). Fig. 2C shows a titration with MgADP in presence of ScCl3 and NaF. As can be seen from the increased affinities, the MgADP·ScFx complex is generated at sites 1 and 2 also in the absence of the βDP/αDP hydrogen-bonding network. The measured Kd values were as follows: for βR394A/βR398A/βQ441A+βY331W mutant F1, Kd1 < 2 nm, Kd2 = 35 nm, Kd3 = 90 μm; for βY331W mutant F1, Kd1 < 2 nm, Kd2 = 58 nm, Kd3 = 66 μm.
Modeling the βR394A/βR398A/βQ441A Mutant—The βR394A/βR398A/βQ441A triple mutant was modeled using either the original structure of bovine mitochondrial F1 with two occupied catalytic sites (7) or the structure with three occupied catalytic sites (20) as template; in both templates, the conformation of the residues forming the βDP/αDP hydrogen-bonding network is very similar. Except for the “mutated” amino acid side chains, the resulting models (not shown) were very similar to the respective template, and therefore to each other. In both models the hydrogen bond between the mainchain oxygen of βL328 and the side chain of αQ399 were preserved. The geometry between the different secondary structural elements carrying the residues that form the network was not affected by the triple alanine substitution.
Conservation of Residues Forming the βDP/αDP Hydrogen-bonding Network—A BLAST (25) search resulted in several thousand sequences each for α and β subunits of ATP synthase. Of the residues forming the βDP/αDP hydrogen-bonding network, βR394 was completely conserved (see Table 2 for selected species). αE402 and αF406 were conserved in >99% of all species. αF406 was replaced by Leu in some plant mitochondrial ATP synthases. In place of αE402 Asp (∼10 cases) and Ser (∼5) were found. αQ399 and βR398 were conserved in ∼90% of all species. In place of βR398 mostly Lys and Phe were found, in place of αQ399 Ala and Ser. In light of the experimental data obtained here, which indicated that the preservation of two possible hydrogen bonds between αQ399 and β might be required for normal function, the latter result was somewhat surprising. Of the residues under investigation here, βQ441 was the most variable. Replacements that can still form a hydrogen bond were found (Asp, Glu, Ser, Asn, and His) but also others where this is no longer possible (Gly, Ala, and Val).
TABLE 2.
Conservation of residues forming the βDP/αDP hydrogen-bonding network Amino acids found in selected species in the position of residues that form the βDP/αDP hydrogen-bonding network in the bovine mitochondrial enzyme (7). Residue numbers refer to the E. coli enzyme.
|
Species |
Position |
|||||
|---|---|---|---|---|---|---|
| β441 | α399 | β398 | α402 | β394 | α406 | |
| Bos taurus | Gln | Gln | Arg | Glu | Arg | Phe |
| Gallus gallus | Gln | Gln | Arg | Glu | Arg | Phe |
| Danio rerio (zebrafish) | Gln | Gln | Arg | Glu | Arg | Phe |
| Drosophila melanogaster | Val | Gln | Arg | Glu | Arg | Phe |
| Caenorhabditis elegans | Val | Gln | Arg | Glu | Arg | Phe |
| Saccharomyces cerevisiae | His | Gln | Arg | Glu | Arg | Phe |
| Neurospora crassa | Gly | Gln | Arg | Glu | Arg | Phe |
| Arabidopsis thaliana | Gln | Gln | Arg | Glu | Arg | Phe |
| Spinacia oleracea | Gln | Gln | Arg | Glu | Arg | Phe |
| Escherichia coli | Gln | Gln | Arg | Glu | Arg | Phe |
| Clostridium acetobutylicum | Ala | Gln | Arg | Glu | Arg | Phe |
| Vibrio alginolyticus | Gln | Ala | Arg | Glu | Arg | Phe |
| Bacillus sp. PS3 | Asp | Ala | Phe | Glu | Arg | Phe |
| Bacillus subtilis | Asp | Ser | Phe | Glu | Arg | Phe |
| Wolinella succinogenes | Asn | Gln | Lys | Glu | Arg | Phe |
| Thermotoga maritima | Gln | Gln | Arg | Glu | Arg | Phe |
| Synechococcus elongatus | Gln | Gln | Lys | Glu | Arg | Phe |
| Methanosarcina barkeri | Gly | Gln | Arg | Glu | Arg | Phe |
DISCUSSION
In the study presented here we analyzed the functional importance of an inter-subunit hydrogen-bonding network that is unique to one of the three β/α interfaces in ATP synthase. This hydrogen-bonding network is located between the C-terminal domains of βDP and αDP. Directly involved in formation of inter-subunit contacts via hydrogen bonds are the main-chain carbonyl oxygen of βL328, the sides chain βQ441, αQ399, βR398, αE402, and βR394, plus, via a cation/π interaction, the phenyl ring of αF406 (Fig. 1). This network is present in all published structures of bovine mitochondrial F1, including the structure with all three catalytic sites occupied by nucleotide (20) and the recent structure obtained in absence of azide (26). Exceptions are the structures that contain the natural inhibitor protein, IF1 (27, 28). IF1 is wedged between the C-terminal domains of βDP and αDP, keeping the interface between both subunits more open (more like at the βTP/αTP interface; see Fig. 1), thus preventing formation of the hydrogen-bonding network (27). Instead, residues αQ399, αE402, αF406, and βR394 appear to contribute to binding of the inhibitor protein (27, 28). In the structure of the yeast mitochondrial F1, the hydrogen-bonding network is present in only one of the three different enzyme conformations; in the two other conformations, the βDP/αDP interface resembled the more open βTP/αTP interface (29). However, as discussed below, there is experimental evidence for the functional importance of some of these residues also in the yeast enzyme. The absence of a comparable hydrogen-bonding network in the recent structure of F1 from a thermoalkaliphilic Bacillus species (30) is not surprising. This structure contains no nucleotide, and all β subunits are in the open conformation, resulting in clefts between the C-terminal domains of β and α at the catalytic interface.
The mutational analysis presented here led to the conclusion that the hydrogen-bonding network is indeed required for normal function of the enzyme. Reduction of the network to a single possible hydrogen bond in the βR394A/βR398A/βQ441A mutant reduced the enzymatic activity to ∼10% of normal. Besides the αQ399A mutant, where no ATP synthase was assembled, the single mutants with the overall lowest activities were αQ399C, βQ441A, αF406A, and αE402A, followed by the three βR394 mutants. These findings indicate that the bonds at either end of the network are more important than the central ones (see Fig. 1). On one end, two hydrogen bonds between αQ399 and β seem to be required for normal function; one of them has to be the bond to βQ441. On the other end, the hydrogen bond between αE402 and βR394 plus the cation/π interaction between βR394 and αF406 appear necessary. Earlier results obtained with an αF406C mutant4 suggest that the cation/π interaction can be replaced by a hydrogen bond.
In general, analysis of the conservation of the residues forming the hydrogen-bonding network confirmed the functional importance of the interactions between αE402 and βR394, and between βR394 and αF406. A possible exception is a group of plant mitochondrial ATPases where αF406 has been replaced by Leu, preventing the cation/π interaction. Unfortunately, the sequences of the β subunits are not known in these cases, making an analysis of the β/α interaction in these enzymes impossible. At the other end of the network, involving positions β441 and α399, a number of cases exist that do not fit into the pattern established here for E. coli ATP synthase (see Table 2). Apparently, some enzymes can compensate for the loss of hydrogen bonds from either of these positions. In the case of the thermophilic Bacillus sp. PS3 ATP synthase, a part of the hydrogen-bonding network appears to be replaced by hydrophobic interactions. Residue β398 is not an Arg, but a Phe, which can interact with the Leu in position α403. Obviously, an Ala in position α399 fits better into this hydrophobic environment than a Gln. We are planning to characterize the βDP/αDP interactions in the PS3 enzyme by mutational analysis as described here.
Although the present study is the first systematic analysis of the βDP/αDP hydrogen-bonding network, the functional importance of some of the participating residues and their capability to form intra-subunit hydrogen bonds had been noticed before. Mutations that suppress ρ0-lethality in the yeast Kluyveromyces lactis were identified in the genes that code for the α, β, and γ subunits of ATP synthase (31). Among these mitochondrial genome integrity, or mgi, mutations (32) found were αF406S,L and βR394G,I,T,V,K (31). None of the mutations abolished the ability of the respective strain to grow on glycerol, and all tested mutations (αF406S and βR394G,K) showed some ATPase activity of isolated mitochondria (31). These results are comparable to those described here for mutations of residues αF406 and βR394. Introduction of a similar set of mutations in Saccharomyces cerevisiae (αF406S and βR394G,I) gave similar results (32). In contrast, another study using S. cerevisiae (33) found that mutations of residue βR394 (in βR394I,T mutants) prevented growth on a nonfermentable lactate medium and eliminated mitochondrial ATPase activity. Mutational analysis of the function of residue βR398 in S. cerevisiae identified the residue as non-essential (33), similar to the results obtained here.
Residue βR398 was identified as necessary to confer E. coli with sensitivity to the antibiotic aurovertin; mutation to His, Cys, and Trp rendered ATPase and ATP synthesis activities resistant to the antibiotic (34, 35). Furthermore, bacterial species where the equivalent residue is phenylalanine seem to be aurovertin-resistant (36). The x-ray structure of bovine mitochondrial F1 complexed with aurovertin (37) showed that two molecules of the antibiotic were bound per F1, one to βTP, the other to βE. In both β subunits residue βR398 contributed to aurovertin binding via hydrogen bond(s). The βDP/αDP hydrogen-bonding network was unperturbed.
After having established the importance of the βDP/αDP hydrogen-bonding network, the question about its role in the mechanism of ATP synthase remains. In the description of the original x-ray structure of F1-ATPase, the fact that the βDP/αDP interface is more closed than the ones at βTP/αTP or βE/αE had led to the proposal that the catalytic site on βDP might be the high affinity site (7). However, using fluorescence resonance energy transfer we could recently demonstrate that the catalytic site on βTP is the high affinity site, whereas the βDP site is the medium affinity site (22). Furthermore, in the present study it is shown that a nearly complete removal of the βDP/αDP hydrogen-bonding network has no influence on nucleotide-binding affinities at any of the three catalytic sites. It is interesting to note in this context that the molecular modeling data suggested that elimination of the hydrogen-bonding network in the βR394A/βR398A/βQ441A mutant does not significantly affect the conformation of the protein backbone of the C-terminal domains of βDP and αDP. Specifically, the βDP/αDP interface in the βR394A/βR398A/βQ441A mutant does not seem to be more “open” than in the wild-type enzyme.
Based on the localization of mgi mutations in K. lactis and their functional consequences, Wang et al. (32) concluded that the respective wild-type residues, including the two contributing to the hydrogen-bonding network, αF406 and βR394, might be “necessary for efficient coupling of ATP synthase, possibly by acting as support to fix the axis of rotation of the central stalk.” Based on the data obtained here, we would like to propose a specific role for the βDP/αDP hydrogen-bonding network in coupling ATP synthesis/hydrolysis and rotation. It is important to note that in the α3β3 ring αDP is opposite of βE. βE carries the low affinity catalytic site, which binds the incoming MgATP during multisite ATP hydrolysis and releases the newly formed MgATP during ATP synthesis (3, 22, 23, 38, 39). It is generally assumed that MgATP binding and release are energy-linked, i.e. the large conformational changes in β accompanying these processes are coupled to rotation of γ (22, 39–42). In ATP hydrolysis direction, MgATP binding and the associated closing of the βE subunit appear to drive the 80° rotation substep; a recent molecular dynamics study supports this notion (43). When βE closes it exerts a force on γ, which has to be converted into a rotary motion of γ, instead of a lateral motion, away from the axis of rotation, which would not allow the catalytic cycle to continue (Fig. 3). As the C terminus of αDP is in the direction of such a lateral motion, hydrogen-bonding it to the C terminus of βDP should make it more rigid, offering greater resistance to a lateral motion of γ (for a more detailed discussion, see the legend to Fig. 3). In ATP synthesis, the situation is similar. The rotating γ subunit, powered by the flow of protons through the F0 subcomplex, has to be kept in a position where its rotation can be translated into opening of the low affinity catalytic site and release of the newly formed MgATP.
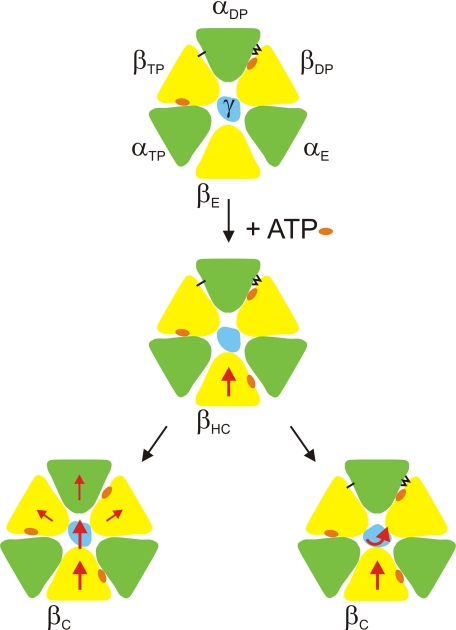
FIGURE 3.
Model of the possible role of the βDP/αDP hydrogen-bonding network in ATP hydrolysis. A transection of F1 at the level of the C-terminal domains ofα andβ is shown, as seen from the direction of the membrane; green, α subunits; yellow, β subunits; blue, γ subunit. A nucleotide bound to the catalytic site in the central domain of the respective β subunit is indicated by an orange oval. The black lines between βDP and αDP indicate the hydrogen-bonding network. The single black line betweenαDP andβTP indicates the possible hydrogenbond(s) between the C-terminal domains of these subunits (see “Discussion”); the functional importance of the latter interaction has not yet been proven. As some of the subunit movements postulated here might be rather subtle, compared with the overall size of the F1 subcomplex, the direction of movement of a specific subunit is indicated by a red arrow. The starting position (top panel) corresponds to the two-nucleotide x-ray structure (7) where the catalytic sites onβTP and βDP are filled with nucleotide, whereas the low affinity nucleotide binding site on βE is empty and the βE subunit is in an open conformation. MgATP binds to the open βE site, and the site starts to close by moving the C-terminal domain toward the pseudosymmetry axis. The β subunit transiently assumes a “half-closed” conformation, “βHC”(middle panel), similar to the conformation observed for the low affinity site in the three-nucleotide structure (20), until it finally reaches the fully closed “βC” conformation (This βC conformation could be very similar to the βTP conformation that the subunit will assume after completion of the 120° rotation step.). Because of steric clashes, γ has to be pushed “out of the way” to allow the β subunit to close completely. In the model, this can occur either by a lateral motion in the same general direction as the “pushing” C-terminal domain of βHC (bottom left-hand panel), or it can be translated into a rotary motion (bottom right-hand panel). The latter movement is the desired one, as only it will allow the overall reaction to proceed. The lateral motion will exert some pressure on the C-terminal domains of αDP, βDP, and/or βTP. Only if these domains can move away from the pseudosymmetry axis (bottom left), the lateral motion can occur (it does not necessarily mean that the enzyme would disintegrate under these circumstances, because it is still held together byα/β interactions in the N-terminal domain and in the nucleotide-binding domain.). According to the model, the βDP/αDP hydrogen-bonding network and, possibly, the hydrogen bonds between αDP and βTP prevent the movement of the C-terminal domains away from the axis, thus forcingγ into a rotary instead of a lateral motion (bottom right).
With the exception of the αQ399A mutant, which did not assemble, none of the mutants with a reduced set of hydrogen bonds was completely inactive. In the context of the model presented here this means that even the most impaired mutants had occasionally a rotation step, probably interspersed with non-productive events where γ moved laterally away from the rotation axis.
The model proposed here is supported by the finding that the bonds at either end of the network are more important than the central ones. The bonds at the end are sufficient to define the geometry of the interactions between αDP and βDP. The central bonds confer additional binding energy, which is, at least under the conditions applied her, not absolutely required; this appears to be another example of “overengineered” subunit-subunit interactions in ATP synthase (44).
The one or two hydrogen bonds that can be formed between the C-terminal domains of αDP and βTP (see Fig. 3) might play a role similar to that of the βDP/αDP hydrogen-bonding network. In the bovine mitochondrial enzyme, the involved residues are αE355 and βS383 (bovine numbers). In E. coli these residues are not conserved but have been replaced by αN358 and βE369, respectively, which could also form a hydrogen bond. Alternatively, a hydrogen bond seems possible between αN358 and βQ365. The function of these residues will be tested in an approach similar to the one described here.
Finally, it should be noted that the functional importance of the βDP/αDP hydrogen-bonding network gives an explanation as to why the medium affinity catalytic site has to be occupied during multisite catalysis (45). So far, only the roles of the other two sites were clearly defined. The high affinity site on βTP performs catalysis, and nucleotide binding and release occur at the low affinity site on βE. Without nucleotide bound to the medium affinity site, the βDP subunit will assume an open conformation, which will prevent formation of the hydrogen-bonding network, therefore impairing catalysis.
This work was supported, in whole or in part, by National Institutes of Health Grant GM071462 (to J. W.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviation used is: AMPPNP, 5′-adenylyl-β,γ-imidodiphosphate.
E. coli residue numbers are used unless indicated otherwise.
S. Nadanaciva, J. Weber, and A. E. Senior, unpublished data.
References
- 1.Leslie, A. G. W., and Walker, J. E. (2000) Philos. Trans. R. Soc. Lond. B Biol. Sci. 355 465-471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Noji, H., and Yoshida, M. (2001) J. Biol. Chem. 276 1665-1668 [DOI] [PubMed] [Google Scholar]
- 3.Weber, J., and Senior, A. E. (2003) FEBS Lett. 545 61-70 [DOI] [PubMed] [Google Scholar]
- 4.Wilkens, S. (2005) Adv. Protein Chem. 71 345-382 [DOI] [PubMed] [Google Scholar]
- 5.Dimroth, P., von Ballmoos, C., and Meier, T. (2006) EMBO Rep. 7 276-282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weber, J. (2006) Biochim. Biophys. Acta 1757 1162-1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abrahams, J. P., Leslie, A. G. W., Lutter, R., and Walker, J. E. (1994) Nature 370 621-628 [DOI] [PubMed] [Google Scholar]
- 8.Bowler, M. W., Montgomery, M. G., Leslie, A. G. W., and Walker, J. E. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 8646-8649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rao, R., Al-Shawi, M. K., and Senior, A. E. (1988) J. Biol. Chem. 263 5569-5573 [PubMed] [Google Scholar]
- 10.Weber, J., Wilke-Mounts, S., Lee, R. S. F., Grell, E., and Senior, A. E. (1993) J. Biol. Chem. 268 20126-20133 [PubMed] [Google Scholar]
- 11.Ahmad, Z., and Senior, A. E. (2004) J. Biol. Chem. 279 31505-31513 [DOI] [PubMed] [Google Scholar]
- 12.Klionsky, D. J., Brusilow, W. S. A., and Simoni, R. D. (1984) J. Bacteriol. 160 1055-1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Senior, A. E., Langman, L., Cox, G. B., and Gibson, F. (1983) Biochem. J. 210 395-403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Senior, A. E., Latchney, L. R., Ferguson, A. M., and Wise, J. G. (1984) Arch. Biochem. Biophys. 228 49-53 [DOI] [PubMed] [Google Scholar]
- 15.Weber, J., and Senior, A. E. (2004) Methods Enzymol. 380 132-152 [DOI] [PubMed] [Google Scholar]
- 16.Perlin, D. S., Cox, D. N., and Senior, A. E. (1983) J. Biol. Chem. 258 9793-9800 [PubMed] [Google Scholar]
- 17.Taussky, H. H., and Shorr, E. (1953) J. Biol. Chem. 202 675-685 [PubMed] [Google Scholar]
- 18.Van Veldhoven, P. P., and Mannaerts, G. P. (1987) Anal. Biochem. 161 45-48 [DOI] [PubMed] [Google Scholar]
- 19.Nadanaciva, S., Weber, J., and Senior, A. E. (2000) Biochemistry 39 9583-9590 [DOI] [PubMed] [Google Scholar]
- 20.Menz, R. I., Walker, J. E., and Leslie, A. G. W. (2001) Cell 106 331-341 [DOI] [PubMed] [Google Scholar]
- 21.Weber, J., Dunn, S. D., and Senior, A. E (1999) J. Biol. Chem. 274 19124-19128 [DOI] [PubMed] [Google Scholar]
- 22.Mao, H. Z., and Weber, J. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 18478-18483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scanlon, J. A. B., Al-Shawi, M. K., Le, N. P., and Nakamoto, R. K. (2007) Biochemistry 46 8785-8797 [DOI] [PubMed] [Google Scholar]
- 24.Nadanaciva, S., Weber, J., and Senior, A. E. (1999) J. Biol. Chem., 274 7052-7058 [DOI] [PubMed] [Google Scholar]
- 25.Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D. J. (1997) Nucleic Acids Res. 25 3389-3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bowler, M. W., Montgomery, M. G., Leslie, A. G. W., and Walker, J. E. (2007) J. Biol. Chem. 282 14238-14242 [DOI] [PubMed] [Google Scholar]
- 27.Cabezon, E., Montgomery, M. G., Leslie, A. G. W., and Walker, J. E. (2003) Nat. Struct. Biol. 10 744-750 [DOI] [PubMed] [Google Scholar]
- 28.Gledhill, J. R., Montgomery, M. G., Leslie, A. G. W., and Walker, J. E. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 15671-15676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kabaleeswaran, V., Puri, N., Walker, J. E., Leslie, A. G. W., and Mueller, D. M. (2007) EMBO J. 25 5433-5442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stocker, A., Keis, S., Vonck, J., Cook, G. M., and Dimroth, P. (2007) Structure 15 904-914 [DOI] [PubMed] [Google Scholar]
- 31.Clark-Walker, G. D., Hansbro, P. M., Gibson, F., and Chen, X. J. (2000) Biochim. Biophys. Acta 1478 125-137 [DOI] [PubMed] [Google Scholar]
- 32.Wang, Y., Singh, U., and Mueller, D. M. (2007) J. Biol. Chem. 282 8228-8236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ichikawa, N., Chisuwa, N., Tanase, M., and Nakamura, M. (2005) J. Biochem. (Tokyo) 138 201-207 [DOI] [PubMed] [Google Scholar]
- 34.Lee, R. S. F., Pagan, J., Satre, M., Vignais, P. V., and Senior, A. E. (1989) FEBS Lett. 253 269-272 [DOI] [PubMed] [Google Scholar]
- 35.Lee, R. S. F., Pagan, J., Wilke-Mounts, S., and Senior, A. E. (1991) Biochemistry 30 6842-6847 [DOI] [PubMed] [Google Scholar]
- 36.Hicks, D. B., and Krulwich, T. A. (1990) J. Biol. Chem. 265 20547-20554 [PubMed] [Google Scholar]
- 37.van Raaij, M. J., Abrahams, J. P., Leslie, A. G. W., and Walker, J. E. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 6913-6917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weber, J., Bowman, C., and Senior, A. E. (1996) J. Biol. Chem. 271 18711-18718 [DOI] [PubMed] [Google Scholar]
- 39.Gao, Y. Q., Yang, W., and Karplus, M. (2005) Cell 123 195-205 [DOI] [PubMed] [Google Scholar]
- 40.Gao, Y. Q., Yang, W., Marcus, R. A., and Karplus, M. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 11339-11344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yasuda, R., Noji, H., Yoshida, M., Kinosita, K., Jr., and Itoh, H. (2001) Nature 410 898-904 [DOI] [PubMed] [Google Scholar]
- 42.Nishizaka, T., Oiwa, K., Noji, H., Kimura, S., Muneyuki, E., Yoshida, M., and Kinosita, K., Jr. (2004) Nat. Struct. Mol. Biol. 11 142-148 [DOI] [PubMed] [Google Scholar]
- 43.Pu, J., and Karplus, M. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 1192-1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weber, J., Wilke-Mounts, S., and Senior, A. E. (2003) J. Biol. Chem. 278 13409-13416 [DOI] [PubMed] [Google Scholar]
- 45.Weber, J., and Senior, A. E. (2001) J. Biol. Chem. 276 35422-35428 [DOI] [PubMed] [Google Scholar]
- 46.Löbau, S., Weber, J., and Senior, A. E. (1998) Biochemistry 37 10846-10853 [DOI] [PubMed] [Google Scholar]
- 47.Weber, J., Wilke-Mounts, S., and Senior, A. E. (2002) J. Biol. Chem. 277 18390-18396 [DOI] [PubMed] [Google Scholar]
- 48.Ketchum, C. J., Al-Shawi, M. K., and Nakamoto, R. K. (1998) Biochem. J. 330 707-712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weber, J., Lee, R. S. F., Grell, E., and Senior, A. E. (1992) Biochemistry 31 5112-5116 [DOI] [PubMed] [Google Scholar]