Abstract
Antibody microarray technology identified Nup88 (nucleoporin 88) as a highly up-regulated protein in response to osmotic stress in inner medullary collecting duct (IMCD3) cells. Changes in expression were verified by Western blot and quantitative PCR for protein and message expression. In mouse and human kidney, Nup88 expression was substantial in the papilla, whereas it was nearly absent in the cortex. Furthermore, the expression of Nup88 increased 410.4 ± 22% in the papilla of mice after 36 h of thirsting. Nup88 protein expression in IMCD3 cells was significantly up-regulated in the first 8 h following exposure to acute osmotic stress, indicating that Nup88 is an early response protein. To define the function of Nup88 in the osmotic stress response, the transcription factor associated with hypertonicity, tonicity enhancer-binding protein (TonEBP), was cloned upstream of the green fluorescent protein. Employing this construct, we demonstrate that silencing Nup88 in IMCD3 cells acutely stressed to hypertonic conditions reduces nuclear retention of TonEBP, resulting in a substantial blunting in transcription of important osmotic stress response target genes and reduced cell viability. Finally, we show that in IMCD3 cells, nuclear export of TonEBP under isotonic conditions involves CRM-1 but under hypertonic stress is CRM1-independent. Our data, therefore, suggest that Nup88 is up-regulated in response to hypertonic stress and acts to retain TonEBP in the nucleus, activating transcription of critical osmoprotective genes.
The cells that inhabit the hypertonic environment of the inner medulla of the kidney possess a number of adaptative mechanisms that allow them to survive in this environment (1-5). The classical osmotic stress response involves the prompt transcription of several target genes by the tonicity enhancer-binding protein (TonEBP),2 also known as NFAT5 (6-9). Under isotonic conditions (300 mosmol/kg H2O), TonEBP is mainly present in the cytosol with only minor localization in the nucleus. However, under hypertonic stress, TonEBP is translocated to the nucleus, where it enhances the transcription of genes that are important in the early osmotic stress response. These genes includes aldose reductase (AR) (10), the sodium-myoinositol transporter (10), the BGT1 (betaine/GABA transporter 1) (11), the taurine transporter (TauT) (12), and heat shock protein 70 (Hsp70) (13) among others. Expression of these target genes results in the accumulation of a number of compatible organic osmolytes (mainly sorbitol, myoinositol, betaine, and taurine) that allow the cell to compensate the extracellular osmotic gradient and, hence, adapt to hypertonic stress (see Refs. 3 and 6-9 for excellent reviews). One of the main mechanisms involved in the regulation of TonEBP activity under hypertonic stress is nucleocytoplasmic trafficking (7, 9, 14, 15).
Our laboratory has employed several proteomic approaches, including two-dimensional difference gel electrophoresis (16) and antibody microarray (17) technologies to evaluate hypertonicity-induced up-regulation of important proteins in inner medullary collecting duct (IMCD3) cells. Here, we describe the marked up-regulation of Nup88 (nucleoporin 88) under hypertonic stress. Nup88 was first identified as an interacting partner of Nup214 (nucleoporin 214) (18). These two proteins are components of a nuclear pore complex in the nuclear membrane and are involved in the nucleocytoplasmic trafficking of different molecules, including transcription factors (19-22).
It was previously demonstrated in Drosophila that Nup88 does not interfere in the nuclear import of NF-κB and instead acts as an inhibitor of CRM1-mediated protein export (20, 22). In experiments with mutated (inactive) Nup88, CRM1 expression is mislocalized in the nucleus and is not present in the nuclear membrane. Interestingly, this mislocalization resulted in a continuous shuttling of the NF-κB Rel-like transcription factor Dorsal in and out of the nucleus (22). These data suggest that CRM1 and, therefore, Nup88 expression are necessary for regulating Dorsal nuclear export. The mechanism by which Nup88 functions is by sequestering CRM1 at the nuclear pore and thereby avoiding its recycling back to the nucleus for another round of export. Similarly, another Rel-like transcription factor, Dif, seems to be regulated by Nup88 expression in Drosophila (20). Likewise, other members of the Rel-like transcription factor class, including TonEBP (10), may be controlled by the expression of Nup88.
The up-regulation of Nup88 under hypertonic stress in the kidney has not been previously described. The present work was undertaken to confirm the observations made by antibody microarray analysis and further characterize the osmotic regulation of expression, half-life, cellular distribution, and in vivo expression. Furthermore, we identify a potential role for Nup88 in the nuclear retention of TonEBP in hypertonically stressed cells.
EXPERIMENTAL PROCEDURES
Materials—Cell culture medium, FCS, and antibiotics were from Invitrogen. Antibodies to Nup88 and GS15 were obtained from Clontech, anti-AR antibody was purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA), anti-Hsp70 was from Stressgen (Ann Arbor, MI), and anti-β-actin was from Cell Signaling (Danvers, MA). All other chemicals were purchased from Sigma.
Antibody Microarray—Antibody microarrays and reagents were purchased from Clontech. Microarrays were processed as per the manufacturer's protocol and as previously described (17). Antibody microarrays were analyzed using a ProScanArray HT scanner (PerkinElmer Life Sciences).
Cell Culture—The established IMCD3 cell line originally developed by Rauchman et al. (23) was provided by Steve Gullans (Rx Gen, Hamden, CT). IMCD3 cells chronically adapted to 600 and 900 mosmol/kg H2O were previously established in our laboratory (24, 25) and were compared with cells grown at isotonic conditions. In experiments involving hypertonic stress, the media in culture dishes were exchanged for that with added NaCl to the specified osmolality, depending on the experiment. Osmolality was determined with a microosmometer (model 3300; Advanced Instruments, Norwood, MA).
Cell Viability and Growth Experiments—Cell viability in tissue culture stress experiments was determined by cell counts following exposure to sublethal osmotic stress. Experiments were initiated once cells reached confluence at isotonic conditions in 24-well flat bottom tissue culture plates (35-3047; Falcon BD Labware, Franklin Lakes, NJ), with each experimental time point after exposure to the sublethal osmotic stress performed in triplicate. In cell growth studies, cell counts were initiated when cultures had reached ∼90% confluence and followed for an additional 80-h period with counts at 24 and 48 h. Growth curves were fit to linear regression using the Prism 4.0 software, and the slopes of the curves were compared for relative differences in growth rate.
Stable Cell Lines Silenced for Nup88 Expression—IMCD3 cultures were transfected with the pSM2 empty vector (v2MM_173601) or the short hairpin RNA vector pSM2-Nup88 (v2HS_152407; Open Biosystems) using Lipofectamine 2000 (Invitrogen) as described by the manufacturer. Stable transfectants (clones) were selected from colonies growing in plates from a 10-fold dilution series in media prepared with 10 μg/ml puromycin antibiotic (Sigma). The absence of Nup88 expression in silenced clones was verified by Western blot of cell lysates obtained from these clones grown in media adjusted to 500 mosmol/kg H2O for 48 h.
Cloning and Transfection of the Construct TonEBP-GFP—The first 800 bp of mouse TonEBP, including the nuclear export signal, the auxiliary export domain, and the nuclear localization signal, was obtained by PCR using the following primers, which contained, respectively, XhoI and EcoRI restriction sites (both underlined) for directional cloning into pAcGFP-N1 vector (Clontech): sense, 5′-GAC CTC GAG ATG CCC TCG GAC TTC ATC TCA TTG CTC-3′; antisense, 5′-GAC GAA TTC GAG GTG CTT TGG CAC TGT CGG CAT CAA-3′. Transfection into IMCD3 cells or into the stable cell lines expressing pSM2-Nup88 vector or pSM2-Empty vector control was performed using Lipofectamine 2000.
Mouse Kidney Tissues—C57/B6 mice were obtained from Jackson Laboratories (Bar Harbor, ME). Mice were subjected to food and ad libitum water or deprived for water for 36 h. Urine samples were collected from the bladder for osmolality analysis. Mice were harvested by cervical dislocation, and kidneys were removed. Papilla and cortex tissues were dissected and homogenized with a glass tissue grinder on ice with mitogen-activated protein kinase lysis buffer for protein and analyzed as described (24).
Human Kidney Tissues—Human kidney tissues from cortex and papilla were obtained under the Colorado Multiple Institutional Review Board and National Institutes of Health Grant U19A10636030 from a kidney that was not suitable for transplantation. Tissues were processed for protein as described above.
RNA Extraction, Analysis, and Message Quantification—Cytosolic RNA was isolated from confluent cell cultures using the RNeasy kit (Qiagen, Valencia, CA) as per the manufacturer's protocol. Before quantitative PCR (QPCR), sample RNA concentration and integrity was assessed by UV spectrometry (absorbance at 260 nm) and by capillary electrophoresis using a bioanalyzer (model 210, Agilent, Foster City, CA; using the 28 S to 18 S rRNA ratio), respectively. RNA was converted to cDNA using the iScript cDNA synthesis kit (Bio-Rad). QPCR was performed using primer pairs identified in supplemental Table 3, designed using Beacon Designer 7.0 (Premier Biosoft, Palo Alto, CA). QPCR runs were performed using the SYBR green JumpStart Taq Readymix QPCR kit (Sigma) on an I-Cycler (Bio-Rad). QPCR runs were analyzed by agarose gel electrophoresis and melt curve to verify that the correct amplicon was produced. β-Actin RNA was used as internal control in all QPCRs, and the amount of RNA was calculated by the comparative CT method.
Protein Extraction and Western Blotting—Cell protein lysates were obtained from confluent cell culture dishes as previously described (24). Protein concentration was determined by the BCA protein assay (Pierce). Seventy micrograms of total protein was loaded per lane for SDS-PAGE (7.5%, w/v) analysis and then transferred to polyvinylidene difluoride membranes. Membranes were incubated with primary antibody and visualized by using an alkaline phosphatase secondary antibody and Lumi-Phos reagent (Pierce) as described by the manufacturer. Chemiluminescence was recorded with an Image Station 440CF (Kodak Digital Science), and results were analyzed with 1D Image Software (Kodak Digital Science). Blots were also analyzed for β-actin as a loading control.
Confocal Fluorescence Microscopy—IMCD3 cells were seeded in 8-well chambers (Nunc) and stained using an anti-Nup88 antibody (1:50 dilution in phosphate-buffered saline; Clontech) or anti-TonEBP antibody (1:400 dilution in PBS) kindly provided by Dr. H. Moo Kwon (University of Maryland). Inmunofluorescence was carried out as previously described (17). Preparations and TonEBP-GFP fusion protein transfections were imaged with a ×40 water immersion objective using a laser-scanning confocal microscope (model LSM510; Zeiss, Thornwood, NY). Data were analyzed with LSM Image analyzer postacquisition software (Zeiss).
Statistics and Data Analysis—All data are presented as the mean ± S.E. Data graphics and statistical analysis were performed using Instat (version 3.0) and Prism 4 (both from GraphPad, San Diego, CA). Independent replicates for each data point (n) are identified in figure legends. p < 0.05 was recognized as statistically significant.
RESULTS
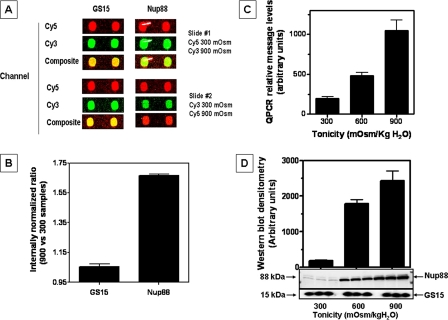
Nup88 Expression Is Up-regulated under Hypertonic Stress—Employing antibody microarray proteomics (Clontech, Mountain View, CA), we found that 5% of the 512 proteins analyzed were up-regulated (17) in IMCD3 cells chronically adapted to hypertonic stress (900 mosmol/kg H2O) as compared with cells grown under isotonic conditions. As shown in Fig. 1A, the Golgi vesicle soluble N-ethylmaleimide-sensitive factor attachment protein receptor protein (GS15) is representative of the greater than 90% of the proteins whose expression did not change under hypertonic conditions. In contrast, Nup88 demonstrated a substantial increase in expression in hypertonically adapted cells. A statistical analysis of four spots for each protein (from two arrays; i.e. dye swap (Fig. 1B)) showed an internal normalization ratio for Nup88 signal intensity of 1.65 (p < 0.001) as compared with 1.05 for GS15. Validation of antibody microarray data was performed by QPCR and Western blot. Data shown in Fig. 1C indicate a substantial increase in Nup88 message levels (7-fold; p < 0.001) in cells adapted to 900 mosmol/kg H2O as compared with isotonic conditions (300 mosmol/kg H2O). This up-regulation was also substantial for protein expression (12.9-fold; p < 0.001) under the same conditions. For comparison, Western blots prepared with anti-GS15 demonstrated equal protein expression in IMCD3 cells at isotonic and hypertonic conditions (Fig. 1D).
FIGURE 1.
Identification of Nup88 as an up-regulated protein in IMCD3 cells in response to osmotic stress by antibody microarray proteomics. A, individual spot signals were analyzed with ScanArrayGx/ProScanarray software. Selected antibody pairs for array 1 are compared with the dye swap array 2 and display pseudocolors for Cy3 (green) and Cy5 (red). Greater than 90% of the array spots demonstrated equal signal intensity (yellow in the composite) as illustrated by the response for Golgi vesicle soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) (GS15) protein. In contrast, spots corresponding to the nucleoporin Nup88 showed a substantial increase in signal intensity from hypertonic adapted cells, as shown in the composite spots for both arrays. B, channel intensity from both arrays was used to calculate the internal normalization ratio for GS15 representing no change (value between 0.82 and 1.15) as compared with a Nup88 internal normalization ratio value of 1.65 (p < 0.001). C, QPCR was performed using specific primers for Nup88 and demonstrated a 7-fold increase in message for IMCD3 cells adapted to 900 versus 300 mosmol/kg H2O(p < 0.001). D, Western blot data reveal a 12-fold increase in Nup88 protein expression in IMCD3 cells chronically adapted to hypertonicity as compared with cells maintained at isotonic conditions. Data represent the average ± S.E. for three independent experiments performed in triplicate (n = 9). Both QPCR and Western blot data were normalized using β-actin message and protein expression, respectively. A representative Western blot is shown including GS15 as a loading control.
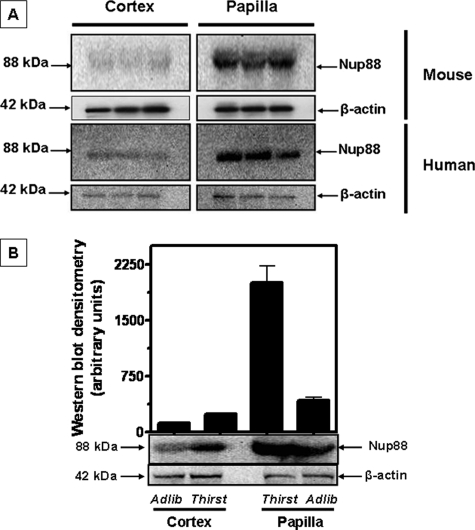
Expression of Nup88 in Renal Cortex and Medulla from Rodents and Human Kidney—To assess whether the changes seen in cultured cells are also observed in vivo, we examined the renal tissues of mice and a human kidney. Western blot data shown in Fig. 2A reveal a near absence of Nup88 protein expression in the cortex of both species. In contrast, substantial Nup88 protein expression was determined in the papilla for both mice and human. These determinations were made in mice on ad libitum water intake. The difference between cortex and papilla is quantitatively depicted in Fig. 2B. We also examined the kidney tissues of mice subjected to 36 h of thirsting (urine osmolality increased from 1,424 ± 211 to 3,105 ± 524 mosmol/kg H2O; n = 6). As shown in Fig. 2B, thirsting increased Nup88 expression by 2-fold (107 ± 12%, p < 0.01) in the cortex and 5-fold (410 ± 22%, p < 0.01) in the papilla tissues.
FIGURE 2.
Nup88 expression in mouse and human kidney tissues. Mouse kidney tissues (cortex and papilla) were harvested from ad libitum water mice (n = 3) and following 36 h of water restriction (thirst, n = 3). Expression of Nup88 protein was substantial in kidney papilla as compared with little or no expression in cortex tissues (A, top). Nup88 protein expression in human cortex and papilla tissues was similar to that observed in mouse tissues as shown in A (bottom). Water restriction in mice led to a 2-fold increase in Nup88 protein expression in the cortex and a 5-fold increase in protein expression in papilla (p < 0.01) (B). Data represent the mean ± S.E. from three Western blots performed in triplicate (n = 9).
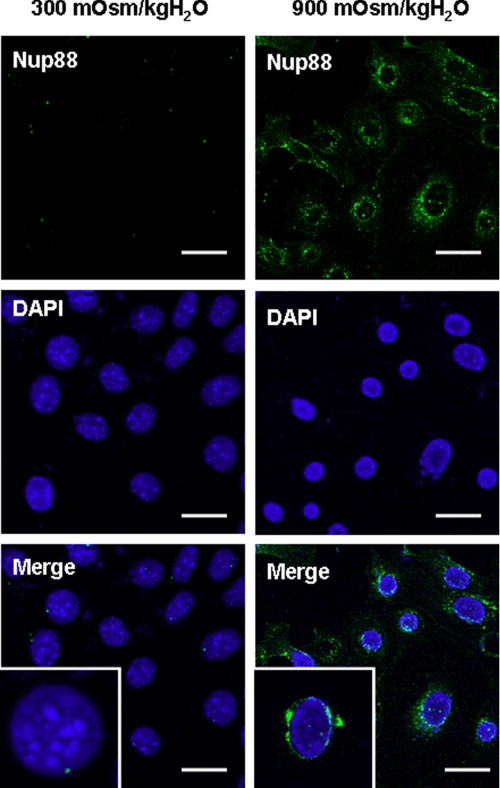
Immunocytochemical Localization of Nup88 in Cells Adapted to Hypertonicity—Immunofluorescence microscopy studies were undertaken to assess the presence and localization of Nup88 protein in IMCD3 cells at isotonic conditions and chronically adapted to hypertonicity (900 mosmol/kg H2O). Fig. 3 shows that Nup88 expression (green) is substantial in IMCD3 cells chronically adapted to 900 mosmol/kg H2Oas compared with isotonic conditions where no Nup88 signal is found. The presence of Nup88 in the nuclear membrane is confirmed by colocalization with the nuclear marker DAPI.
FIGURE 3.
Cellular expression of Nup88 in IMCD3 cells chronically adapted to hypertonicity. Confocal immunohistochemistry analysis of IMCD3 cells grown under isotonic (left) and hypertonic (right) conditions for Nup88 protein expression (300 versus 900 mosmol/kg H2O). Cells grown under isotonic conditions demonstrated little or no expression of Nup88 (green) as compared with cells chronically adapted to hypertonicity. Counter-staining of the nucleus with DAPI demonstrates the localization of Nup88 at the nuclear border. Scale bars, 20 μm.
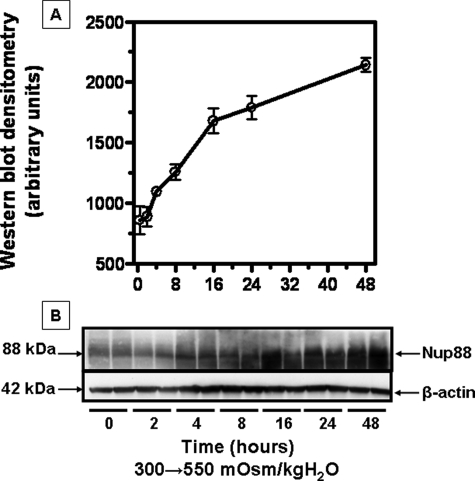
Kinetics of Nup88 Message and Protein Expression with Acute Exposure to Hypertonicity—Nup88 protein expression in IMCD3 cells subjected to acute sublethal osmotic stress was evaluated to determine the timing of expression. To this end, we performed Western blot analysis for protein at numerous time points after exposure to acute sublethal hypertonicity (550 mosmol/kg H2O). Data shown in Fig. 4 demonstrate a significant increase in Nup88 protein expression after 4-6 h of hypertonic stress (p < 0.01). To assess the half-life for Nup88 message and protein, IMCD3 cells were returned to isotonic conditions. Data shown in Fig. 5 were curve-fit for exponential decay, and the half-life was calculated to be 2.8 and 18.4 h for message and protein, respectively. Separately, cells were treated with actinomycin D or cyclohexamide to evaluate potential changes in the stability of Nup88 message and protein. These data were essentially identical to the data shown in Fig. 5 indicating no significant changes in the stability of Nup88 message or protein (data not shown).
FIGURE 4.
Effect of acute sublethal osmotic stress (550 mosmol/kg H2O) on Nup88 protein expression in IMCD3 cells. Cell lysates (0-48 h) were analyzed by Western blot. A, data depict the mean ± S.E. from three Western blots performed in duplicate (n = 6). B, a representative Western blot is shown.
FIGURE 5.
Estimation of half-life for Nup88 mRNA and protein in IMCD3 cells. A, QPCR and Western blot analysis were from three independent experiments performed in duplicate (n = 6). B, a representative Western blot including a β-actin loading control is shown.
Studies on the Osmotic and Ionic Mediators of the Up-Regulation of Nup88 Expression to Hypertonic Stress—To assess whether the effects observed with increasing NaCl levels were unique to this solute, Nup88 protein expression was measured after exposure to other mediators added to the medium to reach the same osmolality (550 mosmol/kg H2O). As depicted in Fig. 6, replacement of sodium by choline or chloride by acetate does not affect the response (p < 0.01 versus isotonic). Other solutes, including sucrose, mannitol, and, interestingly, urea, also caused a marked increase in Nup88 protein expression (p < 0.01 versus isotonic).
FIGURE 6.
Effects of acute osmotic stress using various solutes on MUPP1 protein expression in IMCD3 cells. Solutes were added to increase medium tonicity to 550 mosmol/kg H2O, and cells were exposed for 48 h. Cell lysates were analyzed by Western blot. Data depict the mean ± S.E. from four Western blots performed in duplicate (n = 8). A representative Western blot, including a β-actin loading control is shown.
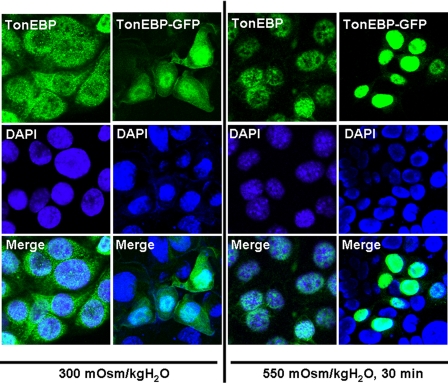
Development and Use of a TonEBP-GFP Construct to Study Trafficking in IMCD3 Cells under Changing Osmotic Stress—Primers for the N terminus of human TonEBP (amino acids 1-267) that include the nuclear export signal, auxiliary export domain, and nuclear localization signal were designed to amplify an 800-bp fragment from cDNA of murine IMCD3 cells chronically adapted at 900 mosmol/kg H2O. Sequence data for the amplicon and its translation are shown as supplemental data (supplemental Tables 1 and 2, respectively). The amplicon was cloned into pAcGFP-N1 (Clontech) upstream of the green fluorescent protein (GFP). To validate the utility of the TonEBP-GFP construct, a comparison was made with antibody labeling of fixed cells. Fig. 7 shows the results obtained from IMCD3 cells transfected with the TonEBP-GFP construct (columns 2 and 4) as compared with immunohistochemistry analysis (columns 1 and 3). In both cases, cells maintained at isotonic conditions (300 mosmol/kg H2O) demonstrated that the majority of the TonEBP signal is present in the cytosol (with some nuclear staining), whereas in cells acutely stressed to 550 mosmol/kg H2O for 30 min, TonEBP is rapidly shifted to the nucleus. Analysis of confocal images reveals that the TonEBP-GFP construct cells demonstrate a more profound and quantifiable image as compared with immunostained cells. The application of the TonEBP-GFP construct also, and more importantly, allows for live imaging of TonEBP trafficking in IMCD3 cells in response to changes in acute osmotic stress. We therefore employed the TonEBP-GFP construct in further experiments.
FIGURE 7.
Comparative analysis of the detection of endogenous TonEBP versus the TonEBP-GFP construct. Confocal analysis of endogenous TonEBP staining (columns 1 and 3) in IMCD3 cells maintained at isotonic conditions (300 mosmol/kg H2O) (column 1) or after 30 min of acute hypertonic stress (550 mosmol/kg H2O) (column 3) shows a similar pattern compared with that obtained employing the TonEBP-GFP construct (columns 2 and 4). TonEBP (green) is primarily localized in the nucleus (DAPI; blue) following acute osmotic stress.
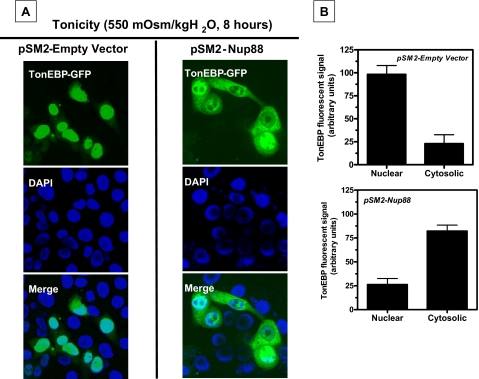
TonEBP Is Exported from the Nucleus under Hypertonic Stress in Cells Silenced for Nup88 Expression—IMCD3 cells were silenced for expression of Nup88 employing a commercial vector pSM2-Nup88 (Open Biosystems). BLAST and alignment analysis were performed to ascertain the selectivity of the silencing sequence with other genes that may induce off-target responses. Analysis reveals a <70% homology of the silencing sequence with well known nucleoporins and exportins. In addition, message levels for nucleoporins and exportins reported to be involved with Nup88, including Nup153 (26), Nup358 (27), Nup214 (28), and CRM1 (18), did not change significantly in Nup88-silenced cells as compared with empty vector controls (data not shown). Stable silenced clones using puromycin (10 μg/ml) were validated as depicted in supplemental Fig. 1A. Two clones (clones 3 and 6) were selected for further experiments, since they showed nearly complete silencing of Nup88 expression under hypertonic stress. Fig. 8A depicts a stable control cell line expressing pSM2-Empty Vector (left) or pSM2-Nup88 clone 3 (right) and transiently transfected with the TonEBP-GFP construct. In cells expressing the empty vector control, TonEBP (green) is present in the nucleus after 8 h of acute sublethal stress (550 mosmol/kg H2O), as demonstrated by its colocalization with DAPI (blue). In contrast, in Nup88-silenced cells (right), the TonEBP is mainly present in the cytosol under hypertonic conditions.
FIGURE 8.
Nup88 retains TonEBP in the nucleus under hypertonic stress. A, the left panel shows the stable cell line obtained by transfection with the pSM2 empty vector control. After 8 h of acute hypertonic stress, TonEBP (green) is substantially present in the nucleus, as demonstrated by colocalization with DAPI (blue). Alternatively, the right panel shows the stable cell line in which Nup88 expression is silenced. TonEBP is primarily present in the cytosol under osmotic stress conditions. B, fluorometry intensity analysis for cells depicted in A (n = 39/6 wells for pSM2-empty vector cells and n = 36/6 wells for pSM2-Nup88 cells from three different experiments). Fluorescent data indicate that 80% of the signal is present in the nucleus of empty vector control cells under hypertonic stress as compared with 25% for the Nup88-silenced cells.
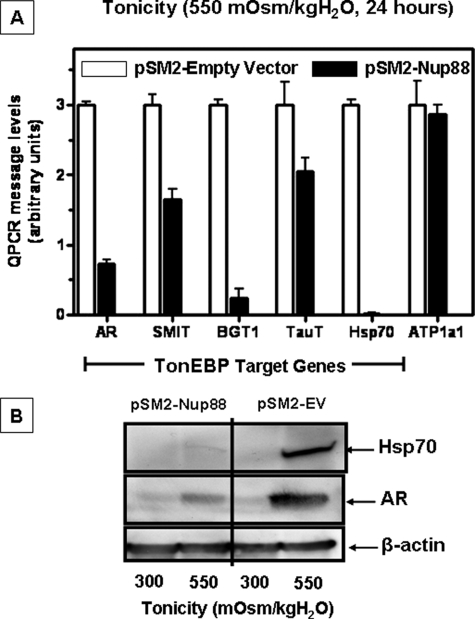
Comparative fluorescence measurement of the TonEBP-GFP signal is shown in Fig. 8B and reveals that 80 ± 4% of the TonEBP signal remains in the nucleus of empty vector control cells, whereas only 25 ± 2% of TonEBP remains in nucleus of cells silenced for Nup88 expression (p < 0.001, n > 35 cells/condition). This finding is supported by a significant decrease in message levels of TonEBP target genes (p < 0.001). As shown in Fig. 9A, levels of AR, sodium-myoinositol transporter, BGT1, TauT, and Hsp70 are lower in Nup88 silenced cells (solid bars) as compared with empty vector control cells (open bars) (p < 0.001). Fig. 9B depicts a representative Western blot for AR and Hsp70 from these cells in which levels of AR protein expression are significantly blunted in cells silenced for Nup88 as compared with empty vector control cells. Interestingly, there is a complete absence of Hsp70 protein expression in cells silenced for Nup88, corroborating the data obtained by QPCR. In contrast, mRNA levels of non-TonEBP target genes, such as the α1 subunit of the Na/K-ATPase (ATP1a1) (29), did not change in Nup88-silenced cells as compared with the empty vector control cells (Fig. 9A).
FIGURE 9.
Effect of silencing Nup88 on TonEBP target gene expression under acute hypertonic stress in IMCD3 cells. A, message levels of TonEBP target genes (AR, BGT1, TauT, and Hsp70) are decreased in the absence of Nup88 under acute sublethal hypertonic stress (550 mosmol/kg H2O, 24 h). Open bars, relative message levels in empty vector control stable cells; solid bars, relative message levels in the stable Nup88-silenced cells. A comparison with message levels for the α1 subunit of Na/K-ATPase (a non-TonEBP target) is shown. B, a representative Western blot for AR and Hsp70 protein expression in empty vector and Nup88-silenced cells treated as above is shown. A β-actin loading control is included.
We also examined the expression of AR and Hsp70 in a clone that was partially silenced, clone 4, and a clone that had almost full Nup88 expression, clone 5 (see supplemental Fig. 1A). As shown in supplemental Fig. 1, B, C, and D, an intermediate phenotype was found for AR, and Hsp70 mRNA (Fig. 1C) and protein (Fig. 1D) expression were found in the partially silenced clone 4. In contrast, no significant difference was found between empty vector control cells and the nonsilenced clone 5.
Silencing Nup88 Expression in IMCD3 Cells Increases Cellular Osmosensitivity to Acute Hypertonic Stress—A comparison of the effect of silencing Nup88 protein expression on the survival of IMCD3 cells during acute sublethal hypertonic stress (575 mosmol/kg H2O) is shown in Fig. 10. Under isotonic conditions, growth rates are comparable for Nup88-silenced cells as compared with empty vector control cells (Fig. 10A). However, under acute hypertonic stress, cells silenced for Nup88 expression demonstrated greater osmosensitivity with a 42% increase in initial cell death over empty vector control cells at 24 h (p < 0.01). Continued incubation at stress resulted in only minor increases in cell numbers in Nup88-silenced cells as compared with complete recovery and substantial growth in the empty vector control cells (Fig. 10B).
FIGURE 10.
Effect of silencing Nup88 expression on IMCD3 cell survival under hypertonic conditions. A, comparison of cell growth rates of IMCD3 clones expressing the empty vector or silencer for Nup88 under isotonic conditions. Data indicate no significant difference in the growth rates over a 72-h period. B, comparison of cell survival and growth for empty vector and Nup88-silenced cells under sublethal hypertonic stress conditions (575 mosmol/kg H2O). A substantial difference in immediate cell viability at 24 h was determined for Nup88-silenced cells as compared with empty vector controls (42%, p < 0.001). In addition, whereas empty vector control cells demonstrated recovery and cell growth over the ensuing 4 days, in Nup88-silenced cultures, no significant cell growth was determined (p < 0.001 between controls and silenced cells). Data represent the mean ± S.E. for three independent experiments performed in triplicate (n = 9).
TonEBP Nuclear Retention by Nup88 under Hypertonic Conditions Is CRM1-Independent—As previously described, Nup88 has been identified to interact with CRM1 (also known as exportin 1) (18). This interaction affects the trafficking of Rel-like transcription factors (20). Since TonEBP is a Rel-like transcription factor, experiments were undertaken to evaluate the potential role of CRM1 in TonEBP trafficking in IMCD3 cells under isotonic and hypertonic stress. Under isotonic conditions, the addition of 10 ng/ml leptomycin B (LMB), a selective inhibitor of CRM-1, resulted in a translocation of TonEBP from the cytosol to the nucleus (supplemental Fig. 2A, right) as compared with cells without LMB addition (left). These data clearly identify that CRM1 is important in the trafficking of TonEBP out of the nucleus under isotonic conditions. However, when IMCD3 cells silenced for Nup88 expression are subjected to hypertonic stress in the presence or absence of LMB (supplemental Fig. 2B, middle and right), TonEBP is found primarily in the cytosol, indicating that CRM1 does not provide the same shuttling under hypertonic stress conditions. These images are compared with normal IMCD3 cells expressing Nup88 (pSM2-empty vector, supplemental Fig. 2B, left) under hypertonic conditions and without LMB addition. These data point to the potential involvement of alternative exportins for shuttling TonEBP under hypertonic stress conditions, as previously suggested in other studies (15).
DISCUSSION
Cellular changes in the genome and proteome are required for cells of the renal inner medulla to adapt to extreme changes in their hypertonic environment. Although a large body of information already exists for changes in expression of proteins involved in the osmotic response (1, 3-5, 30), it is expected that a larger set of genes and proteins yet unknown are also involved that have wide ranging effects of critical importance.
In an approach to elucidate proteins involved in this osmotic stress response, our laboratory is using genomic and proteomic tools, including gene arrays, two-dimensional differential gel electrophoresis (16), and antibody microarrays (17). Although two-dimensional difference gel electrophoresis is a powerful proteomic discovery technique, antibody microarray technology is an excellent complementary approach to two-dimensional difference gel electrophoresis, because it allows for the discovery of changes in expression of hydrophobic as well as low abundance proteins.
The mouse IMCD3 cell line provides a unique tool for studying the adaptative changes in the collecting duct of the kidney. Unlike mouse papilla, IMCD3 cells may be examined under isotonic growth conditions and acutely challenged or chronically adapted to the hypertonic setting (24, 25). In the current work, we have employed antibody microarray proteomics and identified Nup88 as highly up-regulated in IMCD3 cells under hypertonic stress.
Previous researchers have demonstrated that Nup88 associates with nucleoporin 214 to form part of a nuclear pore complex at the nuclear membrane (18). Nuclear pore complexes are involved in the trafficking of several molecules, including RNAs and proteins. Of specific interest are data indicating that Nup88 inhibits the nuclear export of several Rel-like transcription factors through anchoring CRM1 also referred to as exportin 1, to the nuclear membrane, thereby avoiding its return to the nucleus for a new round of export (20). However, other authors (19) propose an opposite role for Nup88, indicating that it enhances nuclear export. We propose that up-regulation of Nup88 in IMCD3 cells in response to hypertonicity acts to retain the Rel-like transcription factor TonEBP in the nucleus. TonEBP is essential for adaptation to hypertonic stress via transcription of target genes necessary for compatible osmolyte accumulation, ion transport, and DNA repair (11, 13, 30-32).
The up-regulation of Nup88 in IMCD3 cells determined by antibody microarray was further validated by both QPCR for message and Western blot for protein. Nup88 protein expression increased rapidly upon exposure to cells to an acute sublethal osmotic stress in a time period (4-6 h after hypertonic stress) similar to early response proteins. Under isotonic conditions, IMCD3 cells were found to express low levels of Nup88 protein, which markedly increased during acute or chronic exposure to hypertonicity. The expression of Nup88 protein in response to hypertonicity was also demonstrated in mice and human kidney tissues with substantial Nup88 protein levels at the hypertonic papilla tissues and nearly absent at the isotonic cortex. It is of interest that Nup88 expression in papilla from mice deprived of water for 36 h was markedly up-regulated as compared with mice that received ad libitum water. Nup88 message and protein half-lives were determined by returning cells chronically adapted to 900 mosmol to isotonic conditions and are relatively short (2.8 and 18.4 h, respectively), possibly due to its proposed role as an inhibitor of TonEBP nuclear export, since it has been demonstrated that TonEBP is rapidly exported out of the nucleus after cells are returned to isotonic conditions (15).
High osmolality induced by ionic solutes as well as mannitol, sucrose, and urea resulted in near equal up-regulation of Nup88. In this regard, up-regulation of Nup88 with urea may be explained by the fact that urea transporter A is a target gene for TonEBP under urea conditions, as shown previously by others (33, 34). Localization of Nup88 protein in IMCD3 cells by confocal fluorescence microscopy demonstrated its presence exclusively at the nuclear membrane, consistent with the protein's potential involvement in nuclear pore complexes (18). In order to better define whether Nup88 is involved in the retention of TonEBP under hypertonic stress, we amplified a fragment of the mouse N terminus of TonEBP containing its nuclear localization signal, its auxiliary export domain, and its nuclear export signal. We cloned this fragment upstream of the GFP to obtain TonEBP-GFP expression. Results obtained with this construct correlate with those obtained from analysis of endogenous TonEBP using a specific antibody. Confocal data indicated essentially identical localization, thereby validating the TonEBP-GFP construct as a useful tool to track the nucleocytoplasmic traffic of TonEBP under hypertonic stress in living cells. We show that TonEBP is present in the cytosol of IMCD3 cells at isotonic conditions and is rapidly shifted to the nucleus in only 30 min of acute sublethal stress. Confocal analysis of TonEBP fluorescent signal showed that 90% of TonEBP is internalized in the nucleus in the first hour of hypertonic stress. As shown in Fig. 4, Nup88 is barely expressed in these cells after 30 min of osmotic stress, indicating that TonEBP import to the nucleus occurs in a Nup88-independent way. This result is confirmed using the stable cell line silenced for Nup88 expression (data not shown).
After 8 h of acute hypertonic stress, the majority of TonEBP is present in the cytosol in cells silenced for Nup88 expression, as compared with empty vector control cells that retain most of the transcription factor in the nucleus. GFP analysis revealed that ∼75% of the TonEBP signal is located in the cytosol of Nup88 silenced cells; in contrast, 80% of the TonEBP is present in the nucleus of nonsilenced cells. The significance of this observation is reflected in the fact that the message levels of well established TonEBP target genes (AR, sodium-myoinositol transporter, BGT1, TauT, and Hsp70) (10, 13, 31, 32) are blunted under acute hypertonic stress in cells silenced for Nup88 as compared with empty vector control cells expressing Nup88. Interestingly, this blunting of mRNA levels of TonEBP target genes under hypertonic stress is similar to that reported in the kidney papilla from TonEBP knock-out mice (35). As confirmation, protein expression for AR and Hsp70 was examined and found to be substantially reduced (66 and 95%, respectively; p < 0.001) in Nup88-silenced cells as compared with control cells under hypertonic stress with an intermediate expression in partially silenced clones. This blunting of expression of critical osmotic stress response genes leads to an increased osmosensitivity in Nup88-silenced cells as compared with controls. In fact, over a 5-day period, little if any cell recovery is determined when Nup88-silenced IMCD3 cells are exposed to sublethal hypertonic stress.
Previous research points to a role for exportin 1 (CRM1) in the localization of Rel-like transcription factors, such as TonEBP. Our data employing the specific inhibitor LMB demonstrate that in IMCD3 cells, CRM1 appears to be involved in the nuclear export of TonEBP under isotonic conditions. In contrast, under hypertonic stress, CRM1 is not involved, since inhibition of CRM1 with LMB did not prevent the loss of TonEBP from the nucleus in Nup88-silenced cells. These results are similar to those previously described by Tong et al. (15).
In conclusion, our data are the first to demonstrate that the nucleoporin protein, Nup88, is up-regulated both in vitro and in vivo in the kidney in response to hypertonic stress. These data also suggest that Nup88 expression serves to retain TonEBP in the nucleus, thus allowing the transcription of target genes essential for cell adaptation to osmotic stress.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants DK-19928 and DK-66544 (to T. B.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1-3 and Figs. 1 and 2.
Footnotes
The abbreviations used are: TonEBP, tonicity enhancer-binding protein; AR, aldose reductase; TauT, taurine transporter; QPCR, quantitative PCR; DAPI, 4′,6-diamidino-2-phenylindole; GFP, green fluorescent protein; LMB, leptomycin B.
References
- 1.Burg, M. B., Kwon, E. D., and Kultz, D. (1997) Annu. Rev. Physiol. 59 437-455 [DOI] [PubMed] [Google Scholar]
- 2.Burg, M. B., Kwon, E. D., and Kültz, D. (1996) FASEB J. 10 1598-1606 [DOI] [PubMed] [Google Scholar]
- 3.Ferraris J. D., and Burg, M. B. (2006) Contrib. Nephrol. 152 125-141 [DOI] [PubMed] [Google Scholar]
- 4.Handler, J. S., and Kwon, H. M. (1996) Kidney Int. 49 1682-1683 [DOI] [PubMed] [Google Scholar]
- 5.Handler, J. S., and Kwon, H. M. (2001) Nephron 87 106-110 [DOI] [PubMed] [Google Scholar]
- 6.Handler, J. S., and Kwon, H. M. (2001) Kidney Int. 60 408-411 [DOI] [PubMed] [Google Scholar]
- 7.Jeon, U. S., Kim, J. A., Sheen, M. R., and Kwon, H. M. (2006) Acta Physiol. (Oxf.) 187 241-247 [DOI] [PubMed] [Google Scholar]
- 8.Woo, S. K., and Kwon, H. M. (2002) Int. Rev. Cytol. 215 189-202 [DOI] [PubMed] [Google Scholar]
- 9.Woo, S. K., Lee, S. D., and Kwon, H. M. (2002) Pflugers Arch. 444 579-585 [DOI] [PubMed] [Google Scholar]
- 10.Miyakawa, H., Woo, S. K., Dahl, S. C., Handler, J. S., and Kwon, H. M. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 2538-2542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miyakawa, H., Woo, S. K., Chen, C. P., Dahl, S. C., Handler, J. S., and Kwon, H. M. (1998) Am. J. Physiol. 274 F753-F761 [DOI] [PubMed] [Google Scholar]
- 12.Ito, T., Fujio, Y., Hirata, M., Takatani, T., Matsuda, T., Muraoka, S., Takahashi, K., and Azuma, J. (2004) Biochem. J. 382 177-182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Woo, S. K., Lee, S. D., Na, K. Y., Park, W. K., and Kwon, H. M. (2002) Mol. Cell. Biol. 22 5753-5760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cha, J. H., Woo, S. K., Han, K. H., Kim, Y. H., Handler, J. S., Kim, J., and Kwon, H. M. (2001) J. Am. Soc. Nephrol. 12 2221-2230 [DOI] [PubMed] [Google Scholar]
- 15.Tong, E. H., Guo, J. J., Huang, A. L., Liu, H., Hu, C. D., Chung, S. S., and Ko, B. C. (2006) J. Biol. Chem. 281 23870-23879 [DOI] [PubMed] [Google Scholar]
- 16.Rivard, C. J., Brown, L. M., Almeida, N. E., Maunsbach, A. B., Pihakaski-Maunsbach, K., Andres-Hernando, A., Capasso, J. M., and Berl, T. (2007) J. Biol. Chem. 282 6644-6652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lanaspa, M. A., Almeida, N. E., Andres-Hernando, A., Rivard, C. J., Capasso, J. M., and Berl, T. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 13672-13677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fornerod, M., van Deursen, J., van Baal, S., Reynolds, A., Davis, D., Murti, K. G., Fransen, J., and Grosveld, G. (1997) EMBO J. 16 807-816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hutten, S., and Kehlenbach, R. H. (2006) Mol. Cell. Biol. 26 6772-6785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roth, P., Xylourgidis, N., Sabri, N., Uv, A., Fornerod, M., and Samakovlis, C. (2003) J. Cell Biol. 163 701-706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uv, A. E., Roth, P., Xylourgidis, N., Wickberg, A., Cantera, R., and Samakovlis, C. (2000) Genes Dev. 14 1945-1957 [PMC free article] [PubMed] [Google Scholar]
- 22.Xylourgidis, N., Roth, P., Sabri, N., Tsarouhas, V., and Samakovlis, C. (2006) J. Cell Sci. 119 4409-4419 [DOI] [PubMed] [Google Scholar]
- 23.Rauchman, M. I., Nigam, S. K., Delpire, E., and Gullans, S. R. (1993) Am. J. Physiol. 265 F416-F424 [DOI] [PubMed] [Google Scholar]
- 24.Capasso, J. M., Rivard, C. J., and Berl, T. (2001) Am. J. Physiol. 280 F768-F776 [DOI] [PubMed] [Google Scholar]
- 25.Capasso, J. M., Rivard, C. J., Enomoto, L. M., and Berl, T. (2003) Ann. N. Y. Acad. Sci. 986 410-415 [DOI] [PubMed] [Google Scholar]
- 26.Kodiha, M., Tran, D., Qian, C., Morogan, A., Presley, J. F., Brown, C. M., and Stochaj, U. (2008) Biochim. Biophys. Acta 1783 405-418 [DOI] [PubMed] [Google Scholar]
- 27.Bernad, R., van der Velde, H., Fornerod, M., and Pickersgill, H. (2004) Mol. Cell. Biol. 24 2373-2384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bernad, R., Engelsma, D., Sanderson, H., Pickersgill, H., and Fornerod, M. (2006) J. Biol. Chem. 281 19378-19386 [DOI] [PubMed] [Google Scholar]
- 29.Capasso, J. M., Rivard, C., and Berl, T. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 13414-13419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dmitrieva, N. I., and Burg, M. B. (2005) Mutat. Res. 569 65-74 [DOI] [PubMed] [Google Scholar]
- 31.Na, K. Y., Woo, S. K., Lee, S. D., and Kwon, H. M. (2003) J. Am. Soc. Nephrol. 14 283-288 [DOI] [PubMed] [Google Scholar]
- 32.Zhang, Z., Ferraris, J. D., Brooks, H. L., Brisc, I., and Burg, M. B. (2003) Am. J. Physiol. 285 F688-F693 [DOI] [PubMed] [Google Scholar]
- 33.Lim, S. W., Ahn, K. O., Sheen, M. R., Jeon, U. S., Kim, J., Yang, C. W., and Kwon, H. M. (2007) J. Am. Soc. Nephrol. 18 421-429 [DOI] [PubMed] [Google Scholar]
- 34.Nakayama, Y., Peng, T., Sands, J. M., and Bagnasco, S. M. (2000) J. Biol. Chem. 275 38275-38280 [DOI] [PubMed] [Google Scholar]
- 35.López-Rodríguez, C., Antos, C. L., Shelton, J. M., Richardson, J. A., Lin, F., Novobrantseva, T. I., Bronson, R. T., Igarashi, P., Rao, A., and Olson, E. N. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 2392-2397 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.