Abstract
The ability to accurately digest and ligate DNA molecules of different origins is fundamental to modern recombinant DNA research. Only a handful of enzymes are capable of recognizing and cleaving novel and long DNA sequences, however. The slow evolution and engineering of new restriction enzymes calls for alternative strategies to design novel and unique restriction enzymes capable of binding and digesting specific long DNA sequences. Here we report on the use of zinc finger nucleases (ZFNs)—hybrid synthetic restriction enzymes that can be specifically designed to bind and cleave long DNA sequences—for the purpose of DNA recombination. We show that novel ZFNs can be designed for the digestion of specific sequences and can be expressed and used for cloning purposes. We also demonstrate the power of ZFNs in DNA cloning by custom-cloning a target DNA sequence and assembling dual-expression cassettes on a single target plasmid, a task that rarely can be achieved using type-II restriction enzymes. We demonstrate the flexibility of ZFN design and the ability to shuffle monomers of different ZFNs for the digestion of compatible recognition sites through ligation of compatible ends and their cleavage by heterodimer ZFNs. Of no less importance, we show that ZFNs can be designed to recognize and cleave existing DNA sequences for the custom-cloning of native target DNA molecules.
Keywords: artificial restriction enzymes, DNA cloning, multigene vectors
DNA cloning is fundamental for nearly every niche of molecular biology research, and site-specific endonucleases are the cornerstones of modern recombinant DNA technology (1, 2). The burden of site-specific DNA digestion falls on a wide variety of restriction enzymes isolated from different organisms. A recent update of the REBASE (enzymes and genes for DNA restriction and modification) database lists >3600 type II restriction enzymes that have been biochemically or genetically characterized (3). But although >600 of these restriction enzymes are currently commercially available, only 262 distinct sequences are targeted (3), most of them 4- to 6-bp palindromic sequences. Thus, not only are many specificities still unavailable, but the number of enzymes capable of recognizing 8- and 10-bp-long sequences is rather small. The situation is even more critical with respect to meganucleases—restriction enzymes capable of targeting very large (>12 bp) sequences. To the best of our knowledge, only six different meganucleases are currently available commercially.
Although many of the 6-bp cutters are widely used for various recombinant DNA applications, their application can be somewhat limited by their high statistical occurrence within various DNA sequences (4). Furthermore, their use for cloning large DNA fragments or for the assembly of complex DNA structures is nearly impossible. Thus, for example, the construction of large artificial DNA molecules (which may contain several reparative elements and/or 6- and 4-bp-long recognition sites) could be facilitated by the introduction of restriction enzymes with long and novel target sequences (e.g., 8- and 10-bp cutters and meganucleases).
Meganucleases (also known as rare cutters) have been particularly useful for in vivo applications. Their high specificity makes them extremely useful for genome engineering in mammalian, animal, and plant cells (5–7) and for novel cloning purposes (8–10). Various groups are engaged in searching for, modifying, and reengineering existing meganucleases for the targeting of novel sequences (8–10). But the extreme specificity of meganucleases (and of other restriction enzymes, for that matter) poses a great challenge for their engineering (11), and we have yet to witness the commercialization and widespread use of some of the recently reported variant and novel meganucleases (8–10).
Recently, zinc finger nucleases (ZFNs), a new type of restriction enzymes, have been developed as a novel tool for genome engineering in living cells (12–14). ZFNs provide an excellent alternative to the tedious and potentially impractical reengineering of existing meganucleases, because they can be engineered to digest virtually any long stretch of DNA sequence (15). ZFNs are modular proteins composed of a specifically engineered zinc finger DNA-binding domain fused to the FokI non-sequence-specific DNA-cleavage domain (shown in Fig. 1A). The zinc finger DNA-binding domain is composed of several fingers, each engineered to bind a specific DNA triplet. Monomers of two ZFNs must be properly aligned and function together to digest a double-stranded target DNA sequence (16, 17). The length of the spacer between the two monomers, where FokI actually digests the target DNA, is rather flexible; with a 6-bp spacer, the nuclease has been proposed to typically digest between the second and fourth nucleotides, leaving a 2-base-long 5′ overhang. Thus, a ZFN dimer, composed of two monomers, each engineered to bind a 9-bp DNA sequence, can be used to target a 20- to 24-bp DNA sequence. It also should be noted that custom-designed ZFNs may bind not only to their target sites, but also to similar 9-bp sequences. But due to the very low likelihood of finding two similar 9-bp-long sequences arranged in reverse orientation in a semipalindromic fashion, it is likely that custom-designed ZFNs will exhibit high specificity toward their target sequence. Furthermore, ZFNs with four or even five zinc fingers possibly can be designed if higher specificity is required. Thus, ZFNs can be classified as supermeganucleases, which have potential uses in various cloning tasks.
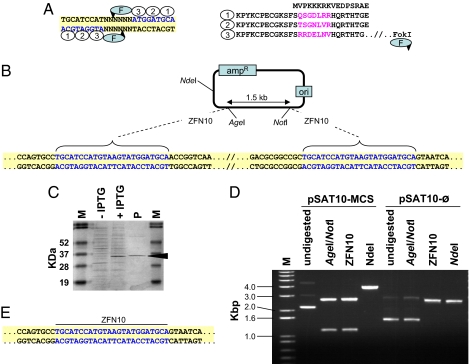
Fig. 1.
Structure and expression of ZFNs and their use for DNA cloning. (A) The structure of a typical 24-bp-long ZFN recognition site and its corresponding zinc finger protein, exemplified by ZFN10. DNA triplets (blue) and their binding zinc fingers (ovals) are numbered correspondingly. The unique amino acid sequences in each zinc finger are in purple. The FokI cleavage domain (F) is linked to the zinc finger protein's carboxyl terminus, and the predicted cleavage sites are indicated by arrowheads. (B) Scheme of the pSAT10-MCS plasmid. Sequences of the ZFN10 palindrome-like recognition sites are in blue. (C) Separation of total crude extract from induced (+IPTG) and noninduced (-IPTG) ZFN10 protein-expressing E. coli cells and of purified ZFN10 protein (P). The 34-kDa band corresponding to the ZFN10 protein is indicated. (D) Restriction analysis of the parental pSAT10-MCS and its progeny plasmid pSAT10- . (E) Sequence analysis of the ZFN10 ligation site in one of the pSAT10-
. (E) Sequence analysis of the ZFN10 ligation site in one of the pSAT10- plasmids. Sequences of the reconstructed ZFN10 palindrome-like recognition sites are in blue.
plasmids. Sequences of the reconstructed ZFN10 palindrome-like recognition sites are in blue.
Recently, several artificially designed ZFNs have been successfully used as powerful tools for gene targeting in various species (12–15). The rules and protocols for the design, assembly, and testing of new ZFNs have been laid out in several recent publications (17–19), but to the best of our knowledge, the application of ZFNs and an evaluation of their potential for in vitro DNA cloning purposes have not yet been reported. In this article, we describe the construction, expression, and purification of several ZFNs and their successful use in recombinant DNA procedures. We demonstrate the power of ZFNs in performing tasks that are nearly impossible using the current repertoire of 6-bp restriction enzymes. We also demonstrate the uniqueness of ZFNs, which lies in the ability to shuffle and combine monomers of different ZFNs for cloning purposes, as well as the potential for digesting and cloning native target DNA molecules. Given that >200,000 different combinations are possible with a 9-bp sequence, the ability to engineer ZFNs to bind to such sequences represents a significant increase in the number of restriction enzymes that possibly can be developed and may be useful for DNA cloning.
Results
ZFN-Digested DNA Ends Can Be Ligated to Each Other.
One of the basic requirements for newly isolated or recombinant restriction enzymes is their ability to accurately and specifically digest donor and recipient DNA molecules. We thus tested whether an artificial ZFN could specifically and accurately digest target DNA molecules, and also whether the products of such digestion could be purified and ligated using common molecular cloning protocols. Toward this end, we first engineered the plasmid pSAT10-MCS (multiple cloning site) to carry two identical 24-bp sequences, each composed of a pair of 9-bp sequences, acting as a palindrome, separated by a 6-bp nonpalindromic sequence (Fig. 1B). We next engineered ZFN10, a ZFN monomer with three zinc fingers capable of binding in a specific orientation to the ATGGATGCA sequences on pSAT10-MCS, as illustrated in Fig. 1A. Because an active ZFN10 site is actually composed of two 9-bp sequences making up a “palindrome-like” recognition site, two ZFN10 proteins were expected to function as a homodimer and cleave within the 6-bp spacer, as reported previously for other ZFNs (20). It is important to note that we designed the spacer sequence such that if digestion occurs between the second and fourth nucleotides, located on the upper and lower strands, respectively (Fig. 1A), then the overhanging parts of the digested molecules can easily ligate to each other.
We expressed ZFN10 in E. coli cells, but the crude protein extract proved useless for accurate digestion of our target pSAT10-MCS plasmid, leading to nearly complete degradation of the sample DNA (data not shown). Thus, we further purified the enzyme by binding it to Ni-NTA agarose beads (Fig. 1C). Indeed, Ni-NTA–purified ZFN10 was able to release a specific ZFN10-flanked 1.2-kb fragment from pSAT10-MCS (Fig. 1D). Similarly, in vitro-expressed ZFN10 also released a well defined ZFN10-flanked fragment from pSAT10-MCS (data not shown). Thus, both E. coli– and in vitro-expressed ZFNs were useful for the efficient and specific digestion of target DNA molecules, and we used them alternately throughout this study.
We next purified the ZFN10-digested plasmid backbone from the gel and subjected it to a simple in vitro ligation procedure. We used commercial T4 ligase in an overnight ligation reaction and then transferred the entire reaction into chemically competent DH5α E. coli cells. We recovered a large number of recombinant colonies and analyzed their structure by both restriction pattern analysis and DNA sequencing. Fig. 1D shows that whereas the digestion of the parental pSAT10-MCS by AgeI and NotI (which are located internal to the ZFN10; Fig. 1B), or solely by ZFN10, released a 1.2-kb fragment, digestion of the self-ligated plasmid's backbone (pSAT- ) was limited to ZFN10. The plasmid no longer could be digested by AgeI or NotI (compare the AgeI/NotI and ZFN10 digestions of pSAT10-MCS and pSAT-
) was limited to ZFN10. The plasmid no longer could be digested by AgeI or NotI (compare the AgeI/NotI and ZFN10 digestions of pSAT10-MCS and pSAT- ; Fig. 1D). Indeed, pSAT-
; Fig. 1D). Indeed, pSAT- was missing the entire MCS fragment and was 1.2 kb shorter than its parental plasmid pSAT10-MCS (compare the NdeI digestions of pSAT10-MCS and pSAT-
was missing the entire MCS fragment and was 1.2 kb shorter than its parental plasmid pSAT10-MCS (compare the NdeI digestions of pSAT10-MCS and pSAT- ; Fig. 1D). This digestion pattern suggests that after removal of the ZFN10-digested 1.2-kb fragment, ligation of the two ZFN10 sites resulted in the accurate reconstruction of a new ZFN10 site at the ligation site. Indeed, DNA sequencing of several recombined clones clearly showed the accurate assembly of a new ZFN10 site (Fig. 1E). Thus, ZFNs not only can be expressed using both E. coli and in vitro expression systems, but also can be used for accurate digestion, and the ZFN-digested DNA molecules can be accurately ligated and recovered using common recombinant DNA protocols.
; Fig. 1D). This digestion pattern suggests that after removal of the ZFN10-digested 1.2-kb fragment, ligation of the two ZFN10 sites resulted in the accurate reconstruction of a new ZFN10 site at the ligation site. Indeed, DNA sequencing of several recombined clones clearly showed the accurate assembly of a new ZFN10 site (Fig. 1E). Thus, ZFNs not only can be expressed using both E. coli and in vitro expression systems, but also can be used for accurate digestion, and the ZFN-digested DNA molecules can be accurately ligated and recovered using common recombinant DNA protocols.
ZFNs Can Be Used to Assemble Large and Complex DNA Constructs.
Once we found that ZFN10 can be used to digest and self-ligate single DNA molecules, we then set out to determine whether ZFNs can be designed and used to recombine DNA fragments from different plasmids. We chose the challenge of assembling two expression cassettes onto a single plasmid. This task cannot typically be achieved using type II restriction enzymes, because of the high occurrence of their recognition sites in the two expression cassettes and the cloned genes (21). One approach to assembling multiple expression cassettes onto a single plasmid is to flank each cassette with novel sequences and use rare-cutters for their digestion and cloning into an acceptor plasmid (21, 22). Basing our strategy on this approach, we decided to construct two plasmids in which each expression cassette was flanked by a different ZFN. We began by constructing pSAT10-YFP-CHS and pSAT11-DsRed2-P, a pair of satellite plasmids (22) carrying plant expression cassettes for the yellow fluorescent protein (YFP)-tagged endoplasmic reticulum protein chalcone synthase (CHS) (23) and the DsRed2-tagged protein P of Sonchus Yellow Net Virus (SYNV) (24), respectively. The YFP-CHS and DsRed2-P expression cassettes were flanked by ZFN10 and ZFN11 24-bp-long recognition sites, respectively (Fig. 2A). We also engineered ZFN11, a ZFN monomer capable of binding to the 9-bp sequences composing the ZFN11 recognition site (data not shown). We constructed pRCS11, a binary plasmid, to carry the ZFN10 and ZFN11 recognition sites on its MCS (Fig. 2B). We successively cloned the YFP-CHS and DsRed2-P expression cassettes from pSAT10-YFP-CHS and pSAT11-DsRed2-P using ZFN10 and ZFN11, respectively, into their corresponding sites on pRCS11. Bombardment of the resultant plasmid, pRCS11[YFP-CHS][DsRed2-P], into tobacco mesophyll cells demonstrated the successful mounting of both expression cassettes onto the acceptor plasmid.
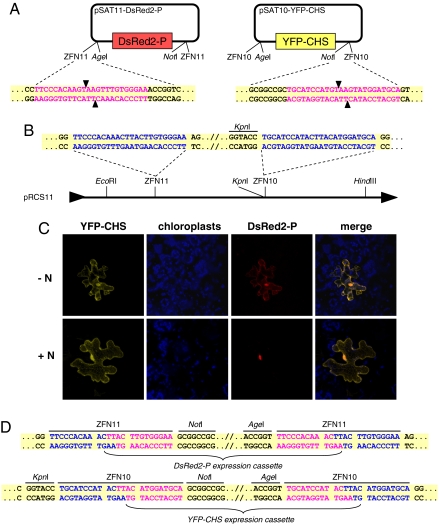
Fig. 2.
Assembly of dual-expression cassettes by ZFNs. (A) Schemes of pSAT10-YFP-CHS and pSAT11-DsRed2-P. Sequences of ZFN10 and ZFN11 recognition sites are in purple, and the predicted cleavage sites in each sequence are indicated by arrowheads. (B) Scheme of the pRCS11 acceptor plasmid. Sequences of ZFN10 and ZFN11 recognition sites in the plasmid's MCS are in blue. (C) Detection of YFP-CHS and DsRed2-P expression from pRCS11[YFP-CHS][ DsRed2-P]-bombarded plant cells in the presence (+N) or absence (-N) of the N protein of SYNV. Expression of YFP-CHS, DsRed-P, and chloroplast autofluorescence is shown in yellow, blue, and red, respectively. The images are projections of several confocal sections. (D) Sequence analysis of the ZFN10 and ZFN11 ligation sites in pRCS11[YFP-CHS][DsRed2-P]. ZFN10 and ZFN11 sequences derived from the acceptor plasmid pRCS11 are in blue, and those derived from the inserted expression cassettes are in purple.
Consistent with previous observations (23, 24), DsRed2-P was found in both the cell nucleus and the cytoplasm, whereas YFP-CHS decorated the rough endoplasmic reticulum and the nucleus (Fig. 2C). Furthermore, when we cobombarded pRCS11[YFP-CHS][DsRed2-P] with pSAT3-N (22), a plasmid expressing free SYNV N protein, the DsRed2-P accumulated predominantly in the nucleus (compare the expression pattern of DsRed2-P in the presence and absence of N; Fig. 2C), in accordance with the N protein's function: translocating the P protein into the host cell nucleus (24).
We next analyzed the ligation junction site sequences, and found that ZFN-digested cassettes were accurately ligated into their corresponding cloning sites on pRCS11 (Fig. 2D). Collectively, our data suggest that ZFNs can be used successfully for the accurate digestion and recombination of DNA fragments from different sources and can be useful for unique tasks, such as the assembly of complex and multigene transformation vectors.
ZFNs Can Be Used to Clone Native DNA Molecules.
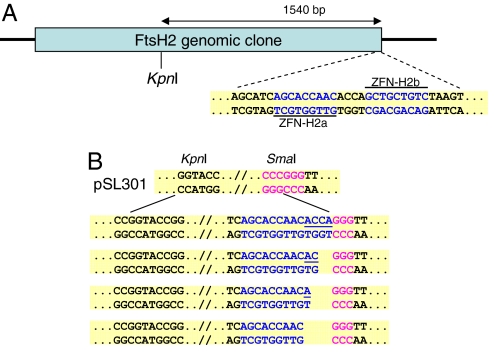
The potential use of ZFNs for cloning purposes goes far beyond the cloning of artificially assembled DNA sequences. An important feature of ZFNs is their ability to recognize and cleave existing native sequences. We chose the Arabidopsis ATP-dependent chloroplast-protease-encoding FtsH2 gene as a model sequence to demonstrate how ZFNs can be designed and used to assist in the cloning of existing DNA molecules. To further demonstrate the flexibility of ZFN design (i.e., targeting various lengths of sequences), we selected a 22-bp-long target sequence and constructed a pair of new ZFN monomers designated ZFN-H2a and ZFN-H2b, capable of binding two target sequences on our model genomic sequence separated by a 4-bp-long spacer (Fig. 3A). We next used ZFN-H2a and ZFN-H2b as a heterodimer ZFN to digest their target and linearize a plasmid carrying a genomic fragment containing the FtsH2 gene. The linearized plasmid was gel-purified and further digested with KpnI, which released the expected 1.5-kb fragment. Because digestion with ZFNs leaves a 5′ overhang, we used Klenow fragment to fill in the recessed 3′ ends of the digested site, and cloned the semiblunted fragment into the SmaI and KpnI sites of our target plasmid, pSL301. Sequence analyses of the ligation junction between the FtsH2 fragment and the SmaI site on pSL301 (Fig. 3B) in recombinant molecules revealed the different cleavage point made by the ZFN-H2a/ZFN-H2ba pair digested at the 4-bp spacer between the 9-bp DNA-binding recognition sites (Fig. 3B). These observations are in line with a previous report (20) showing that ZFN digestion patterns may depend on the length of the spacer between the ZFN monomers' binding sites. Thus, our data demonstrate that ZFNs can be designed to facilitate in vitro cloning of not only artificial, but also native DNA sequences.
Fig. 3.
Custom-cloning of a target DNA sequence. (A) Outline of the Arabidopsis FtsH2 genomic clone. Sequences of the ZFN-H2a and ZFN-H2b binding sites are shown in blue. (B) Sequence analysis of ZFN-H2a-ZFN-H2b/SmaI and KpnI/KpnI junctions in various pSL301-AtFtsH2 clones. Sequences derived from the SmaI site on acceptor plasmid pSL301 are in purple, and those derived from the inserted DNA are in blue. Filled-in nucleotides are underlined.
ZFN Monomers Can Be Shuffled for Digestion Compatibility.
The unique structure of a typical ZFN recognition site allows for flexible design in producing ZFNs with compatible restriction patterns. With the proper spacer sequence, pairs of ZFNs can be designed to function as compatible enzymes during the ligation of two DNA molecules (similar to the compatibility existing between, e.g., BamHI and BglII). To demonstrate this feature for artificially designed ZFNs, we linearized pSAT12.1-CHRD-RFP, a plasmid carrying a ZFN-H2a recognition sequence (Fig. 4A) and the RFP-tagged chloroplast ChrD protein expression cassette, and cloned it into the ZFN10 recognition site on pRCS11 (Fig. 4B). Bombardment of the resultant plasmid, pRCS11[10/12.1-CHRD-RFP], into tobacco mesophyll cells demonstrated the functionality of the CHRD-RFP expression cassette, as evidenced by a distinct red signal in the chloroplasts (data not shown). More importantly, sequence analyses of the ligation-junction sites revealed accurate ligation between the ZFN-H2a- and ZFN10-digested DNA fragments (Fig. 4C).
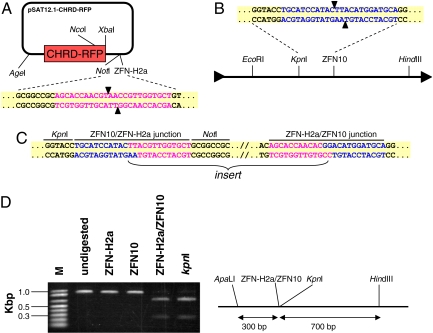
Fig. 4.
DNA cloning using compatible ZFNs. (A) Scheme of pSAT12.1-CHRD-RFP. The ZFN-H2a recognition-site sequence is in purple, and the predicted cleavage sites are indicated by arrowheads. (B) Scheme of the pRCS11 acceptor plasmid. The ZFN10 recognition site in the plasmid's MCS is in blue. (C) Sequence analysis of the ZFN10/ZFN-H2a junctions at the ligation sites in pRCS11[10/12.1-CHRD-RFP]. The ZFN10 recognition sequences derived from the acceptor plasmid pRCS11 are in blue, and the ZFN-H2a sequences derived from pSAT12.1-CHRD-RFP are in purple. (D) Restriction analysis of an ≈1-kb-long ApaLI-HindIII fragment from pRCS11[10/12.1-CHRD-RFP]. The fragment's restriction map, which includes a ZFN-H2a/ZFN10 ligation junction, is on the right.
Naturally, because the palindrome-like ZFN10 and ZFN-H2a recognition sequences at the ligation sites were lost after the ligation (Fig. 4C), we predicted that ZFN10 and ZFN-H2a would not be able to digest any of these sites on the recombined plasmid. To examine this hypothesis, we isolated a 1-kb ApaLI-HindIII fragment containing a ZFN-H2a/ZFN10 recombination site from pRCS11[10/12.1-CHRD-RFP] and redigested it with either ZFN10 or ZFN-H2a. Indeed, as shown in Fig. 4D, although the fragment could be digested by KpnI, an enzyme with recognition sites located near the ZFN-H2a/ZFN10 junction recognition site, neither ZFN10 nor ZFN-H2a alone was capable of digesting the recombined site. More importantly, because both ZFN10 and ZFN-H2a could each still bind to their corresponding 9-bp sites and because they share a similar structure typical of all ZFNs, they potentially could function as heterodimers and digest at the junction site. Indeed, digestion of the 1-kb ApaLI-HindIII fragment with a combination of ZFN10 and ZFN-H2a resulted in a digestion pattern similar to that after digestion with KpnI alone (Fig. 4D), suggesting that the ZFN10-ZFN-H2a heterodimer indeed is capable of binding and digesting the ZFN10/ZFN-H2a junction recognition site. Thus, our data demonstrate that ZFN monomers can be shuffled and matched for digestion, cloning, and analysis of recombined molecules.
Discussion
Novel restriction enzymes with unique specificities, particularly enzymes that are capable of recognizing and cleaving long sequences, are extremely useful for genome analysis and manipulation, assembly of complex DNA structures, and genome editing (25–27). Thus, although other molecular techniques, such as overlap extension polymerase chain reaction (PCR), SfiI-based ligation, multiGATEWAY recombination, and uracil-DNA glycosylase, can be used for the construction of complex DNA structures (21, 28), the search continues for novel and rare-cutting restriction enzymes that can facilitate the cloning of DNA molecules. Various approaches have been taken to creating artificial rare cutters, most involving engineering existing enzymes for unique specificities. For example, a rational approach combined with a cell-based high-throughput assay was recently used to identify functional variants of the naturally occurring rare cutter I-CreI (9). In another approach, a unique positive and negative selection system was developed to direct the in vivo evolution of a I-SceI mutant with altered DNA-cleavage specificities (29). Although such approaches potentially can be used to develop additional variants and novel restriction enzymes, they have proven quite challenging. In fact, most of these methods require special expertise (11, 30), and they rarely if ever help in the prediction or assembly of new restriction enzymes with predetermined novel target sequences. Thus, for example, although Lanio et al. (31) used a structure-guided approach to alter the recognition sequence of EcoRV and produced several variants with altered selectivity, none had the desired (and predicted) selectivity. Because the structure of many restriction enzymes is not modular by nature (in contrast to, e.g., zinc finger transcription factors), reengineering such enzymes to bind novel sequences while maintaining their specific enzymatic activity is a challenge (30).
The development of ZFNs as a tool for genome engineering (15) laid the foundation for the design and assembly of novel restriction enzymes. Important advantages of ZFNs as rare-cutting restriction enzymes include the clear separation between their DNA-binding and catalytic domains, and the ability to design them from the ground up. The rules and protocols for the design and assembly of novel ZFNs have been laid out in several recent publications (17–19, 32–33), and a collection of ZFNs already has been assembled and successfully used in gene-targeting experiments in various species (12–15). During the course of this study, we successfully used publicly available knowledge and information for the design and construction of four novel ZFNs, all of which were then successfully used for cloning purposes. We used basic protein expression and purification methods for the production of our proteins. These protocols are similar to those used for the development and analysis of other ZFN proteins (14, 17, 19, 33); thus, we suggest that similar strategies can be used to produce novel ZFNs suitable for in vitro DNA digestion in cloning tasks. We also used routine molecular biology procedures for digestion, purification, and ligation of our target and vector DNA molecules, further supporting the notion that self-produced ZFNs can be easily implemented into existing recombinant DNA protocols.
The biochemical properties and digestion characteristics of ZFNs have been the subject of several reports (20, 33). When properly aligned on their target DNA, two ZFN monomers will act together to digest spacers of different lengths and will leave various lengths of 5′ overhang (17, 20, 33). This phenomenon allows for great flexibility in the design and use of ZFN monomers for the cloning of native sequences, and also in the construction of artificial complex DNA constructs. We demonstrated this flexibility during the cloning of the FtsH2 genomic clone (Fig. 3) and the assembly of the CHRD-RFP expression cassette onto a plant-transformation vector (Fig. 4). We showed that although ZFN-H2a was originally designed to function together with ZFN-H2b in digesting a unique 22-bp-long target site in the FtsH2 genomic sequence, it also could be used to target a 24-bp palindrome-like recognition site carrying a 6-bp spacer. Furthermore, we showed that different ZFN monomers potentially can be shuffled to act together, because the product of ZFN-H2a was ligated into a ZFN10-digested vector, and product could then be digested by a mixture of ZFN-H2a and ZFN10 (Fig. 4D).
ZFN-H2a and ZFN-H2b were designed to target a native genomic sequence and were intentionally located only a short (4-bp) distance from each other (Fig. 3A). This design allowed us to demonstrate that, in line with the notion that the ZFN digestion pattern depends on the length of the target spacer (20), digestion of the 4-bp-long sequence indeed occurred at different positions on the spacer (Fig. 3B). Although we cannot rule out the possibility that the digestion of a 6-bp-long spacer also produces various 5′ overhang products, a sequence analysis of the ZFN-H2a/ZFN10 ligation junctions indicates that at least in the cases that we examined, digestion of both the target and vector sequences occurred between the third and fourth nucleotides of the spacer (Fig. 4C). Rare cutters' ability to achieve site-specific digestion of such long spacers allows for another important use, in directional cloning (Fig. 2). Directional cloning is particularly important during the assembly of complex multigene transformation vectors, where the direction of several expression cassettes in relation to others may affect the expression patterns of the cloned genes. Whereas the assembly of multigene expression cassettes possibly can be acheived by various means, methods that are based on the use of rare-cutting restriction enzymes, are expected to be more relevant to the assembly of multigene transformation vectors (21) and other complex DNA structures. The latter approaches certainly will benefit from the introduction of a much larger collection of novel rare cutters. Although in this article we have described the use of only four new rare cutters for cloning purposes, the previous descriptions of many others (13, 14) along with the relative ease of design and assembly of such enzymes, support the notion that various novel ZFNs can be constructed and introduced for recombinant DNA applications. Furthermore, because ZFN monomers with four or more fingers potentially can be designed to target sequences of 12 bp or longer, they can be used to produce ZFNs with extremely high specificity.
To conclude, here we have shown for the first time that ZFNs can be used for accurate in vitro cloning of DNA molecules of both artificial and native DNA sequences. Furthermore, we have shown that ZFNs can be used in conjunction with commercially available restriction enzymes, and that pairs of ZFNs can be designed for compatibility. Because ZFNs can be specifically tailored to suit individual needs (17, 18, 33), they represent a powerful and novel addition to the molecular biologist's toolbox.
Materials and Methods
Molecular Assembly of ZFNs.
The ZFN DNA-binding regions (i.e., zinc finger protein domains) for ZFN10, ZFN11, ZFN-H2a, and ZFN-H2b were designed based on a zinc-finger framework consensus sequence developed by Desjarlais and Berg (34). The detailed recognition helices are given in supporting information (SI) Table S1. Each zinc finger protein was assembled from sets of overlapping backbone oligos (BBOs) and sequence-dependent oligos (SDOs) as described previously (33), using a high-fidelity Pfu DNA polymerase (Stratagene). A comprehensive list of the BBO and SDO primers used for the assembly of ZFN10, ZFN11, ZFN-H2a, and ZFN-H2b is given in Table S2. Each PCR was composed of 5 pM BBO and SDO primers and 200 pM 5′CCGCTCGAGCTGAAAAACCTTACAAGTGTCC3′ and 5′GGACTAGTCCTCCAGTATGAGTACGTTGATG3′ primers and was carried out for 35 cycles. PCR products were cloned into the XhoI and SpeI sites of a modified pET28 vector, resulting in fusion of the zinc finger protein with the FokI endonuclease domain from pHS::QQR-QEQ/2300 (12) and a 6xHis-tag at the C terminus of the protein driven by the T7 promoter.
Assembly of Modified pSAT and pRCS Plasmids.
To construct pSAT10-MCS, pSAT11-MCS, and pSAT12.1-MCS, the entire plasmid backbone of pSAT6-MCS (22) was PCR-amplified using the appropriate set of primers (Table S3). The PCR product was digested by AgeI and NotI, and ligated with the AgeI-NotI 1.2-kb fragment from pSAT6-MCS (22). The YFP-CHS, DsRed2-P, and CHRD-RFP expression cassettes were cloned as AgeI-NotI fragments from pSAT6-EYFP-C1-CHS, pSAT4-DsRed2-P, and pSAT6(A)-CHRD-mRFP-N1, respectively, producing pSAT10-YFP-CHS, pSAT11-DsRed2-P, and pSAT12.1-CHRD-RFP. pRCS11 was constructed by modifying the MCS of pRCS2 (22); this was done by subsequent cloning of self-annealed pairs of primers (Table S3) encoding the ZFN10 and ZFN11 sites into the KpnI and SmaI sites, respectively, of pRCS2.
Expression of ZFN Protein.
For E. coli expression, ZFN expression plasmids (i.e., pET28-ZFN10, pET28-ZFN11, pET28-ZFN-H2a, and pET28-ZFN-H2b) were transformed into BL21 GOLD (DE3) PlyS cells (Stratagene). The cells were cultured in 100 ml of Luria broth medium supplemented with 50 μg/μl of kanamycin and 100 μM ZnCl2 and grown at 22°C. At an OD600 of 0.6, ZFN expression was induced by 0.7 mM IPTG for 3 h at 22°C. Cells were harvested by centrifugation; resuspended in 35 ml of 25 mM Tris·HCl [pH 7.5], 300 mM NaCl, 5% (vol/vol) glycerol, and 100 μM ZnCl2; and lysed twice using a French press. Proteins were loaded on 0.5 ml of Ni-NTA agarose beads (Qiagen) and eluted with 1 ml of buffer containing 500 mM imidazole. Eluted proteins were stored at −20°C in 50% glycerol. Alternatively, the Expressway in vitro protein synthesis system (Invitrogen) was used for in vitro expression of ZFN proteins. Crude in vitro-produced proteins, with or without further purification through Ni-NTA agarose beads, were used for in vitro digestion experiments.
Digestion, Ligation, and Transformation into E. coli.
Digestion of ≈200 ng of plasmid DNA was carried out in 10 mM Tris [pH 8.8], 50 mM NaCl, 1 mM DTT, 100 μM ZnCl2, 50 μg/ml of BSA, and 100 μg/ml of tRNA in a total reaction volume of 20–30 μl. Typically, 0.05–1 μl of purified enzyme was added to the reaction, gently mixed, and preincubated for 30 min at room temperature, to allow the enzyme to bind to its target sequences. MgCl2 was added next, to a final concentration of 5 mM, and then the reaction was incubated for 2–40 min at room temperature. Cleaved fragments were separated by gel electrophoresis and purified using a GFX Gel Band Purification Kit (Amersham). Compatible ends in the target plasmids were dephosphorylated using shrimp alkaline phosphatase (Fermentas), and fragments were ligated with T4 ligase (NEB) and then transferred to chemically competent DH5α E. coli cells using standard molecular biology protocols.
Microbombardment and Confocal Microscopy.
For biolistic delivery, 50 μg of DNA was adsorbed onto 10 mg of 1-μm gold particles (Bio-Rad) according to the manufacturer's instructions, and then microbombarded onto the leaf epidermis of greenhouse-grown Nicotiana tabacum cv. Turk plants. For cobombardment of two constructs, the plasmids were mixed at a 1:1 molar ratio before the adsorption onto gold beads. The microbombardment was performed at a pressure of 150 psi using a portable Helios gene gun system (PDS-1000/He; Bio-Rad). After incubation for 16–24 h at 25°C to allow expression of the transfected DNA, the plant tissues were viewed directly under a confocal laser-scanning microscope (TCS SP5; Leica). Enhanced YFP was excited by an argon laser at 514 nm, and fluorescence was monitored between 520 and 540 nm. DsRed2 was excited by a helium-neon laser at 543 nm, and fluorescence was monitored between 570 and 630 nm. Chlorophyll fluorescence was monitored >660 nm. All experiments were repeated at least three times, with 10–20 cells exhibiting fluorescent signal examined in each independent experiment.
Supplementary Material
Acknowledgments.
We thank Dr. G. N. Drews for the gift of pHS::QQR-QEQ/2300. We also thank Dan Weinthal for his instrumental advice and support. This work was supported by University of Michigan startup funds.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0803618105/DCSupplemental.
References
- 1.Roberts RJ. How restriction enzymes became the workhorses of molecular biology. Proc Natl Acad Sci USA. 2005;102:5905–5908. doi: 10.1073/pnas.0500923102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brownlee C. Danna and Nathans: Restriction enzymes and the boon to modern molecular biology. Proc Natl Acad Sci USA. 2005;102:5909. doi: 10.1073/pnas.0502760102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roberts RJ, Vincze T, Posfai J, Macelis D. REBASE: Enzymes and genes for DNA restriction and modification. Nucleic Acids Res. 2007;35:D269–D270. doi: 10.1093/nar/gkl891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pingoud A, Fuxreiter M, Pingoud V, Wende W. Type II restriction endonucleases: Structure and mechanism. Cell Mol Life Sci. 2005;62:685–707. doi: 10.1007/s00018-004-4513-1. [DOI] [PubMed] [Google Scholar]
- 5.Puchta H, Dujon B, Hohn B. Two different but related mechanisms are used in plants for the repair of genomic double-strand breaks by homologous recombination. Proc Natl Acad Sci USA. 1996;93:5055–5060. doi: 10.1073/pnas.93.10.5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thermes V, et al. I-SceI meganuclease mediates highly efficient transgenesis in fish. Mech Dev. 2002;118:91–98. doi: 10.1016/s0925-4773(02)00218-6. [DOI] [PubMed] [Google Scholar]
- 7.Windbichler N, et al. Homing endonuclease-mediated gene targeting in Anopheles gambiae cells and embryos. Nucleic Acids Res. 2007;35:5922–5933. doi: 10.1093/nar/gkm632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fajardo-Sanchez E, Stricher F, Paques F, Isalan M, Serrano L. Computer design of obligate heterodimer meganucleases allows efficient cutting of custom DNA sequences. Nucleic Acids Res. 2008;36:2163–2173. doi: 10.1093/nar/gkn059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arnould S, et al. Engineering of large numbers of highly specific homing endonucleases that induce recombination on novel DNA targets. J Mol Biol. 2006;355:443–458. doi: 10.1016/j.jmb.2005.10.065. [DOI] [PubMed] [Google Scholar]
- 10.Smith J, et al. A combinatorial approach to create artificial homing endonucleases cleaving chosen sequences. Nucleic Acids Res. 2006;34:e149. doi: 10.1093/nar/gkl720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jeltsch A, Wenz C, Wende W, Selent U, Pingoud A. Engineering novel restriction endonucleases: Principles and applications. Trends Biotechnol. 1996;14:235–238. doi: 10.1016/0167-7799(96)10030-5. [DOI] [PubMed] [Google Scholar]
- 12.Lloyd A, Plaisier CL, Carroll D, Drews GN. Targeted mutagenesis using zinc-finger nucleases in. Arabidopsis. Proc Natl Acad Sci USA. 2005;102:2232–2237. doi: 10.1073/pnas.0409339102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Urnov FD, et al. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- 14.Doyon Y, et al. Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat Biotechnol. 2008;26:702–708. doi: 10.1038/nbt1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Porteus MH, Carroll D. Gene targeting using zinc finger nucleases. Nat Biotechnol. 2005;23:967–973. doi: 10.1038/nbt1125. [DOI] [PubMed] [Google Scholar]
- 16.Durai S, et al. Zinc finger nucleases: Custom-designed molecular scissors for genome engineering of plant and mammalian cells. Nucleic Acids Res. 2005;33:5978–5990. doi: 10.1093/nar/gki912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carroll D, Morton JJ, Beumer KJ, Segal DJ. Design, construction and in vitro testing of zinc finger nucleases. Nat Protoc. 2006;1:1329–1341. doi: 10.1038/nprot.2006.231. [DOI] [PubMed] [Google Scholar]
- 18.Wright DA, et al. Standardized reagents and protocols for engineering zinc finger nucleases by modular assembly. Nat Protoc. 2006;1:1637–1652. doi: 10.1038/nprot.2006.259. [DOI] [PubMed] [Google Scholar]
- 19.Cathomen T, Segal DJ, Brondani V, Muller-Lerch F. Generation and functional analysis of zinc finger nucleases. Methods Mol Biol. 2008;434:277–290. doi: 10.1007/978-1-60327-248-3_17. [DOI] [PubMed] [Google Scholar]
- 20.Smith J, et al. Requirements for double-strand cleavage by chimeric restriction enzymes with zinc finger DNA-recognition domains. Nucleic Acids Res. 2000;28:3361–3369. doi: 10.1093/nar/28.17.3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dafny-Yelin M, Tzfira T. Delivery of multiple transgenes to plant cells. Plant Physiol. 2007;145:1118–1128. doi: 10.1104/pp.107.106104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tzfira T, et al. pSAT vectors: A modular series of plasmids for autofluorescent protein tagging and expression of multiple genes in plants. Plant Mol Biol. 2005;57:503–516. doi: 10.1007/s11103-005-0340-5. [DOI] [PubMed] [Google Scholar]
- 23.Saslowsky D, Winkel-Shirley B. Localization of flavonoid enzymes in Arabidopsis roots. Plant J. 2001;27:37–48. doi: 10.1046/j.1365-313x.2001.01073.x. [DOI] [PubMed] [Google Scholar]
- 24.Goodin MM, Dietzgen RG, Schichnes D, Ruzin S, Jackson AO. pGD vectors: Versatile tools for the expression of green and red fluorescent protein fusions in agroinfiltrated plant leaves. Plant J. 2002;31:375–383. doi: 10.1046/j.1365-313x.2002.01360.x. [DOI] [PubMed] [Google Scholar]
- 25.Caccio S, Camilli R, La Rosa G, Pozio E. Establishing the Cryptosporidium parvum karyotype by NotI and SfiI restriction analysis and Southern hybridization. Gene. 1998;219:73–79. doi: 10.1016/s0378-1119(98)00376-x. [DOI] [PubMed] [Google Scholar]
- 26.Shen W, Huang Y, Tang Y, Liu DP, Liang CC. A general method to modify BACs to generate large recombinant DNA fragments. Mol Biotechnol. 2005;31:181–186. doi: 10.1385/MB:31:3:181. [DOI] [PubMed] [Google Scholar]
- 27.Paques F, Duchateau P. Meganucleases and DNA double-strand break-induced recombination: Perspectives for gene therapy. Curr Gene Ther. 2007;7:49–66. doi: 10.2174/156652307779940216. [DOI] [PubMed] [Google Scholar]
- 28.Lu C, Mansoorabadi K, Jeffries T. Comparison of multiple gene assembly methods for metabolic engineering. Appl Biochem Biotechnol. 2007;137–140:703–710. doi: 10.1007/s12010-007-9090-y. [DOI] [PubMed] [Google Scholar]
- 29.Doyon JB, Pattanayak V, Meyer CB, Liu DR. Directed evolution and substrate specificity profile of homing endonuclease I-SceI. J Am Chem Soc. 2006;128:2477–2484. doi: 10.1021/ja057519l. [DOI] [PubMed] [Google Scholar]
- 30.Collins CH, Yokobayashi Y, Umeno D, Arnold FH. Engineering proteins that bind, move, make and break DNA. Curr Opin Biotechnol. 2003;14:665. doi: 10.1016/j.copbio.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 31.Lanio T, Jeltsch A, Pingoud A. On the possibilities and limitations of rational protein design to expand the specificity of restriction enzymes: A case study employing EcoRV as the target. Protein Eng. 2000;13:275–281. doi: 10.1093/protein/13.4.275. [DOI] [PubMed] [Google Scholar]
- 32.Liu Q, Xia Z, Zhong X, Case CC. Validated zinc finger protein designs for all 16 GNN DNA triplet targets. J Biol Chem. 2002;277:3850–3856. doi: 10.1074/jbc.M110669200. [DOI] [PubMed] [Google Scholar]
- 33.Mani M, Kandavelou K, Dy FJ, Durai S, Chandrasegaran S. Design, engineering, and characterization of zinc finger nucleases. Biochem Biophys Res Commun. 2005;335:447–457. doi: 10.1016/j.bbrc.2005.07.089. [DOI] [PubMed] [Google Scholar]
- 34.Desjarlais JR, Berg JM. Use of a zinc-finger consensus sequence framework and specificity rules to design specific DNA-binding proteins. Proc Natl Acad Sci USA. 1993;90:2256–2260. doi: 10.1073/pnas.90.6.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.