Abstract
The differential gain and loss of genes from homologous gene families represents an important source of functional variation among the genomes of different species. Differences in gene content between species are primarily attributable to lineage-specific gene gains via duplication and lineage-specific losses via deletion or inactivation. Here, we use a comparative genomic approach to investigate this process of gene turnover in the β-globin gene family of placental mammals. By analyzing genomic sequence data from representatives of each of the main superordinal clades of placental mammals, we were able to reconstruct pathways of gene family evolution during the basal radiation of this physiologically and morphologically diverse vertebrate group. Our analysis revealed that an initial expansion of the nonadult portion of the β-globin gene cluster in the ancestor of placental mammals was followed by the differential loss and retention of ancestral gene lineages, thereby generating variation in the complement of embryonic globin genes among contemporary species. The sorting of ε-, γ-, and η-globin gene lineages among the basal clades of placental mammals has produced species differences in the functional types of hemoglobin isoforms that can be synthesized during the course of embryonic development.
Keywords: β-globin gene family, gene duplication, gene family evolution, genome evolution, hemoglobin
Efforts to identify genetic changes that underlie phenotypic differences among species traditionally focus on nucleotide divergence between orthologous genes. The differential gain and loss of genes from homologous gene families represents a less widely appreciated source of functional variation among the genomes of different species (1–6). Differences in the complement of genes between species are primarily attributable to lineage-specific gene gains via duplication and lineage-specific losses via deletion or inactivation. The β-globin gene cluster of mammals represents an especially good model for investigating mechanisms and processes of genome evolution, because it is one of the most intensively studied multigene families from the standpoint of molecular genetics and phylogenetic history (7–9). The β-globin gene cluster of mammals contains a set of developmentally regulated genes that are arranged in their temporal order of expression (10–12). The ε-, γ-, and η-globin genes (HBE, HBG, and HBH, respectively) are expressed in embryonic erythroid cells and are descended from an ancestral HBE gene. The δ- and β-globin genes (HBD and HBB, respectively) are expressed in fetal and adult erythroid cells and are descended from an ancestral HBB gene. There are some exceptions to these general patterns of stage-specific expression, because duplicated copies of HBG genes have been recruited for fetal expression in anthropoid primates (8) and duplicated copies of HBB genes have been recruited for exclusive fetal expression in some cetartiodactyls (13).
In contrast to the diverse repertoire of β-like globin genes in eutherian (placental) mammals that have been studied to date, monotremes and marsupials possess a single pair of β-like genes: an early-expressed 5′ copy and an ontogenetically later-expressed 3′ copy (14–18). Remarkably, the early- and late-expressed β-like globin genes in monotremes and therian mammals (marsupials and eutherians) are the products of independent duplications of a proto β-globin gene in each of these two lineages (19). Whereas the β-globin gene cluster of marsupials has retained the ancestral two-gene structure, the addition of new early- and late-expressed genes to the β-globin gene cluster of eutherian mammals is attributable to several successive rounds of duplication and divergence after the eutherian/marsupial split, which is thought to have occurred ≈170 Mya (20). Because the HBD gene is either weakly expressed or completely nonfunctional in the majority of eutherian mammals, the increased functional diversity of the β-globin gene cluster in eutherian mammals is mainly attributable to the expansion of the “nonadult” HBE-HBG-HBH portion of the gene cluster.
Thanks to recent advances in the molecular systematics of eutherian mammals, we now have a solid phylogenetic framework for reconstructing pathways of gene family evolution in this morphologically and physiologically diverse vertebrate group. Eutherian mammals are classified into four superordinal groups: Afrotheria (which includes elephants, hyraxes, manatees, aardvarks, tenrecs, and allies), Xenarthra (which includes sloths, armadillos, and anteaters), Laurasiatheria (which includes bats, eulipotyphlans, pangolins, carnivores, perrisodactyls, and cetartiodactyls), and Euarchontoglires (which includes primates, tree shrews, colugos, rabbits, and rodents). Recent phylogenomic studies have demonstrated that a clade (Atlantogenata) composed of Afrotheria and Xenarthra is the sister group of all remaining members of the eutherian crown group (Boreoeutheria) (21, 22). The inferred structure of the β-globin gene cluster in the common ancestor of Boreoeutheria is: 5′-ε-γ-η-δ-β-3′ (13, 14, 16, 23). Because the genomic structure of the β-globin gene cluster has not been previously characterized in any representatives of Atlantogenata, the full complement of β-like globin genes in the crown group ancestor of eutherian mammals has yet to be elucidated.
Here, we report the results of a comparative genomic analysis of the β-globin gene cluster in a diverse set of species that encompasses the four main superordinal clades of eutherian mammals. Results of our analysis demonstrate that the initial expansion of the nonadult portion of the gene cluster in the ancestor of eutherian mammals was followed by differential retention of ancestral gene lineages among different clades, thereby generating variation in the complement of embryonic globin genes among contemporary species. By using a phylogenetic approach to reconstruct pathways of gene family evolution during the basal diversification of eutherian mammals, we found that (i) all eutherian species examined have retained an HBE gene at the 5′ end of the cluster; (ii) most representatives of Xenarthra, Afrotheria, and Euarchontoglires have lost the HBH gene while retaining the HBG gene; and (iii) most representatives of Laurasiatheria have lost the HBG gene while retaining the HBH gene.
Results
Genomic Structure of the Mammalian β-Globin Gene Cluster.
We obtained genomic sequences that spanned the β-globin gene clusters of 21 eutherian and 3 metatherian species. Comparison of the β-globin gene clusters among the eutherian species in our study revealed considerable variation in the size and membership composition of the gene family (Fig. 1). The number of putatively functional genes in the cluster ranged from 2 in the pig to 8 in the rat. As is generally the case in the globin gene clusters of vertebrates (11, 12), the nonadult genes—HBE, HBH, and HBG—were located upstream of the late-expressed HBD and HBB genes. The only exceptions involved en bloc duplications in the goat and cow, where HBE and HBH genes in the 3′ duplication blocks were located downstream of the HBB gene in the 5′ blocks (13).
Fig. 1.
Genomic structure of the β-globin gene cluster in therian mammals. Phylogenetic relationships are based on a loose consensus of recent studies (21, 22, 49, 50). Diagonal slashes indicate gaps in genomic coverage. Segments containing such gaps were not drawn to scale. The orientation of the cluster is from 5′ (on the left) to 3′ (on the right).
We found that all eutherian species possess one to two copies of the HBE gene at the 5′ end of the cluster, and the vast majority of species possess a functional copy of one additional embryonic gene: either HBG or HBH, but never both. The variation in gene family size is mainly attributable to variation in the number of HBB genes at the 3′ end of the cluster. Whereas myomorph rodents (e.g., Mus and Rattus) possess two to four HBB genes and the tenrec possesses four HBB genes, most species possess a single copy. The two eulipotyphlan species in our dataset, the African pygmy hedgehog and the Eurasian shrew, have lost HBB altogether (Fig. 1). In these two species it appears that paralogous copies of the HBD gene are solely responsible for synthesizing the β-chain subunits of adult hemoglobin. The variation in membership composition of the β-globin gene family is mainly attributable to the differential loss of the embryonic HBH and HBG genes and the late-expressed HBD gene. Below we assess this variation in the complement of β-like globin genes in a phylogenetic framework. Because the genomic structure of the β-globin gene family has been characterized previously in primates, rodents, and rabbits (all members of the superorder Euarchontoglires), here we focus on resolving orthologous relationships of β-like globin genes among species in Afrotheria, Xenarthra, and Laurasiatheria. The genomic structure of the β-globin gene cluster has not been previously characterized in any species from the former two groups.
Genomic Structure and Orthologous Relationships in Atlantogenata.
Among atlantogenatan species, we obtained complete coverage of the β-globin gene cluster in one xenarthran species, the nine-banded armadillo, and one afrotherian species, the lesser hedgehog tenrec. In the nonadult portion of the cluster, both species possess a single copy of HBE and a single copy of HBG, although HBG is present only as a pseudogene in the armadillo (Fig. 1). We did not find a functional HBH gene in either of the atlantogenatan species examined. The only trace of HBH was a fragment (spanning intron 2, exon 3, and the 3′ untranslated region) in the armadillo gene cluster that appears to be orthologous to the human HBHps pseudogene (=ψη) [supporting information (SI) Fig. S1]. In the adult portion of the cluster, the armadillo possesses single copies of the HBD and HBB genes, whereas the gene cluster of the tenrec contains no trace of HBD, but contains four copies of HBB (Fig. 1).
In the nonadult portion of the cluster, phylogeny reconstructions of flanking and intronic sequences strongly suggest that the HBE genes in the armadillo and tenrec are 1:1 orthologs of the HBE gene in humans (Fig. 2). Phylogeny reconstructions based on upstream flanking sequence and intron 2 sequence also indicate that the HBG genes in the two atlantogenatan species are orthologous to the duplicated pair of HBG genes in humans (Gγ- and Aγ-globin) (Fig. 2).
Fig. 2.
Maximum likelihood phylograms depicting relationships among β-like globin genes in species of the superorder Atlantogenata based on 1 kb of 5′ flanking sequence (Left), intron 2 (Center), and 1 kb of 3′ flanking sequence (Right). Bootstrap support for the relevant nodes was evaluated by using 1,000 pseudoreplicates.
In the adult portion of the cluster, phylogeny reconstructions based on flanking and intronic sequence clearly show that the HBD genes of armadillo and human are 1:1 orthologs, as are the HBB genes of these same two species (Fig. 2). By contrast, in the same phylogenetic trees, monophyly of the four HBB paralogs of the tenrec indicates that this set of genes originated via three successive rounds of lineage-specific gene duplication (Fig. 2). Curiously, the four HBB paralogs of the tenrec have downstream flanking sequences that exhibit strong affinities to HBD-like sequences of other species (Fig. 2 and Fig. S1). One possible explanation for this pattern is that the coding region and upstream flanking region of an ancestral, single-copy HBD gene were completely converted by an HBB donor gene that has since been deleted in the tenrec lineage. Subsequent rounds of duplication then produced a total of four HBB-like gene copies that have each retained an unconverted HBD-like downstream flanking sequence.
Genomic Structure and Orthologous Relationships in Laurasiatheria.
Among laurasiatherian species, we obtained complete coverage of the β-globin gene cluster in two eulipotyphlans, two bats, two carnivores, one perissodactyl, and one cetartiodactyl. In the nonadult portion of the cluster, we found that the African pygmy hedgehog (order Eulipotyphla) is the only mammalian species that possesses two copies of the HBE gene (Fig. 1). The coding regions of these two HBE paralogs were distinguished by a total of two synonymous substitutions, which suggests two possibilities: (i) the two genes are the products of a relatively recent, lineage-specific duplication event, or (ii) the two genes have a more ancient origin, but have undergone a relatively recent, lineage-specific gene conversion event. Although the horse possesses an HBGps pseudogene, no trace of the HBG gene was found in any of the other laurasiatherian species. All bats and carnivores in our sample possess a single HBH gene and the horse possesses an HBH gene in addition to an HBHps pseudogene. The HBH gene has been inactivated or lost in the remaining laurasiatherian species (Fig. 1).
For the nonadult portion of the cluster, phylogeny reconstructions of flanking and intronic sequence show that the HBE genes in laurasiatherian species are 1:1 orthologs of the HBE gene in humans (or coorthologs in the case of the HBE-T1 and HBE-T2 genes in the hedgehog) (Fig. 3 and Fig. S2). Phylogeny reconstructions also indicated that the HBGps pseudogene of horse is sister to the two HBG paralogs of humans, and that the HBH genes of all laurasiatherian species are 1:1 orthologs of the HBHps pseudogene in humans (or coorthologs in the case of the HBH gene and the HBHps pseudogene in the horse) (Fig. 3). In combination with the results for the atlantogenatan taxa (see above) and marsupials (16, 19), these results indicate that the HBE genes of therian mammals originated via duplication of a proto-HBB gene after the therian/prototherian split (≈220 Mya), and that HBG and HBH are the products of two successive rounds of duplication that occurred after the eutherian/metatherian split (≈170 Mya). For the adult portion of the cluster, phylogeny reconstructions of flanking and intronic sequence demonstrated that the HBB genes of laurasiatherian species are 1:1 orthologs of the HBB gene in humans, and likewise for the HBD genes (Fig. 3). However, we did identify several cases in which recombinational exchanges between HBB and HBD affected upstream flanking sequence (the three HBD paralogs of the African pygmy hedgehog, HBD-T2 of dog, and HBB of cat) and intron 2 sequence (the HBDps pseudogene of pig) (Fig. 3).
Fig. 3.
Maximum likelihood phylograms depicting relationships among β-like globin genes in species of the superorder Laurasiatheria based on 1 kb of 5′ flanking sequence (Left), intron 2 (Center), and 1 kb of 3′ flanking sequence (Right). Bootstraps support for the relevant nodes was evaluated by using 1,000 pseudoreplicates.
Discussion
Results of our comparative genomic analysis revealed that the initial expansion of the nonadult portion of the β-globin gene cluster in the ancestor of eutherian mammals was followed by the differential loss and retention of ancestral gene lineages, thereby generating partially overlapping inventories of embryonic globin genes among contemporary species. All eutherian species have retained at least one copy of the HBE gene at the 5′ end of the gene cluster. However, as a result of the sorting of HBG and HBH gene lineages among the four main clades of eutherian mammals, contemporary species possess either HBE paired with HBG (e.g., lesser hedgehog tenrec, and the majority of species in Euarchontoglires) or HBE paired with HBH (the majority of species in Laurasiatheria) (Fig. 1). There are also several eutherian species that have independently lost both HBG and HBH (e.g., armadillo, African pygmy hedgehog, Eurasian shrew, guinea pig, and pig). In fact, as a result of independent inactivations and deletions, the β-globin gene clusters of the pig and the guinea pig have independently reverted to the ancestral 5′-HBE-HBB-3′ structure seen in marsupials. The armadillo, African pygmy hedgehog, and Eurasian shrew also approximate this ancestral state except that they have two or more copies of each early- and late-expressed paralog (Fig. 1).
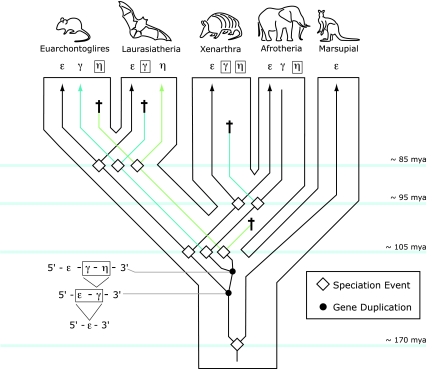
We inferred the differential loss of the embryonic HBG and HBH genes by using a phylogenetic approach to reconstruct pathways of gene family evolution in the four main groups of eutherian mammals: Afrotheria, Xenarthra, Laurasiatheria, and Euarchontoglires. Our model for the evolution of the nonadult portion of the mammalian β-globin gene cluster is graphically depicted in Fig. 4. According to this model, two successive duplications of a proto-HBE gene gave rise to the HBG and HBH genes in the ancestor of eutherian mammals after divergence from marsupials (see also refs. 9, 23, and 24). Consequently, the full complement of embryonic globin genes—HBE-HBG-HBH—was present in the common ancestor of the two main clades of eutherian mammals, Boreoeutheria (comprising Euarchontoglires and Laurasiatheria) and Atlantogenata (comprising Xenarthra and Afrotheria). Subsequent to the Boreoeutheria–Atlantogenata split (≈105 Mya), the HBH gene was lost in the common ancestor of xenarthrans and afrotherians, and subsequent to the divergence of the two latter groups (≈95 Mya), the HBG gene was lost in xenarthrans. The ancestral three-gene set was also present in the common ancestor of Euarchontoglires and Laurasiatheria. Subsequent to the divergence of these two groups (≈85 Mya), the HBG gene was lost in Laurasiatheria and the HBH gene was lost in Euarchontoglires. Although HBH was deleted altogether in the gene clusters of rabbits and rodents, a HBHps pseudogene has been retained in nearly all primates that have been examined (9, 25). As shown in Fig. 1, the HBH gene has also been independently lost in several laurasiatherian taxa (e.g., African pygmy hedgehog, Eurasian shrew, and pig).
Fig. 4.
A model describing the evolution of the β-globin gene family in eutherian mammals. According to this model, two successive duplications of a proto-HBE gene gave rise to the HBG and HBH genes in the ancestor of eutherian mammals after divergence from marsupials. Consequently, the full complement of embryonic globin genes—HBE-HBG-HBH—was present in the common ancestor of the two main clades of eutherian mammals, Boreoeutheria (comprising Euarchontoglires and Laurasiatheria) and Atlantogenata (comprising Xenarthra and Afrotheria). Subsequent to the Boreoeutheria–Atlantogenata split (≈105 Mya), the HBH gene was deleted in the common ancestor of xenarthrans and afrotherians, and the HBG gene was lost in xenarthrans after divergence from the afrotherian lineage (≈95 Mya). The ancestral three-gene set was also present in the common ancestor of Euarchontoglires and Laurasiatheria. Subsequent to the divergence of these two groups (≈85 Mya), the HBG gene was lost in laurasiatherians and the HBH gene was lost in Euarchontoglires. Latin crosses denote lineage-specific gene losses, either via deletion or inactivation.
These species differences in the complement of β-like globin genes are associated with differences in the functional diversity of prenatally expressed hemoglobin isoforms. In bats, cats, dogs, and horses, prenatal hemoglobins incorporate β-chain products of HBE and HBH. By contrast, in representatives of Afrotheria (tenrec) and Euarchontoglires (rabbits, myomorph rodents, and prosimian primates), prenatal hemoglobins incorporate β-chain products of HBE and HBG. The functional significance of this hemoglobin isoform diversity remains to be elucidated.
Mode of Gene Family Evolution.
Results of our analysis demonstrate that the genomic structure of the mammalian β-globin gene family has been shaped by a mixed process of concerted evolution and birth-and-death evolution. However, concerted evolution appears to have been largely restricted to tandemly duplicated copies of the same paralogous type (e.g., between HBB-T1 and HBB-T2 of mouse or between HBG-T1 and HBG-T2 of anthropoid primates; refs. 26, 27). In the adult portion of the gene cluster, ectopic recombination between HBB and HBD paralogs has created chimeric β/δ fusion genes in multiple, independent lineages (9, 23, 28–30). In the nonadult portion of the gene cluster, ectopic recombination between HBE and HBG has created a chimeric γ/ε fusion gene in myomorph rodents (24, 31). Aside from this one exception in rodents, we found no evidence of recombinational exchange among the HBE, HBG, and HBH paralogs in any other placental mammals. There are no pronounced differences in levels of interparalog divergence between species that possess an HBE–HBG gene pair (tenrec and most representatives of Euarchontoglires) vs. those that possess an HBE-HBH gene pair (most representatives of Laurasiatheria). In species that possessed HBE and HBG in tandem, levels of amino acid sequence divergence between the two paralogs ranged from 20.5% to 25%, and in species that possessed HBE and HBH in tandem, levels of interparalog divergence ranged from 19 to 22%. In comparison with the HBB genes, the nonadult genes are characterized by higher levels of sequence conservation, which presumably reflects a higher level of functional constraint (16, 23).
Gene Duplication, Functional Redundancy, and Evolutionary Innovation.
The differential loss of HBG and HBH genes among different lineages of eutherian mammals may have been a purely stochastic process such that the particular complement of genes inherited by a given species was simply a matter of chance. However, the particular complement of genes inherited by a given species may help steer the trajectory of physiological evolution. The possession of multiple, functionally redundant gene copies may provide increased scope for evolutionary innovation because it allows duplicated genes to take on new functions or divide up ancestral functions (32–34).
Whereas monotremes and marsupials possess a single pair of early- and late-expressed β-like globin genes, the majority of eutherian mammals possess a more functionally diverse repertoire of globin genes. The expanded gene complement of eutherian mammals may allow for a greater degree of evolutionary versatility. For example, in anthropoid primates, duplicate copies of HBG have been recruited for fetal expression. It has been argued that the acquisition of fetally expressed hemoglobin played an important role in the life history evolution of anthropoid primates because it facilitated an extended duration of fetal development (8). In New World monkeys, HBG-T1 is expressed in nucleated erythroid cells derived from the embryonic yolk sac (the ancestral condition), but HBG-T2 is expressed in enucleated erythroid cells derived from the fetal liver. In Old World monkeys and apes, both HBG-T1 and HBG-T2 are fetally expressed (35). The cooption of HBG for fetal expression in anthropoid primates was probably facilitated by the fact that redundant or semiredundant copies of the HBE and HBG genes continued to perform their ancestral functions during the early stages of embryogenesis. The evolutionary pathway that led to the acquisition of fetally expressed hemoglobin would not have been accessible if the ancestor of anthropoid primates had possessed only a single embryonic gene, as in contemporary monotremes, marsupials, and some eutherian species like the guinea pig. In a similar fashion, the cooption of the embryonic αD-globin gene for expression during postnatal life appears to have played an important role in the evolution of hypoxia tolerance in Old World vultures and other birds that fly at high altitude (36).
In conclusion, two successive rounds of gene duplication and divergence produced a set of three embryonic β-like globin genes in the ancestor of eutherian mammals. The differential loss and retention of these genes during the subsequent diversification of this group generated variation in the complement of embryonic globin genes among contemporary species and variation in the functional types of hemoglobin isoforms that can be synthesized during the course of prenatal development.
Materials and Methods
Nomenclature for β-Like Globin Genes.
Following the nomenclature of Aguileta et al. (37), we refer to the embryonic ε-, γ-, and η-globin genes as HBE, HBG, and HBH, respectively, and we refer to the late-expressed δ- and β-globin genes, as HBD and HBB, respectively. Because mammalian β-globin genes have undergone multiple rounds of duplication that have resulted in tandemly repeated sets of paralogous gene copies in many species, we index each duplicated gene with the symbol T followed by a number that corresponds to the linkage order in the 5′ to 3′ orientation.
DNA Sequence Data and Gene Identification.
The genomic structure of the β-globin gene cluster has been characterized for cow (Bos taurus), goat (Capra hircus), and several species in the superorder Euarchontoglires (e.g., primates, rodents, and lagomorphs; refs. 12, 13, 30, 38–41). For this subset of taxa, we annotated genomic sequences using information from the database records. For the remaining taxa, we obtained genomic sequences from the High Throughput Genomic Sequences database (HTGS). We characterized the genomic structure of the β-globin gene cluster of one xenarthran species (nine-banded armadillo, Dasypus novemcinctus), one afrotherian species (lesser hedgehog tenrec, Echinops telfairi), and eight laurasiatherian species, including two eulipotyphlans (African pygmy hedgehog, Atelerix albiventris, and Eurasian shrew, Sorex araneus), two bats (little brown bat, Myotis lucifugus, and greater horseshoe bat, Rhinolophus ferrumequinum), two carnivores (cat, Felis catus, and dog, Canis lupus familiaris), one perissodactyl (horse, Equus caballus), and one cetartiodactyl (pig, Sus scrofa) (Table S1). For these species, we identified globin genes in unannotated genomic sequences by using the program Genscan (42) and by comparing known exon sequences to genomic contigs using the program BLAST 2, version 2.2 (43).
Structure of the β-Globin Gene Cluster and Orthologous Relationships.
The genomic structure of the β-globin gene cluster in afrotherian, xenarthran, and laurasiatherian species was investigated by using pairwise analyses of sequence similarity. In each case we included 5 kb of upstream sequence flanking the most 5′ gene copy and 5 kb of downstream sequence flanking the most 3′ gene copy. When comparing gene families among species, it is often difficult to assign orthologous relationships because interparalog gene conversion can obscure the true history of gene duplication and species divergence. Because interparalog gene conversion is typically restricted to the coding regions of globin genes (26, 31, 44–46), we used phylogeny reconstructions of noncoding sequences (flanking regions and intron 2) to assign orthologous relationships among β-like globin genes. All analyses were based on two independent alignments, one containing laurasiatherian species and another containing atlantogenatan species. In both cases the human sequence was included as an outgroup.
Sequence alignments were carried out by using the program MUSCLE (47) as implemented in the Berkeley Phylogenomics Group web server (http://phylogenomics.berkeley.edu). Phylogeny reconstructions were based on coding sequence, 1 kb of upstream flanking sequence, 1 kb of downstream flanking sequence, and intron 2 (1,451 bp in the atlantogenata sequence alignment and 2486 bp in the laurasiatheria sequence alignment). Phylogenetic relationships were inferred in a maximum likelihood framework by using Treefinder, version January 2008 (48) and support for the nodes was assessed with 1,000 bootstrap pseudoreplicates. For analyses based on the atlantogenata sequence alignment, we used an HKY + Γ model of nucleotide substitution. For analyses based on the laurasiatheria sequence alignment, we used the HKY + Γ model (upstream flanking sequence) and a GTR + I + Γ model (intron 2 and downstream flanking sequence).
Supplementary Material
Acknowledgments.
We thank M. Goodman and three anonymous reviewers for helpful comments and suggestions. This work was supported by National Institutes of Health Grant R01 HL087216–01A2 (to J.F.S.), National Science Foundation Grant DEB-0614342 (to J.F.S.), the Nebraska Research Council (J.F.S.), and a University of Nebraska postdoctoral fellowship (to F.G.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0804392105/DCSupplemental.
References
- 1.Demuth JP, De Bie T, Stajich JE, Cristianini N, Hahn MW. The evolution of mammalian gene families. PLoS ONE. 2006;1:e85. doi: 10.1371/journal.pone.0000085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hahn MW, Demuth JP, Han SG. Accelerated rate of gene gain and loss in primates. Genetics. 2007;177:1941–1949. doi: 10.1534/genetics.107.080077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hahn MW, Han MV, Han SG. Gene family evolution across 12 Drosophila genomes. PLoS Genet. 2007;3:e197. doi: 10.1371/journal.pgen.0030197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nam J, Nei M. Evolutionary change of the numbers of homeobox genes in bilateral animals. Mol Biol Evol. 2005;22:2386–2394. doi: 10.1093/molbev/msi229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Niimura Y, Nei M. Evolutionary dynamics of olfactory and other chemosensory receptor genes in vertebrates. J Hum Genet. 2006;51:505–517. doi: 10.1007/s10038-006-0391-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang J, Dyer KD, Rosenberg HF. Evolution of the rodent eosinophil-associated RNase gene family by rapid gene sorting and positive selection. Proc Natl Acad Sci USA. 2000;97:4701–4706. doi: 10.1073/pnas.080071397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Forget BG. Molecular genetics of the human globin genes. In: Steinberg MH, Forget BG, Higgs D, Nagel R, editors. Disorders of Hemoglobin Genetics, Pathophysiology, and Clinical Management. Cambridge, UK: Cambridge Univ Press; 2001. pp. 117–130. [Google Scholar]
- 8.Goodman M, Czelusniak J, Koop BF, Tagle DA, Slightom JL. Globins: A case study in molecular phylogeny. Cold Spring Harb Symp Quant Biol. 1987;52:875–890. doi: 10.1101/sqb.1987.052.01.096. [DOI] [PubMed] [Google Scholar]
- 9.Goodman M, Koop BF, Czelusniak J, Weiss ML. The η-globin gene. Its long evolutionary history in the β-globin gene family of mammals. J Mol Biol. 1984;180:803–823. doi: 10.1016/0022-2836(84)90258-4. [DOI] [PubMed] [Google Scholar]
- 10.Hardison R. Hemoglobins from bacteria to man: Evolution of different patterns of gene expression. J Exp Biol. 1998;201:1099–1117. doi: 10.1242/jeb.201.8.1099. [DOI] [PubMed] [Google Scholar]
- 11.Hardison R. Organization, evolution, and regulation of the globin genes. In: Steinberg M, Forget B, Higgs D, Nagel R, editors. Disorders of Hemoglobin: Genetics, Pathophysiology, and Clinical Management. Cambridge, UK: Cambridge Univ Press; 2001. pp. 95–115. [Google Scholar]
- 12.Collins FS, Weissman SM. The molecular genetics of human hemoglobin. Prog Nucleic Acid Res Mol Biol. 1984;31:315–462. doi: 10.1016/s0079-6603(08)60382-7. [DOI] [PubMed] [Google Scholar]
- 13.Townes TM, Fitzgerald MC, Lingrel JB. Triplication of a four-gene set during evolution of the goat β-globin locus produced three genes now expressed differentially during development. Proc Natl Acad Sci USA. 1984;81:6589–6593. doi: 10.1073/pnas.81.21.6589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cooper SJ, Hope RM. Evolution and expression of a β-like globin gene of the Australian marsupial Sminthopsis crassicaudata. Proc Natl Acad Sci USA. 1993;90:11777–11781. doi: 10.1073/pnas.90.24.11777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cooper SJ, Murphy R, Dolman G, Hussey D, Hope RM. A molecular and evolutionary study of the beta-globin gene family of the Australian marsupial Sminthopsis crassicaudata. Mol Biol Evol. 1996;13:1012–1022. doi: 10.1093/oxfordjournals.molbev.a025651. [DOI] [PubMed] [Google Scholar]
- 16.Koop BF, Goodman M. Evolutionary and developmental aspects of two hemoglobin β-chain genes (εM and βM) of opossum. Proc Natl Acad Sci USA. 1988;85:3893–3897. doi: 10.1073/pnas.85.11.3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wheeler D, et al. An orphaned mammalian β-globin gene of ancient evolutionary origin. Proc Natl Acad Sci USA. 2001;98:1101–1106. doi: 10.1073/pnas.98.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wheeler D, Hope RM, Cooper SJ, Gooley AA, Holland RA. Linkage of the beta-like omega-globin gene to alpha-like globin genes in an Australian marsupial supports the chromosome duplication model for separation of globin gene clusters. J Mol Evol. 2004;58:642–652. doi: 10.1007/s00239-004-2584-0. [DOI] [PubMed] [Google Scholar]
- 19.Opazo JC, Hoffmann FG, Storz JF. Genomic evidence for independent origins of beta-like globin genes in monotremes and therian mammals. Proc Natl Acad Sci USA. 2008;105:1590–1595. doi: 10.1073/pnas.0710531105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Rheede T, et al. The platypus is in its place: Nuclear genes and indels confirm the sister group relation of monotremes and therians. Mol Biol Evol. 2006;23:587–597. doi: 10.1093/molbev/msj064. [DOI] [PubMed] [Google Scholar]
- 21.Hallstrom BM, Kullberg M, Nilsson MA, Janke A. Phylogenomic data analyses provide evidence that Xenarthra and Afrotheria are sister groups. Mol Biol Evol. 2007;24:2059–2068. doi: 10.1093/molbev/msm136. [DOI] [PubMed] [Google Scholar]
- 22.Wildman DE, et al. Genomics, biogeography, and the diversification of placental mammals. Proc Natl Acad Sci USA. 2007;104:14395–14400. doi: 10.1073/pnas.0704342104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hardison RC. Comparison of the β-like globin gene families of rabbits and humans indicates that the gene cluster 5′-ε-γ-δ-β-3′ predates the mammalian radiation. Mol Biol Evol. 1984;1:390–410. doi: 10.1093/oxfordjournals.molbev.a040326. [DOI] [PubMed] [Google Scholar]
- 24.Hill A, et al. Two mouse early embryonic β-globin gene sequences. Evolution of the nonadult β-globins. J Biol Chem. 1984;259:3739–3747. [PubMed] [Google Scholar]
- 25.Bailey WJ, et al. Reexamination of the African hominoid trichotomy with additional sequences from the primate β-globin gene cluster. Mol Phylogenet Evol. 1992;1:97–135. doi: 10.1016/1055-7903(92)90024-b. [DOI] [PubMed] [Google Scholar]
- 26.Storz JF, et al. Complex signatures of selection and gene conversion in the duplicated globin genes of house mice. Genetics. 2007;177:481–500. doi: 10.1534/genetics.107.078550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Slightom J, Koop B, Xu P, Goodman M. Rhesus fetal globin genes: Concerted gene evolution in the descent of higher primates. J Biol Chem. 1988;263:12427–12438. [PubMed] [Google Scholar]
- 28.Hardies SC, Edgell MH, Hutchison CA. Evolution of the mammalian β-globin gene cluster. J Biol Chem. 1984;259:3748–3756. [PubMed] [Google Scholar]
- 29.Hutchison CA, Hardies SC, Padgett RW, Weaver S, Edgell MH. The mouse globin pseudogene βh3 is descended from a premammalian δ-globin gene. J Biol Chem. 1984;259:12881–12889. [PubMed] [Google Scholar]
- 30.Tagle DA, et al. The β-globin gene cluster of the prosimian primate. Galago crassicaudatus: Nucleotide sequence determination of the 41-kb cluster and comparative sequence analyses. Genomics. 1992;13:741–760. doi: 10.1016/0888-7543(92)90150-q. [DOI] [PubMed] [Google Scholar]
- 31.Hardison R, Miller W. Use of long sequence alignments to study the evolution and regulation of mammalian globin gene clusters. Mol Biol Evol. 1993;10:73–102. doi: 10.1093/oxfordjournals.molbev.a039991. [DOI] [PubMed] [Google Scholar]
- 32.Ohno S. Evolution by Gene Duplication. New York: Springer; 1970. p. 160. [Google Scholar]
- 33.Force A, et al. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151(4):1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hughes AL. The evolution of functionally novel proteins after gene duplication. Proc Biol Sci. 1994;256:119–124. doi: 10.1098/rspb.1994.0058. [DOI] [PubMed] [Google Scholar]
- 35.Johnson RM, et al. Phylogenetic comparisons suggest that distance from the locus control region guides developmental expression of primate β-type globin genes. Proc Natl Acad Sci USA. 2006;103:3186–3191. doi: 10.1073/pnas.0511347103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoffmann FG, Storz JF. The αD-globin gene originated via duplication of an embryonic α-like globin gene in the ancestor of tetrapod vertebrates. Mol Biol Evol. 2007;24:1982–1990. doi: 10.1093/molbev/msm127. [DOI] [PubMed] [Google Scholar]
- 37.Aguileta G, Bielawski JP, Yang Z. Proposed standard nomenclature for the α- and β-globin gene families. Genes Genet Syst. 2006;81:367–371. doi: 10.1266/ggs.81.367. [DOI] [PubMed] [Google Scholar]
- 38.Margot JB, Demers GW, Hardison RC. Complete nucleotide sequence of the rabbit β-like globin gene cluster. Analysis of intergenic sequences and comparison with the human β-like globin gene cluster. J Mol Biol. 1989;205:15–40. doi: 10.1016/0022-2836(89)90362-8. [DOI] [PubMed] [Google Scholar]
- 39.Shehee WR, et al. Nucleotide sequence of the BALB/c mouse β-globin complex. J Mol Biol. 1989;205:41–62. doi: 10.1016/0022-2836(89)90363-x. [DOI] [PubMed] [Google Scholar]
- 40.Efstratiadis A, et al. The structure and evolution of the human β-globin gene family. Cell. 1980;21:653–668. doi: 10.1016/0092-8674(80)90429-8. [DOI] [PubMed] [Google Scholar]
- 41.Schimenti JC, Duncan CH. Structure and organization of the bovine β-globin genes. Mol Biol Evol. 1985;2:514–525. doi: 10.1093/oxfordjournals.molbev.a040369. [DOI] [PubMed] [Google Scholar]
- 42.Burge C, Karlin S. Prediction of complete gene structures in human genomic DNA. J Mol Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- 43.Tatusova TA, Madden TL. BLAST 2 Sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol Lett. 1999;174:247–250. doi: 10.1111/j.1574-6968.1999.tb13575.x. [DOI] [PubMed] [Google Scholar]
- 44.Hardison RC, Gelinas RE. Assignment of orthologous relationships among mammalian alpha-globin genes by examining flanking regions reveals a rapid rate of evolution. Mol Biol Evol. 1986;3:243–261. doi: 10.1093/oxfordjournals.molbev.a040392. [DOI] [PubMed] [Google Scholar]
- 45.Hoffmann FG, Opazo JC, Storz JF. Rapid rates of lineage-specific gene duplication and deletion in the α-globin gene family. Mol Biol Evol. 2008;25:591–602. doi: 10.1093/molbev/msn004. [DOI] [PubMed] [Google Scholar]
- 46.Storz JF, Hoffmann FG, Opazo JC, Moriyama H. Adaptive functional divergence among triplicated α-globin genes in rodents. Genetics. 2008;178:1623–1638. doi: 10.1534/genetics.107.080903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jobb G, von Haeseler A, Strimmer K. TREEFINDER: A powerful graphical analysis environment for molecular phylogenetics. BMC Evol Biol. 2004;4:18. doi: 10.1186/1471-2148-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 49.Murphy WJ, et al. Resolution of the early placental mammal radiation using Bayesian phylogenetics. Science. 2001;294:2348–2351. doi: 10.1126/science.1067179. [DOI] [PubMed] [Google Scholar]
- 50.Murphy WJ, Pringle TH, Crider TA, Springer MS, Miller W. Using genomic data to unravel the root of the placental mammal phylogeny. Genome Res. 2007;17:413–421. doi: 10.1101/gr.5918807. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.