Abstract
Bacteroides species are the most abundant Gram-negative bacteria of the human colonic microbiota. These endogenous organisms are unique in that they synthesize an extensive number of phase-variable surface polysaccharides. Pathogenic bacteria phase vary expression of surface molecules for immune evasion, but the importance of the synthesis of multiple phase-variable polysaccharides to these commensal bacteria is unknown. We previously showed that a Bacteroides fragilis mutant unable to synthesize 4 of the 8 capsular polysaccharides and unable to glycosylate proteins properly is rapidly outcompeted by the wild-type strain for colonization of the gnotobiotic mouse intestine. In the present study, we constructed mutants defective only in capsule polysaccharide synthesis to define better the importance of these surface molecules to intestinal colonization. We discovered a key enzymatic activity required for synthesis of 7 of the 8 capsular polysaccharides. Deletion of its gene resulted in the first B. fragilis mutant able to synthesize only one phase-variable polysaccharide, and further mutation resulted in a stable acapsular mutant. We show that the acapsular mutant is rapidly outcompeted, but synthesis of a single polysaccharide is sufficient for the organism to colonize the gnotobiotic intestine competitively. These data demonstrate that initial colonization of the gnotobiotic mouse intestine by B. fragilis requires that the organism synthesize only a single polysaccharide and suggest that the synthesis of multiple phase-variable polysaccharides is important for the bacteria's long-term maintenance in the normally complex and competitive ecosystem.
Keywords: acapsular, microbiota, phase variation, polysaccharide, glycoprotein
The human intestinal tract is home to a vast and diverse alliance of microbes comprising one of the densest microbial ecosystems in the world and one that provides functions essential to human health. Many species of the order Bacteroidales are abundant members of the human intestinal microbiota and have served as models for studying mutualistic bacterial–host relationships in this ecosystem (1, 2). Despite their importance, relatively little is known about the biology of the predominant members of this ecosystem. Deciphering how these mutualistic microorganisms successfully and stably colonize the human intestine may help us understand how they are tolerated, how different members interact with one another, what additional benefits the microbiota provide us, and how microbiota-associated diseases are triggered in the susceptible host.
The intestinal Bacteroidales synthesize a vast repertoire of surface glycans. A single strain of B. fragilis synthesizes 8 distinct capsular polysaccharides (3), an extracellular polysaccharide (4), and numerous glycoproteins (5). The synthesis of extensive numbers of glycosylated molecules is a general property of the intestinal Bacteroidales (6–8). We estimate that the genome of B. fragilis 9343 encodes ≈80 glycosyltransferases and dedicates at least 215,000 bp of DNA (≈4% of its chromosome) to the synthesis and regulation of glycosylated molecules.
All intestinal Bacteroidales species analyzed to date synthesize numerous capsular polysaccharides, most of which undergo phase variation; however, other non-intestinal members of this order, such as those in the oral cavity, do not (8). This conserved characteristic among the intestinal Bacteroidales seems to highlight its importance to colonization and persistence in the mammalian intestine. B. fragilis Δgmd-fclΔfkp is outcompeted rapidly by wild-type B. fragilis in a gnotobiotic mouse colonization model (5). This mutant is defective in both the de novo and salvage GDP-fucose pathways and cannot synthesize 4 of the 8 capsular polysaccharides [polysaccharide B (PSB), polysaccharide C (PSC), polysaccharide D (PSD), and polysaccharide E (PSE)] nor incorporate fucose into glycoproteins. We thus were unable to determine if this mutant's colonization deficiency was caused by its defect in capsular polysaccharide synthesis, in glycoprotein synthesis, or a combination of both. In this study, we determine the specific importance of capsular polysaccharide synthesis to the organism's ability to colonize the gnotobiotic mouse. Contrary to a recent report (9), our data indicate that synthesis of a single capsular polysaccharide is sufficient for competitive colonization of the gnotobiotic mouse intestine. Our data suggest that the ability to synthesize multiple phase-variable polysaccharides probably is an important biological property for long-term survival of these organisms in the complex and competitive intestinal ecosystem.
Results
Construction of Mutants Attenuated for Capsular Polysaccharide Synthesis.
To create a mutant that was stably and irreversibly attenuated for polysaccharide synthesis, we sought to delete gene(s) encoding key product(s) necessary for the synthesis of multiple polysaccharides. Such genes often are located outside the polysaccharide biosynthesis loci in conserved areas of the genome to ensure that synthesis of a polysaccharide is not dependent on a product encoded by another phase-variable locus (5). The structures of B. fragilis 9343 polysaccharide A (PSA) and PSB are known (10), and both contain complex di- and tri-deoxy monosaccharides: PSA contains 2-acetamido-4-amino-2,4,6-trideoxy-d-galactose (AATGal), and PSB contains d- and l-N-acetylquinovosamine (d- and l-QuiNAc). The synthesis of such sugars should first require conversion of UDP-N-acetylglucosamine (UDP-GlcNAc) to a UDP-2-acetamido-4-keto-2,4,6-trideoxy-d-glucose intermediate by a UDP-GlcNAc 4,6-dehydratase [supporting information (SI) Fig. S1]. None of the 8 polysaccharide biosynthesis loci of B. fragilis 9343 contain a putative UDP-GlcNAc 4,6-dehydratase gene. We therefore considered such a gene a good candidate for mutation, because it probably would reside in a conserved area of the chromosome and be required for synthesis of at least PSA and PSB.
We searched the B. fragilis 9343 proteome for orthologs of WbpM, a characterized Pseudomonas aeruginosa UDP-GlcNAc 4,6-dehydratase (11), and found 2 candidates that we designated “UngD1” and “UngD2,” respectively, encoded by genes BF1706 and BF2848 (GenBank accession nos. EU682264 and EU682265). These genes are not located near any of the 8 polysaccharide biosynthesis loci (Fig. 1A). UngD1 and UngD2 have very hydrophobic N-termini, each with 4 predicted membrane-spanning domains, as does WbpM (11). Exclusive of the N-terminal transmembrane regions, UngD1 and UngD2 are 64% and 62% similar to WbpM, respectively, and are 91% similar to each other. UngD1 and UngD2 each contain the Ser-Met-Lys catalytic triad, the GxxGxxG nucleotide co-factor binding motif, and the conserved glutamate residue characteristic of this branch of the short-chain dehydrogenase/reductase family of enzymes (12). To determine if ungD1 and ungD2 encode UDP-GlcNAc 4,6 dehydratase activity, we assessed the ability of each to complement a wbpM mutant strain of P. aeruginosa PAO1 (13). Each was able to restore expression of serotype O5 LPS to P. aeruginosa wbpM::Gm (Fig. 1B).
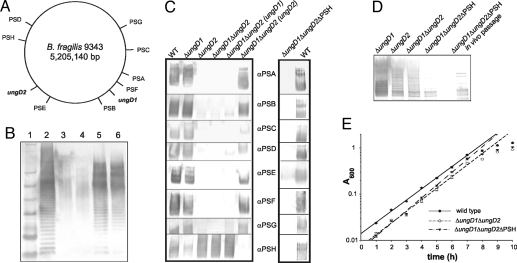
Fig. 1.
Functional and genetic analyses of ungD1 and ungD2. (A) Schematic diagram of the B. fragilis 9343 chromosome with the locations of the 8 capsule loci (PSA–PSH), ungD1, and ungD2. (B) Ability of ungD1 and ungD2 to complement a P. aeruginosa wbpM mutant. Lane 1: SeeBlue Plus2 prestained protein ladder (Invitrogen). Lane 2: P. aeruginosa PAO1. Lane 3: P. aeruginosa PAO1 wbpM::Gm. Lane 4: P. aeruginosa PAO1 wbpM::Gm pUCP18 (vector control). Lane 5: P. aeruginosa PAO1 wbpM::Gm pLEC216 (ungD1). Lane 6: P. aeruginosa PAO1 wbpM::Gm pLEC217 (ungD2). (C) Production of each of the 8 capsular polysaccharides by wild type, ΔungD1, ΔungD2, and ΔungD1ΔungD2 mutants and mutants with ungD1 and ungD2 in trans and by the acapsular mutant (ΔungD1ΔungD2ΔPSH). (D) Whole-cell lysates of B. fragilis mutants probed with antiserum generated to wild-type B. fragilis. Lane 5 is blank. The ΔungD1ΔungD2ΔPSH in vivo sample is bacteria isolated from the feces of mice monoassociated for 3 days and passaged in vitro once before analysis. (E) Growth comparison of B. fragilis 9343 wild type, ΔungD1ΔungD2, and ΔungD1ΔungD2ΔPSH in supplemented basal medium. The average absorbance at 600 nm of 5 replicate cultures is shown.
Single-deletion mutants of ungD1 and ungD2 and an ungD1 and ungD2 double mutant were created to assess the role of these genes in capsular polysaccharide synthesis. All 8 polysaccharides are expressed by the ΔungD1 mutant (Fig. 1C), but deletion of ungD2 abrogates synthesis of 7 of the 8 capsular polysaccharides; only polysaccharide H (PSH) is expressed by this mutant (Fig. 1C). To obtain a completely acapsular mutant, we deleted a large region of the PSH biosynthesis locus from the ΔungD1ΔungD2 mutant. This mutant, ΔungD1ΔungD2ΔPSH, does not synthesize any capsular polysaccharides (Fig. 1 C and D). Multiple passages and phenotypic screenings confirmed that these phenotypes are stable properties of ΔungD1ΔungD2 and ΔungD1ΔungD2ΔPSH. Synthesis of PSA–PSG is restored by providing ungD2 in trans to the ΔungD1ΔungD2 mutant. Because very slight expression of a few polysaccharides was observed when ungD1 was provided in trans to ΔungD1ΔungD2, we continued to use the ΔungD1ΔungD2 double mutant rather than the ΔungD2 single mutant for subsequent studies to ensure that synthesis of all 7 polysaccharides was abrogated.
Glycoprotein Analysis of the ΔungD1, ΔungD1ΔungD2 and Acapsular Mutants.
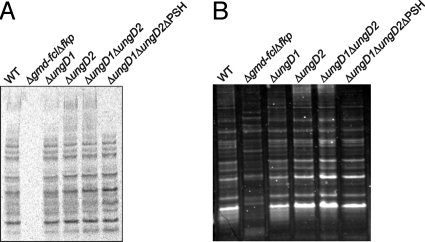
Two different assays, 3H-l-fucose incorporation and glycostain analysis, were used to determine if glycoprotein expression was affected in these mutants. Unlike Δgmd-fclΔfkp, which has an altered glycoprotein profile, the glycoproteins of the ΔungD1, ΔungD2, ΔungD1ΔungD2, and ΔungD1ΔungD2ΔPSH mutants are not altered compared with wild type when analyzed by incorporation of 3H-l-fucose (Fig. 2A) or with a nonspecific glycoprotein stain (Fig. 2B); thus we were able to analyze the distinct contribution of capsular polysaccharides to intestinal colonization.
Fig. 2.
Analysis of glycoprotein synthesis by ungD mutants. (A) Phosphoimager scan of the blot of a PAGE gel of whole-cell lysates of B. fragilis mutants grown in the presence of 3H-l-fucose. (B) Glycostain analysis of whole-cell lysates separated by PAGE.
In Vitro Growth Characteristics of the ΔungD1ΔungD2 and Acapsular Mutants.
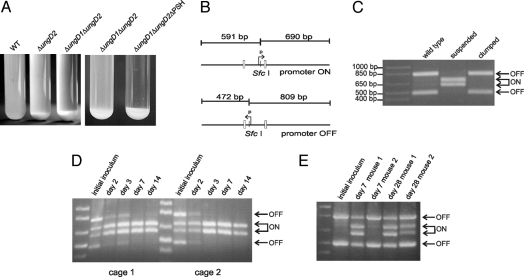
When grown in liquid culture, all the ΔungD1ΔungD2ΔPSH cells settled as an aggregated mass at the bottom of the tube (Fig. 3A). Most ΔungD2 and ΔungD1ΔungD2 cells displayed the same phenotype; only a small percentage of bacteria grew in suspension. The biphasic nature of these ΔungD2 and ΔungD1ΔungD2 cultures suggested that the aggregated cells are acapsular because of the PSH promoter being off, and that the suspended cells have the PSH promoter on. These populations were separated from an actively growing ΔungD1ΔungD2 culture, and the PSH promoter orientations in each were assessed quantitatively by a PCR-digestion assay (Fig. 3B). The results confirmed that the aggregated organisms have the PSH promoter oriented off and therefore are acapsular, whereas suspended cells have the PSH promoter oriented on and are encapsulated (Fig. 3C).
Fig. 3.
Phase variation of PSH in ΔungD1ΔungD2 mutant. (A) Appearance of cultures after overnight growth demonstrating that most of the bacteria with deletions of ungD2 are settled at the bottom of the tube compared with the wild-type culture and that the acapsular mutant grows as a completely aggregated population. (B) PCR-digestion scheme used to determine quantitatively the portions of a given population with the PSH promoter in the on and off orientations. (C) PCR-digestion results for the PSH promoter from the aggregated bacteria versus those growing in suspension for the ΔungD1ΔungD2 mutant. (D) PSH promoter orientations of bacteria from fecal samples of mice monoassociated with ΔungD1ΔungD2 for various intervals. (E) PSH promoter orientations of bacteria from fecal samples of 2 mice monoassociated with wild type for 7 or 28 days.
Based on the aggregative phenotype of the acapsular mutant, gene expression levels of the wild type and the acapsular mutant were examined using whole-genome microarrays to detect substantial alterations in the acapsular mutant (GEO accession numbers GSM284753, GSM284777, GSM284778, GSM284779, GSM284780, and GSM284781). Surprisingly, the ΔungD1ΔungD2ΔPSH expression profile is unremarkable compared with wild type (Tables S1 and S2), suggesting that the physiology of the acapsular mutant strain is not significantly different from that of wild type. Of the 91 genes identified as differentially expressed, 20 (22.0%) are capsular polysaccharide genes from phase-variable loci, which often demonstrate drastic expression differences even in wild-type cultures, and another 44 (48.4%) are annotated only as hypothetical proteins. The cluster of orthologous groups analysis is instructive as well: many categories expected to be heavily represented if growth of ΔungD1ΔungD2ΔPSH were significantly altered compared with wild type (e.g., translation, transcription, replication, recombination and repair, and chaperones) contained very few or no differentially expressed genes.
Based on these microarray data, we did not expect to find a significant alteration in the growth rate of the acapsular mutant despite its aggregative phenotype. Indeed, we were unable to detect any in vitro growth defect in the ΔungD1ΔungD2 or the acapsular strains (Fig. 1E).
Mouse Monoassociation Experiments.
The ability of ΔungD1ΔungD2 and ΔungD1ΔungD2ΔPSH to colonize the intestinal tract of gnotobiotic mice in the absence of competing organisms was analyzed separately. In these monoassociation assays, each strain colonized at ≈3 × 1010 bacteria/g feces, equivalent to the level attained by wild-type bacteria (5). Bacteria from the stool of mice monoassociated with ΔungD1ΔungD2ΔPSH for 3 days were cultured to ensure that the acapsular mutant did not regain the ability to synthesize a capsule following in vivo passage. These bacteria do not revert to an encapsulated state but rather are stably acapsular (Fig. 1D).
The ΔungD1ΔungD2 mutant grows as a mixed population; some bacteria are encapsulated (PSH promoter on), whereas others are acapsular (PSH promoter off). A PCR-digestion assay was used to analyze the PSH promoter orientation of bacteria in fecal samples of mice monoassociated with ΔungD1ΔungD2 to detect if either population has an advantage for intestinal colonization. The in vitro–grown inoculum used to initially colonize the mice had > 60% of the bacterial population with the PSH promoter oriented off (Fig. 3D). A rapid and drastic population shift favoring bacteria with the PSH promoter on was observed by day 2, and by day 7 nearly all of the ΔungD1ΔungD2 analyzed from mouse fecal samples had the PSH promoter oriented on, suggesting that bacteria expressing PSH have a colonization advantage over those that are acapsular.
To investigate whether the PSH promoter has a natural tendency to shift on in the mouse intestine, bacteria in fecal samples from mice monoassociated with wild-type organisms were examined for PSH promoter orientation. These analyses revealed that although the population of bacteria with the PSH promoter on does increase in vivo, the shift is very gradual, and the promoter remains off in > 30% of the wild-type population, even by day 28 (Fig. 3E). These data further suggest that encapsulated bacteria have a survival advantage in the mouse intestine.
Competitive Colonization Assays.
Competitive colonization assays were performed to detect whether the acapsular mutant is less fit to survive in the gnotobiotic mouse intestine, as suggested by the ΔungD1ΔungD2 monoassociation data, and to determine whether a strain expressing only a single polysaccharide can compete with wild-type organisms in this model.
The initial competitive colonization assay used an inoculum consisting of 34% wild type and 66% ΔungD1ΔungD2. After 15 days, the ΔungD1ΔungD2 mutant comprised 60% and 79% of the bacteria in the feces of mice from 2 cages (Table 1) and thus exhibited no colonization defect. The experiment was repeated using an inoculum in which ΔungD1ΔungD2 was reduced to 38%. The ΔungD1ΔungD2 continued to persist for the 15-day duration of the experiment, with the percentage of reduction possibly representing selection against the acapsular population of ΔungD1ΔungD2. In contrast, our previous competitive colonization experiments with Δgmd-fclΔfkp showed that this mutant was almost completely outcompeted by wild type in 3 days (5).
Table 1.
Percentage of ΔungD1ΔungD2 in fecal samples during competitive colonization assays with wild type
| Source | ΔungD1ΔungD2 present (%) at day |
|||
|---|---|---|---|---|
| Zero* | Three | Seven | Fifteen | |
| Cage 1 | 66 | 54 | 40 | 60 |
| Cage 2 | 66 | – | – | 79 |
| Cage 1 | 38 | 10 | 21 | 18 |
| Cage 2 | 38 | 6 | 10 | 12 |
*Day zero represents the ratio used to initially inoculate mice.
Similar competitive colonization assays were performed with wild type and the acapsular mutant. In two separate experiments using a mixed culture initial inoculum in which ΔungD1ΔungD2ΔPSH comprised 81% or 84% of total bacteria, we noted a consistent and rapid decrease in the number of mutant bacteria detected so that by day 7 no acapsular bacteria were detected in the fecal samples from mice in 3 of the 4 cages (Table 2).
Table 2.
Percentage of ΔungD1ΔungD2ΔPSH in fecal samples during competitive colonization assays with wild type
| Source | ΔungD1ΔungD2ΔPSH present (%) at day |
||||
|---|---|---|---|---|---|
| Zero* | Two | Three | Seven | Fourteen | |
| Cage 1 | 81 | 33 | 18 | 1 | 8 |
| Cage 2 | 81 | 11 | 3 | 0 | 0 |
| Cage 1 | 84 | 12 | 0 | 0 | 0 |
| Cage 2 | 84 | 2 | 0 | 0 | 0 |
*Day zero represents the ratio used to initially inoculate mice.
Survival of Acapsular Mutant Under Conditions Simulating the in Vivo Environment.
The rapid decline in the acapsular population in the presence of wild type suggests that host factors contribute to this selection. There are numerous host factors that could limit the growth and survival of acapsular bacteria. We first tested whether the acapsular mutant is as able as wild type to survive at low pH, a condition these organisms must survive to transit the stomach (pH minimum of 2). The wild-type, ΔungD1ΔungD2, and ΔungD1ΔungD2ΔPSH strains all grew normally in supplemented basal media adjusted to pH 5.5 and above and failed to grow at a pH of 5.0 or below. Although the bacteria failed to grow, they still were viable after exposure to pH 4.0 for 24 h, pH 3.5 for 6 h, and pH 3.0 for 2 h. Importantly, no differences were noted in the susceptibilities of these 3 strains to acidic pH.
The ability to resist killing by bile and cationic detergents also is important for intestinal survival. We tested the 3 strains for differences in their sensitivity to the cationic detergents hexadecyltrimethylammonium bromide (CTAB), benzalkonium chloride (BZAC), deoxycholic acid (a bile acid and an anionic detergent), bile salts, and porcine bile extract. All 3 strains grew at 0.0000781% (wt/vol, ≈2.1 mM) CTAB and at 0.0003125% (≈8.5 mM) BZAC and below and were killed by concentrations of bile salts >0.02% (≈473.3 μM), and by deoxycholic acid levels >0.01% (≈241.2 μM). All 3 strains also grew well in porcine bile extract at concentrations ≤ 1% wt/vol. Thus, there were no differences in the ability of these 3 strains to grow under these conditions.
Many innate immune factors are present in the gut, and some selectively target specific bacterial populations (14–16). The synthesis of surface polysaccharides is correlated directly with the ability of many bacteria to resist the bactericidal effects of a particular innate immune factor, complement (17–19). Complement has been studied largely as a serum factor, but components of the complement cascade are synthesized by normal colonic epithelial cells (20, 21) and may play a role in maintaining homeostasis in the gut. Based on the importance of surface polysaccharides of other bacteria in resisting complement-mediated killing, we tested the ability of the acapsular strain to survive in the presence of complement. We found that wild-type bacteria are resistant to the bactericidal effects of normal human serum but that the acapsular strain is susceptible, demonstrating a 30-fold reduction in viable counts after 1 h (Fig. S2).
Discussion
The construction of defined capsular polysaccharide mutants allows us to investigate the role of these molecules in intestinal colonization. The creation of a stable acapsular mutant and one that synthesizes only a single polysaccharide in a normal phase-variable manner was not achieved by other mutational strategies (9, 22). Because we targeted genes whose products are involved in the synthesis of capsular polysaccharides rather than in their regulation, we obtained mutants with stable phenotypes. We found that UngD2 is required for the synthesis of 7 of the 8 capsular polysaccharides of B. fragilis 9343. The complementation and mutational data support the homology and motif-based analyses that UngD2 is an UDP-GlcNAc 4,6-dehydratase and provides an intermediate derived from UDP-GlcNAc necessary for synthesis of many N-acetylated di- or tri-deoxy monosaccharides. These data further suggest that PSC, PSD, PSE, polysaccharide F (PSF), and polysaccharide G (PSG) all contain N-acetylated di- or tri-deoxy monosaccharides similar to those of PSA and PSB. Based on the lack of apparent alterations in the glycoproteins of any of the ungD mutants, these molecules may contain less complex monosaccharides than the capsular polysaccharides.
Because all 8 of the B. fragilis capsular polysaccharides undergo phase variation in wild-type cells, one might expect there would be an acapsular portion in the population in which all of these loci were phased off simultaneously. The acapsular state, however, does not occur naturally in a wild-type population because the PSC promoter does not invert. Moreover, PSC is not expressed when other polysaccharide promoters are locked on (22), and therefore PSC probably is the “fail-safe” polysaccharide, expressed only when needed to ensure that the organism always produces at least one capsule. The aggregative growth phenotype and colonization deficiency exhibited by the acapsular mutant suggest the importance of this fail-safe mechanism to the organism.
Our experimental data contradict some conclusions of a recent study (9). This report states that production of at least one capsular polysaccharide is required for viability and that the acapsular mutant is not tolerated and must revert to express capsular polysaccharides. Our acapsular mutant is very stable and grows comparably to wild type in vitro (Fig. 1E). Additionally, we saw no propensity for ΔungD1ΔungD2ΔPSH to become encapsulated, even after passage through the mouse intestine. Moreover, our single polysaccharide-producing mutant is not notably defective for intestinal colonization for the period analyzed. We suspect differences in the nature of the mutants used in the two studies contributed to these disparities. Because the earlier study deleted regulatory genes rather than genes involved in capsule biosynthesis, the regulatory mutant strain was able to revert. The mutant used in the earlier study was deleted not only for the gene encoding the global polysaccharide promoter DNA invertase Mpi but also for the adjacent gene tsr19. Mpi mediates the inversion not only of the 7 invertible PS biosynthesis loci promoters but also of 6 other promoter regions that transcribe regions encoding products with unknown functions (22). Therefore, deletion of mpi affects more than just polysaccharide biosynthesis. In addition, Tsr19 is a tyrosine-family site-specific recombinase that inverts a promoter governing extracellular polysaccharide expression (4). Because the contribution of extracellular polysaccharide expression to intestinal colonization has not yet been determined, colonization deficiencies observed using mpi-tsr19 deletion mutants cannot be attributed solely to differences in capsular polysaccharide expression.
It also is possible that heterogeneous polysaccharides of B. fragilis are not functionally equivalent, and some may not bestow upon the organism the ability to compete competitively with wild type. The mutants of the prior study expressed only PSB or PSC. Our mutant expressing a single polysaccharide synthesized only PSH. Therefore, further studies are necessary to determine the ability of each of the 8 polysaccharides to impart competitive colonization properties to the organism.
The results of our study allow us to conclude that the Δgmd-fclΔfkp mutant's inability to compete with wild type in the gnotobiotic mouse intestine is not caused by its inability to synthesize PSB, PSC, PSD, and PSE, because the ΔungD1ΔungD2 mutant is unable to synthesize these 4 polysaccharides and 3 others (PSA, PSF, and PSG), and it still persists in competition with wild-type organisms. Rather, it is likely that the Δgmd-fclΔfkp mutant's glycoprotein deficiency accounts for its colonization defect in this model. Based on the data in this study and on earlier data showing that a large number of proteins are glycosylated in B. fragilis (5), further analysis of these glycoproteins will be important to understand their role in intestinal colonization.
The ΔungD1ΔungD2 unencapsulated PSH phase variant and the stable acapsular mutant cannot compete for intestinal colonization in this animal model, strongly suggesting that synthesis of at least one capsular polysaccharide is necessary for B. fragilis to colonize its niche. However, constitutive synthesis of the same capsular polysaccharide probably would be disastrous for the organism over time in the complex intestinal ecosystem. Defensive products produced by the host, such as antibody, or offensive products produced by other members of the microbiota target surface polysaccharides (23–25). Thus, the capacity to produce multiple phase-variable capsular polysaccharides creates diverse populations with regard to surface architecture, including some that would be less susceptible to attack by deleterious products. Indeed, many pathogenic bacteria and parasites have evolved mechanisms by which they alter their surface antigenicity to persist in the host (26–28).
Direct demonstration that the synthesis of multiple phase-variable polysaccharides confers a survival advantage probably will require more complex animal models and extended periods of colonization. However, the widespread presence of systems devoted to generating surface variability throughout intestinal species of the Bacteroidales order argues strongly that such an advantage is operable. Factors such as change in diet, microbial content, phage exposure, and health of the host should be addressed to assess better the importance of this unique and conserved feature of the intestinal Bacteroidales.
Experimental Procedures
Bacterial Strains and Growth Conditions.
B. fragilis 9343 was the parental strain of all mutants and was grown as previously described (29). Labeling with l-[5,6-3H] fucose (1 mCi/ml, specific activity 50 Ci/mmol, American Radiolabeled Chemicals) followed a published method (5). P. aeruginosa strains were grown in L-broth or on L-agar plates supplemented where appropriate with gentamicin (300 μg/ml, Invitrogen) and carbenicillin (500 μg/ml, Invitrogen). ΔungD1ΔungD2ΔPSH was agitated vigorously before each dilution and spectrophotometer reading.
Construction of ΔungD1, ΔungD2, ΔungD1ΔungD2, and ΔungD1ΔungD2ΔPSH.
Creation of deletion mutants involved PCR amplification (primer sequences are provided in Table S3) of DNA flanking each side of the region to be deleted, digestion of these products with restriction enzymes using sites engineered into the primers, and 3-way ligation into the Bacteroides conjugal suicide vector pNJR6 (30) or pJST55 (31). After conjugal transfer to B. fragilis, cointegrates were selected by resistance to erythromycin, passaged, plated on nonselective medium, and replica plated to medium containing erythromycin. Erythromycin-sensitive colonies were screened by PCR to detect those acquiring the mutant genotypes.
The ΔungD1 (BF1706) mutant has 1810 bp of ungD1 deleted, whereas the ΔungD2 (BF2848) mutant lacks 1816 bp of the ungD2 gene. The ΔungD1ΔungD2 double mutant was engineered by using the ΔungD1 mutant as the recipient for the ΔungD2 plasmid construct. The acapsular ΔungD1ΔungD2ΔPSH mutant was created using the ΔungD1ΔungD2 double-mutant strain as a recipient and has a 4929 bp deletion of the PSH locus affecting 6 genes: BF3454–BF3459.
Plasmid Constructs for Complementation Studies.
pMCL54 and pLEC216 were created for ungD1 trans-complementation experiments in B. fragilis and P. aeruginosa, respectively. Similarly, pLEC61 and pLEC217 allowed ungD2 complementation analysis in B. fragilis and P. aeruginosa. PCR products of ungD1 and ungD2 were cloned into the BamHI site of the Bacteroides expression vector pFD340 (32) or the Pseudomonas shuttle vector pUCP18 (33). pMCL54 and pLEC61 were introduced into the appropriate B. fragilis strains by mobilization from Escherichia coli using the conjugal helper plasmid RK231 as described in ref. 22. pLEC216 and pLEC217 were transformed into RbCl2-competent (34) P. aeruginosa PAO1 wbpM::Gm recipients.
Analysis of Products Separated by SDS/PAGE.
SDS/PAGE and Western blotting were performed essentially as described in ref. 29. Glycoproteins were stained in gel with the Pro-Q Emerald 300 glycoprotein stain (Invitrogen). Radioactively labeled cultures separated by SDS/PAGE were transferred to polyvinylidine fluoride membrane and exposed to a tritium storage phosphor screen (Amersham Biosciences) and scanned at a pixel size of 100 μm with a Typhoon 9410 variable mode imager (Amersham Biosciences).
Gene Expression Analysis.
Microarray procedures are provided as SI Materials and Methods.
Analysis of the PSH Promoter Orientation.
PSH promoter orientation ratios of various samples were measured quantitatively by a PCR digestion protocol similar to that described in ref. 3. Chromosomal DNA was isolated from fecal samples using the ExtractMaster Fecal DNA Extraction Kit (Epicentre Biotechnologies). Chromosomal DNA from bacteria grown in vitro or from mouse feces was PCR amplified using primers that anneal outside of and on opposite sides of the invertible PSH promoter region, producing a 1281-bp product (Fig. 3B, Table S3). The PCR products were purified and digested with SfcI (New England Biolabs), which cleaves asymmetrically within the PSH invertible promoter region. Digestion fragments of 690 bp and 591 bp result if the PSH promoter is oriented on, and fragments of 809 bp and 472 bp result if the promoter is oriented off.
Mouse Colonization Experiments.
Mouse studies were approved by the Harvard Medical Area Standing Committee on Animals. Swiss–Webster germ-free mice (male, 3–5 weeks old) were purchased from Taconic. Mice were housed in gnotobiotic isolators (2 mice per cage, 2 cages per experiment) and were monoassociated with ΔungD1ΔungD2 or ΔungD1ΔungD2ΔPSH by spreading bacteria on the face and fur. Fresh fecal samples were collected and pooled from both mice in each cage 2, 3, 7, and 14 days after colonization. For competitive colonization experiments, the inocula contained varying ratios of wild-type and mutant bacteria. Fresh fecal samples were collected, diluted in PBS, and plated. Wild-type and mutant colonies were enumerated by PCR amplification using primers that yield a 2291-bp product from wild type and a 473-bp product from mutants (Table S1).
Assessment of in Vitro Growth Under Various Conditions.
These methods are provided as SI.
Complement Bactericidal Assay.
One ml bacteria (≈1 × 108 cfu) was centrifuged, washed, and resuspended in 1 ml PBS containing 5 mM magnesium chloride, and 90-μl volumes were aliquoted to tubes. Ten microliters of normal human serum (NHS) or of heat-inactivated NHS were added, and the samples were incubated anaerobically at 37°C for 1 h and plated to determine viable counts. The fold-change is reported as the decrease in the number of cfu from wild type or mutants incubated in NHS compared with the same strain incubated in heat-inactivated NHS.
Supplementary Material
Acknowledgments.
We thank Joseph Lam for providing P. aeruginosa PAO1 wbpM::Gm, G. Priebe for rabbit antisera to P. aeruginosa PAO1, and V. Carey and C.M. Fletcher for helpful discussions. This work was supported by National Institutes of Health/National Institute of Allergy and Infectious Diseases Grants AI044193 and AI053694.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0804220105/DCSupplemental.
References
- 1.Stappenbeck TS, Hooper LV, Gordon JI. Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells. Proc Natl Acad Sci USA. 2002;99:15451–15455. doi: 10.1073/pnas.202604299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 3.Krinos CM, et al. Extensive surface diversity of a commensal microorganism by multiple DNA inversions. Nature. 2001;414:555–558. doi: 10.1038/35107092. [DOI] [PubMed] [Google Scholar]
- 4.Chatzidaki-Livanis M, Coyne MJ, Roche-Hakansson H, Comstock LE. Expression of a uniquely regulated extracellular polysaccharide confers a large capsule phenotype to Bacteroides fragilis. J Bacteriol. 2008;190(3):1020–1026. doi: 10.1128/JB.01519-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coyne MJ, Reinap B, Lee MM, Comstock LE. Human symbionts use a host-like pathway for surface fucosylation. Science. 2005;307:1778–1781. doi: 10.1126/science.1106469. [DOI] [PubMed] [Google Scholar]
- 6.Fletcher CM, Coyne MJ, Bentley DL, Villa OF, Comstock LE. Phase-variable expression of a family of glycoproteins imparts a dynamic surface to a symbiont in its human intestinal ecosystem. Proc Natl Acad Sci USA. 2007;104:2413–2418. doi: 10.1073/pnas.0608797104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu J, et al. Evolution of symbiotic bacteria in the distal human intestine. PLoS Biol. 2007;5:e156. doi: 10.1371/journal.pbio.0050156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coyne MJ, Comstock LE. Niche-specific features of the intestinal Bacteroidales. J Bacteriol. 2008;190:736–742. doi: 10.1128/JB.01559-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu CH, Lee SM, Vanlare JM, Kasper DL, Mazmanian SK. Regulation of surface architecture by symbiotic bacteria mediates host colonization. Proc Natl Acad Sci USA. 2008;105:3951–3956. doi: 10.1073/pnas.0709266105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baumann H, Tzianabos AO, Brisson JR, Kasper DL, Jennings HJ. Structural elucidation of two capsular polysaccharides from one strain of Bacteroides fragilis using resolution NMR spectroscopy. Biochem. 1992;31:4081–4089. doi: 10.1021/bi00131a026. [DOI] [PubMed] [Google Scholar]
- 11.Creuzenet C, Lam JS. Topological and functional characterization of WbpM, an inner membrane UDP-GlcNAc C6 dehydratase essential for lipopolysaccharide biosynthesis in Pseudomonas aeruginosa. Mol Microbiol. 2001;41:1295–1310. doi: 10.1046/j.1365-2958.2001.02589.x. [DOI] [PubMed] [Google Scholar]
- 12.Creuzenet C, Urbanic RV, Lam JS. Structure-function studies of two novel UDP-GlcNAc C6 dehydratases/C4 reductases: Variation from the SYK dogma. J Biol Chem. 2002;277:26769–26778. doi: 10.1074/jbc.M202882200. [DOI] [PubMed] [Google Scholar]
- 13.Burrows LL, Charter DF, Lam JS. Molecular characterization of the Pseudomonas aeruginosa serotype O5 (PAO1) B-band lipopolysaccharide gene cluster. Mol Microbiol. 1996;22:481–495. doi: 10.1046/j.1365-2958.1996.1351503.x. [DOI] [PubMed] [Google Scholar]
- 14.Hooper LV, Stappenbeck TS, Hong CV, Gordon JI. Angiogenins: A new class of microbicidal proteins involved in innate immunity. Nat Immunol. 2003;4:269–273. doi: 10.1038/ni888. [DOI] [PubMed] [Google Scholar]
- 15.Cash HL, Whitham CV, Behrendt CL, Hooper LV. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science. 2006;313:1126–1130. doi: 10.1126/science.1127119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Uemura K, et al. L-MBP is expressed in epithelial cells of mouse small intestine. J Immunol. 2002;169:6945–6950. doi: 10.4049/jimmunol.169.12.6945. [DOI] [PubMed] [Google Scholar]
- 17.Geoffroy MC, Floquet S, Métais A, Nassif X, Pelicic V. Large-scale analysis of the meningococcus genome by gene disruption: Resistance to complement-mediated lysis. Genome Res. 2003;13:391–398. doi: 10.1101/gr.664303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iadarola MJ, Reckseidler-Zenteno SL, DeVinney R, Woods DE. The capsular polysaccharide of Burkholderia pseudomallei contributes to survival in serum by reducing complement factor C3b deposition. Infect Immun. 2005;73:1106–1115. doi: 10.1128/IAI.73.2.1106-1115.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Russo TA, et al. The effects of Escherichia coli capsule, O-antigen, host neutrophils, and complement in a rat model of Gram-negative pneumonia. FEMS Microbiol Lett. 2003;226:355–361. doi: 10.1016/S0378-1097(03)00636-0. [DOI] [PubMed] [Google Scholar]
- 20.Andoh A, et al. Detection of complement C3 and factor B gene expression in normal colorectal mucosa, adenomas and carcinomas. Clin Exp Immunol. 1998;111:477–483. doi: 10.1046/j.1365-2249.1998.00496.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giacomin PR, Wang H, Gordon DL, Botto M, Dent LA. Loss of complement activation and leukocyte adherence as Nippostrongylus brasiliensis develops within the murine host. Infect Immun. 2005;73:7442–7449. doi: 10.1128/IAI.73.11.7442-7449.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coyne MJ, Weinacht KG, Krinos CM, Comstock LE. Mpi recombinase globally modulates the surface architecture of a human commensal bacterium. Proc Natl Acad Sci USA. 2003;100:10446–10451. doi: 10.1073/pnas.1832655100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nesper J, Kapfhammer D, Klose KE, Merkert H, Reidl J. Characterization of Vibrio cholerae O1 antigen as the bacteriophage K139 receptor and identification of IS1004 insertions aborting O1 antigen biosynthesis. J Bacteriol. 2000;182:5097–5104. doi: 10.1128/jb.182.18.5097-5104.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dupont K, Janzen T, Vogensen FK, Josephsen J, Stuer-Lauridsen B. Identification of Lactococcus lactis genes required for bacteriophage adsorption. Appl Environ Microbiol. 2004;70:5825–5832. doi: 10.1128/AEM.70.10.5825-5832.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stummeyer K, et al. Evolution of bacteriophages infecting encapsulated bacteria: Lessons from Escherichia coli K1-specific phages. Mol Microbiol. 2006;60:1123–1135. doi: 10.1111/j.1365-2958.2006.05173.x. [DOI] [PubMed] [Google Scholar]
- 26.McCulloch R. Antigenic variation in African trypanosomes: Monitoring progress. Trends Parasitol. 2004;20:117–121. doi: 10.1016/j.pt.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 27.Khamri W, et al. Variations in Helicobacter pylori lipopolysaccharide to evade the innate immune component surfactant protein D. Infect Immun. 2005;73:7677–7686. doi: 10.1128/IAI.73.11.7677-7686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barbour AG, Dai Q, Restrepo BI, Stoenner HG, Frank SA. Pathogen escape from host immunity by a genome program for antigenic variation. Proc Natl Acad Sci USA. 2006;103:18290–18295. doi: 10.1073/pnas.0605302103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weinacht KG, et al. Tyrosine site-specific recombinases mediate DNA inversions affecting the expression of outer surface proteins of Bacteroides fragilis. Mol Microbiol. 2004;53:1319–1330. doi: 10.1111/j.1365-2958.2004.04219.x. [DOI] [PubMed] [Google Scholar]
- 30.Stevens AM, Shoemaker NB, Salyers AA. The region of a Bacteroides conjugal chromosomal tetracycline resistance element which is responsible for production of plasmidlike forms from unlinked chromosomal DNA might also be involved in transfer of the element. J Bacteriol. 1990;172:4271–4279. doi: 10.1128/jb.172.8.4271-4279.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thompson JS, Malamy MH. Sequencing the gene for an imipenem-cefoxitin-hydrolyzing enzyme (CfiA) from Bacteroides fragilis TAL2480 reveals strong similarity between CfiA and Bacillus cereus β-lactamase II. J Bacteriol. 1990;172:2584–2593. doi: 10.1128/jb.172.5.2584-2593.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith CJ, Rogers MB, McKee ML. Heterologous gene expression in Bacteroides fragilis. Plasmid. 1992;27:141–154. doi: 10.1016/0147-619x(92)90014-2. [DOI] [PubMed] [Google Scholar]
- 33.Schweizer HP. Escherichia-Pseudomonas shuttle vectors derived from pUC18/19. Gene. 1991;97:109–121. doi: 10.1016/0378-1119(91)90016-5. [DOI] [PubMed] [Google Scholar]
- 34.Hanahan D, Jessee J, Bloom FR. Plasmid transformation of E coli and other bacteria. Methods Enzymol. 1991;204:63–113. doi: 10.1016/0076-6879(91)04006-a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.