Abstract
Since cdc48 mutants were isolated by the first genetic screens for cell division cycle (cdc) mutants in yeast, the requirement of the chaperone-like ATPase Cdc48/p97 during cell division has remained unclear. Here, we discover an unanticipated function for Caenorhabditis elegans CDC-48 in DNA replication linked to cell cycle control. Our analysis of the CDC-48UFD−1/NPL−4 complex identified a general role in S phase progression of mitotic cells essential for embryonic cell division and germline development of adult worms. These developmental defects result from activation of the DNA replication checkpoint caused by replication stress. Similar to loss of replication licensing factors, DNA content is strongly reduced in worms depleted for CDC-48, UFD-1, and NPL-4. In addition, these worms show decreased DNA synthesis and hypersensitivity toward replication blocking agents. Our findings identified a role for CDC-48UFD−1/NPL−4 in DNA replication, which is important for cell cycle progression and genome stability.
Keywords: ATL-1/ATR, C. elegans, CDC-48/p97, genome stability
Many biological processes including development and cell division are tightly controlled by ubiquitin-mediated protein degradation. A central factor for mobilizing and targeting ubiquitylated substrates to the 26S proteasome is Cdc48/p97 (Cdc48 in yeast, CDC-48 in C. elegans, p97 in mammals), a chaperone-like AAA ATPase (1). Its activity is modulated by alternative adaptor proteins, which determine recruitment and processing of specific substrates. Cdc48/p97 forms a complex with the cofactors Ufd1 and Npl4 that is involved in endoplasmic reticulum (ER)-associated protein degradation (ERAD) (2), membrane fusion and cell cycle progression (3, 4). Temperature sensitive cdc48 mutants have already been isolated by early cdc-screens (5) in Saccharomyces cerevisiae. However, the essential role of Cdc48 during cell cycle progression remained elusive.
Meanwhile, different activities of Cdc48/p97 in mitosis have been addressed by several studies. Early observations in yeast and recent findings using Xenopus egg extracts suggested that Cdc48/p97 regulates spindle disassembly during exit from mitosis (6, 7). For example, spindle regulators such as the Polo-like kinase Plx remain attached and probably stabilize the spindle in the absence of p97Ufd1/Npl4. However, contradictory evidence exists concerning a specific role of the p97Ufd1/Npl4 complex in spindle dynamics (8, 9). Beside spindle function, p97 together with its Ufd1-Npl4 cofactor is important for nuclear envelope assembly (10). Interestingly, it has been shown that p97 stimulates nucleus reformation after mitosis by extracting and thereby inactivating the mitotic progression kinase Aurora B from chromatin (11). Together, these diverse processes involving Cdc48/p97 suggest the existence of multiple substrates that need to be regulated during mitosis.
Recently, we found that the C. elegans Cdc48/p97 homologues CDC-48.1 and CDC-48.2 form an evolutionarily conserved complex with UFD-1 and NPL-4 important for the degradation of misfolded proteins from the ER. In addition, downregulation of cdc-48.1/cdc-48.2, ufd-1, or npl-4 results in embryonic lethality, indicating an essential role for the CDC-48UFD−1/NPL−4 complex (12). Here, we identified CDC-48UFD−1/NPL−4 to be crucial for DNA replication. Consequently, inactivation of the whole complex leads specifically to a replication checkpoint dependent delay in S phase progression; however, mitosis time is not affected. Our data show that similar to loss of the replication licensing factors CDT-1 and CDC-6, DNA content is strongly reduced in worms depleted for CDC-48, UFD-1, and NPL-4. Together, these findings provide a link between DNA replication and the chaperone-like CDC-48UFD−1/NPL−4 complex, which is important for cell cycle progression and genome stability.
Results and Discussion
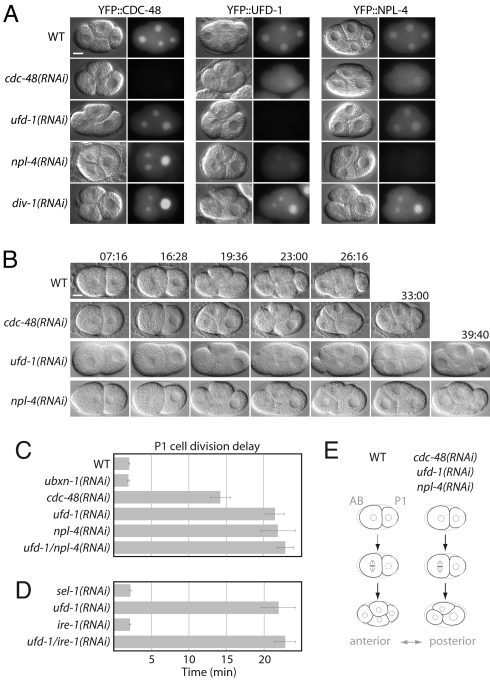
To determine the function of the CDC-48UFD−1/NPL−4 complex in early development, we analyzed its subcellular embryonic localisation using yellow fluorescent protein (YFP) fusions. In contrast to a cytosolic distribution during mitosis, YFP::CDC-48, YFP::UFD-1, and YFP::NPL-4 all similarly accumulated in the nucleus after its reformation [Fig. 1A, supporting information (SI) Fig. S1, and Movie S1]. Codepletion of CDC-48.1 and CDC-48.2 by RNA interference (RNAi) (hereafter referred to as CDC-48 depletion or cdc-48(RNAi)) diminished the nuclear localisation of UFD-1 and NPL-4, whereas CDC-48 localisation remained unchanged in the converse experiment (Fig. 1A). These data suggest CDC-48UFD−1/NPL−4 complex formation in early embryos and identify CDC-48 as the major determinant for its nuclear localisation.
Fig. 1.
Depletion of CDC-48UFD−1/NPL−4 delays cell cycle progression. (A) DIC and fluorescent images of wild-type (WT), cdc-48(RNAi), ufd-1(RNAi), npl-4(RNAi), and div-1(RNAi) embryos expressing YFP::CDC-48, YFP::UFD-1, and YFP::NPL-4. (B) Selected images from time-lapse DIC microscopy of wild-type (WT), cdc-48(RNAi), ufd-1(RNAi), and npl-4(RNAi) embryos. (C and D) Quantification of the division delay between AB and P1 cells. (E) Cartoon illustrating major differences between wild-type (WT) embryos and those depleted for CDC-48UFD−1/NPL−4, exhibiting a persistent three-cell stage. Anterior is to the left. In time-lapse analyses, time 0 is defined by nuclear envelope breakdown (NEBD) of the P0 cell. Error bars represent s.e.m. (Scale bar, 10 μm).
The first C. elegans embryonic division of the P0 zygote is asymmetric and generates an anterior AB cell, and a smaller posterior P1 cell. These cells have different developmental fates and division timing, with AB dividing ≈2 min before P1 (Fig. 1E) (13). Time-lapse differential interference contrast (DIC) microscopy identified that downregulation of cdc-48, ufd-1, and npl-4 increases the cell division delay of P1 in comparison to AB, leading to a prolonged three-cell stage (Fig. 1 A–C and E, Fig. S1, and Movies S2–S5). In line with CDC-48UFD−1/NPL−4 acting as a functional unit (12), the delay in P1 cell division was not significantly enhanced in codepleted ufd-1/npl-4(RNAi) embryos (Fig. 1C and Table S1). This phenotype was not caused by depletion of the p47 homologue UBXN-1, an alternative cofactor of CDC-48/p97 (12, 14), indicating that CDC-48 cooperates specifically with the UFD-1/NPL-4 adaptor in cell cycle progression (Fig. 1C and Table S1). Moreover, this cell cycle function of the CDC-48UFD−1/NPL−4 complex is not a consequence of general ER stress conditions as downregulation of the ERAD or unfolded protein response (UPR) regulators sel-1 or ire-1 (15) neither recapitulated nor suppressed the P1 cell division delay phenotype (Fig. 1D and Table S1).
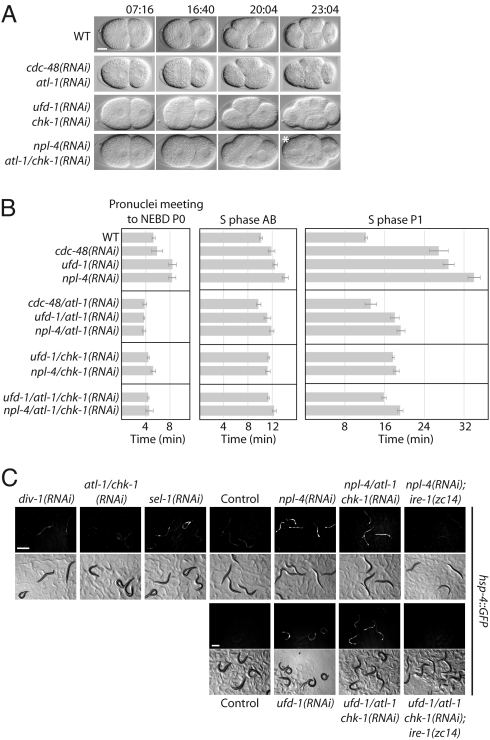
In early C. elegans embryonic cell cycles, S and M phases rapidly alternate without apparent gap phases (16). In contrast to mitosis, the progression of S phase was significantly delayed in P0, AB, and to a larger extent in P1 cells of cdc-48(RNAi), ufd-1(RNAi), or npl-4(RNAi) embryos (Fig. 2B, Fig. S2A, and Table S1). Intriguingly, this phenotype is reminiscent of defects in the replication machinery, for example caused by depletion of the DNA polymerase α-subunit DIV-1 (17, 18). Nonetheless, div-1(RNAi) did not affect the nuclear localisation of CDC-48, UFD-1, and NPL-4 (Fig. 1A). The cell cycle delay of div-1 downregulation depends on the DNA replication checkpoint kinases ATL-1/ATR and CHK-1/Chk1, which are required for the cellular response to stalled DNA replication forks and UV-induced DNA damage (17, 19). We tested whether activation of the DNA replication checkpoint is responsible for the delayed S phase progression associated with the depletion of a functional CDC-48UFD−1/NPL−4 complex. Indeed, downregulation of atl-1 and/or chk-1 suppressed the P1 cell division delay phenotype of cdc-48, ufd-1, and npl-4 RNAi embryos (Fig. 2 A and B, Table S1, and Movie S6). In contrast, RNAi-mediated depletion of ATM-1, related to the mammalian DNA double-strand break sensing ATM kinase (19, 20), had no effect (Fig. S3 and Table S1). Similarly, downregulation of the mitotic regulator genes mdf-1 and air-2 (21, 22), which encode homologues of the spindle checkpoint factor MAD1 and the mitotic progression kinase Aurora B, did not suppress the cell division delay (Fig. S2 B and C, and Table S1).
Fig. 2.
Downregulation of the DNA replication checkpoint suppresses the S phase arrest caused by CDC-48UFD−1/NPL−4 depletion. (A) Selected images from time-lapse DIC microscopy of wild-type (WT), cdc-48/atl-1(RNAi), ufd-1/chk-1(RNAi), and npl-4/atl-1/chk-1(RNAi) embryos. The picture marked with an asterisk was taken at time point 26:24 min. (Scale bar, 10 μm). (B) Quantification of S phase duration in P0, AB, and P1 cells. Error bars represent s.e.m. (C) Monitoring UPR induction by expression of GFP under the control of the hsp-4 promoter. (Scale bar, 500 μm).
We addressed the importance of ATL-1 and CHK-1 for the previously described ERAD function of UFD-1 and NPL-4 (12), by monitoring ER stress conditions with the UPR-inducible hsp-4::gfp reporter (15). The expression level of the ER-resident chaperone HSP-4 is directly related to the accumulation of misfolded proteins. Unlike the cell cycle phenotype, depletion of atl-1 or chk-1 did not suppress ERAD defects caused by ufd-1(RNAi) or npl-4(RNAi) (Fig. 2C). The UPR was also not significantly induced in div-1(RNAi) worms, indicating that ER quality control pathways are not influenced by DNA replication defects (Fig. 2C).
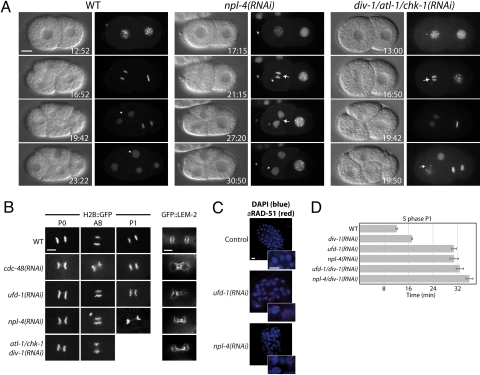
To further investigate S phase defects of cdc-48, ufd-1, and npl-4 RNAi embryos, we studied chromatin structure and chromosome segregation by following GFP-tagged histone H2B (H2B::GFP) by time-lapse microscopy. In wild-type, DNA replication generates two sets of sister chromatids that segregate toward opposite spindle poles during mitosis (16). In contrast, in cdc-48(RNAi), ufd-1(RNAi), and npl-4(RNAi) embryos the separating chromatids often remain connected via chromosome bridges. This phenotype is reported to be associated with S phase defects (17, 23), and occurs throughout the divisions of P0, AB, and P1 cells (Fig. 1E, Fig. 3 A and B, and Movie S8). We tested whether downregulation of atl-1 and/or chk-1 is able to restore correct sister chromatid separation. In contrast to suppression of delayed S phase progression (Fig. 2 A and B, Table S1, and Movie S6), these chromosomal abnormalities were enhanced in ufd-1(RNAi) embryos as a consequence of DNA replication checkpoint inactivation (Table S2), which is comparable to embryos carrying div-1 temperature-sensitive (ts) alleles (17). Therefore, activation of the DNA replication checkpoint in embryos lacking a functional CDC-48UFD−1/NPL−4 complex seems to result from replication defects.
Fig. 3.
Embryos depleted for CDC-48UFD−1/NPL−4 display abnormal chromatin structures. (A) Images of time-lapse movies showing wild-type (WT), npl-4(RNAi), and div-1/atl-1/chk-1(RNAi) embryos expressing H2B::GFP. Arrows point at chromatin abnormalities. (B) Close up of fluorescence images of wild-type (WT), cdc-48(RNAi), ufd-1(RNAi), npl-4(RNAi), and div-1/atl-1/chk-1(RNAi) embryos expressing H2B::GFP or GFP::LEM-2. (Scale bar, 5 μm). (C) Merged images of ufd-1(RNAi) and npl-4(RNAi) treated embryos stained with DAPI (blue) and anti-RAD-51 antibody (red). In contrast to Fig. 4D, DAPI signals were adjusted to visualize the DNA content. (Scale bar, 5 μm). (D) Quantified S phase in P1 cells for epistatic analysis of div-1 with ufd-1 or npl-4. Error bars represent s.e.m.
Since in vitro studies suggested a role for p97 in nuclear envelope assembly (10), we expressed a GFP-tagged version of the inner nuclear membrane protein MAN1 (24) (GFP::LEM-2) in embryos depleted for CDC-48, UFD-1, and NPL-4. The nuclear envelope integrity is not affected in these depleted embryos lacking chromosome bridges (Fig. S4), which is consistent with previous reports (25). In contrast, chromosome bridges seem to cause defects in nuclear envelope formation, since they are also detectable in embryos lacking DIV-1 (Fig. 3B). Similar secondary effects caused by chromosome bridges might be reflected by chromatin condensation problems (Fig. 3A, Fig. S5, and Movie S7 and Movie S8).
These data provide evidence that in embryos depleted for the CDC-48UFD−1/NPL−4 complex, the DNA replication checkpoint is activated as a consequence of replication defects. Evidence for stalled replication forks can be obtained cytologically by assessing the accumulation of RAD-51 foci (26) that assembles on processed single-stranded DNA at stalled or collapsed replication forks. In contrast to wild-type, ufd-1(RNAi) and npl-4(RNAi) embryos formed RAD-51 foci, indicating the existence of replication stress (Fig. 3C). In addition to embryonic defects, npl-4(RNAi) adult hermaphrodite worms displayed RAD-51 positive arrested nuclei in the proliferative zone of the gonad, demonstrating that the replication checkpoint is also activated in mitotic germ cells (Fig. S6). Codepletion of DIV-1 together with UFD-1 or NPL-4 neither significantly suppressed nor enhanced the P1 cell division delay observed in single RNAi experiments, indicating that they act in a common genetic pathway (Fig. 3D). These observations suggest a general role of the CDC-48UFD−1/NPL−4 complex in DNA replication of mitotic/proliferating cells, required for embryonic and germline development.
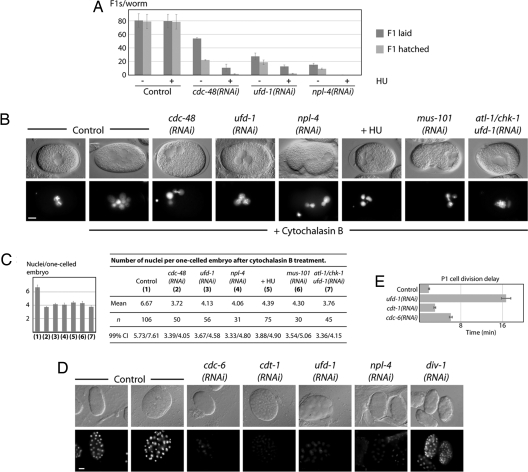
We expected that replication defects caused by downregulation of CDC-48UFD−1/NPL−4 combined with sublethal doses of the replication blocking drug HU would result in synthetic effects (27). Indeed, worms depleted for cdc-48, ufd-1, or npl-4 exhibited enhanced sensitivity for HU, resulting in embryonic lethality and sterile worms (Fig. 4A). To assess a direct requirement of the CDC-48UFD−1/NPL−4 complex for DNA replication, we blocked cytokinesis with cytochalasin B and quantified the number of nuclei in single-celled embryos expressing H2B::GFP (23). In contrast to cytokinesis, DNA replication remained unaffected and consequently wild-type embryos accumulated large amounts of DNA after 2–3 h (Fig. 4 B and C). The production of multinucleated cells depends on DNA synthesis because it was dramatically reduced in the presence of the replication inhibitor HU. Comparable to HU treatment, embryos depleted for CDC-48, UFD-1, NPL-4, or the replication factor MUS-101 (23) exhibited significantly less nuclei, indicating the requirement for DNA replication (Fig. 4 B and C, and Table S3). Depletion of ATL-1/CHK-1 did not increase the number of nuclei in ufd-1(RNAi) embryos, which showed that the replication defect is the primary consequence of UFD-1 depletion not influenced by checkpoint activation (Fig. 4 B and C, and Table S3). This observation is in line with the enhanced accumulation of chromosome bridges in ufd-1/atl-1/chk-1(RNAi) embryos (Table S2). In contrast to DIV-1 depletion, cdc-48, ufd-1, and npl-4 RNAi embryos (≈30 to 100 cell stage) contain only trace amounts of DNA, which implies a function at early steps of DNA replication rather than elongation. In fact depletion of the key replication licensing factors CDT-1 and CDC-6 shows a similar block in DNA synthesis (Fig. 4D). Consequently, their downregulation leads to a P1 cell division delay similar to loss of CDC-48, UFD-1, and NPL-4 (Fig. 4E). Once per cell cycle CDT-1 and CDC-6 are required to form the prereplication complex at replication origins. To avoid re-replication, CDT-1 is targeted for degradation by the CUL-4 E3 ligase (28). However, ubiquitin-dependent regulation of CDT-1 protein levels does not involve CDC-48, UFD-1, and NPL-4 (Fig. S7). These observations suggest that the CDC-48UFD−1/NPL−4 complex is crucial for DNA replication initiation, which might not involve a role in CDT-1 turnover.
Fig. 4.
Depletion of CDC-48UFD−1/NPL−4 results in replication stress sensitivity and DNA replication defects. (A) Developmental defects of cdc-48(RNAi), ufd-1(RNAi), and npl-4(RNAi) embryos exposed to (5 mM) hydroxyurea (HU). (B) DIC and fluorescence images of cdc-48(RNAi), ufd-1(RNAi), npl-4(RNAi), ufd-1/chk-1(RNAi), mus-101(RNAi), and HU-treated one-cell stage embryos expressing H2B::GFP after cytochalasin B exposure. (Scale bar, 10 μm). (C) Quantification of nuclear division/replication efficiency shown in Fig. 4B. Error bars represent s.e.m. Confidence interval at 99% indicated as 99% CI. (D) DIC and fluorescence images of cdt-1(RNAi), cdc-6(RNAi), ufd-1(RNAi), npl-4(RNAi), and div-1(RNAi) embryos stained with DAPI. Fluorescence images were acquired in the same conditions. In contrast to Fig. 3C, adjustments of DAPI signals were equally performed for all photographs. (Scale bar, 10 μm). (E) Quantification of the division delay between AB and P1 cells.
Several previous studies have described different activities of Cdc48/p97 in mitotic events, such as spindle disassembly and nuclear envelope reformation (6, 7, 10, 11). Here, we identified a role for CDC-48UFD−1/NPL−4 in DNA replication, which is important for cell cycle progression (Fig. S8). Inactivation of the complex leads to hypersensitivity toward HU treatment, decreased DNA synthesis, and to a replication checkpoint dependent delay in S phase progression. Several lines of evidence suggest that the requirement of CDC-48UFD−1/NPL−4 for DNA replication is independent of its roles in spindle dynamics and nuclear assembly. First, in contrast to the S phase delay, depletion of the whole complex did not affect mitosis time. Second, downregulation of the homologues of the spindle checkpoint factor MAD1 and the mitotic progression kinase Aurora B did not suppress the S phase delay of cdc-48, ufd-1, and npl-4 RNAi embryos. Third, the integrity of the nuclear envelope is not affected in embryos depleted for CDC-48UFD−1/NPL−4 that lack chromosome bridges.
Consistent with our findings, yeast cdc48 (ts) alleles arrest at G2/M phase (5, 6), a phenotype reminiscent of S phase replication defects (29). The recently documented arrest of cdc-48-depleted worm oocytes in meiosis I might also be linked to DNA replication problems (9), which would be in line with NPL-4 being important for S phase progression of mitotic germ cells (Fig. S6). Given that the CDC-48UFD−1/NPL−4 complex is required for embryonic cell division and germline development of adult worms together with its ubiquitous expression (30), we propose a general role in DNA replication of mitotic cells. At present it is unclear how CDC-48 affects replication but recent studies reported that p97 binds to DNA replication factors (31, 32). Interestingly, p97 associates with the human Werner RecQ helicase (33) and C. elegans CDC-48.1 with HIM-6, a Bloom Syndrome helicase homolog (34). As both RecQ helicases are involved in DNA replication to prevent accelerated aging, it is conceivable to speculate about an evolutionarily conserved requirement of CDC-48/p97 in DNA synthesis linked to cancer and longevity.
Although the exact molecular mechanism remains unclear, it is likely that the ubiquitin-related function of the CDC-48UFD−1/NPL−4 complex is important for its role in DNA replication. Thus, based on the link between the general DNA replication machinery and CDC-48/p97-dependent regulation, it will be of great interest to identify critical substrates.
Materials and Methods
Strains.
Worms were handled according to standard procedures and grown at 15°C unless otherwise stated (35). Mutations and transgenes used in this study are listed by chromosomes as follows: Mutations and transgenes used in this study are listed as follows: unc-119(ed3) ruls32[unc-119(+) pie-1::GFP::H2B]III, unc-119(ed3) qaIs3507[unc-119(+) pie-1::GFP::lem-2]III, zcIs4[hsp-4::GFP]V, ire-1(zc14)II; zcIs4[hsp-4::GFP]V. The C. elegans Bristol strain N2 was used as wild-type strain.
Cloning Procedures.
Standard molecular biology protocols were used (36). div-1 cDNA was amplified by PCR and cloned into pPD129.36 (pPD129.36-DIV-1), pPD129.36-UFD-1 (pPD129.36-DIV-1/UFD-1), and pPD129.36-NPL-4, (pPD129.36-DIV-1/NPL-4). ufd-1 cDNA was subcloned into pPD129.36-NPL-4 (pPD129.36-UFD-1/NPL-4). air-2 cDNA subcloned into pPD129.36 (pPD129.36-AIR-2) was obtained from the C. elegans ORFeome-library (Geneservice Ltd). mdf-1 and mus-101 cloned into pPD129.36 (pPD129.36-MDF-1; pPD129.36-MUS101) were obtained from the C. elegans RNAi library (Geneservice Ltd). atl-1, chk-1, and atl-1/chk-1 cloned into pPD129.36 (pPD129.36-ATL-1, pPD129.36-CHK-1, pPD129.36-ATL-1/CHK-1) were kindly provided by Pierre Gönczy (ISREC, Lausanne, Switzerland). sel-1 cloned into pPD129.36 (pPD129.36-SEL-1) was kindly provided by David Ron (NYU Medical Center, New York). Other constructs used here have been published previously (12).
Expression Constructs and Generation of Transgenic Animals.
The YFP translational fusion genes YFP::cdc-48, YFP::ufd-1, and YFP::npl-4 were constructed by PCR amplification of the corresponding cDNAs and cloning into the pAZ-YFP(N-terminal) bombardement vector, which contains the pie-1 promoter for maternal expression and the unc-119(+) marker for selection of transgenic worms. The constructs were bombarded into unc-119(ed4) mutants as described previously (37). Fluorescence images were taken with an Axioplan2 Imaging microscope mounted with Axiocam HR camera (Carl Zeiss) or with a fluorescent stereo microscope SZX12 (Olympus), outfitted with a Colorview camera (Soft Imaging System) and processed with analysis software (Soft Imaging Solutions). Chromosome bridges in Table S2 were documented with a TCS SP2 confocal microscope and Leica Control (Leica Camera).
RNAi.
RNA interference was performed using the feeding method (38). L4 larvae were placed on IPTG-containing plates seeded with Escherichia coli [HT115(DE3)] expressing double-stranded RNA as described in ref. 12. RNAi-depleted embryos that were able to divide at least until the four-cell stage were analyzed. For strains expressing fluorescent proteins, RNAi was performed at 20°C or 25°C. In RNAi control experiments, bacteria only contained the empty vector pPD129.36.
Time-Lapse Microscopy.
C. elegans eggs were extruded in M9 buffer from dissected adult worms and mounted on 2% agarose pads. Recordings in Fig. 1 B and D, Fig. 2 A and B, Fig. 3D, Fig. S2, Fig. S3, and Table S1 were acquired at 4-s intervals with AxioCam HR or AxioCam MRc cameras mounted on Axioplan2 Imaging or Axiophot microscopes, respectively, equipped with differential interference contrast (DIC) optics (100×/1.3 Plan-Neofluar; Carl Zeiss). Movement or meeting of pronuclei, nuclear envelope breakdown (NEBD), and the end of cytokinesis were estimated visually. To quantify the P1 cell division delay phenotype, the time separating cytokinesis of AB from that of P1 was measured. Since early embryonic cells of C. elegans alternate between S and M phase (16), the time separating NEBD from cytokinesis corresponds to M phase, while that separating cytokinesis from NEBD in the subsequent cell cycle corresponds to S phase. Recordings in Fig. S1, Fig. 3 A and B (H2B::GFP), and Fig. S5 were acquired in 20- or 10-second intervals (2 × 2 binning) with an Orca ER 12-bit digital camera (Hamamatsu) mounted on a wide-field microscope (Axioplan2, 40×/1.3 Plan-Apochromat objective; Carl Zeiss) or a spinning disk confocal microscope (Axioplan, 63×/1.4 Plan-Apochromat objective; Carl Zeiss; and Yokogawa disk head). Image processing was done with AxioVision (Carl Zeiss) or MetaMorph software (Universal Imaging).
Immunotechniques.
For DNA staining, worms were dissected onto polylysine (Sigma) coated microscope slides, and embryos were freeze-cracked in liquid nitrogen for at least 5 min before fixing in a methanol bath at −20°C for 10 min, followed by an acetone bath at −20°C for 10 min. Embryos were rehydrated in decreasing ethanol concentration steps (from 90% to 10%), 5 min each. A last washing with PBS-T (PBS containing 0.5% Tween-20) for 5 min was performed before applying DAPI stain diluted 1:10,000 in PBS-T buffer for 5 min at room temperature. Finally embryos were mounted with mounting medium for fluorescence (Vector Laboratories Inc.). RAD-51 staining was performed by fixation of worms with 3.7% paraformaldehyde for 5 min, incubation in liquid nitrogen for 15 min, and permeabilisation with ethanol in PBS containing 0.1% Triton. An anti-RAD-51 specific antibody (39) was applied at 1:200 in PBS containing 0.5% BSA.
Replication Assay and Drug Treatment.
To detect replication defects wild-type or RNAi treated adult hermaphrodite worms expressing H2B::GFP were soaked in M9 buffer containing 10 μg/ml cytochalasin B and incubated 2–3 h with continuous shaking before observation. Soaked worms were dissected and single-celled embryos mounted on 2% agarose pads were analyzed randomly. The number of distinct nuclei in these embryos was estimated by fluorescence microscopy. Hydroxyurea (HU) was applied to M9 buffer for the replication assay or NGM agar plates for growth-sensitivity tests at a final concentration of 20 mM or 5 mM, respectively.
Statistical Analysis.
Statistical significance was accessed for parametric data sets by two-tailed paired student's t test. For non-parametric data sets the confidence intervals are indicated. Values and statistics of time-lapse movies are described in Table S1.
Supplementary Material
Acknowledgments.
We thank P. Gönczy, A. Fire, E. T. Kipreos, Y. Kohara, D. Ron, M. Vidal, the Caenorhabditis Genetics Center (funded by the NIH Center for Research Resources), and the Dana–Farber Cancer Institute and Geneservice Ltd for antibodies, plasmids, cDNAs, and strains; and S. Ernst for technical help. We also thank S. Jentsch, J. Kim, A. Segref, and O. Stemmann for critical reading of the manuscript. This work is supported by grants of the Deutsche Forschungsgemeinschaft and the EMBO Young Investigator Program to T.H., a Cancer Research U.K. CDA grant to A.G., the Max Planck Society, A.A.H. and C.H., and a postdoctoral fellowship of the Schering Foundation to C.H.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0805944105/DCSupplemental.
References
- 1.Richly H, et al. A Series of Ubiquitin Binding Factors Connects CDC48/p97 to Substrate Multiubiquitylation and Proteasomal Targeting. Cell. 2005;120:73–84. doi: 10.1016/j.cell.2004.11.013. [DOI] [PubMed] [Google Scholar]
- 2.Meusser B, Hirsch C, Jarosch E, Sommer T. ERAD: the long road to destruction. Nat Cell Biol. 2005;7:766–772. doi: 10.1038/ncb0805-766. [DOI] [PubMed] [Google Scholar]
- 3.Ye Y. Diverse functions with a common regulator: Ubiquitin takes command of an AAA ATPase. J Struct Biol. 2006;156:29–40. doi: 10.1016/j.jsb.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 4.Meyer HH, Shorter JG, Seemann J, Pappin D, Warren G. A complex of mammalian ufd1 and npl4 links the AAA-ATPase, p97, to ubiquitin and nuclear transport pathways. EMBO J. 2000;19:2181–2192. doi: 10.1093/emboj/19.10.2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moir D, Stewart SE, Osmond BC, Botstein D. Cold-sensitive cell-division-cycle mutants of yeast: isolation, properties, and pseudoreversion studies. Genetics. 1982;100:547–563. doi: 10.1093/genetics/100.4.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frohlich KU, et al. Yeast cell cycle protein CDC48p shows full-length homology to the mammalian protein VCP and is a member of a protein family involved in secretion, peroxisome formation, and gene expression. J Cell Biol. 1991;114:443–453. doi: 10.1083/jcb.114.3.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao K, Nakajima R, Meyer HH, Zheng Y. The AAA-ATPase Cdc48/p97 regulates spindle disassembly at the end of mitosis. Cell. 2003;115:355–367. doi: 10.1016/s0092-8674(03)00815-8. [DOI] [PubMed] [Google Scholar]
- 8.Heubes S, Stemmann O. The AAA-ATPase p97-Ufd1-Npl4 is required for ERAD but not for spindle disassembly in Xenopus egg extracts. J Cell Sci. 2007;120:1325–1329. doi: 10.1242/jcs.006924. [DOI] [PubMed] [Google Scholar]
- 9.Sasagawa Y, Yamanaka K, Nishikori S, Ogura T. Caenorhabditis elegans p97/CDC-48 is crucial for progression of meiosis I. Biochem Biophys Res Commun. 2007;358:920–924. doi: 10.1016/j.bbrc.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 10.Hetzer M, et al. Distinct AAA-ATPase p97 complexes function in discrete steps of nuclear assembly. Nat Cell Biol. 2001;3:1086–1091. doi: 10.1038/ncb1201-1086. [DOI] [PubMed] [Google Scholar]
- 11.Ramadan K, et al. Cdc48/p97 promotes reformation of the nucleus by extracting the kinase Aurora B from chromatin. Nature. 2007;450:1258–1262. doi: 10.1038/nature06388. [DOI] [PubMed] [Google Scholar]
- 12.Mouysset J, Kaehler C, Hoppe T. A conserved role of Caenorhabditis elegans CDC-48 in ER-associated protein degradation. J Struct Biol. 2006;156:41–49. doi: 10.1016/j.jsb.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 13.Pellettieri J, Seydoux G. Anterior–posterior polarity in C. elegans and Drosophila–PARallels and differences. Science. 2002;298:1946–1950. doi: 10.1126/science.1072162. [DOI] [PubMed] [Google Scholar]
- 14.Kondo H, et al. p47 is a cofactor for p97-mediated membrane fusion. Nature. 1997;388:75–78. doi: 10.1038/40411. [DOI] [PubMed] [Google Scholar]
- 15.Urano F, et al. A survival pathway for Caenorhabditis elegans with a blocked unfolded protein response. J Cell Biol. 2002;158:639–646. doi: 10.1083/jcb.200203086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van den Heuvel S. Cell-cycle regulation. WormBook. 2005:1–16. doi: 10.1895/wormbook.1.28.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brauchle M, Baumer K, Gonczy P. Differential activation of the DNA replication checkpoint contributes to asynchrony of cell division in C. elegans embryos. Curr Biol. 2003;13:819–827. doi: 10.1016/s0960-9822(03)00295-1. [DOI] [PubMed] [Google Scholar]
- 18.Encalada SE, et al. DNA replication defects delay cell division and disrupt cell polarity in early Caenorhabditis elegans embryos. Dev Biol. 2000;228:225–238. doi: 10.1006/dbio.2000.9965. [DOI] [PubMed] [Google Scholar]
- 19.Abraham RT. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 2001;15:2177–2196. doi: 10.1101/gad.914401. [DOI] [PubMed] [Google Scholar]
- 20.Garcia-Muse T, Boulton SJ. Distinct modes of ATR activation after replication stress and DNA double-strand breaks in Caenorhabditis elegans. EMBO J. 2005;24:4345–4355. doi: 10.1038/sj.emboj.7600896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Encalada SE, Willis J, Lyczak R, Bowerman B. A spindle checkpoint functions during mitosis in the early Caenorhabditis elegans embryo. Mol Biol Cell. 2005;16:1056–1070. doi: 10.1091/mbc.E04-08-0712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oegema K, Desai A, Rybina S, Kirkham M, Hyman AA. Functional analysis of kinetochore assembly in Caenorhabditis elegans. J Cell Biol. 2001;153:1209–1226. doi: 10.1083/jcb.153.6.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holway AH, Hung C, Michael WM. Systematic, RNA-interference-mediated identification of mus-101 modifier genes in Caenorhabditis elegans. Genetics. 2005;169:1451–1460. doi: 10.1534/genetics.104.036137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee KK, Gruenbaum Y, Spann P, Liu J, Wilson KL. C. elegans nuclear envelope proteins emerin, MAN1, lamin, and nucleoporins reveal unique timing of nuclear envelope breakdown during mitosis. Mol Biol Cell. 2000;11:3089–3099. doi: 10.1091/mbc.11.9.3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poteryaev D, Squirrell JM, Campbell JM, White JG, Spang A. Involvement of the actin cytoskeleton and homotypic membrane fusion in ER dynamics in Caenorhabditis elegans. Mol Biol Cell. 2005;16:2139–2153. doi: 10.1091/mbc.E04-08-0726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haaf T, Golub EI, Reddy G, Radding CM, Ward DC. Nuclear foci of mammalian Rad51 recombination protein in somatic cells after DNA damage and its localization in synaptonemal complexes. Proc Natl Acad Sci USA. 1995;92:2298–2302. doi: 10.1073/pnas.92.6.2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Woodward AM, et al. Excess Mcm2–7 license dormant origins of replication that can be used under conditions of replicative stress. J Cell Biol. 2006;173:673–683. doi: 10.1083/jcb.200602108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhong W, Feng H, Santiago FE, Kipreos ET. CUL-4 ubiquitin ligase maintains genome stability by restraining DNA-replication licensing. Nature. 2003;423:885–889. doi: 10.1038/nature01747. [DOI] [PubMed] [Google Scholar]
- 29.Weinert TA, Kiser GL, Hartwell LH. Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes Dev. 1994;8:652–665. doi: 10.1101/gad.8.6.652. [DOI] [PubMed] [Google Scholar]
- 30.Yamauchi S, Yamanaka K, Ogura T. Comparative analysis of expression of two p97 homologues in Caenorhabditis elegans. Biochem Biophys Res Commun. 2006;345:746–753. doi: 10.1016/j.bbrc.2006.04.160. [DOI] [PubMed] [Google Scholar]
- 31.Yamada T, et al. p97 ATPase, an ATPase involved in membrane fusion, interacts with DNA unwinding factor (DUF) that functions in DNA replication. FEBS Lett. 2000;466:287–291. doi: 10.1016/s0014-5793(99)01673-7. [DOI] [PubMed] [Google Scholar]
- 32.Indig FE, et al. Werner syndrome protein directly binds to the AAA ATPase p97/VCP in an ATP-dependent fashion. J Struct Biol. 2004;146:251–259. doi: 10.1016/j.jsb.2003.11.009. [DOI] [PubMed] [Google Scholar]
- 33.Partridge JJ, Lopreiato JO, Jr, Latterich M, Indig FE. DNA damage modulates nucleolar interaction of the Werner protein with the AAA ATPase p97/VCP. Mol Biol Cell. 2003;14:4221–4229. doi: 10.1091/mbc.E03-02-0111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Caruso ME, et al. GTPase-mediated regulation of the Unfolded Protein Response in C. elegans is dependent on the AAA+ ATPase CDC-48. Mol Cell Biol. 2008;28:4261–4274. doi: 10.1128/MCB.02252-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Habor, NY: Cold Spring Habor Laboratory Press; 1989. [Google Scholar]
- 37.Praitis V, Casey E, Collar D, Austin J. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics. 2001;157:1217–1226. doi: 10.1093/genetics/157.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Timmons L, Fire A. Specific interference by ingested dsRNA. Nature. 1998;395:854. doi: 10.1038/27579. [DOI] [PubMed] [Google Scholar]
- 39.Alpi A, Pasierbek P, Gartner A, Loidl J. Genetic and cytological characterization of the recombination protein RAD-51 in Caenorhabditis elegans. Chromosoma. 2003;112:6–16. doi: 10.1007/s00412-003-0237-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.